Abstract
Objective:
Given the potential of real-world evidence (RWE) to inform understanding of the risk–benefit profile of next-generation sequencing (NGS)–based testing, we undertook a study to describe the current landscape of whether and how payers use RWE as part of their coverage decision making and potential solutions for overcoming barriers.
Methods:
We performed a scoping literature review of existing RWE evidentiary frameworks for evaluating new technologies and identified barriers to clinical integration and evidence gaps for NGS. We synthesized findings as potential solutions for improving the relevance and utility of RWE for payer decision-making.
Results:
Payers require evidence of clinical utility to inform coverage decisions, yet we found a relatively small number of published RWE studies, and these are predominately focused on oncology, pharmacogenomics, and perinatal/pediatric testing. We identified 3 categories of innovation that may help address the current undersupply of RWE studies for NGS: (1) increasing use of RWE to inform outcomes-based contracting for new technologies, (2) precision medicine initiatives that integrate clinical and genomic data and enable data sharing, and (3) Food and Drug Administration reforms to encourage the use of RWE. Potential solutions include development of data and evidence review standards, payer engagement in RWE study design, use of incentives and partnerships to lower the barriers to RWE generation, education of payers and providers concerning the use of RWE and NGS, and frameworks for conducting outcomes-based contracting for NGS.
Conclusions:
We provide numerous suggestions to overcome the data, methodologic, infrastructure, and policy challenges constraining greater integration of RWE in assessments of NGS.
Keywords: coverage policies, decision making, next-generation sequencing, payers, real-world data, real-world evidence, reimbursement
Introduction
Next-generation sequencing (NGS)–based tests (multigene panels, whole-exome sequencing, whole-genome sequencing) have started to transform the clinical approach to prenatal testing, cancer treatment, diagnosis of rare disorders, and predisposition testing for chronic diseases.1–4 Nevetheless, 2 interrelated factors—inadequate evidence base and lack of coverage by public and private payers—are issues that must be addressed before NGS becomes part of routine care. Payer coverage policies often highlight evidence deficiencies in the clinical validity of the test (result is clinically meaningful) or the clinical utility of the test (result is clinically useful) as reasons for denying coverage. Clinical utility is the evidentiary standard used by most payers when evaluating tests for coverage decision making, a standard that has been affirmed for Medicare by the courts in their determination that the Centers for Medicare and Medicaid services may consider health outcomes and patient management when deciding whether to cover a diagnostic test.5
From a regulatory perspective in the United States, NGS tests are governed by the Food and Drug Administration’s (FDA’s) Center for Devices and Radiological Health’s evidence requirements for in vitro diagnostics (IVDs), which focuses on the demonstration of analytic validity (technical efficacy of the test) and clinical validity. The absence of regulatory requirements for premarket evidence of clinical utility and complexity of NGS (tests examine multiple genes and produce multiple results, each with distinct clinical implications) makes conventional randomized controlled trials (RCTs) more challenging and expensive to conduct. Also, the clinical actionability of NGS results can evolve over time as new information regarding the relationship between genetic variants and disease risk or drug response becomes available. Thus, observational data may be particularly useful for informing estimates of the effectiveness of testing in different disorders, patient subgroups, and clinical settings.
Although payers may prefer direct evidence of clinical utility from randomized trials of the impact of NGS tests on provider behavior and patient health outcomes, they frequently must rely on indirect evidence when addressing coverage determinations. In the context of IVDs, indirect evidence is obtained by extrapolating from robust clinical validity studies and treatment outcome studies, constructing a chain of evidence linking test results to patient health outcomes.6 Use of real-world data (RWD) from registries, surveys, and observational studies is a practical alternative for building indirect evidence of the clinical impact of NGS tests, also referred to as real-world evidence (RWE; Box 1).
BOX 1.
Real world Data (RWD):
data relating to patient health states and/or delivery of health care routinely collected from a variety of sources, including electronic health records, claims and billing data, product and disease registries and data gathered through personal devices and health applications.7
Real world evidence (RWE):
the analysis of RWD in a study designed with a high degree of pragmatism, regardless of study type.7
Importantly, the FDA has recognized that RWE can be used as valid scientific evidence to support regulatory claims for new devices. For example, RWE can be used to support expanded indications, conduct postmarketing surveillance, as a control group, and as evidence to identify, demonstrate, or support the clinical validity of a biomarker.8 Although decision making for regulatory approval of a new test is distinct from payer coverage determinations regarding the test, improvements in the quality of RWD and usefulness of RWE can be seen as “a rising tide that lifts all boats,” such that there should be a downstream benefit to payer decision making.9 One caveat is that the FDA can choose to exercise enforcement discretion for IVDs, allowing laboratory-developed tests to enter the market without FDA approval. Nevertheless, there are ongoing efforts to modernize federal oversight of laboratory-developed tests based on a risk-based approach10 in addition to recent evidence that the FDA is stepping up its regulation of pharmacogenetic testing (PGx) in particular.11 Our objective was to describe the challenges and opportunities for payers, researchers, and test developers to capitalize on the growing availability and applicability of RWE for coverage decisions and develop potential solutions to support greater use of RWE in the context of NGS.
Previous studies12–15 focused on pharmaceuticals have outlined the relevance of RWE for payers; however, there is a similar and growing interest regarding how RWE may be used to demonstrate the clinical utility of IVDs, a critical evidentiary threshold for payers. The analytic framework for evaluating the clinical utility of IVDs is well established16,17 and makes clear that tests must first have demonstrated analytic and clinical validity before clinical utility can be considered. Once the relationship between test use in a defined patient group changes in provider and patient behavior, and health outcomes are established, RWD from observational studies are frequently used to provide indirect evidence of clinical utility. RWD is also essential in the development of decision-analytic models to evaluate the cost-effectiveness of NGS tests,18 information that is often relevant for private payer coverage determinations. RWD collection may also occur as part of a coverage with evidence development study or as part of outcomes-based contract evaluations.12,19 The former refers to a situation in which a payer provides provisional coverage of certain items or services conditional on further collection of population-level evidence from a prespecified study20 and the latter to contracts intended to lessen the financial risks to payers for expensive treatments by measuring the actual value delivered to patients.19,21
Nevetheless, whether and how payers use RWE for NGS coverage decision making is not well understood, and diagnostics present unique methodological challenges for the use of RWE.22 In particular, both evidence developers and decision makers have underscored the need to adapt evidentiary frameworks and the use of RWE for NGS-based tests in clinical areas such as oncology, an area of intensive activity for NGS.23,24 There is a similar need to understand the current landscape of how RWD is being used to elucidate the risk–benefit profile of NGS in a broader range of clinical contexts and what factors can accelerate greater uptake of RWE by payers. We briefly describe approaches that have been used to support the use of RWE in general by payers, then focus on identifying trends in the literature, policy environment, and data ecosystem that could affect the use of RWE for coverage decision making for NGS specifically. This information fills a critical gap in the RWE literature and forms the basis of potential solutions for enabling the evidence-based use of NGS by payers over time. The development of potential solutions that can be endorsed and applied by NGS stakeholders is the focus of this study.
Methods
Scoping Review
We undertook a scoping review as opposed to a systematic review to rapidly map key concepts underlying this research topic, identify evidence gaps, and synthesize knowledge within policy and practice contexts.25,26 Scoping reviews are particularly useful for examining the extent of research activity in a particular area and determining the value of undertaking a full systematic review in the future. We limited our review to the United States because of significant variability internationally in how healthcare is financed and new technologies are evaluated for approval and payment. For this review, PubMed was used to search peer-reviewed, scientific literature from January 2013 to November 2019 in an attempt to answer 3 research questions: (1) How has RWE been used to support public and private payer decision making in the United States? (2) What are specific examples of RWE uses related to NGS? and (3) How has RWE been used to support decision making by stakeholders that influence payers? (eg, FDA, clinical guideline developers, health technology assessment). The search strategy and inclusion and exclusion criteria are described in Appendix 1 in Supplemental Materials found at https://doi.org/10.1016/j.jval.2020.02.001.
To locate gray literature, we searched Google and GenomeWeb websites as of November 2019 using the following keywords: “Real World Evidence AND Coverage,” “Real World Evidence AND Reimbursement,” “Real World Evidence AND NGS,” “Real World Evidence AND payer,” and “real world evidence” as exact phrases. We also searched Google for gray literature on oncology data-sharing initiatives referenced in articles (see Appendix 1 in the Supplemental Materials found at https://doi.org/10.1016/j.jval.2020.02.001) using the terms “initiative name” and “coverage or reimbursement,” “clinical utility,” “performance-based risk sharing arrangement,” and “payers.” In addition, we reviewed the press releases on company websites known to be sponsors of RWE initiatives (see Appendix 1 in the Supplemental Materials found at https://doi.org/10.1016/j.jval.2020.02.001) to identify additional gray literature. Data abstraction forms were created in Microsoft Excel and used to summarize information from the included peer-reviewed and gray literature articles (see Appendix 1 in the Supplemental Materials found at https://doi.org/10.1016/j.jval.2020.02.001). Two of the authors (P.A.D. and M.P.D.) extracted data independently from the selected documents; the findings were reviewed jointly with discrepancies resolved by consensus.
Finally, we hand searched the National Human Genome Research Institute’s list of accomplishments in genomic medicine since 201127 and the peer-reviewed and gray literature reference lists of included articles. Search results from scientific (title and abstract) and gray literature were independently screened by 2 authors according to inclusion and exclusion criteria (see Appendix 1 in the Supplemental Material found at https://doi.org/10.1016/j.jval.2020.02.001). Discrepancies were resolved by consensus and review by a third author. Data from final versions of the abstraction forms were synthesized into themes organized by research question and evidence gaps highlighted.
Results
Literature Review
The PubMed search yielded a total of 426 articles, whereas the various gray literature searches produced 183 hits. From the total of 609 articles or hits, we excluded 529 articles based on the following reasons (see Fig. 1): title/abstract review (509), international studies (22), and background studies (19). Hand searching of manuscript reference lists added an additional 53 articles for a total of 69 peer-reviewed studies and 43 gray literature articles included for analysis.
Figure 1.
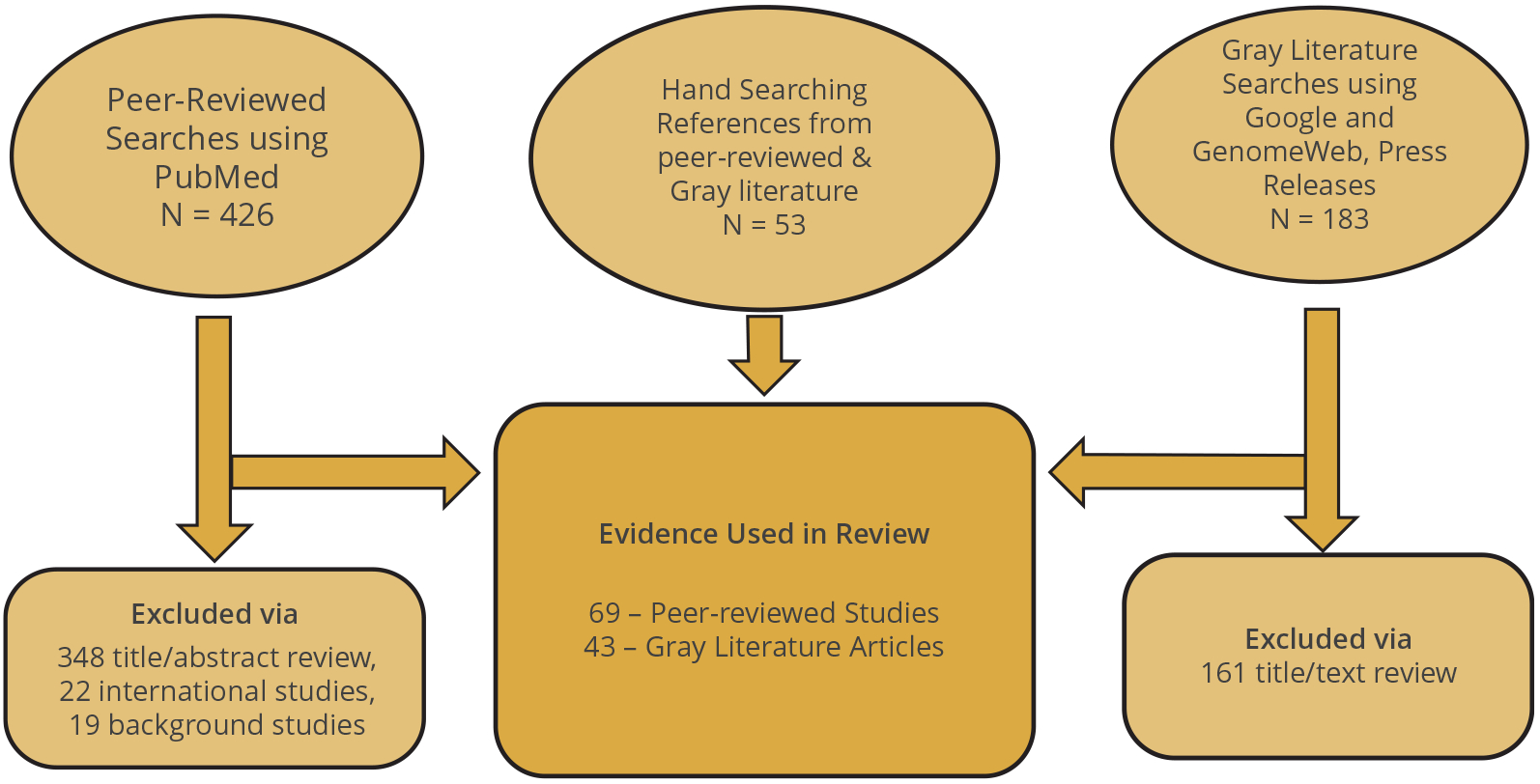
Flow diagram of included and excluded articles and sources.
Many articles touted the potential uses of RWE by payers; however, the vast majority of the published literature focused on drugs, not diagnostics.12,13,28–30 When researchers critically evaluated how payers used RWE for drug decision-making, RWE was infrequently cited in pharmacy and therapeutics materials, even among therapeutic class reviews, in which RWE is more readily available and studies are of high quality.14 One reason why RWE has not had a bigger impact is that payers lack confidence in the rigor of study designs and the validity of study conclusions (Table 1). This concern by payers exists despite numerous guidance documents published by researchers and other stakeholders regarding good research practices for conducting and reporting RWE studies.31–38
Table 1.
RWE limitations.
| Payer concerns about RWE studies |
|---|
|
HIPAA indicates Health Insurance Portability and Accountability Act; RWD, real-world data; RWE, real-world evidence.
The situation becomes more complicated with respect to NGS because researchers attempt to capture probabilistic information as binary results, false-positives/-negatives are not addressed, testing and reporting standards are in flux, data silos prevent data sharing, reclassification of variants is common,39 and clinical outcomes data are lacking.40,41 A major hurdle for clinical laboratories is deciding which genes have sufficient evidence to support use in clinical care. A method to evaluate the strength of evidence for a gene’s role in a given disease has been developed by ClinGen,42 which oversees the only FDA-recognized public variant database to support clinical validity claims for genetic tests.43
We identified several RWE studies that evaluated the use of NGS to guide oncology therapy in clinical practice44 and the impact of NGS tumor profiling on the health outcomes45,46 and economic outcomes47–49 of cancer patients. Although tumor profiling using NGS has been gaining coverage if the test has received FDA approval50 or there are clinical guidelines supporting test use,51 there is widespread recognition of the need for RWE to drive new frameworks for payer coverage policy development.52 These frameworks include coverage with evidence development and real-world performance-based risk-sharing arrangements (also referred to as “outcomes-based contracts”). There is also a growing body of studies evaluating the clinical utility of NGS for inherited conditions such as hereditary breast and ovarian cancer, familial hypercholesterolemia (FH), and Lynch syndrome (increased risk for colorectal, endometrial, and ovarian cancers), which have resulted in the Centers for Disease Control and Prevention’s assigning these conditions “tier 1” status.53 This designation refers to conditions for which there is some reasonable evidence supporting implementation such as a clinical guideline based on systematic reviews, Centers for Medicare & Medicaid Services (CMS) coverage of testing, or FDA labeling.54 Most public and private payers cover testing for hereditary breast and ovarian cancer and Lynch syndrome55; however, coverage for FH is highly variable despite recognition by professional associations of the clinical utility of genetic testing. One of the reasons cited for this coverage gap is the lack of cost-effectiveness data supporting FH testing,56 which requires RWD to populate economic models.
Pharmacogenomics previously focused on the analysis of variants in a single gene to predict drug response; however, there has recently been much more emphasis on the analysis of multiple pharmacogenes using NGS panels, ideally in a preemptive fashion before the prescription of any targeted drug.57 In this scenario, pharmacogenomic results are stored in the electronic health record along with a clinical decision support (CDS) system that alerts the prescriber when an affected drug is prescribed for a patient with variant genetics. The clinical and economic impacts of implementing a preemptive PGx strategy for antiplatelet agents, statins, and warfarin has been demonstrated through modeling and application to a health system cohort.58 The improvements were only modest and are consistent with the observation that payers do not reimburse for preemptive PGx panel testing. The strongest evidence from RWE concerns the relationship between variants in CYP2C19 and antiplatelet therapy such as clopidogrel,59 HLA B and carbamazepine,60 and TMTP genotypes and thiopurines,61 findings further confirmed by a review of 44 economic evaluations of a PGx-informed strategy.62 Nevertheless, lack of reimbursement of PGx testing remains a major barrier for the field and appears less related to lack of RWE and more related to implementation barriers, for example, the inability to represent genomic data in the electronic health record and the need to update CDS recommendations to reflect changes in variant interpretation.63
Some of the most definitive work using RWE has examined the clinical utility and cost-effectiveness of NGS in critically ill infants and pediatric patients with suspected monogenic disorders.64–68 Correspondingly, a recent qualitative study of payer decision-making revealed that 71% of payers representing 170 million insured lives cover pediatric exome sequencing, primarily because there are available interventions or to end the diagnostic odyssey.69 In the specific context of neurodevelopmental disorders in pediatric patients, a study of private payer coverage policies of whole-exome sequencing demonstrated a trend toward more favorable coverage decisions over time, correlated with a larger evidence base, including RWE studies.70 Another study demonstrated that payers relied on modeled evidence of clinical utility when affirmatively covering noninvasive prenatal testing.1 Finally a review of 55 coverage policies for NGS tests compared with coverage policies for other interventions such as drugs or diagnostic imaging revealed that most clinical studies cited as supporting evidence relied on RCTs; however, some policies did cite health technology assessments and cost-effectiveness assessments, which presumably included RWE.71 Notably, NGS tests had a weaker evidence base than other technologies, confirming the perception that there is an undersupply of clinical utility data for this type of intervention.
There are also studies documenting the growing importance of RWD in the setting of outcomes-based contracting (OBC) for new technologies, primarily concerning drugs.19,21,72 In addition, there are incentives in bundled care and value-based payment reimbursements mandated by federal legislation73 that are based on outcomes best assessed using RWD. This emphasis on value-based payments is a critical factor reshaping how RWE is used for payer decision-making. The limitation is that the results of OBCs are rarely disclosed publicly. The only publicly available description of an OBC related to NGS is the example of Illumina working with Harvard Pilgrim to offer noninvasive prenatal testing to women at average risk while also committing to third-party evaluation of the impact of risk sharing on clinical outcomes and costs and publication of the results. The investigators demonstrated an increase in noninvasive prenatal testing use, modest increases in total testing and diagnostic expenditures, and a decrease in invasive procedures compared with the baseline year when testing was covered only for high-risk pregnancies.74
Numerous data-sharing initiatives have been organized to overcome the data limitations inhibiting the assessment of the real-world impact of NGS. To date, many of these initiatives are specific to oncology, but there are a growing number of population and public health–focused efforts (Table 2).45,75–96 These information networks have enabled greater use of RWD for various types of NGS evaluations, including studies of clinical and economic impact that have the potential to be useful to payers.45,47,48,76,77 Nevertheless, we were unable to find publicly available evidence that these study results have informed positive or negative coverage decisions to date, perhaps because of their relatively recent formation. Similarly, there are no published examples of coverage with evidence development for NGS, although this strategy is frequently described as a valuable option for payers and manufacturers when there is uncertainty regarding clinical utility.
Table 2.
Illustrative US-based RWD networks including NGS.
| Cancer related | Population based |
|---|---|
|
|
NGS indicates next-generation sequencing; RWD, real-world data.
NGS testing currently in planning phase.
A residual data barrier to developing RWE for NGS is the fact that genomic data are not represented in a structured format in the electronic health record (EHR), and test billing codes are too nonspecific to be able to rely solely on claims data.97,98 To facilitate the use of RWD and overcome the need for manual curation, artificial intelligence–based methods such as natural language processing, machine learning, and deep learning are increasingly being applied to process and analyze unstructured data from the EHR and patient-generated data.99 Critical to the success of these methods will be transparency regarding data sources and analytic methods so that payers will understand and trust the results. In addition, systematic analyses of NGS implementation efforts such as the IGNITE (Implementing Genomics in Pratice) network demonstrate that sustainability drivers include infrastructure (EHR, CDS, laboratories, manufacturers, community), evidence of clinical effectiveness, economic measures, workforce and workflow impact, provider and patient education, regulatory/legal updates specific to NGS, and stakeholder engagement in research. After a priority-setting exercise, the top 3 sustainability constructs were provider education, availability of genomic-focused CDS/EHR tools, and reimbursement.100 Addressing these constructs is necessary to ensure the quality and availability of RWD and RWE.
Finally, there are several policy developments that are also endorsing greater use of RWE by stakeholders that influence payers. In 2017, the Center for Devices and Radiologic Health and the Center for Biologics Evaluation and Research at the FDA issued a guidance document describing the use of RWE to support regulatory decision making for medical devices.8 The goal is to incentivize the creation of a system for characterizing, aggregating, and analyzing data from all uses of medical devices so that innovative and accurate tests are made available to patients as efficiently as possible. Related efforts include FDA guidance on the use of public human genetic variant databases to support claims of clinical validity for genomic-based IVD and considerations for the development of evidence of analytic validity for NGS-based IVDs for suspected germline diseases.9,43,101
The FDA has also helped to establish the Medical Device Innovation Consortium (MDIC) in 2012, a public-private partnership focused solely on advancing medical device regulatory science. The MDIC engages a wide variety of stakeholders, including representatives of the FDA, National Institutes of Health, CMS, industry, nonprofits, and patient organizations to improve medical technology development and review processes. The group recently convened FDA, industry, and payer representatives to build a framework to help IVD manufacturers develop credible evidence of analytical and clinical validity and clinical utility. The section on clinical utility specifically describes the use of RWD as a source of evidence that can be used to support regulatory and reimbursement decision-making.6 The FDA has also provided funding to establish the The National Evaluation System for health Technology Coordinating Center (https://nestcc.org/about/faqs/), a coordinating center for a voluntary network of data partners that act as a national evaluation system for devices. This network emphasizes the use of RWE in the evaluation of its test cases, which include an IVD test panel for lung cancer focused on developing evidence of clinical utility.102,103 Although all of these efforts are primarily focused on improving regulatory and clinical decision making about device access and safety, the relevance to payer decision making is that RWE studies that meet robust methodological standards in the regulatory science arena should also prove useful to payers in as much as they include information about clinical utility.
Patient advocacy groups,104 professional societies,105 and nonprofit health and research agencies106,107 have all become engaged in developing more comprehensive and accurate RWD to support decision makers, including patients and families. The expectation is that over time, patients will become key drivers of use of RWE by payers, particularly as there is greater availability of patient-generated data as part of clinical care.
The scoping review revealed a number of evidence gaps for use of RWE in coverage decision-making for NGS. First, most studies are in oncology, PGx, and diagnosis of suspected genetic disorders in the perinatal or pediatric period, rather than the full spectrum of clinical genomic applications. Second, a wide variety of methods are used to conduct RWE studies, and we could find no examples in which investigators cited adherence to methodologic guidance documents or best practices for RWE studies. Third, published analyses of payer coverage policies do not distinguish RWE from RCT data when evaluating the relationship between clinical studies and coverage determinations. Finally, it is difficult to describe how RWE specifically can inform payer decision making based on publicly available data without an in-depth review of coverage policies and their evolution over time.
Potential Solutions
We developed potential solutions (Table 3) for overcoming the challenges limiting payer uptake of RWE in the setting of NGS based on our team’s experience and findings from the scoping review. The suggestions spanned a spectrum, from the importance of following RWE methodological best practices, to building transparency and relevance of study methods, to collaborating with existing expert groups addressing issues of data standardization, data sharing, and assessment of value.
Table 3.
Potential solutions for advancing use of RWE.
| Area of focus | Challenges | Opportunities | Potential solutions |
|---|---|---|---|
| I. Relevance and rigor of RWE for payers |
|
|
|
| II. Incentives for RWE development |
|
|
|
| III. Educational needs regarding both RWE and NGS |
|
|
|
| IV. Standards for testing and reporting NGS data |
|
|
|
| V. Standards for genomic data representation in EHR |
|
|
|
| VI. Standards for NGS evidence review by payers |
|
|
|
| VII. Partnerships |
|
|
|
| VIII. Role of RWD in support of OBCs for NGS |
|
|
|
| IX. Narrow definition of how value of NGS is measured |
|
|
|
CDS indicates clinical decision support; CMS, Centers for Medicare & Medicaid Services; EHR, electronic health record; FDA, US Food and Drug Administration; MDIC, Medical Device Innovation Consortium; NGS, next-generation sequencing; OBC, outcomes-based contracting; PGx, pharmacogenetic testing; RWD, real-world data; RWE, real-world evidence.
Health Information Technology for Economic and Clinical Health, Medicare Access and CHIP Reauthorization Act of 2015.
College of American Pathology, Association for Molecular Pathology, American College of Genetics and Genomics.
International Society for Pharmacoeconomics and Outcomes Research.
The first 3 solutions address the need to develop processes to promote the relevance and rigor of RWE for payers. Payers must be able to trust study findings and understand how to apply the results in coverage decision-making. Efforts at payer engagement and tailoring existing RWE best practices and evaluation tools to NGS are best pursued as part of multistakeholder initiatives that are already working to advance use of RWE with payers generally. The next 4 potential solutions (4–7) target the lack of incentives for test developers to conduct RWE studies and undersupply of published research. There is an increasing number of curated data networks and integrated healthcare delivery systems focused on genomics that can facilitate robust RWE studies and initiatives supported by the federal government to encourage greater use of RWE in policy decisions, including coverage. Progress could be accelerated if stakeholders advocated for including genomic data in meaningful use requirements. The next 2 solutions (8 and 9) tackle the widespread problem that payers lack the requisite skills to review and apply RWE studies in their local context but through the lens of NGS-related decisions.
There is widespread recognition that a lack of standards is hampering development of clinical utility; these include standards for testing and reporting NGS data, representing clinic-genomic data in the HER and payer requirements for NGS evidence review. There are 7 potential solutions (10–16) that address these interrelated gaps; however, they vary in the level of effort that will be required to advance these tactics. Until these issues are solved in a scalable way, it will be very difficult for researchers and test developers to capitalize on RWD sources. These solutions also address gaps in capturing the necessary clinical, digital, and patient-reported data to conduct RWE studies by focusing on artificial intelligence–based methods to reduce the need for manual curation.
The need for public–private partnerships is addressed by the next 2 solutions (17 and 18), as pooling of infrastructure resources, patient populations, and expertise will be required if the clinical utility of NGS will be demonstrated for genomic conditions, many of which are relatively rare. The next 2 potential solutions (19 and 20) take on the need to overcome barriers to using RWD to support OBCs for NGS, recognizing that although all of the obstacles are not specific to NGS, the remedies should be targeted to how OBCs can be structured and evaluated for NGS specifically. Finally, the last 3 solutions (21–23) focus on the importance of including the patient and other relevant perspectives in value-based frameworks focused on NGS. It is also important to note that for most of these proposed solutions, the policy and data environments are in flux; therefore, each effort will need to be evaluated and recalibrated in response to changing trends.
Discussion
To our knowledge, this is the first article to directly examine payer use of RWE for NGS and what new approaches are needed. Although the topic of RWE as a valuable source of information for clinical, regulatory, and payer decision makers has been addressed by numerous authors and policy makers,7,8,12–15,30,40,104,105,108–116 we explored whether and how RWE could play a similar role for NGS specifically in the context of coverage decisions. In the scoping review, we identified numerous examples of how RWE was likely used in oncology, PGx, and pediatric or perinatal settings to bridge the existing evidence gaps for payers, as there is concurrent evidence of increasingly positive coverage decisions for NGS tests in these clinical contexts. These applications included developing empiric evidence of current test and treatment use patterns, assessing real-world cost implications, demonstrating the incremental value of testing additional genes when compared with standard-of-care single-gene tests, supplementing RCT data, creating efficiencies and greater certainty in coverage policy development, and enabling OBC.
Nevertheless, it appears that payers still primarily rely on clinical guidelines and RCTs as opposed to RWE as the type of evidence cited as justification for their decisions. The reasons for this disconnect are multifactorial, including challenges related to RWD quality and comprehensiveness, difficulties representing genomic data in a standardized manner in the EHR, and lack of payer engagement in study development, resulting in results that are not relevant for coverage decision-making. Payers also lack familiarity with RWE study methods and continue to prefer RCT data over observational data. They also have concerns about the lack of transparency regarding data sources and analytic methods, resulting in a lack of trust, particularly regarding studies conducted by industry. Generally, payers are not sure how to use RWE within their current coverage processes and do not fully understand all the relevant questions that RWE could answer. Nevertheless, there is a confluence of policy, technology, and data infrastructure trends that are likely to enable greater use of RWE for NGS, for example, greater receptivity to RWE by the FDA, artificial intelligence–based methods to facilitate data analyses, and a proliferation of data networks built and curated to support RWD studies involving the use of NGS in people with and without known disease.
The data and infrastructure-related barriers to conducting RWE studies for NGS are being addressed by healthcare providers, federal research funders, and private companies. Currently, there is a growing number of learning healthcare systems focused on implementing and evaluating population-based NGS and numerous precision medicine programs focused on oncology.45,75–82 Each of these organizations has a vested interest in developing the data infrastructure to support collection of high-quality RWD and to publicize the results of RWE studies. By engaging payers in the design of these studies, there is a real opportunity to provide RWE that can inform coverage decision making in an efficient and timely manner. Nevertheless, to ensure that these studies have the desired impact, researchers must follow methodologic best practices for conducting and reporting RWD studies. Fortunately, there are many existing consensus statements from expert groups to guide researcher efforts and reassure payers that the study results are valid. What is missing is agreement about which guidelines are best to follow for NGS. Similarly, existing tools for evaluating the quality and applicability of RWE need to be tailored to NGS for payers to more consistently evaluate these studies. Both efforts will require multistakeholder groups to discuss options, explicitly address conflicts of interest, and agree on a path forward for using RWE to inform payers and other decision makers about the clinical utility of NGS. Although conflicts of interest are ubiquitous, agreeing how these conflicts will be identified and managed is possible, as demonstrated by groups such as the MDIC, a public–private partnership working to promote patient access to innovative medical technologies such as NGS.
Lack of standards is a major stumbling block for using RWE to evaluate the clinical utility of NGS. This refers not only to a lack of standards for analyzing and reporting NGS results, and a lack of standards for representing genomic data in the EHR, but also to a lack of standards for payer assessment of clinical utility data for NGS. Although individual labs or health systems may attempt to reduce variation in their NGS testing practices or NGS-related CDS tools, the definitive solutions require multistakeholder groups to ensure interoperability and efficiency as patients move from one clinical setting to another. Similarly, multistakeholder groups need to define the methodological standards for developing clinical utility evidence for NGS that will address payer requirements for coverage decision-making. Although this has been successfully done for molecular diagnostics in oncology,117 RWE has the potential to address the lack of evidence of clinical utility of NGS, provided that payers agree on how RWE can be used for coverage determinations. Although affirmative coverage decisions are necessary, they are not sufficient to ensure clinical uptake of NGS unless there is simultaneous attention to NGS implementation requirements.
There are also reimbursement-focused enablers that will require access to robust RWD, such as the growing use of OBCs between manufacturers and payers to ensure realization of promised benefits of new interventions. Nevertheless, there are significant barriers to implementing OBCs, and many preliminary discussions fail to result in actual contracts. These challenges include difficulties obtaining accurate data, lack of outcome measures, concerns regarding patient data privacy, and costs of data collection. There is an opportunity for an honest broker to convene payers and manufacturers to develop a framework to assess the suitability/desirability of OBCs for NGS and also guide the negotiation and implementation processes to increase the likelihood of success. Greater transparency regarding the results of OBCs is desirable, but the proprietary nature of contract terms makes this goal unlikely.
Nonprofit groups such as the Personalized Medicine Coalition, the Innovation and Value Initiative, and patient advocacy groups are highlighting the importance of putting the patient perspective at the center of any assessment of the value of new technologies such as NGS. Development of patient-centered outcome measures would advance these efforts and address the lack of standard outcome measures limiting RWE studies and OBCs. Because each group is engaging relevant stakeholders to develop suggestions or consensus statements for a path forward, we recommend following a similar approach for NGS-specific adaptation or evaluating the feasibility of joint efforts.
There are several limitations to this study. There is no Medical Subject Headings term for either RWD or RWE118; therefore, we may have missed relevant articles. We made several efforts to overcome this limitation by searching reference lists and asking experts to share seminal articles. We did not examine specific payer coverage policies, so our conclusions regarding the impact of RWE on payer decision making is likely an underestimate of the true impact. We also limited our evaluation to US payers and data networks. We are aware that there are comparable payer information needs and data initiatives in other countries but determined these to be out of scope for our literature and policy analyses. Future studies could characterize the RWE landscape in other countries and determine whether our recommendations could be generalized.
This study assessed the opportunities and challenges surrounding use of RWE by payers to inform decision making regarding NGS and found a growing number of published RWE studies, particularly in oncology, PGx, and perinatal or pediatric genomic testing, that have played either a direct or supportive role given the state of published coverage analyses. We also identified many examples of policy and data network enablers designed to address the current undersupply of RWE for NGS. For RWE to become a game changer for payers, there must be multifaceted efforts to raise awareness of data and methods advances and publicly available examples of robust studies that are relevant for payer decision making. We present 20 potential solutions that, if pursued collaboratively with stakeholders over time, represent promising steps toward ensuring greater and more effective use of RWE by payers.
Supplementary Material
Acknowledgments
We gratefully acknowledge comments and input on earlier drafts from the Global Economics and Evaluation of Clinical Sequencing Working Group (GEECS).
Source of financial support: This study was funded by an unrestricted consulting agreement with Illumina (no number) and grants from the National Human Genome Research Institute (U01 HG009599) and from the National Cancer Institute (R01 CA221870).
Footnotes
Supplemental Material
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.jval.2020.02.001.
Conflict of interest: All authors (P.A.D., M.P.D., K.A.P.) received consulting fees from Illumina to support the research conducted for this publication. All authors (P.A.D., M.P.D., K.A.P.) received travel support from Illumina to attend a health economist work group (GEECS) meeting to obtain expert feedback on project findings.
REFERENCES
- 1.Dervan AP, Deverka PA, Trosman JR, et al. Payer decision making for next-generation sequencing-based genetic tests: insights from cell-free DNA prenatal screening. Genet Med. 2017;19:559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freedman AN, Klabunde CN, Wiant K, et al. Use of next-generation sequencing tests to guide cancer treatment: results from a nationally representative survey of oncologists in the United States. JCO Precis Oncol. 2018:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Splinter K, Adams DR, Bacino CA, et al. Effect of genetic diagnosis on patients with previously undiagnosed disease. N Engl J Med. 2018;379: 2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalayinia S, Goodarzynejad H, Maleki M, et al. Next generation sequencing applications for cardiovascular disease. Ann Med. 2018;50:91–109. [DOI] [PubMed] [Google Scholar]
- 5.Burwell Kort v. Civil No. 1:14-cv-01519 (APM). Washington, DC: United States District Court for the District of Columbia; 2016. [Google Scholar]
- 6.Medical Device Innovation Consortium. Developing clinical evidence for regulatory and coverage assessments in in vitro diagnostics (IVDs). https://mdic.org/wp-content/uploads/2019/08/Clinical-Evidence-IVD-Framework-FINAL.pdf. Accessed November12, 2019.
- 7.Jarow JP, LaVange L, Woodcock J. Multidimensional evidence generation and FDA regulatory decision making: defining and using “real-world” data. JAMA. 2017;318:703–704. [DOI] [PubMed] [Google Scholar]
- 8.Use of real-world evidence to support regulatory decision-making for medical devices: guidance for industry and food and drug administration staff. https://www.fda.gov/media/99447/download. Accessed May29, 2019.
- 9.O’Neill T, Miksad R, Miller D, et al. ISPOR, the FDA, and the evolving regulatory science of medical device products. Value Health. 2019;22:754–761. [DOI] [PubMed] [Google Scholar]
- 10.What are in vitro diagnostic tests, and how are they regulated? https://www.pewtrusts.org/en/research-and-analysis/issue-briefs/2019/05/what-are-in-vitro-diagnostic-tests-and-how-are-they-regulated. Accessed November12, 2019.
- 11.Ray T FDA stepping up actions against PGx testing, forcing some labs to stop reporting drug information. https://www.360dx.com/regulatory-news-fdaapprovals/fda-stepping-actions-against-pgx-testing-forcing-some-labs-stop#.XcrUmJpKiUk. Accessed November12, 2019.
- 12.Garrison LP Jr, Neumann PJ, Erickson P, et al. Using real-world data for coverage and payment decisions: the ISPOR Real-World Data Task Force report. Value Health. 2007;10:326–335. [DOI] [PubMed] [Google Scholar]
- 13.Hampson G, Towse A, Dreitlein WB, et al. Real world evidence for coverage decisions: opportunities and challenges. https://icer-review.org/wp-content/uploads/2018/03/ICER-Real-World-Evidence-White-Paper-03282018.pdf. Accessed May, 2019. [DOI] [PubMed]
- 14.Hurwitz JT, Brown M, Graff JS, et al. Is real-world evidence used in P&T monographs and therapeutic class reviews? J Manag Care Spec Pharm. 2017;23:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malone DC, Brown M, Hurwitz JT, et al. Real-world evidence: useful in the real world of US Payer decision making? How? When? And what studies? Value Health. 2018;21:326–333. [DOI] [PubMed] [Google Scholar]
- 16.Fryback DG, Thornbury JR. The efficacy of diagnostic imaging. Med Decis Making. 1991;11:88–94. [DOI] [PubMed] [Google Scholar]
- 17.Methods Guide for Medical Test Reviews. AHRQ Publication No. 12-EC017. www.effectivehealthcare.ahrq.gov/reports/final.cfm. Accessed November19, 2019.
- 18.Rational Integration of Clinical Sequencing (RISE), Vanderbilt University Medical Center. http://grantome.com/grant/NIH/R01-HG009694-01. Accessed November19, 2019.
- 19.Carlson JJ, Chen S, Garrison LP Jr. Performance-based risk-sharing arrangements: an updated international review. Pharmacoeconomics. 2017;35:1063–1072. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Medicare and Medicaid Services. Coverage with evidence development. https://www.cms.gov/medicare/coverage/coverage-withevidence-development/. Accessed May29, 2019.
- 21.Mahendraratnam N, Sorenson C, Richardson E, et al. Value-based arrangements may be more prevalent than assumed. Am J Manag Care. 2019;25: 70–76. [PubMed] [Google Scholar]
- 22..Shore C, Gee AW, Kahn B, et al. , eds. Examining the Impact of Real-World Evidence on Medical Product Development: Proceedings of a Workshop Series. Washington, DC: The National Academies Press; 2019. [PubMed] [Google Scholar]
- 23.Conley RB, Dickson D, Zenklusen JC, et al. Core clinical data elements for cancer genomic repositories: a multi-stakeholder consensus. Cell. 2017;171:982–986. [DOI] [PubMed] [Google Scholar]
- 24.Green Park Collaborative. Initial medical policy and model coverage guidelines for clinical next generation sequencing in oncology: report and recommendations. http://www.cmtpnet.org/docs/resources/Full_Release_Version_August_13__2015.pdf. Accessed May29, 2019.
- 25.Anderson S, Allen P, Peckham S, et al. Asking the right questions: scoping studies in the commissioning of research on the organisation and delivery of health services. Health Res Policy Syst. 2008;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arksey H, O’Malley L. Scoping studies: towards a methodological framework. Int J Soc Res Methodol. 2008;8:19–32. [Google Scholar]
- 27.National Human Genome Research Institute. Accomplishments in genomic medicine. https://www.genome.gov/health/Genomics-and-Medicine/accomplishments. Accessed November19, 2019.
- 28.Pearson SD, Dreitlein WB, Towse A, et al. A framework to guide the optimal development and use of real-world evidence for drug coverage and formulary decisions. J Comp Eff Res. 2018;7:1145–1152. [DOI] [PubMed] [Google Scholar]
- 29.Green Park Collaborative. RWE decoder framework: a practical tool for assessing relevance and rigor of real world evidence. http://www.cmtpnet.org/docs/resources/RWE_Decoder_Framework.pdf. Accessed May29, 2019. [Google Scholar]
- 30.Makady A, Ham RT, de Boer A, et al. Policies for use of real-world data in health technology assessment (HTA): a comparative study of six HTA agencies. Value Health. 2017;20:520–532. [DOI] [PubMed] [Google Scholar]
- 31.Velentgas P, Dreyer NA, Nourjah P, et al. , eds. Developing a Protocol for Observational Comparative Effectiveness Research: A User’s Guide. Rockville, MD: Agency for Healthcare Research and Quality; 2013. [PubMed] [Google Scholar]
- 32.ENCePP checklist for study protocols (revision 3). http://www.encepp.eu/encepp/openAttachment/methodologicalDeclaration/19034. Accessed May29, 2019.
- 33.US Food and Drug Administration. Good pharmacovigilance practices and pharmacoepidemiologic assessment. https://www.fda.gov/regulatoryinformation/search-fda-guidance-documents/good-pharmacovigilance-practicesand-pharmacoepidemiologic-assessment. Accessed May29, 2019.
- 34.Berger ML, Mamdani M, Atkins D, et al. Good research practices for comparative effectiveness research: defining, reporting and interpreting nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—part I. Value Health. 2009;12:1044–1052. [DOI] [PubMed] [Google Scholar]
- 35.Cox E, Martin BC, Van Staa T, et al. Good research practices for comparative effectiveness research: approaches to mitigate bias and confounding in the design of nonrandomized studies of treatment effects using secondary data sources: the International Society for Pharmacoeconomics and Outcomes Research Good Research Practices for Retrospective Database Analysis Task Force Report—part II. Value Health. 2009;12:1053–1061. [DOI] [PubMed] [Google Scholar]
- 36.Johnson ML, Crown W, Martin BC, et al. Good research practices for comparative effectiveness research: analytic methods to improve causal inference from nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—part III. Value Health. 2009;12:1062–1073. [DOI] [PubMed] [Google Scholar]
- 37.Dreyer NA, Bryant A, Velentgas P. The GRACE checklist: a validated assessment tool for high quality observational studies of comparative effectiveness. J Manag Care Spec Pharm. 2016;22:1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patient Centered Outcomes Research Insitute (PCORI). methodology standards. https://www.pcori.org/research-results/about-our-research/researchmethodology/pcori-methodology-standards. Accessed May29, 2019.
- 39.Liu P, Meng L, Normand EA, et al. Reanalysis of clinical exome sequencing data. N Engl J Med. 2019;380:2478–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickson DJ, Pfeifer JD. Real-world data in the molecular era: finding the reality in the real world. Clin Pharmacol Ther. 2016;99:186–197. [DOI] [PubMed] [Google Scholar]
- 41.Rosenman MB, Decker B, Levy KD, et al. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health. 2017;20:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strande NT, Riggs ER, Buchanan AH, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Food US and Adminstration Drug. Use of public human genetic variant databases to support clinical validity for genetic and genomic-based in vitro diagnostics. https://www.fda.gov/media/99200/download. Accessed May29, 2019.
- 44.Velcheti V, Patwardhan PD, Liu FX, et al. Real-world PD-L1 testing and distribution of PD-L1 tumor expression by immunohistochemistry assay type among patients with metastatic non-small cell lung cancer in the United States. PLoS One. 2018;13:e0206370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Presley CJ, Tang D, Soulos PR, et al. Association of broad-based genomic sequencing with survival among patients with advanced non-small cell lung cancer in the community oncology setting. JAMA. 2018;320:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321:1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haslem DS, Chakravarty I, Fulde G, et al. Precision oncology in advanced cancer patients improves overall survival with lower weekly healthcare costs. Oncotarget. 2018;9:12316–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steuten L, Goulart B, Meropol NJ, Pritchard D, Ramsey SD. Cost-effectiveness of multigene panel sequencing for patients with advanced non-small-cell lung cancer. JCO Clin Cancer Inform. 2019;3:1–10. [DOI] [PubMed] [Google Scholar]
- 49.Signorovitch J, Zhou Z, Ryan J, et al. Budget impact analysis of comprehensive genomic profiling in patients with advanced non-small cell lung cancer. J Med Econ. 2019;22:140–150. [DOI] [PubMed] [Google Scholar]
- 50.CMS and commercial insures extend coverage of Oncomine Dx Target Test to more than 160 million U.S. lives. http://thermofisher.mediaroom.com/2018-03-19-CMS-and-Commercial-Insurers-Extend-Coverage-of-Oncomine-Dx-Target-Test-to-more-than-160-Million-U-S-Lives. Accessed November19, 2019.
- 51.Lu CY, Loomer S, Ceccarelli R, et al. Insurance coverage policies for pharmacogenomic and multi-gene testing for cancer. J Pers Med. 2018;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trosman JR, Weldon CB, Gradishar WJ, et al. From the past to the present: insurer coverage frameworks for next-generation tumor sequencing. Value Health. 2018;21:1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dotson WD, Douglas MP, Kolor K, et al. Prioritizing genomic applications for action by level of evidence: a horizon-scanning method. Clin Pharmacol Ther. 2014;95:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Centers for Disease Control and Prevention. Introducing the CDC tierclassified database. https://blogs.cdc.gov/genomics/2019/07/16/introducingthe-cdc-tier/. Accessed November18, 2019.
- 55.American Society of Clinical Oncology. Genetic testing coverage & reimbursement. https://www.asco.org/practice-guidelines/cancer-care-initiatives/genetics-toolkit/genetic-testing-coverage-reimbursement. Accessed November19, 2019. [Google Scholar]
- 56.Hendricks-Sturrup RM, Lu CY. Understanding implementation challenges to genetic testing for familial hypercholesterolemia in the United States. J Pers Med. 2019;9(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y, Graves JA, Garbett SP, et al. A decision-theoretic approach to panel-based, preemptive genotyping. MDM Policy Pract. 2019;4:2381468 319864337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin J, Williams AK, Klein MD, et al. Frequency and clinical outcomes of CYP2C19 genotype-guided escalation and de-escalation of antiplatelet therapy in a real-world clinical setting. Genet Med. 2020;22(1):160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volpi S, Bult CJ, Chisholm RL, et al. Research directions in the clinical implementation of pharmacogenomics: an overview of US programs and projects. Clin Pharmacol Ther. 2018;103:778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manzi SF, Fusaro VA, Chadwick L, et al. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration: experience from a pediatric tertiary care facility. J Am Med Inform Assoc. 2017;24:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verbelen M, Weale ME, Lewis CM. Cost-effectiveness of pharmacogenetic guided treatment: are we there yet? Pharmacogenomics J. 2017;17:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet. 2019;394:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monroe GR, Frederix GW, Savelberg SM, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18:949–956. [DOI] [PubMed] [Google Scholar]
- 66.Lazaridis KN, Schahl KA, Cousin MA, et al. Outcome of whole exome sequencing for diagnostic odyssey cases of an individualized medicine clinic: the Mayo Clinic experience. Mayo Clin Proc. 2016;91:297–307. [DOI] [PubMed] [Google Scholar]
- 67.Clark MM, Stark Z, Farnaes L, et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clark MM, Hildreth A, Batalov S, et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med. 2019;11(489). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trosman JR, Weldon CB, Slavotinek A, et al. Perspectives of US private payers on insurance coverage for pediatric and prenatal exome sequencing: results of a study from the Program in Prenatal and Pediatric Genomic Sequencing (P3EGS). Genet Med. 2020;22(2):283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Douglas MP, Parker SL, Trosman JR, et al. Private payer coverage policies for exome sequencing (ES) in pediatric patients: trends over time and analysis of evidence cited. Genet Med. 2019;21:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chambers JD, Saret CJ, Anderson JE, et al. Examining evidence in U.S. payer coverage policies for multi-gene panels and sequencing tests. Int J Technol Assess Health Care. 2017;33:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duhig AM, Saha S, Smith S, et al. The current status of outcomes-based contracting for manufacturers and payers: an AMCP membership survey. J Manag Care Spec Pharm. 2018;24:410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Centers for Medicare and Medicaid Services. Medicare Access and CHIP Reauthorization Act of 2015 (MACRA). https://www.cms.gov/medicare/quality-initiatives-patient-assessment-instruments/value-based-programs/macra-mips-and-apms/macra-mips-and-apms.html. Accessed May29, 2019.
- 74.McQueen R, Schroeder B, Wright G, et al. Evaluating coverage expansion for NIPT through a perfomrance-based risk sharing agreement. Denmark: ISPOR Europe Copenhagen; 2019. [Google Scholar]
- 75.Illumina Media Relations . Illumina and Harvard Pilgrim Partner on value-based contract. https://www.illumina.com/company/news-center/featurearticles/illumina-harvard-pilgrim-health-care-partner-on-value-based-con.html. Accessed May29, 2019.
- 76.Farnaes L, Hildreth A, Sweeney NM, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 2018;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwartz MLB, McCormick CZ, Lazzeri AL, et al. A model for genome-first care: returning secondary genomic findings to participants and their healthcare providers in a large research cohort. Am J Hum Genet. 2018;103:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Renown Institute for Health Innovation. Healthy Nevada Project. https://healthynv.org/about/. Accessed May29, 2019.
- 80.Philanthropy News Digest. Sanford Health receives $25 million for precision medicine initiative. https://philanthropynewsdigest.org/news/sanfordhealth-receives-25-million-for-precision-medicine-initiative. Accessed May29, 2019.
- 81.Ray T Precision lung cancer drugs on pricing evaluation group’s agenda. https://www.genomeweb.com/cancer/precision-lung-cancer-drugs-pricingevaluation-groups-agenda. Accessed May29, 2019.
- 82.Pecora AL, Norden AD, Hervey J, et al. Development of a precise, clinically relevant, digital classification schema for cancer. JCO Clin Cancer Inform. 2018;2:1–10. [DOI] [PubMed] [Google Scholar]
- 83.Tempus Collaborations. https://www.tempus.com/collaborations/. Accessed May 2019, 2019.
- 84.The White House. Fact sheet: President Obama’s precision medicine initiative. https://obamawhitehouse.archives.gov/the-press-office/2015/01/30/fact-sheetpresident-obama-s-precision-medicine-initiative. Accessed May29, 2019.
- 85.CancerLinQ LLC. Collaborating with the American College of Medical Genetics and Genomics. https://www.asco.org/about-asco/press-center/news-releases/cancerlinq-llc-collaborating-american-college-medical-genetics. Accessed May 2019, 2019.
- 86.Foundation Medicine. Foundation Medicine Launches Precision Medicine Exchange Consortium™ (PMEC) to advance the integration of molecular information in clinical oncology and accelerate adoption of precision care. https://investors.foundationmedicine.com/news-releases/news-release-details/foundation-medicine-launches-precision-medicine-exchange. Accessed May 2019.
- 87.M2GEN. https://m2gen.com/. Accessed May29, 2019.
- 88.Information Exchange and Data Transformation (INFORMED). https://www.fda.gov/about-fda/oncology-center-excellence/information-exchange-anddata-transformation-informed. Accessed May29, 2019. [Google Scholar]
- 89.VITAL Innovation. https://vitalinnovation.com/. Accessed May29, 2019.
- 90.SetYale School of Medicine. Digging deep into data. https://medicine.yale.edu/people/about/article.aspx?id=12041. Accessed May29, 2019.
- 91.McKesson. iKnowMed: Oncology practice EHR system. https://www.mckesson.com/specialty/oncology-electronic-health-records/. Accessed May29, 2019.
- 92.American Association for Cancer Research. American Association for Cancer Research’s Genomics Evidence Neoplasia Information Exchange (GENIE). https://www.aacr.org/Research/Research/Pages/aacr-project-genie.aspx. Accessed November, 2019.
- 93.Health Resources and Services Administration. National Coordinating Center for the Regional Genetics Network. https://nccrcg.org/. Accessed November20, 2019.
- 94.Michigan Department of Health and Human Services. https://www.michigan.gov/mdhhs/0,5885,7-339-73971_4911_4916-85137-,00.html. Accessed November20, 2019.
- 95.Alabama Genomic Health Initiative (AGHI). https://healthitanalytics.com/news/alabama-recruits-for-statewide-population-health-genomics-program. Accessed November20, 2019.
- 96.Syapse Learning Health Network. https://www.syapse.com/offerings/syapselearning-health-network. Accessed November21, 2019.
- 97.Phillips KA, Deverka PA, Hooker GW, et al. Genetic test availability and spending: where are we now? Where are we going? Health Aff (Millwood). 2018;37:710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boussios C Gliklich R Artificial intelligence for real-world evidence. https://www.healtheconomics.com/blog/2018/01/artificial-intelligence-for-real-world-evidence/. Accessed May29, 2019.
- 100.Levy KD, Blake K, Fletcher-Hoppe C, et al. Opportunities to implement a sustainable genomic medicine program: lessons learned from the IGNITE Network. Genet Med. 2019;21:743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.U. S. Food and Drug Administration. Considerations for design, development, and analytical validation of next generation sequencing (NGS)—based in vitro diagnostics (IVDs) intended to aid in the diagnosis of suspected germline diseases. https://www.fda.gov/media/99208/download. Accessed May29, 2019.
- 102.Fleurence RL, Shuren J. Advances in the use of real-world evidence for medical devices: an update from the National Evaluation System for Health Technology. Clin Pharmacol Ther. 2019;106:30–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.NEST Coordinating Center. Test-Cases. https://nestcc.org/test-cases/. Accessed November19, 2019.
- 104.National Health Council. Patient perspectives on real-world evidence: a roundtable to gather views, needs, and recommendations. https://www.nationalhealthcouncil.org/sites/default/files/Patient%20Perspectives%20on%20Real-World%20Evidence.pdf. Accessed May29, 2019.
- 105.Reinke T Real-world evidence faces some real-world challenges. https://www.managedcaremag.com/archives/2017/5/real-world-evidence-facessome-real-world-challenges. Accessed May29, 2019. [PubMed] [Google Scholar]
- 106.Forsythe LP, Szydlowski V, Murad MH, et al. A systematic review of approaches for engaging patients for research on rare diseases. J Gen Intern Med. 2014;29(suppl 3):S788–S800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ray T BCBS “Evidence Street” group enabling engagement with MDx players. https://www.genomeweb.com/molecular-diagnostics/bcbs-evidence-street-group-enabling-engagement-mdx-players#.XO6vZYhKiUk. Accessed May29, 2019. [Google Scholar]
- 108.Center for Medicaid and Medicare Services. CMS finalizes coverage of next generation sequencing tests, ensuring enhanced access for cancer patients. https://www.cms.gov/newsroom/press-releases/cms-finalizes-coveragenext-generation-sequencing-tests-ensuring-enhanced-access-cancerpatients. Accessed May29, 2019. [Google Scholar]
- 109.Guardant Health. Guardant Health announces Medicare coverage for the Guardant360 assay in non-small cell lung cancer. https://www.prnewswire.com/news-releases/guardant-health-announces-medicare-coverage-for-theguardant360-assay-in-non-small-cell-lung-cancer-300680330.html. Accessed May29, 2019.
- 110.Ray T Myriad pushing ahead with payors on GeneSight as data from large randomized study is published. https://www.genomeweb.com/reimbursement/myriad-pushing-ahead-payors-genesight-data-largerandomized-study-published#.XPAyVohKiUk. Accessed May29, 2019.
- 111.Genomic Health. Multiple Oncotype DX study presentations at the 2018 San Antonio Breast Cancer Symposium reinforce real-world value of the Oncotype DX Breast Recurrence Score® test in patients regardless of age or race. https://investor.genomichealth.com/news-releases/news-release-details/multiple-oncotype-dx-study-presentations-2018-san-antonio-breast. Accessed May29, 2019.
- 112.MacLean E, Cisar L, Mehle K, et al. Real-world axitinib use in the United States: a retrospective study using linked datasets. J Manag Care Spec Pharm. 2016;22:723–732u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hess LM, Kern DM, Carter GC, et al. Real-world treatment sequences and outcomes among patients with non-small cell lung cancer (RESOUNDS) in the United States: study protocol. JMIR Res Protoc. 2017;6:e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heng DY, Signorovitch J, Swallow E, et al. Comparative effectiveness of second-line targeted therapies for metastatic renal cell carcinoma: a systematic review and meta-analysis of real-world observational studies. PLoS One. 2014;9:e114264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Davis B, Morgan J, Shah S. Getting real with real-world evidence. https://www2.deloitte.com/content/dam/Deloitte/us/Documents/life-sciences-healthcare/us-ls-2017-real-world-evidence-survey-031617.pdf. Accessed May29, 2019.
- 116.Ostrovsky L Implications of real-world data and pharmacoeconomics for managed care. Am Health Drug Benefits. 2016;9:151–155. [PMC free article] [PubMed] [Google Scholar]
- 117.Deverka P, Messner DA, McCormack R, et al. Generating and evaluating evidence of the clinical utility of molecular diagnostic tests in oncology. Genet Med. 2016;18:780–787. [DOI] [PubMed] [Google Scholar]
- 118.Mullins CD. Evolving use of real-world evidence for devices: good for patients, good for policy makers. Value Health. 2019;22:751–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


