Abstract
Background
Carbapenemase-producing Escherichia coli are increasing worldwide. In recent years, an increase in OXA-244-producing E. coli isolates has been seen in the national surveillance of carbapenemase-producing organisms in Denmark.
Aim
Molecular characterisation and epidemiological investigation of OXA-244-producing E. coli isolates from January 2016 to August 2019.
Methods
For the epidemiological investigation, data from the Danish National Patient Registry and the Danish register of civil registration were used together with data from phone interviews with patients. Isolates were characterised by analysing whole genome sequences for resistance genes, MLST and core genome MLST (cgMLST).
Results
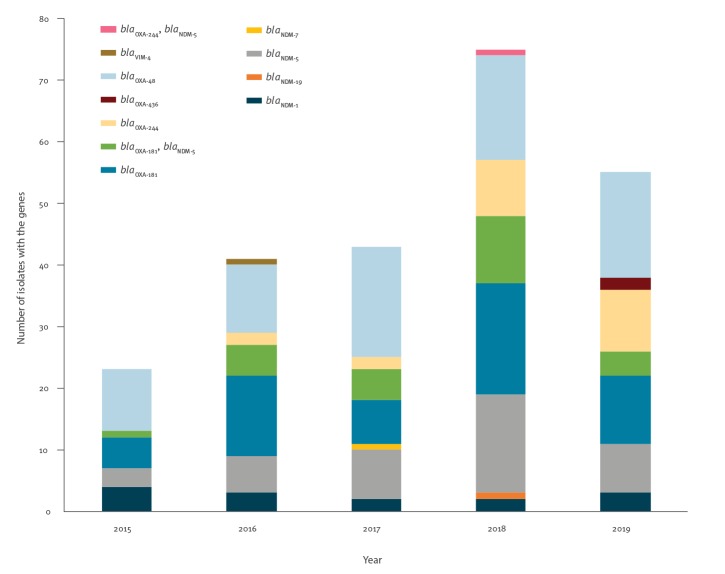
In total, 24 OXA-244-producing E. coli isolates were obtained from 23 patients. Among the 23 patients, 13 reported travelling before detection of the E. coli isolates, with seven having visited countries in Northern Africa. Fifteen isolates also carried an extended-spectrum beta-lactamase gene and one had a plasmid-encoded AmpC gene. The most common detected sequence type (ST) was ST38, followed by ST69, ST167, ST10, ST361 and ST3268. Three clonal clusters were detected by cgMLST, but none of these clusters seemed to reflect nosocomial transmission in Denmark.
Conclusion
Import of OXA-244 E. coli isolates from travelling abroad seems likely for the majority of cases. Community sources were also possible, as many of the patients had no history of hospitalisation and many of the E. coli isolates belonged to STs that are present in the community. It was not possible to point at a single country or a community source as risk factor for acquiring OXA-244-producing E. coli.
Keywords: OXA-244, carbapenemase, cgMLST, MLST, Escherichia coli
Introduction
Carbapenems are used for treatment of infections with multi-resistant Gram-negative bacteria, e.g. extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. Carbapenemase production can be caused by the presence of various carbapenemases. One of the frequently detected carbapenemases in Europe is OXA-48. Besides OXA-48, 17 other variants belong to the OXA-48 carbapenemase group) [1] including OXA-244. In comparison to OXA-48, OXA-244 has a single amino acid substitution (Arg-222-Gly) and has reduced carbapenemase activity [2]. It was first described in Spain from a Klebsiella pneumoniae isolate in 2013 [2]. Subsequently, OXA-244-producing E. coli isolates were reported from patients in the United Kingdom (UK), France and Egypt, from a healthy person in Germany, from a Dutch traveller and her spouse visiting Indonesia, from river water in Algeria and from an estuary in Lebanon [3-8]. OXA-244-producing E. coli can be a challenge for clinical laboratories as they may not grow on selective media used for detection of carbapenemase producers or may not be detected by carbapenemase-specific tests [4]. Furthermore, OXA-244-producing E. coli isolates can have low minimal inhibitory concentrations (MIC) for temocillin and meropenem [4,9].
On 18 February 2020, the European Centre for Disease Prevention and Control (ECDC) published a Rapid Risk Assessment (RRA) on the increase in OXA-244-producing E. coli, including in Denmark [10]. Before the invitation to contribute data to the RRA, the national reference laboratory at Statens Serum Institute (SSI), Copenhagen had already detected an increase in OXA-244-producing E. coli in Denmark, whereas bla OXA-244 was not detected from other carbapenemase-producing Enterobacterales.
In this study, we characterised by whole genome sequencing (WGS), all OXA-244-producing E. coli isolates submitted to SSI during 2016 through July 2019. Data from the Danish National Patient Registry were used for epidemiological investigation together with data from phone interviews with patients infected/colonised with OXA-244-producing E. coli isolates from January 2018 through July 2019. Our aim was to investigate if the increase in OXA-244-producing E. coli was related to hospital outbreaks or was related to travel from abroad.
Methods
Setting
Before 2018, Danish departments of clinical microbiology (DCM) submitted on voluntary basis, carbapenemase-producing organisms (CPO) for verification and genotyping at the national reference laboratory at SSI [11]. On 5 September 2018, the Danish Health Authority made CPO notifiable and published a national guideline to prevent the spread of CPO, including criteria for screening for CPO colonisation after hospitalisation abroad [12]. The CPO isolates are both from screening, e.g. patients travelling abroad or during outbreak investigations, and clinical samples from hospitals and primary care facilities detected during routine susceptibility testing.
Screening isolates are obtained using faecal material from a faecal swabs added to a selective Brain heart infusion (BHI) broth with 0.25 mg/L meropenem. After overnight incubation (18-24 hours) at 35°C, 10 µl from the BHI broth are plated in a selective agar e.g. chromID CARBA SMART (bioMérieux, Ballerup, Denmark) and incubated for 18–24 h at 35°C [13]. Isolates (from clinical and screenings samples) suspected to be carbapenemase-producing according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [14,15] are submitted to SSI from the Danish DCM [11].
Samples
From January 2016 to August 2019, 24 OXA-244-producing E. coli isolates from 23 patients were detected in Denmark as part of the national surveillance of CPO. For patients from whom more than one OXA-244-producing E. coli isolate was reported within a rolling 12-month period, only the first isolate was included in the study. From a single patient, two isolates were included as the second isolate carried an additional carbapenemase gene. The numbers of OXA-244 producing E. coli isolates were compared with the total numbers of carbapenemase-producing E. coli during the same period.
Epidemiological data and sources
Available data on travel history abroad 6 months before detection of the OXA-244-producing E. coli isolates were, as part of the national surveillance, reported by the DCM or by the responsible clinician/physician. Information on age, sex and postal code for residence was collected from the Danish Civil Registration System. Furthermore, patients found positive with OXA-244-producing E. coli during January 2018 to August 2019 (or their parents if children under 15 years of age) were interviewed by phone if possible during the summer of 2019, using a questionnaire created for this study containing questions on travelling within 6 months before date of sampling, food consumption and animal contact.
For investigation of possible nosocomial transmission in Denmark, hospitalisation data were retrieved from the DNPR for all patients during the period November 2015 to February 2019. The DNPR data included administration data, diagnoses, examinations and treatment procedures. Administration data included among others; hospital and department codes, dates of admission and discharge on patient level [16]. On 1 February 2019, the coding procedure for DNPR changed. Therefore, DNPR hospital data were only available until 1 February 2019 for epidemiological analysis in the present study.
Whole genome sequencing and in silico analysis
The genomic DNA was extracted (DNeasy Blood and Tissue Kit, Qiagen, Copenhagen, Denmark), with subsequent library construction (Nextera Kit, Illumina, Little Chesterford, UK) and finally sequenced (MiSeq or Nextseq, Illumina) according to the manufacturer’s instructions to obtain paired-end reads of 2x250 or 2X150 bp in length. Quality control was performed on the raw reads, using the Bifrost pipeline at SSI (https://github.com/ssi-dk/bifrost) with accepted average coverage of 30.00x or above. The WGS data were either used as raw data or de novo assembled using the assemblies generated with SKESA in Bifrost. The raw reads of all 24 isolates were assembled into draft genomes using SKESA version 2.2 in the Bifrost pipeline.
Sequences are available from GenBank under accession number PRJEB36710.
Resistance genes were identified with ResFinder version 2.1 [17] (included in the Bifrost pipeline), using a threshold of 100% identities for identifying genes encoding beta-lactamases and carbapenemases, and 98.00% ID for all other genes encoding transferable antimicrobial resistance. Two different MLST schemes were used, i.e. the Achtman scheme (MLST 1) [18] and the Pasteur scheme (MLST 2) [19].
Phylogenetic analyses
The contigs of the 24 OXA-244-producing E. coli in this study were uploaded to SeqSphere+ version 5.1.0 (Ridom GmbH, Münster, Germany (http://www.ridom.de/seqsphere/)) using the Enterobase E. coli cgMLST scheme based on 2,513 genes with a cluster distance threshold of ≤ 10 allele differences.
Ethical statement
Data on cases were collected as part of the national CPO surveillance according to guidelines for the prevention of spread of CPO by the Danish Health Authority according to which, both patients and treating medical doctor may be contacted for investigation [12]. All data were anonymised and cannot be inferred directly or indirectly to a person.
Results
Patients with OXA-244-producing Escherichia coli
From January 2016 to August 2019, 24 OXA-244-producing E. coli isolates were detected in 23 patients in Denmark (Figure 1). From one of the patients, an OXA-244-producing E. coli and an OXA-244/NDM-5-producing E. coli were obtained from the same faecal sample; both isolates were included in the study (Table 1). The remaining 22 OXA-244-producing E. coli isolates were obtained from blood samples (n = 2), urine samples (n = 18) and faecal samples (n = 2).
Figure 1.
Carbapenemase-producing Escherichia coli, Denmark, January 2015–August 2019 (n = 237)
Table 1. Sample information, demographic data and travel history for patients with OXA-244-producing Escherichia coli, Denmark, January 2016–August 2019 (n = 23).
| Patient number | Sample date | Sample | Sample site | Region | Age group (years)a | Sex | Travelb |
|---|---|---|---|---|---|---|---|
| 1 | May 2016 | Urine | General practitioner | Capital | 31–50 | Female | No travel |
| 2 | Nov 2016 | Urine | Emergency department | Zealand | 51–65 | Male | No information |
| 3 | Jun 2017 | Urine | Hospital | Southern Denmark | 31–50 | Female | No travel |
| 4 | Dec 2017 | Urine | General practitioner | Southern Denmark | 51–65 | Female | No information |
| 5 | Feb 2018 | Blood | Hospital | Southern Denmark | 51–65 | Female | Greece |
| 6 | Feb 2018 | Urine | General practitioner | Zealand | > 65 | Female | No |
| 7 | May 2018 | Faeces | Hospital | Capital | > 65 | Female | Egypt |
| 8 | Jun 2018 | Urine | General practitioner | Capital | 3–10 | Female | No information |
| 9 | Jun 2018 | Urine | Hospital | Central Denmark | > 65 | Female | No travel |
| 10 | Aug 2018 | Urine | General practitioner | Southern Denmark | > 65 | Female | Tunisia |
| 11 | Sep 2018 | Blood | Hospital | Zealand | > 65 | Male | Poland, Austria |
| 12 | Sep 2018 | Faeces | Emergency department | Capital | 31–50 | Female | Egypt, Turkey |
| 13 | Oct 2018 | Urine | General practitioner | North Denmark | 31–50 | Female | No travel |
| 14 | Feb 2019 | Urine | Hospital | Capital | > 65 | Female | Egypt |
| 15 | Mar 2019 | Faeces | Emergency department | Capital | > 65 | Male | India |
| 16 | Apr 2019 | Urine | General practitioner | Central Denmark | > 65 | Female | Egypt, Turkey, Italy |
| 17 | May 2019 | Urine | Hospital | Capital | 3–10 | Female | Sweden, Portugal |
| 18 | May 2019 | Urine | General practitioner | Southern Denmark | 60 | Female | Germany |
| 19 | Jun 2019 | Urine | General practitioner | Southern Denmark | 31–50 | Female | Egypt |
| 20 | May 2019 | Urine | General practitioner | Central Denmark | > 65 | Female | No |
| 21 | Jun 2019 | Urine | General practitioner | Southern Denmark | > 65 | Female | Egypt |
| 22 | Jun 2019 | Urine | Hospital | Central Denmark | 51–65 | Female | No information |
| 23 | Jul 2019 | Urine | Hospital | Southern Denmark | > 65 | Female | Spain |
a The age of the patients ranged from 5 to 79 years, with a median of 58 years.
b Travel 6 months before the OXA-244-producing Escherichia coli was detected.
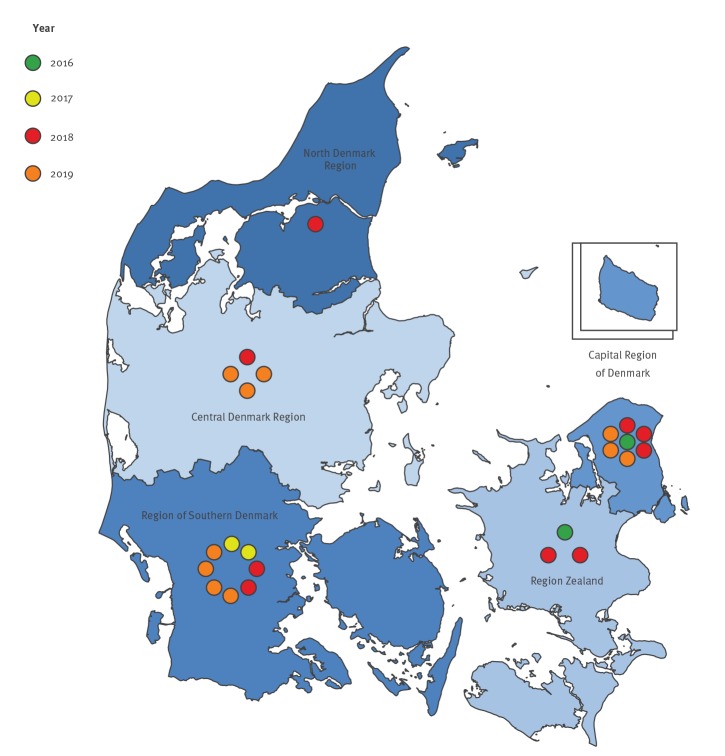
The 24 isolates were collected from hospitals and primary healthcare facilities across all five Danish regions (Figure 2, Table 1).
Figure 2.
OXA-244-producing Escherichia coli by date of detection and region, Denmark, January 2016–August 2019 (n = 24 isolates)
The age of the patients ranged from 5 to 79 years, with a median of 58 years, 20 were female and three were male. In comparison, during the study period, 63.5% (136/214) of patients with carbapenemase-producing E. coli were female in the Danish CPO surveillance, while the majority of the patients with OXA-244-producing E. coli in our study were female (p < 0.05).
Of the 23 patients, 13 had been travelling 6 months before detection of the OXA-244-producing E. coli isolates, six had no reported history of recent travel and for the remaining four patients, travel information was unavailable (Table 1). Of the 13 patients reporting travel abroad within 6 months before detection of the OXA-244-producing E. coli isolate, seven had been to countries in Northern Africa (six to Egypt, one to Tunisia).
During the study period, Carbapenemase-producing E. coli isolates were detected from 214 patients; of these, 21 patient samples were taken at a general practitioner, 11 of these patients had OXA-244-producing E. coli isolates. Furthermore, OXA-244-producing E. coli isolates were detected from the samples from three patients visiting an emergency department and from nine patients hospitalised at different hospital wards.
Data from DNPR were available for the 13 patients with positive samples before February 2019. None of the 13 patients had been hospitalised at the same time at the same hospitals. Among these 13 patients, seven had not been hospitalised in Denmark, from January 2016 to February 2019, and their samples were either from a visit at the general practitioner or at the emergency department.
Questionnaire of food consumption and animal contact
Fifteen of 19 patients with OXA-244-producing E. coli isolates during January 2018 to August 2019 were interviewed on the phone, while four patients were unavailable.
All 15 patients were eating meat and eggs. Four patients had been in contact with cats, 12 with dogs and two with wild birds. None of the patients had been in contact with livestock, hunting animals or reptiles.
Phylogenetic analysis
The 24 OXA-244-producing E. coli isolates belonged to six different STs; ST10 (n = 1), ST38 (n = 13), ST69 (n = 6), ST167 (n = 2), ST361 (n = 1) and ST3268 (n = 1) (Table 2), and 15 had an ESBL gene (bla CTX-M-14b, bla CTX-M-15 or bla CTX-M-27) and one had a plasmid-encoded AmpC gene (bla CMY-2). Genes encoding colistin resistance, i.e. mcr genes, were not detected in any of the isolates. In four isolates, bla OXA-244 was the only detected resistance gene (Table 2).
Table 2. Typing data for OXA-244-producing Escherichia coli isolates, Denmark, January 2016–August 2019 (n = 24)a .
| MLST1b | MLST2c | Patient number | Sample date | Cluster number | Resistance genes (other than bla OXA-244) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta-lactam | Aminoglycoside | Fluoroquinolone | Phenicol | Sulphonamide | Tetracycline | Trimethoprim | |||||
| ST10 | ST2 | 3 | Jun 2017 | – | bla TEM-1B | strA, strB | – | – | sul2 | tet(A) | dfrA14 |
| ST38 | ST8 | 4 | Dec 2017 | 1 | bla CTX-M-27 | aph(3”)-Ib, aph (6)-Id, aadA5 | – | – | sul1,sul2 | tet(A) | dfrA17 |
| ST38 | ST8 | 5 | Feb 2018 | 1 | bla CTX-M-27 | aph (6)-Id, aph(3”)-Ib, aadA5 | – | – | sul1, sul2 | tet(A) | dfrA17 |
| ST38 | ST8 | 14 | Feb 2019 | 1 | bla CTX-M-27 | aph (6)-Id, aph(3”)-Ib | – | – | sul2 | tet(A) | – |
| ST38 | ST8 | 16 | Apr 2019 | 1 | bla CTX-M-27 | aph (6)-Id, aph(3”)-Ib | – | – | sul2 | tet(A) | – |
| ST38 | ST8 | 17 | May 2019 | 1 | – | – | – | – | – | – | – |
| ST38 | ST8 | 1 | May 2016 | 2 | bla CTX-M-14b, bla TEM-1B | aadA1,aph(3')-Ia, strA, strB | – | – | sul2 | – | dfrA1 |
| ST38 | ST8 | 8 | Jun 2018 | 2 | bla CTX-M-14b, bla TEM-1B | aadA1,aph(3')-Ia, strA, strB | – | – | sul2 | – | dfrA1 |
| ST38 | ST8 | 19 | Jun 2019 | – | bla CTX-M-27 | aph (6)-Id, aph(3”)-Ib | – | – | sul2 | tet(A) | – |
| ST38 | ST8 | 6 | Feb 2018 | – | bla CTX-M-14b, bla TEM-1B | aph(3”)-Ib,aph (6)-Id, addA1 | – | catA1 | sul2 | tet(D) | dfrA1 |
| ST38 | ST8 | 10 | Aug 2018 | – | bla CMY-2, bla TEM-1B | aph (6)-Id, aph(3”)-Ib | – | – | sul2 | – | dfrA8 |
| ST38 | ST8 | 13 | Oct 2018 | – | bla CTX-M-14b, bla TEM-1B | aph (6)-Id, aph(3')-Ia, aph(3”)-Ib | – | – | sul2 | – | dfrA1 |
| ST38 | ST8 | 18 | May 2019 | – | bla CTX-M-14b, bla TEM-1B | – | – | catA1 | – | tet(D) | dfrA1 |
| ST38 | ST8 | 23 | Jul 2019 | – | bla CTX-M-14b, bla TEM-1B | aph (6)-Id-like, aph(3')-Ia, aph(3”)-Ib-like | qnrS1 | – | sul2 | tet(D) | dfrA14, dfrA1 |
| ST69 | ST3 | 11 | Sep 2018 | 3 | bla TEM-1B | aph (6)-Id, aph(3”)-Ib | QnrS1 | – | sul2 | tet(A) | dfrA14 |
| ST69 | ST3 | 21 | Jun 2019 | 3 | – | – | – | – | – | – | – |
| ST69 | ST3 | 22 | Jun 2019 | 3 | – | – | – | – | – | – | – |
| ST69 | ST3 | 9 | Jun 2018 | – | – | – | qnrS1 | – | – | tet(A) | dfrA14 |
| ST69 | ST3 | 15 | Mar 2019 | – | – | – | – | – | – | – | – |
| ST69 | ST3 | 2 | Nov 2016 | – | – | – | QnrB19 | – | – | tet(A), tet(U) | – |
| ST167 | ST2 | 12 | Sep 2018 | – | bla NDM-5, bla CTX-M-15, bla OXA-1 | aadA2, aac(6')Ib-cr | aac(6')Ib-cr | catB4 | sul1 | – | dfrA12, dfrA14 |
| ST167 | ST437 | 12 | Sep 2018 | – | bla CTX-M-15, bla TEM-176, bla OXA-1 | aph (6)-Id, aph(3')-Ia, aph(3”)-Ib, aadA5, aac(6')Ib-cr | aac(6')Ib-cr, QnrS1 | catB4, floR | sul2, sul1 | tet(A) | dfrA17, dfrA14 |
| ST361 | ST650 | 7 | May 2018 | – | bla CTX-M-14b, bla TEM-1B | aph(3')-Ia,aph(3”)-Ib, aph (6)-Id | – | – | sul2 | – | dfrA14 |
| ST3268 | ST535 | 20 | May 2019 | – | bla CTX-M-15, bla TEM-1B | aph (6)-Id, aph(3”)-Ib | qnrS1 | – | sul2 | tet(A) | dfrA14 |
One patient had both an OXA-244-producing ST167 E. coli (CPO20180141) and an OXA-244/NDM-5-producing ST167 E. coli (CPO20180142), but the two isolates did not cluster together in the cgMLST analysis and they had different resistance gene profiles (Table 2).
Three phylogenetic clusters were detected by cgMLST for the 24 isolates from this study (Table 2).
Core-genome multilocus sequence typing clusters
The five isolates in Cluster 1 belonged to ST38 (Table 2). The samples were obtained from December 2017 to May 2019 from three different regions in Denmark. Data from DNPR until February 2019 did not detect any overlapping hospitalisations between the first three patients. Travel information was missing for the first patient in this cluster. Six months prior to detection of the OXA-244- producing E. coli isolates, the second patient had been to Greece, the third patient had visited Egypt, the fourth patient had travelled to Egypt, Turkey as well as Italy, and the last patient had been to Sweden and Portugal. There were no geographical associations for place of residence among the patients in Cluster 1.
Both isolates in Cluster 2 belonged to ST38 (Table 2). The isolates were sampled in May 2016 and June 2018 in the same region. The first patient had not been travelling 6 months before detection of the ST38 OXA-244-producing E. coli isolate, whereas travel information was missing for the other patient. No epidemiologic links regarding hospitalisation or place of residence were detected between the patients.
The three isolates in Cluster 3 belonged to ST69 (Table 2). The samples were from three different regions and the patients also lived in different parts of the country. Unfortunately, DNPR data were not available for these patients as they all were detected after February 2019. The first patient had been to Poland and Austria 6 months before detection and the second patient had been to Egypt, while travel information was unavailable for the third patient.
Discussion
In recent years, several hospital-related outbreaks with CPO have been observed in Denmark [11]. The increase in OXA-244-producing E. coli in Denmark described here did not appear to be related to Danish hospital outbreaks. Unfortunately, travel information was not available for all patients. Despite this, repeated import from abroad of the OXA-244-producing E. coli isolates appears likely. Many of the patients had been travelling 6 months before detection of the OXA-244-producing E. coli, predominantly to Northern Africa destinations. Intestinal persistence of CPO for more than 6 months has been reported, so a limitation in our study could by the lack of information about travel prior to 6 months [20]. It could be speculated that the OXA-244-producing E. coli could have been acquired during travel abroad longer than 6 months ago (e.g. a single patient reported travel to Egypt 5 years ago, data not shown).
Among the 15 patients interviewed by phone, 12 reported contacts to dogs and four to cats. To our knowledge, OXA-244-producing E. coli isolates have neither been reported from dogs nor cats, but a recent study reported on OXA-181-producing E. coli isolates obtained from dogs and cats after hospitalisation in a veterinary clinic in Switzerland [21].
All 15 patients interviewed by phone were eating meat, but none of them had been in direct contact with livestock. OXA-244-producing E. coli have neither been reported from meat nor from livestock. More generally, CPO are very rarely reported from livestock and meat in Europe. In contrast to Europe, dissemination of CPO among livestock are more common in China, India and Northern Africa [22].
Community sources for the OXA-244-producing E. coli isolates seem possible since many of the patients had not been hospitalised and the E. coli isolates belonged to STs that are present in the community [23-32]. The ECDC RRA identified one main geographically dispersed ST38 OXA-244-producing E. coli cluster with chromosomal-encoded bla OXA-244 present in all 10 countries, including Denmark, that submitted data for the RRA [10]. The exact location (plasmid or chromosomal) of bla OXA-244 was not investigated in our study as is would require utilisation of long read sequencing techniques (e.g. MinION and PacBio). The bla OXA-244 gene can be part of the Tn51098 transposon [33]. This transposon has been reported at the same location of the chromosomal position among different STs [34].
In conclusion, an increase in OXA-244-producing E. coli has been detected in Denmark and other countries in Europe. Import of OXA-244 E. coli isolates from travelling abroad seems most likely for the majority of cases. Community sources of infections are also possible, as many of the patients had no history of hospitalisation. Furthermore, many of the E. coli isolates belonged to STs that are present in the community. It was not possible to point at a single country or a community source as risk factor for acquiring OXA-244-producing E. coli isolates.
Acknowledgements
Karin Sixhøj Pedersen, Pia Thurø Hansen, Alexandra Medina, Lone Ryste Hansen Kildevang Mette Bar Ilan, Luise Müller and the Interview Center at Infectious Disease Preparedness, SSI are thanked for excellent assistance.
Conflict of interest: None declared.
Authors’ contributions: Lone Jannok Porsbo, Frank Hansen, Louise Roer, Hülya Kaya, Anna Henius, Karina Lauenborg Møller, Ulrik S Justesen, Lillian Søes, Bent Røder, Philip K Thomsen, Mikala Wang, Turid Snekloth Søndergaard, Barbara Juliane Holzknecht, Claus Østergaard, Anne Kjerulf, Brian Kristensen and Henrik Hasman, contributed to the revision of the manuscript and approved the final version. Louise Roer, Frank Hansen, Hülya Kaya, Anette M Hammerum and Henrik Hasman did the molecular analysis. Ulrik S Justesen, Lillian Søes, Bent L Røder, Philip K Thomsen, Mikala Wang, Turid Snekloth Søndergaard, Barbara Juliane Holzknecht, Claus Østergaard detected CPOs at the DCMs in Denmark. Lone Jannok Porsbo and Anette M Hammerum drafted the questionnaire. Lone Jannok Porsbo, Anette M Hammerum, Henrik Hasman, Anna Henius, Karina Lauenborg Møller, Anne Kjerulf and Brian Kristensen were involved in the epidemiology analysis. Anette M Hammerum drafted the manuscript, incorporated comments, additions and feedback throughout the revision.
References
- 1. Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, et al. Beta-lactamase database (BLDB) - structure and function. J Enzyme Inhib Med Chem. 2017;32(1):917-9. 10.1080/14756366.2017.1344235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oteo J, Hernández JM, Espasa M, Fleites A, Sáez D, Bautista V, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother. 2013;68(2):317-21. 10.1093/jac/dks383 [DOI] [PubMed] [Google Scholar]
- 3. Findlay J, Hopkins KL, Loy R, Doumith M, Meunier D, Hill R, et al. OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J Antimicrob Chemother. 2017;72(5):1340-9. 10.1093/jac/dkx012 [DOI] [PubMed] [Google Scholar]
- 4. Hoyos-Mallecot Y, Naas T, Bonnin RA, Patino R, Glaser P, Fortineau N, et al. OXA-244-producing Escherichia coli isolates, a challenge for clinical microbiology laboratories. Antimicrob Agents Chemother. 2017;61(9):e00818-17. 10.1128/AAC.00818-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valenza G, Nickel S, Pfeifer Y, Eller C, Krupa E, Lehner-Reindl V, et al. Extended-spectrum-β-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob Agents Chemother. 2014;58(2):1228-30. 10.1128/AAC.01993-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Hattem JM, Arcilla MS, Bootsma MC, van Genderen PJ, Goorhuis A, Grobusch MP, et al. Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol. 2016;11(7):857-64. 10.2217/fmb.16.18 [DOI] [PubMed] [Google Scholar]
- 7. Tafoukt R, Touati A, Leangapichart T, Bakour S, Rolain J-M. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017;120:185-9. 10.1016/j.watres.2017.04.073 [DOI] [PubMed] [Google Scholar]
- 8. Diab M, Hamze M, Bonnet R, Saras E, Madec J-Y, Haenni M. Extended-spectrum beta-lactamase (ESBL)- and carbapenemase-producing Enterobacteriaceae in water sources in Lebanon. Vet Microbiol. 2018;217:97-103. 10.1016/j.vetmic.2018.03.007 [DOI] [PubMed] [Google Scholar]
- 9. Potron A, Poirel L, Dortet L, Nordmann P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. Int J Antimicrob Agents. 2016;47(1):102-3. 10.1016/j.ijantimicag.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control (ECDC). Rapid risk assessment: increase in OXA-244-producing Escherichia coli in the European Union/European Economic Area and the UK since 2013. Stockholm: ECDC; 2020. Available from: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-increase-oxa-244-producing-escherichia-coli-eu-eea
- 11.National Food Institute, Technical University of Denmark, Statens Serum Institut. DANMAP 2018 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Lyngby, Copenhagen: National Food Institute, Statens Serum Institut; 2019.Available from: https://www.danmap.org/-/media/arkiv/projekt-sites/danmap/danmap-reports/danmap-2018/danmap_2018.pdf?la=en
- 12.Sundhedsstyrelsen. [Danish Health Authority]. Vejledning om forebyggelse af spredning af CPO. [Guidance on preventing the spread of CPO]. Copenhagen: Sundhedsstyrelsen; 2018. Danish. Available from: https://www.sst.dk/-/media/Udgivelser/2018/CPO/Vejledning-om-forebyggelse-af-spredning-af-CPO.ashx?la=da&hash=060943943A71EA7E2AA6B51C229577B87E5937A3
- 13.Wang M, Hansen DS, Littauer P, Schumacher H, Hammerum A. Undersøgelse for Carbapenemase Producerende Organismer (CPO) bærertilstand - en metodevejledning. [Carbapenemase Producing Organism (CPO) Carrier Condition Study - A Method Guide]. Copenhagen: Dansk Selskab for Klinisk Mikrobiologi [Danish Society for Clinical Microbiology]; 2016. Danish. Available from: https://dskm.dk/wp-content/uploads/2016/10/Unders%c3%b8gelse-for-Carbapenemase-Producerende-Organismer-okt-2016.pdf
- 14.European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance version 1.0. Växjö: EUCAST; 2013. Available from: http://www.eucast.org/resistance_mechanisms/
- 15.European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance version 2.0. Växjö: EUCAST; 2017. Available from: http://www.eucast.org/resistance_mechanisms/
- 16. Schmidt M, Schmidt SAJ, Sandegaard JL, Ehrenstein V, Pedersen L, Sørensen HT. The Danish National Patient Registry: a review of content, data quality, and research potential. Clin Epidemiol. 2015;7:449-90. 10.2147/CLEP.S91125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640-4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136-51. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics. 2008;9(1):560. 10.1186/1471-2164-9-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lázaro-Perona F, Ramos JC, Sotillo A, Mingorance J, García-Rodríguez J, Ruiz-Carrascoso G, et al. Intestinal persistence of a plasmid harbouring the OXA-48 carbapenemase gene after hospital discharge. J Hosp Infect. 2019;101(2):175-8. 10.1016/j.jhin.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 21. Nigg A, Brilhante M, Dazio V, Clément M, Collaud A, Gobeli Brawand S, et al. Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018. Euro Surveill. 2019;24(39):1900071. 10.2807/1560-7917.ES.2019.24.39.1900071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. 2018;24(12):1241-50. 10.1016/j.cmi.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 23. Grönthal T, Österblad M, Eklund M, Jalava J, Nykäsenoja S, Pekkanen K, et al. Sharing more than friendship - transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 2018;23(27):1700497. 10.2807/1560-7917.ES.2018.23.27.1700497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tarlton NJ, Moritz C, Adams-Sapper S, Riley LW. Genotypic analysis of uropathogenic Escherichia coli to understand factors that impact the prevalence of β-lactam-resistant urinary tract infections in a community. J Glob Antimicrob Resist. 2019;19:173-80. 10.1016/j.jgar.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hornsey M, Betts JW, Mehat JW, Wareham DW, van Vliet AHM, Woodward MJ, et al. Characterization of a colistin-resistant Avian Pathogenic Escherichia coli ST69 isolate recovered from a broiler chicken in Germany. J Med Microbiol. 2019;68(1):111-4. 10.1099/jmm.0.000882 [DOI] [PubMed] [Google Scholar]
- 26. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32(3):e00135-18. 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu L, Wang P, Cheng J, Qin S, Xie W. Characterization of a novel bla NDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect Drug Resist. 2019;12:511-9. 10.2147/IDR.S192998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang P, Xie Y, Feng P, Zong Z. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58(12):7548-52. 10.1128/AAC.03911-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeng X, Chi X, Ho BT, Moon D, Lambert C, Hall RJ, et al. Comparative genome analysis of an extensively drug-resistant isolate of avian sequence type 167 Escherichia coli strain Sanji with novel In Silico serotype O89b:H9. mSystems. 2019;4(1):e00242-18. 10.1128/mSystems.00242-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Atterby C, Börjesson S, Ny S, Järhult JD, Byfors S, Bonnedahl J. ESBL-producing Escherichia coli in Swedish gulls-A case of environmental pollution from humans? PLoS One. 2017;12(12):e0190380. 10.1371/journal.pone.0190380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freitag C, Michael GB, Kadlec K, Hassel M, Schwarz S. Detection of plasmid-borne extended-spectrum β-lactamase (ESBL) genes in Escherichia coli isolates from bovine mastitis. Vet Microbiol. 2017;200:151-6. 10.1016/j.vetmic.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 32. Zogg AL, Zurfluh K, Schmitt S, Nüesch-Inderbinen M, Stephan R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Vet Microbiol. 2018;216:79-84. 10.1016/j.vetmic.2018.02.011 [DOI] [PubMed] [Google Scholar]
- 33. Turton JF, Doumith M, Hopkins KL, Perry C, Meunier D, Woodford N. Clonal expansion of Escherichia coli ST38 carrying a chromosomally integrated OXA-48 carbapenemase gene. J Med Microbiol. 2016;65(6):538-46. 10.1099/jmm.0.000248 [DOI] [PubMed] [Google Scholar]
- 34. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1):e00102-19. 10.1128/CMR.00102-19 [DOI] [PMC free article] [PubMed] [Google Scholar]