Abstract
The MADS-domain transcription factor SEEDSTICK (STK) controls several aspects of plant reproduction. STK is co-expressed with CESTA (CES), a basic Helix-Loop-Helix (bHLH) transcription factor-encoding gene. CES was reported to control redundantly with the brassinosteroid positive signaling factors BRASSINOSTEROID ENHANCED EXPRESSION1 (BEE1) and BEE3 the development of the transmitting tract. Combining the stk ces-4 mutants led to a reduction in ovule fertilization due to a defect in carpel fusion which, caused the formation of holes at the center of the septum where the transmitting tract differentiates. Combining the stk mutant with the bee1 bee3 ces-4 triple mutant showed an increased number of unfertilized ovules and septum defects. The transcriptome profile of this quadruple mutant revealed a small subset of differentially expressed genes which are mainly involved in cell death, extracellular matrix and cell wall development. Our data evidence a regulatory gene network controlling transmitting tract development regulated directly or indirectly by a STK-CES containing complex and reveal new insights in the regulation of transmitting tract development by bHLH and MADS-domain transcription factors.
Keywords: bHLH, cell death, cell wall, extracellular matrix, MADS-box, transmitting tract
Introduction
The plant life cycle in Angiosperms is characterized by the alternation of diploid sporophyte and haploid gametophyte generations. The sporophyte produces spores, which then develop into gametophytes. The gametophytes produce either the male or the female gametes. Sexual reproduction requires the delivery of sperm nuclei, via the pollen tube, to the embryo sac, where fertilization occurs, and the new diploid sporophyte is formed.
The growing of the pollen tube through the female reproductive organ tissues in Angiosperms is a crucial step in plant sexual reproduction. In Arabidopsis thaliana, the pistil or gynoecium results from the fusion of two carpels. The apical part of the pistil is formed by the stigma and the stigma papillae, which are needed to attach the pollen. The stigma is connected by the style to the ovary, in which lodge the ovules. The septum divides the Arabidopsis ovary into two locules. At the center of the septum, as in the style, the transmitting tract tissue develops. Pollen tubes must travel through several distinct tissues before reaching ovules, including the stigma and the transmitting tract both in the style and in the septum (Lord and Russell, 2002; Roeder and Yanofsky, 2006).
The transmitting tract differentiates from the carpel margin meristem (CMM), a meristematic tissue that develops as two internal crests that, fuse when they reach each other in the middle of the young pistil at developmental stage 9. The transmitting tract is fully developed at stage 12 of pistil development (Bowman et al., 1999; Roeder and Yanofsky, 2006; Reyes-Olalde et al., 2013). The cells composing the transmitting tract secrete an extracellular matrix (ECM), a complex mixture of polysaccharides, glycoproteins, and glycolipids that accompany the growing pollen tubes (Lennon et al., 1998). Indeed, the pollen tubes grow faster, longer and more precisely in vivo than in vitro, suggesting the importance of the ECM for pollen tube growth in response to female signal factors (Johnson and Preuss, 2002; Johnson and Lord, 2006; Kim et al., 2006). Moreover, programmed cell death (PCD) in the transmitting tract is critical for pollen tube growth as well. The PCD of the transmitting tract starts before anthesis and can be accelerated by pollination (Crawford et al., 2007).
A complex genetic network controls the development of the transmitting tract. The zinc-finger transcription factor NO TRANSMITTING TRACT (NTT) has pivotal roles in this process. In the ntt mutant, the PCD of the cells composing the transmitting tract during fertilization, which is also influenced by the ECM composing cell wall, is severely compromised (Crawford et al., 2007). The changes in PCD and ECM in the ntt mutant cause a partial failure in pollen tube growth, with a high percentage of unfertilized ovules from the middle to the bottom part of the siliques (Crawford et al., 2007). NTT acts upstream of three genes that work redundantly in the control of the style and transmitting tract development, that are CESTA (CES), also named HALF FILLED (HAF), a bHLH-encoding transcription factor, and two phylogenetically related genes BRASSINOSTEROID ENHANCED EXPRESSION1 (BEE1) and BEE3 (Crawford and Yanofsky, 2011; Poppenberger et al., 2011).
BEEs expression and activity are induced in response to brassinosteroids (Friedrichsen et al., 2002). BRASSINOSTEROID INSENSITIVE 2 (BIN2) directly represses CES activity by phosphorylating it and thus promoting its degradation, restricting its subcellular localization and inhibiting the DNA binding activity of this transcription factor (Vert et al., 2005; Kim and Wang, 2010; Poppenberger et al., 2011). In response to brassinosteroids, phosphorylation of CES is inhibited and the unphosphorylated isoform accumulates promoting its SUMOylation (Khan et al., 2014). It is hypothesized that the brassinosteroid-induced SUMOylation of CES may facilitate the activation of some target genes involved in plant resistance to freezing stress (Eremina et al., 2016).
The triple mutant ces (haf) bee1 bee3 shows similar defects in transmitting tract development as Crawford and collaborators previously observed in the ntt mutant (Crawford and Yanofsky, 2011). Furthermore, ectopic expression of CES (HAF) increases the diameter of the transmitting tract allowing more pollen tubes to pass through it simultaneously (Crawford and Yanofsky, 2011).
Recently, NTT was found to interact with the MADS-domain transcription factor SEEDSTICK (STK) to control transmitting tract development (Herrera-Ubaldo et al., 2019). STK was previously identified for its role in ovule development and acts redundantly with other two MADS-box transcription factors SHATTERPROOF1 (SHP1) and SHP2 (Pinyopich et al., 2003).
NTT and STK have similar expression patterns in the medial domain of the pistil (Herrera-Ubaldo et al., 2019). At anthesis, the ntt stk double mutant presents a number of cells composing the septum similar to wild type (Herrera-Ubaldo et al., 2019). However, in the ntt stk mutant siliques, there is a partial failure in septum fusion, and it appears to have holes. The fact that the cell number composing the septum has not changed suggests that the abnormal fusion is not related to early stages of development but rather to later defects during epidermis formation (Herrera-Ubaldo et al., 2019). The partial failure of pollen tube growth in the ntt stk mutant is also caused by the absence of degeneration and collapse of the cells composing the septum and consequently the transmitting tract (Herrera-Ubaldo et al., 2019).
Mendes et al. (2016) performed a bioinformatic analysis to identify genes with expression patterns similar to STK, using Arabidopsis Affymetrix microarray data from 2000 independent experiments. From this experiment CES was identified as one of the best candidates to participate in the same processes as STK.
Here we report a genetic and molecular analysis which revealed interactions between the AGAMOUS family members STK, SHP1, and SHP2, the bHLH gene CES and the closely related genes BEE1 and BEE3 in the control of transmitting tract development in Arabidopsis. We further performed a transcriptome analysis of the bee1 bee3 stk ces-4 quadruple mutant uncovering new downstream players, involved in transmitting tract differentiation.
Materials and Methods
Plant Material and Growth Conditions
All plants used in this study were in the Columbia (Col-0) ecotype. For all analyses, Arabidopsis thaliana seeds were incubated for 2 days at 4°C after sowing. Plants were grown at 22°C under long day conditions (LD). The ces-4 mutant was obtained from NASC (SAIL_674_A01 lines). This line phenocopy the ces-3 (haf), ces-1 and ces-2 mutant alleles that have been previously characterized (Crawford and Yanofsky, 2011; Eremina et al., 2016). The stk, shp1, and shp2 mutants were provided by M. Yanofsky (Pinyopich et al., 2003), the bee1 bee2 bee3 triple mutant was provided by J. Chory (Friedrichsen et al., 2002). The other mutant combinations analyzed were obtained by crossing. The pCES::GUS line was provided by Prof. B. Poppenberger (Poppenberger et al., 2011).
Mutant Genotyping
Genotyping of the mutants was done by PCR analysis. All the primers used in this study are listed in Supplementary Table S11.
In situ Hybridization Analysis
Arabidopsis flowers and siliques were fixed and embedded in paraffin as previously described in Dreni et al. (2011). Tissue sections (8 μm) were hybridized with a CES specific antisense RNA probe and observed with a Zeiss Axiophot D1 microscope. Images were captured on an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1).
Gus-Staining Analysis
GUS staining of pCES::GUS was performed as described in Balanzà et al. (2016).
Silique Analysis
The percentage of unfertilized ovules per fruit was determined by opening at least 20 mature siliques for each genotype, counting the number of unfertilized ovules and the total number of developed seeds per silique under a stereomicroscope. The statistical significance of the differences in unfertilized ovules was analyzed using an Anova test followed by the Tukey HSD test (∗∗p < 0.01). All the experiments were repeated four times. For each experiment 5 siliques were collected from 4 independent plant lines of the same genotype.
Scanning Electron Microscope (SEM) Pictures
Biological samples were collected and fixed overnight at 4°C in FAA solution (3.7% formaldehyde, 5% acetic acid and 50% ethanol). Fixed tissues were washed with water and post-fixed with aqueous 2% osmium tetroxide for 2 h at room temperature. Tissues were rinsed several times in deionized water and dehydrated in a graded series of ethanol for 15 min per rinse. This step was followed by critical point drying with liquid CO2 and sputter-coating with gold in a Nanotech sputter coater. Specimens were analyzed using a LEO 1430 Scanning Electron Microscope.
Pollen Tube Guidance Analysis
Experiments of pollen tube guidance were performed as previously described by Mendes et al. (2016) and images were captured on an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1).
Bimolecular Fluorescence Complementation Assay (BiFC)
Coding Sequences (CDS) of CES and STK were amplified by PCR with the primers indicated in Supplementary Table S11 and first cloned into pDONOR207 (Life Technologies) and subsequently into the pYFPN43 and pYFPC43 vectors.1 BiFC experiments were conducted as previously described in Balanzà et al. (2016). Essentially, the abaxial surfaces of tobacco leaves were observed 2–5 days after agroinfiltration and the interactions were monitored using a Leica TCS confocal microscope.
Yeast Two-Hybrid Assays
The two-hybrid assays were performed at 22°C in the yeast strain AH109 (Clontech), using the cotransformation technique (Egea-Cortines et al., 1999). The coding sequences of CESTA, STK, and BEE1 were cloned in the Gateway vector GAL4 system (pGADT7 and pGBKT7; Clontech) passing through pDONOR207 (Life Technologies). Yeast two-hybrid interaction assays were performed on selective yeast synthetic dropout medium lacking Leu, Trp, Adenine, and His supplemented with different concentrations of 3-aminotriazole (1, 2.5, 5, 10, and 15 mM of 3-AT).
RNA Extraction and qRT-PCR Analysis
For the qRT-PCR experiment of ces-4 (knockout validation) total RNA was extracted from whole inflorescences using the LiCl method as previously described (Verwoerd et al., 1989). First-strand cDNA was synthesized using an IMProm-II TM Reverse Transcription System (Promega). Enrichments fold were detected using a SYBER Green assay (Bio-Rad).2 The qRT-PCR assay was performed in triplicate using a Bio-Rad iCycler iQ optical system (software version 3.0a). The qRT-PCR assay was repeated three times. For each experiment the RNA was extracted from three independent plant lines.
RNA Sequencing
Total RNA was extracted from three biological replicates (1 gr, obtained from a pool of 5 plants for each replicate) from both wild-type and bee1 bee3 stk ces-4 mutant inflorescences till stage 12, using the Macherey Nagel “Nucleospin RNA Plant” according to the manufacturer’s instructions. RNA concentrations and integrity were determined using Qubit Fluorometer and the QubitTM RNA XR Assay Kit (Thermo Fisher Scientific). Sequencing libraries were prepared using the NEBNext Ultra II Directional RNA library Prep Kit for Illumina (NEB) according to the manufacture’s instruction and sequenced on the HiSeq Illumina platform. Reads were mapped on the reference Arabidopsis thaliana genome (TAIR, version 10) using the bowtie2 program (Langmead and Steven, 2012; Supplementary Table S4). Estimation of gene expression levels was performed using RSEM (Li and Dewey, 2011). Identification of differentially expressed genes was performed by the quasi-likelihood F-test as implemented by edgeR (Robinson et al., 2009). A False Discovery Rate (FDR) cut-off value of 0.05 was applied for the identification of significantly differentially expressed genes. Graphical representation of the data was performed by means of the gplots R library. To validate the data obtained from the RNAseq experiment, we extracted total RNA from wild type and bee1 bee3 stk ces-4 mutant inflorescences till stage 12, using the Macherey Nagel “Nucleospin RNA Plant” according to the manufacturer’s instructions. The cDNA and qRT-PCR were performed as described in the previous paragraph “RNA extraction and qRT-PCR analysis.” The qRT-PCR assay to validate RNAseq experiment was repeated three times. For each experiment the RNA was extracted from three independent plant lines that were used for RT-PCR analysis.
TFBS Analysis
Enrichment analysis of Transcription Factor Binding Sites (TFBS) was performed by means of the Pscan tool, using the “non-redundant” set of Jaspar TFBS matrices (Fornes et al., 2019) and considering a region spanning from the TSS to 500 bp upstream (default settings). Archetype consensus sites for the W-box, MADS-box and G-box elements were established as the Jaspar TFBS matrix showing the highest enrichment in our dataset according to Pscan. To identify genes with high confidence W-box, MADS-box and G-box elements in their promoters, the complete set of 33239 Arabidopsis transcripts according to the TAIR10 annotation were scanned using the highly significantly enriched TFBS matrices as defined above. Distribution of scores were established for each element, and an Expectation-Maximization algorithm as implemented in the R EM-cluster package was used to fit a 2 Gaussian mixture model. Based on the EM model promoters were partitioned in “high” and “low” scoring. Only promoters that were confidently assigned to high scoring clusters (confidence > 95%) were considered to have a bona fide TFBS element. Similarly, to our previous analyses promoters were defined as regions spanning 500 bp upstream of the TSS. P-values for the enrichment of genes with W-box, MADS-box and G-box elements in their promoters were performed by means of the hypergeometric distribution test as implemented in the R stats package.
Accession Numbers of the Genes
The GenBank/EMBL accession numbers for the genes shown in this study are the following: BEE1, At1g18400; BEE2, At4g36540; BEE3, At1g73830; CES, At1g25330; SHP1, At3g58780; SHP2, At2g42830; STK, At4g09960. The accession number of the genes identified in the transcriptomic analysis are indicated in Supplementary Tables S5, S6.
Results
SEEDSTICK and CESTA Are Co-expressed
In order to identify new targets, partners and/or regulators of STK, a bioinformatic analysis was previously performed in the laboratory to discover genes co-expressed with STK (Mendes et al., 2016). This kind of analysis assumes that correlation between the patterns of gene expression in a large range of different experimental conditions could indicate a functional relationship (Aoki et al., 2007). As already described, the procedure to identify genes that have expression profiles that correlate with STK was proven successful since among the top listed genes SHP1 and SHP2 were found, which act redundantly with STK in the control of ovule identity (Pinyopich et al., 2003). Furthermore, top rated were VERDANDI (VDD) and VALKYRIE (VAL), targets of the STK-SEPALLATA3 (SEP3) complex, which both act in the control of the death of the synergid cell that is receptive of the pollen tube (Mendes et al., 2016). CES (At1g25330) rated in position 14 of the correlators, having an expression correlation with STK of p(LIN) 0.759 and p(LOG) 0.505 (Mendes et al., 2016).
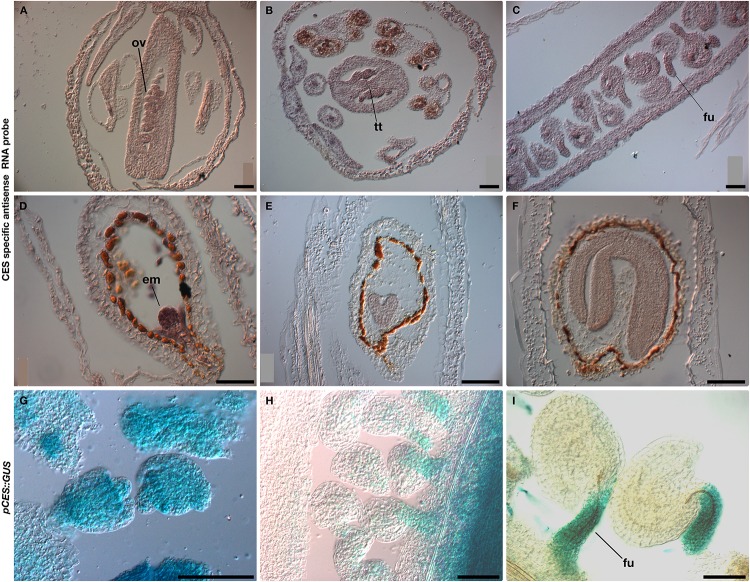
Previously, it has been reported that CES is expressed from stage 8 to 15 of flower development, first in CMM and later in the stigma, septum and funiculi (Crawford and Yanofsky, 2011; Poppenberger et al., 2011). Since STK is expressed during female reproductive organ development (mainly septum, transmitting tract, replum, funiculus and ovule/seed) (Pinyopich et al., 2003; Mizzotti et al., 2014; Herrera-Ubaldo et al., 2019; Di Marzo et al., 2020), we investigated in more detail the spatio-temporal expression pattern of CES during pistil, ovule and seed development. We performed both RNA in situ hybridization and the analysis of the pCES::GUS reporter gene construct (Figure 1). The RNA in situ analysis experiments showed that CES was expressed in ovules during early stages of development until mature ovule formation and its transcripts were also present in the transmitting tract before fertilization (Figures 1A–C). In mature ovules, a high expression was detected in the funiculus (Figure 1C). After fertilization and during embryo development CES mRNA was present at high levels in the early globular stage, and at heart and torpedo stages mRNA levels were lower (Figures 1D–F). Analysis of the pCES::GUS plants confirmed the in situ data (Figures 1G–I) and these experiments showed that STK and CES partially overlap in their expression patterns during ovule and transmitting tract development (Pinyopich et al., 2003; Crawford and Yanofsky, 2011; Poppenberger et al., 2011; Herrera-Ubaldo et al., 2019).
FIGURE 1.
Analysis of CES expression. In situ hybridization (A–F) and GUS activity of the pCES::GUS marker line (G–I) showed CES expression in ovules (A–C,G–I), transmitting tract (B), and in the embryos of the developing seeds (D–F). Em, embryo; fu, funiculus; ov, ovule; tt, transmitting tract. Scale bars: 50 μm.
Arabidopsis Plants With stk, shp, ces, and bee Mutant Combinations Are Partially Sterile
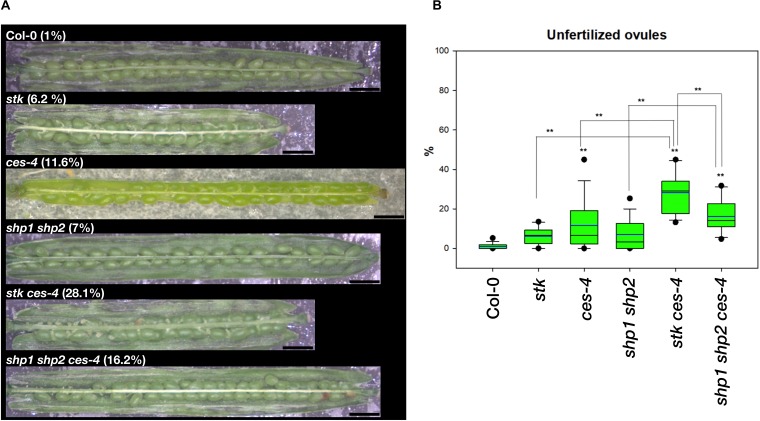
To further analyze the role of CES, we characterized a CES loss-of-function allele that we obtained from the SAIL collection (Alonso and Stepanova, 2003). This allele, that we called ces-4, has a T-DNA insertion in the second exon, differently from ces-1, ces-2, and ces-3 (haf), previously published T-DNA knockout insertion alleles (Crawford and Yanofsky, 2011; Poppenberger et al., 2011; Eremina et al., 2016). The qRT-PCR analysis performed to check CES expression in the ces-4 mutant indicated that this is also a null allele (Supplementary Figure S1). Analysis of ces-4 homozygous plants did not reveal any general phenotype at the level of the entire plant, which was consistent with previous reports for the ces (haf) mutant alleles (Crawford and Yanofsky, 2011; Poppenberger et al., 2011; Eremina et al., 2016). We crossed the ces-4 mutant with stk to obtain the stk ces-4 double mutant. The double mutant was indistinguishable from wild type plants apart from a large number of unfertilized ovules at the lower part of the siliques. In wild type plants on average 1.0% of the ovules in a silique remained unfertilized. In the stk mutant we observed on average 6.2% unfertilized ovules, and in the ces-4 single mutant 11.6% (∗∗p < 0.01 in respect to wild type; Figure 2 and Supplementary Table S1). In the stk ces-4 double mutant this phenotype was further increased to 28.1% of unfertilized ovules (Figure 2 and Supplementary Table S1). Like in the ces-4 single mutant, also in the stk ces-4 double mutant the unfertilized ovules were located always at the lower part of the siliques (Figure 2 and Supplementary Table S1).
FIGURE 2.
Unfertilized ovules analysis in multiple mutant combination of stk, ces-4, and shp1 shp2. (A) Stereomicroscope images of opened siliques of the stk and ces-4 single mutants, the stk ces-4 and the shp1 shp2 double mutants, the shp1 shp2 ces-4 triple mutant and the wild type Col-0; Scale bars: 1 mm. (B) Box plots of all the mutants analyzed in comparison to wild type Col-0. Statistical analysis was performed using Anova followed by Tukey HSD test (**p < 0.01). The experiment was repeated four times. For each experiment 5 siliques were collected from 4 independent plant lines of the same genotype to obtain a total of 20 analyzed siliques.
Since SHP1 and SHP2 are closely related to STK and have shown to control together with STK ovule identity redundantly (Pinyopich et al., 2003), we investigated whether they also play a role together with CES in the control of ovule fertilization. Therefore, the shp1 shp2 double mutant was crossed with the ces-4 mutant to obtain the shp1 shp2 ces-4 triple mutant. The shp1 shp2 double mutant showed on average 7.0% unfertilized ovules (Figure 2 and Supplementary Table S1). The shp1 shp2 ces-4 triple mutant showed a clear increase to 16.2%, also in this case the unfertilized ovules were localized at the lower part of the siliques (∗∗p < 0.01 in respect to wild type and shp1 shp2; Figure 2 and Supplementary Table S1). Unfortunately we could not analyze the stk shp1 shp2 triple mutant, because the triple mutant does not develop normal ovules (Pinyopich et al., 2003). These results suggest that the MADS-domain transcription factor STK, SHP1 and SHP2 have a role together with CES in the control of ovule fertilization.
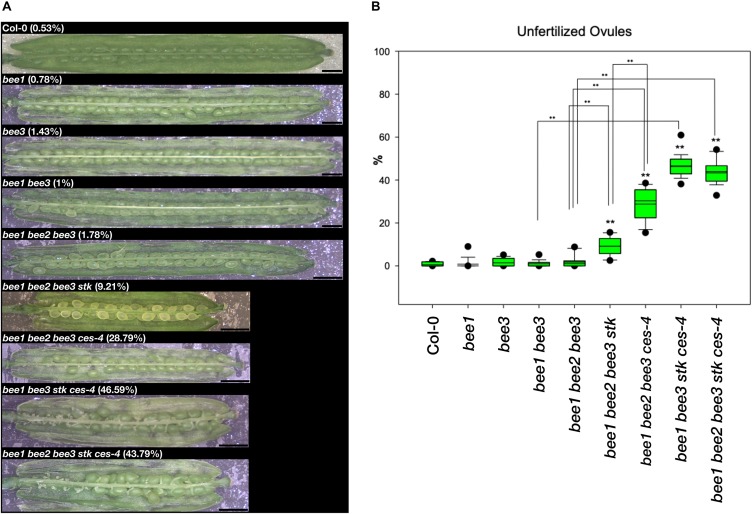
The contribution of the BEE genes to this process was also investigated (Figure 3 and Supplementary Table S2). CES is phylogenetically related to BEE1 and BEE3, which have been described as redundant with BEE2 (Friedrichsen et al., 2002). Moreover, it has been shown that CES together with BEE1 and BEE3 redundantly control the development of the transmitting tract in the pistil (Crawford and Yanofsky, 2011; Poppenberger et al., 2011). The single mutants bee1 and bee3 and the multiple mutant combinations bee1 bee3 and bee1 bee2 bee3 did not show any obvious phenotype (Figure 3 and Supplementary Table S2). To investigate redundancy between BEE genes and STK, we obtained the bee1 bee2 bee3 stk quadruple mutant, that presented 9.2% of unfertilized ovules, with ∗∗p < 0.01 when compared to wild type and the triple mutant bee1 bee2 bee3, which again were preferentially positioned at the lower part of the siliques (Figure 3 and Supplementary Table S2). This percentage is lower than the bee1 bee2 bee3 ces-4 quadruple mutant, which presented in average 28.7% of unfertilized ovules (Figure 3 and Supplementary Table S2). Despite the weaker phenotype, our results demonstrated that STK and BEEs also have a role together in ovule fertilization. To investigate if combining the ces-4 mutant could further enhance the unfertilized ovule phenotype, we generated the bee1 bee3 stk ces-4 quadruple and the bee1 bee2 bee3 stk ces-4 quintuple mutants. The results of this analysis showed that bee1 bee3 stk ces-4 had on average 46.6% unfertilized ovules, and a similar result was obtained in the bee1 bee2 bee3 stk ces-4 (44.0%) (Figure 3 and Supplementary Table S2), showing that the addition of the bee2 allele did not further enhance the phenotype. The unfertilized ovules in these cases are localized from the middle to the bottom part of the siliques. Altogether, these results indicated that the MADS-domain transcription factors function not only together with CES, but also with the BEEs in the control of the fertilization of all the ovules within the Arabidopsis fruit.
FIGURE 3.
Unfertilized ovules analysis in multiple mutant combination of bee1, bee2, bee3, stk, and ces-4. (A) Stereomicroscope images of opened siliques of the bee1 and bee3 single mutants, the bee1 bee3 double mutant, the bee1 bee2 bee3 triple mutant, the bee1 bee2 bee3 stk, the bee1 bee2 bee3 ces-4, and bee1 bee3 stk ces-4 quadruple mutants, the bee1 bee2 bee3 stk ces-4 quintuple mutants and the wild type Col-0; Scale bars: 1 mm. (B) Box plots of all the mutants analyzed in comparison to wild type Col-0. Statistical analysis was performed using Anova followed by Tukey HSD test (**p < 0.01). The experiment was repeated four times. For each experiment 5 siliques were collected from 4 independent plant lines of the same genotype to obtain a total of 20 analyzed siliques.
STK and CES Regulate Transmitting Tract Development
To better understand the defect that caused the presence of a high percentage of unfertilized ovules at the lower part of the siliques in the different mutant combinations, we performed reciprocal crosses between wild type and stk ces-4 or shp1 shp2 ces-4 mutant plants, to analyze if the female, male or both parts were defective. The outcome of the reciprocal crosses shown in Supplementary Figure S2, suggested that the unfertilized-ovules phenotype was only present when the mutant genotypes were used as mothers. This result indicates that the defects resides only in the female reproductive organ.
To further investigate the defect present in the female reproductive organs, which caused the high number of unfertilized ovules at the lower part of the siliques in the multiple mutant combinations, we analyzed the septum morphology by Scanning Electron Microscope (SEM), at stage 17-B of fruit development, when the siliques reach their maximum size (Roeder and Yanofsky, 2006).
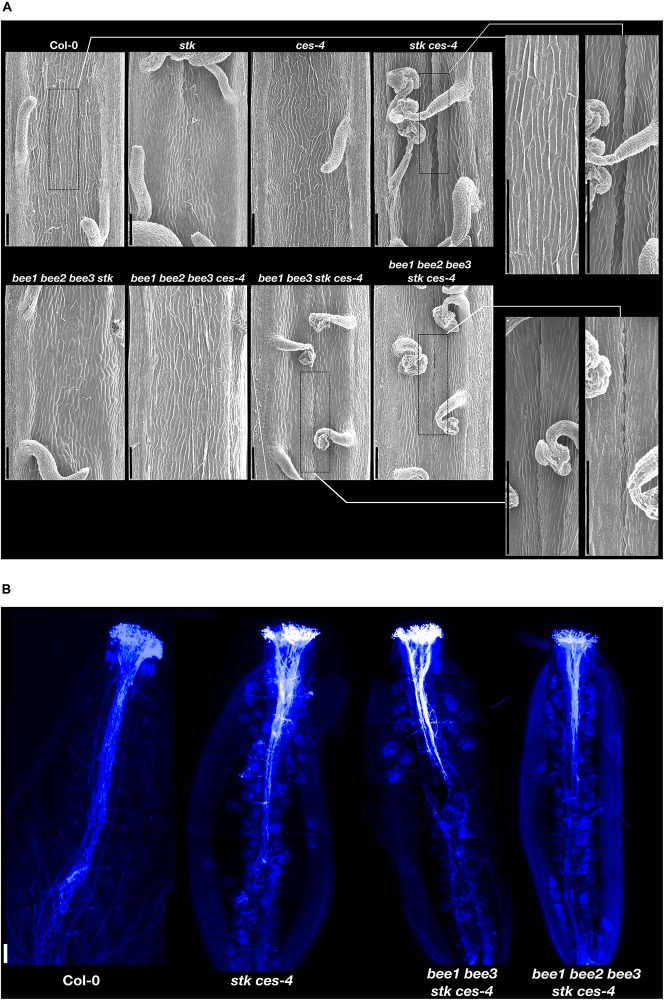
The stk and ces-4 single mutants presented a septum similar to wild type, while the double mutant stk ces-4 disclosed a defect in septum morphology (Figure 4A). The double mutant was characterized by a partial failure of septum fusion in the central portion of the septum in conjunction with the medium portion of the ovary (Figure 4A). The quadruple mutant combinations bee1 bee2 bee3 stk and bee1 bee2 bee3 ces-4 did not display defects in septum morphology when compared to wild type (Figure 4A). However, the damage of the septum appears stronger in the quadruple mutant bee1 bee3 stk ces-4 and in the quintuple mutant bee1 bee2 bee3 stk ces-4 where a hole in the central portion of the septum was seen in conjunction with the medium region of the ovary (Figure 4A).
FIGURE 4.
Septum analysis and pollen tube growth. (A) Scanning Electron Microscope pictures of the septum of opened siliques of stk, ces-4, stk ces-4, bee1 bee2 bee3 stk, bee1 bee2 bee3 ces-4, bee1 bee3 stk ces-4 and bee1 bee2 bee3 stk ces-4 and wild type Col-0; Scale bars: 100 μm. (B) Aniline blue staining of stk ces-4, bee1 bee3 stk ces-4 and bee1 bee2 bee3 stk ces-4 and wild type Col-0; Scale bar: 50 μm.
The septum morphological analysis at stage 17-B suggests a transmitting tract defect caused by a failure of the carpel fusion which happens from stage 9 to 12 (Roeder and Yanofsky, 2006). At these stages, fusion occurs within the two crests of the CMM leading to septum and transmitting tract formation (Bowman et al., 1999; Roeder and Yanofsky, 2006; Reyes-Olalde et al., 2013).
To confirm that the defects found in the septum and transmitting tract formation, described for stk ces-4, bee1 bee3 stk ces-4, and bee1 bee2 bee3 stk ces-4 caused a defective growth of the pollen tubes through the ovary transmitting tract, pistils were hand pollinated with wild type pollen and an aniline blue staining was performed after 36 h (Figure 4B). In the wild-type-hand-pollinated control pistils all the pollen tubes reached the bottom part of the ovary. However, pollen tube growth was compromised in the three mutant combinations. In the stk ces-4 double mutant the pollen tubes did not grow further than the middle region of ovary length (Figure 4B). A stronger phenotype was observed in bee1 bee3 stk ces-4 quadruple and in the bee1 bee2 bee3 stk ces-4 quintuple mutants, where the pollen tubes stopped growing at the upper part of the ovary (Figure 4B).
The differences in septum morphology and in pollen tube growth in the double mutant stk ces-4, in the quadruple mutant bee1 bee3 stk ces-4 and in the quintuple mutant bee1 bee2 bee3 stk ces-4 clearly indicate that the fusion of the carpels, that permits the correct development of the septum and consequently of the transmitting tract, was partially disturbed in these mutants and the cause that the ovules at the lower part of the siliques remained unfertilized.
SEEDSTICK and CESTA Interact in Yeast and Planta
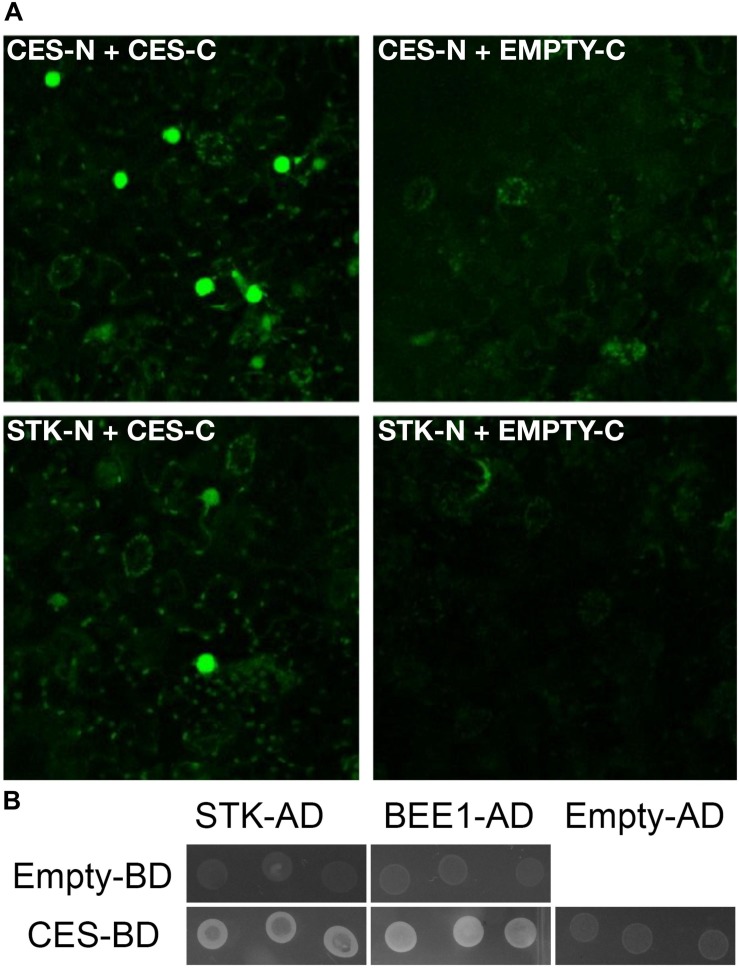
The experiments described above showed a genetic interaction between STK and CES. To investigate if there is also a physical interaction between these proteins, Bimolecular Fluorescence Complementation (BiFC) was performed. As a positive control, cells were co-transformed with CES (YC-CES) and (YN-CES), as CES was shown to homodimerize in yeast (Poppenberger et al., 2011; Figure 5A). Reconstitution of YFP fluorescence in the nuclei of co-transformed cells confirmed that CES is able to interact with STK in vivo (Figure 5A), supporting our idea that CES and STK form a complex that controls transmitting tract formation. The interaction between STK and CES was also confirmed by Y2H experiments (Figure 5B). As positive control in these experiments we used the previously published interaction between BEE1 and CES (Poppenberger et al., 2011; Figure 5B). Furthermore, we observed that STK and BEE1 were not able to interact. The complete list of vector combinations, interactions and growth media used in this Y2H assay are described in Supplementary Table S3.
FIGURE 5.
STK, CES and BEE1 interaction assays in yeast and in planta. (A) BiFC assays showing the interaction of STK with CES. STK-N or CES-N: STK or CES fusion with the N-terminal part of the split YFP; STK-C or CES-C: STK or CES fusion with the C-terminal part of the split YFP. CES-N CES-C interaction was used as positive control. STK-N EMPTY-C and CES-N EMPTY-C were used as negative control. (B) Yeast two-hybrid assays testing interactions between STK-AD, BEE1-AD, and CES-BD; interactions were considered positive when growth on -W-L-H +15 mM of 3-AT selective media was observed. The experiments were performed at 22°C. The CES-BEE1 interaction was chosen as a positive control (Poppenberger et al., 2011), the growth of transformants carrying empty AD or BD vectors were used as negative controls of the experiments.
Transcriptome Analysis of the bee1 bee3 stk ces-4 Quadruple Mutant Reveals Target Genes Involved in Transmitting Tract Development
To obtain a snapshot of the regulatory network controlled by STK, CES, and BEE proteins, in the development of the septum/transmitting tract, transcriptional profiles of the quadruple mutant bee1 bee3 stk ces-4, which displayed the most severe phenotype, were studied by RNA sequencing. RNA was extracted from the quadruple mutant and wild type inflorescences until stage 12 of pistil development, before fertilization since the transmitting tract is fully developed at this stage (Bowman et al., 1999; Roeder and Yanofsky, 2006; Reyes-Olalde et al., 2013).
For the identification of significantly differentially expressed genes, a FDR cut-off value of 0.05 was applied and a total of 202 differentially expressed genes were identified. A total of 31 genes were upregulated in the mutant (Supplementary Table S5) while 171 genes were downregulated (Supplementary Table S6). Furthermore, functional enrichment analyses revealed that gene ontology categories such as glycosinolate, glycoside and carbohydrate biosynthetic and metabolic processes were enriched for the upregulated genes (Supplementary Table S7), while the downregulated genes were involved in the cell death, cell wall, plant-type cell wall biosynthesis components and response to hormone stimulus (Supplementary Table S8).
Remarkably, among the downregulated genes, three genes were previously described as direct targets of STK (Herrera-Ubaldo et al., 2019): AT3G26140, AT1G28710, and AT1G06080. The AT3G26140 gene which encodes a glycosyl hydrolase (GH5_11) is expressed in CMM during pistil development (Aspeborg et al., 2012; Herrera-Ubaldo et al., 2019). The GH5 enzymes are all mannan endo-beta-1,4-mannosidases which are known to be involved in cell wall remodeling processes (Aspeborg et al., 2012; Lombard et al., 2014; Herrera-Ubaldo et al., 2019). AT1G28710 encodes for a nucleotide diphospho-sugar transferase protein which is involving in the biosynthesis of polysaccharides, that are fundamental components of the ECM (Lennon et al., 1998; Tung et al., 2005; Herrera-Ubaldo and de Folter, 2018). AT1G06080 [DELTA 9 DESATURASE1 (ADS1)], encodes for a fatty acid desaturase which is involved in lipid biosynthesis, expressed in flowers (Fukuchi-Mizutani et al., 1998; Yao et al., 2003; Heilmann et al., 2004; Table 1, see also Figure 6 for heatmap description). Downregulation of these three genes was also confirmed by qRT-PCR (Supplementary Figure S3).
TABLE 1.
RNAseq data of candidate genes regulated by STK-CES protein complex and involved in transmitting tract development.
| TAIR ID | Gene name | log FC | FDR | Gene function |
| AT4G14400 | ACD6 | –4.0117 | 2.33E-05 | Cell death |
| AT1G06080 | ADS1 | –1.0338 | 2.17E-05 | Cell wall and ECM biosynthesis |
| AT1G35230 | AGP5 | –2.1455 | 0.0145 | Cell death and ECM biosynthesis |
| AT5G65390 | AGP7 | –1.3013 | 0.0024 | Cell death and ECM biosynthesis |
| AT5G56540 | AGP14 | –0.7840 | 0.0473 | Cell death and ECM biosynthesis |
| AT1G72290 | ATWSCP | –6.5142 | 1.42E-07 | Cell death and pollen tube growth |
| AT5G09730 | BXL3 | –2.0805 | 1.71E-06 | Cell wall |
| AT1G75890 | GDSL family of serine esterases/lipases | 0.8845 | 0.0038 | Cell wall and lipid metabolic process |
| AT4G28780 | GDSL family of serine esterases/lipases | 0.6227 | 0.0393 | Cell wall and lipid metabolic process |
| AT4G16230 | GDSL family of serine esterases/lipases | 1.0662 | 0.0127 | Cell wall and lipid metabolic process |
| AT3G26140 | Glycosyl hydrolase family 5 | –2.7099 | 3.13E-06 | Cell wall and ECM biosynthesis |
| AT1G53130 | GRI | –7.9981 | 4.16E-09 | Cell death |
| AT4G14560 | IAA1 | –0.7727 | 0.0387 | AUX signaling |
| AT3G23030 | IAA2 | –0.6711 | 0.0393 | AUX signaling |
| AT4G14550 | IAA14 | –0.9381 | 0.0265 | AUX signaling |
| AT1G04250 | IAA17 | –1.2215 | 0.0037 | AUX signaling |
| AT2G38530 | LTP2 | –0.8702 | 0.0257 | Lipid transport, cell death, cuticle biosynthesis |
| AT1G28710 | Nucleotide diphospho-sugar transferase protein | –1.2338 | 0.00008 | Cell wall and ECM biosynthesis |
| AT5G49180 | PME58 | –0.6750 | 0.0459 | Cell wall |
| AT4G36110 | SAUR9 | –1.2233 | 0.0447 | Auxin response |
| AT2G21220 | SAUR12 | –0.7960 | 0.0239 | Auxin response |
Column 1, Gene ID according to the TAIR10 annotation of the A. thaliana genome; Column 2, Gene name; Column 3, Log Fold Change of expression between bee1 bee3 stk ces-4 and wild type plants; Column 4, FDR: Benjamini Hochberg adjusted P-value for differential expression. A Significance cut-off of 0.05 is applied; Column 5, Brief description of the gene according to TAIR10 (if available). Total RNA was extracted from three biological replicates (1 gr, obtained from a pool of 5 plants for each replicate) from both wild-type and bee1 bee3 stk ces-4 mutant inflorescences.
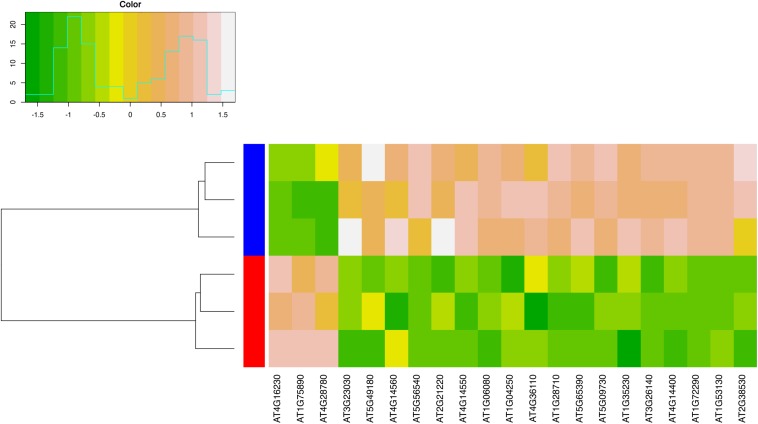
FIGURE 6.
Heatmap of differentially expressed genes described in Table 1. Expression levels are shown for each of the 6 independent biological replicates analyzed in this study. Genes are represented in the columns and the biological replicates in the rows. To facilitate comparison, the expression values were standardized with respect to the average expression of each gene. Dark green indicates low expression, light brown indicates high expression. In the columns, red is used to mark biological replicates for the quadruple mutant, blue is used to indicate biological replicates for wild type plants.
PCD is a key process in the transmitting tract for ovule fertilization (Crawford et al., 2007). Interestingly, several genes involved in the regulation of cell death were downregulated in the quadruple mutant (Table 1 and Figure 6). One of the most important is GRIM REAPER (GRI, AT1G53130) that encodes for a small protein with an extracellular localization that is able to positively regulate cell death (Wrzaczek et al., 2009, 2015; Table 1 and Figure 6). Moreover, we observe that ACCELERATED CELL DEATH 6 (ACD6, AT4G14400) another positive regulator of cell death is also downregulated (Rate et al., 2007; Table 1 and Figure 6).
Additionally, we notice the downregulation of three genes encoding for the Arabinogalactan proteins (AGPs) ARABINOGALACTAN PROTEIN 5 (AGP5, AT1G35230), AGP7 (AT5G65390), and AGP14 (AT5G56540). AGPs are structurally complex plasma membrane and cell wall proteoglycans that are implicated in diverse developmental processes, including plant sexual reproduction (Lennon et al., 1998; Coimbra et al., 2007, 2009). Importantly, these protein are able to regulate cell death in Arabidopsis cell culture (Gao and Showalter, 1999; Guan and Nothnagel, 2004). Intriguingly, we also detect a significant downregulation of ATWSCP (AT1G72290), which encodes for a Kunitz-type protease inhibitor, member of a small family of proteins, that are water-soluble chlorophyll proteins, and LIPID TRANSFER PROTEIN 2 (LTP2, AT2G38530) (Table 1 and Figure 6). ATWSCP is expressed in the septum and transmitting tract during its development, and after fertilization the protein is located in the medial domain of the silique where is able to regulate cell death and pollen tube growth (Bektas et al., 2012; Boex-Fontvieille et al., 2015). The LTP2 gene encodes for a lipid transfer protein necessary for the movement of hydrophobic wax components through the hydrophilic cell wall matrix to reach the cuticle (Kunst and Samuels, 2003). LTP2 mediates cell wall loosening in vitro (Nieuwland et al., 2005).
Numerous genes involved in cell wall biosynthesis processes are mis-regulated in the quadruple mutant, these include BETA-XYLOSIDASE 3 (BXL3, AT5G09730), which encodes for an enzyme involved in arabinan and xylan catabolic process (Minic et al., 2006), and PECTIN METHYLESTERASE 58 (PME58, AT5G49180), encoding for a pectin methylesterases that contributes to cell wall modification (Turbant et al., 2016; Table 1 and Figure 6); both genes were downregulated. Among the upregulated genes, there are three genes of the GDSL family of serine esterases/lipases, AT1G75890, AT4G28780, and AT4G16230 (Table 1 and Figure 6). These genes are involved in numerous aspect of plant development, but in particular the acetylxylan esterase activity of these enzymes could contribute in the degradation of cell walls in association with xylanases, cellulases, and mannanases (Akoh et al., 2004).
Moreover, some genes involved in the auxin signaling pathway were downregulated. These include genes encoding for Aux/IAAs proteins, INDOLE-3-ACETIC ACID INDUCIBLE 1 (IAA1) (AT4G14560), IAA2 (AT3G23030), IAA14 (AT4G14550), and IAA17 (AT1G04250) (Table 1 and Figure 6). The Aux/IAA proteins are the early auxin response proteins and participate in auxin signaling through interacting, as repressors, with AUXIN RESPONSE FACTOR (ARF) proteins (Lavy and Estelle, 2016). In addition, the SMALL AUXIN UPREGULATED RNA 9 (SAUR9) and SAUR12 were also downregulated. The SAURs are small transcripts induced by auxin, that are localized in elongated tissues (McClure and Guilfoyle, 1987; Knauss et al., 2003). SAUR9 and SAUR12 are part of the SAUR10-clade (van Mourik et al., 2017).
Finally, we decided to explore the enriched binding sites from our RNAseq data of the quadruple mutant in order to find possible direct target genes of the complex formed by STK, CES, BEE1, and BEE3. The binding sites used by the MADS-domain and bHLH transcription factors have been characterized: MADS factors bind to CArG boxes (Egea-Cortines et al., 1999; de Folter and Angenent, 2006) while bHLHs recognize E-boxes, being G-boxes the most common form (Quail, 2000; Toledo-Ortiz et al., 2003), in order to regulate the activity of their target genes. On the other hand, W-box is the binding site used by the WRKY transcription factors (Ciolkowski et al., 2008).
Transcription factor binding sites enrichment analyses, performed by means of the Pscan tool (Zambelli et al., 2009) suggest a strong enrichment of G-box and G-box like elements in the upregulated genes (Supplementary Table S9), while a significant enrichment of W-box and MADS-box elements is observed in the downregulated genes (Supplementary Table S10). Notably, according to our analyses G-box elements are identified in 35% (p = 4E-05) of the upregulated genes, while W-box elements and MADS box elements are identified in 17 and 19 downregulated genes (p-values 9.3E-04 and 0.035), respectively. Unsurprisingly, considering the limited number of genes that are differentially expressed in our experimental settings, we observe that only one gene has both a G-box and a CArG-box element in its promoter (AT3G49270, upregulated), only one gene has both a W-box and a MADS-box element (AT1G05450, downregulated), and none of the differentially expressed genes have a W-box and a G-box.
Discussion
Genetic Redundancy Between MADS-Domain and bHLH Transcription Factors in Transmitting Tract Development
In Arabidopsis, the gynoecium is formed by the fusion of two carpels, which fuse vertically at their margins and give rise to the CMM. From this meristem the transmitting tract develops, an essential tissue to facilitate the growing pollen tubes to reach the ovules and fertilize them.
Recently, a role for STK in relation with NTT was shown during the development of transmitting tract (Herrera-Ubaldo et al., 2019). Here we reveal that STK may also act in the downstream pathway of NTT in a protein complex with CES. Our genetic analysis evidence the role and redundancies between the MADS-box genes STK, SHP1, and SHP2, the bHLH transcription factor gene CES and the closely related bHLH transcription factors BEE1 and BEE3 in the development of the transmitting tract in Arabidopsis thaliana. The CES gene was selected based on high correlation of its expression pattern with STK (Mendes et al., 2016) but also because this gene was shown to play a role in transmitting tract development (Crawford and Yanofsky, 2011; Poppenberger et al., 2011). CES, BEE1, and BEE3 were shown to be expressed with distinct but overlapping patterns within the reproductive tract (Crawford and Yanofsky, 2011; Poppenberger et al., 2011). In the ces (haf) bee1 bee3 triple mutant ECM biosynthesis and PCD fail to occur within this tissue (Crawford and Yanofsky, 2011).
The stk mutant did not present any type of reproductive defect (no statistical differences when the number of unfertilized ovules was compared with wild type), while the single mutant ces-4 is characterized by a small but significative percentage of unfertilized ovules (Figure 2). However, the stk ces-4 double mutant had 28.1% of unfertilized ovules, which were almost exclusively found at the basal part of the silique (Figure 2). This phenotype suggests, similar to the ces bee1 bee3 triple mutant (Crawford and Yanofsky, 2011; Poppenberger et al., 2011), a defect in transmitting tract development.
Interestingly, we also observed a genetic interaction between STK and BEE1 and BEE3 in transmitting tract development since the bee1 bee2 bee3 stk quadruple mutant showed 9.21% unfertilized ovules at the lower part of the silique, while stk had only 6.2% and the triple mutant bee1 bee2 bee3 displayed 1.8% of unfertilized ovules (Figure 3). In the light of the fact that this phenotype was weaker than that observed for stk ces-4 (Figure 2), we suggest that CES plays a more predominant role in transmitting tract formation together with STK. To understand a possible role of SHP1 and SHP2 in the formation of the transmitting tract, the triple mutant shp1 shp2 ces-4 was obtained. This triple mutant showed 16.2% of unfertilized ovules which is also a milder phenotype when compared to stk ces-4 (Figure 2). This suggests that STK is important for transmitting tract development, although is not able to cover for 100% SHPs function since the shp1 shp2 ces-4 triple mutant presents a considerable percentage of unfertilized ovules.
Recently, it has been reported that in Arabidopsis thaliana the correct accomplishment of ovule fertilization directly influences the final length of the fruit. In the ntt mutant, in which the seeds only develop in the upper part of the fruit (Crawford et al., 2007), the authors described by using a fruit live imaging platform (FLIP) that in the upper part of the ntt siliques (where fertilization happens) the cells composing the valves elongate properly like in the wild type control while in the bottom part (where no fertilization occurs) the cells do not elongate properly (Ripoll et al., 2019). The quadruple mutant bee1 bee3 stk ces-4, which, similarly, to the ntt mutant displayed the highest percentage of unfertilized ovules from the middle to the bottom part of the fruit (Crawford et al., 2007), was characterized by a clear decrease in fruit length as can be seen in Figure 3, confirming the importance of ovule fertilization for fruit size.
The different degree of unfertilized ovule phenotypes obtained in the different mutant combinations points to redundancy between MADS-box, bHLH, and the BEEs transcription factor encoding genes in a dosage-dependent manner. The unfertilized ovules phenotype was only observed when different mutant alleles were combined and was more pronounced in the quadruple and quintuple mutants, suggesting an additive effect. Such a dosage dependent effect was already described for other MADS-box genes, like FLOWERING LOCUS C (FLC) that acts, in a dosage-dependent manner, as a potent repressor of the floral transition (Bastow et al., 2004; Michaels and Amasino, 2007; Sheldon et al., 2007).
Y2H and BIFC experiments showed that STK is able to interact with CES, but not with BEE1 (Figure 5 and Supplementary Table S3). This result suggests that CES might bridge the interaction between MADS and BEE proteins to form a hypothetical multimeric complex that controls correct transmitting tract formation. Cooperative action of MADS-domain and bHLH transcription factors has been previously described in mice. Molkentin et al. (1995) showed that MEF2 factors, a family of MADS-domain proteins expressed in muscle cells and other cell types, acted as coregulators of myogenic bHLH proteins to activate muscle genes (Molkentin et al., 1995). Moreover, the authors show that these two types of transcription factors physically interact, and that either factor can interact with the other when one is bound to DNA (Molkentin et al., 1995). Interestingly, the authors present this as a general mechanism for the regulation of transcription in specific cell types (Molkentin et al., 1995). This might also be a mechanism for the combinatorial control of MADS-box (with STK as a main player) and bHLH (CES as principal component) transcription factors for the formation of the transmitting tract tissue in Arabidopsis. Studies to understand the nature of a multimeric complex formed by MADS-box and bHLH proteins would really be a very interesting field for future studies.
STK and CES Play a Role in Carpels Fusion
When we discovered, by backcrossing, that the male part was not compromised in the stk ces-4 and shp1 shp2 ces-4 multiple mutants (Supplementary Figure S2), we decided to perform a detailed analysis of the septum morphology of the mutants displaying the most stronger phenotype related to the percentage of unfertilized ovules. The single mutants stk and ces-4 possess a similar septum respect to the wild type (Figure 4). However, when STK was mutated together with CES the fruits showed a septum fusion defect in the lower middle part in concomitance with the medial region of the ovary. The defective septum was found in the double mutant stk ces-4, in the quadruple mutant bee1 bee3 stk ces-4 and in the quintuple mutant bee1 bee2 bee3 stk ces-4. Notably, no defective carpel fusion was revealed for the quadruple mutants bee1 bee2 bee3 stk and bee1 bee2 bee3 ces-4 (Figure 4) which have a significant percentage of unfertilized ovules (Figure 3), confirming the hypothesis that only when STK and CES are mutated simultaneously the fusion of the carpels was in part compromised. Gynoecium defects were previously described only in the ntt stk double mutant, where there is a failure of the fusion with a septum that has a hole (Herrera-Ubaldo et al., 2019). The organ fusion defects were not disclosed for the single mutants stk and ntt and for the triple mutant ces bee1 bee3 (Crawford and Yanofsky, 2011; Herrera-Ubaldo et al., 2019). In the ces bee1 bee3 and probably in our quadruple mutant bee1 bee2 bee3 ces-4 the unfertilized ovule phenotype is due to defects, as previously described by Crawford and Yanofsky (2011), in ECM components synthases and PCD failure in transmitting tract. In conclusion, only when the dimer STK-CES is not formed, the carpels fusion is compromised.
These phenotypes could be explained by mis-regulation of the genes involved in cuticle formation. The cuticle is a lipidic alteration of the cell wall (Yeats and Rose, 2013). The alteration in cuticle biosynthesis, deposition and wax component transports could cause organ fusion defects in plants, but in particular in flowers and carpels (Nawrath, 2006; Sieber et al., 2007; Panikashvili et al., 2010). Intriguingly transcriptional profiles of the quadruple mutant suggest a strong downregulation of LTP2 (Table 1 and Figure 6), a gene involved in lipid transport and/or wax transport to form the cuticle (Kunst and Samuels, 2003). Notably, the LTP5 gene, expressed in the transmitting tract, which belongs to the same gene family as LTP2, and which did not show significantly altered expression in the quadruple mutant according to our analysis, has previously been shown to be involved in pollen tube guidance (Kunst and Samuels, 2003; Chae et al., 2009). We believe that also LTP2 might be involved in the correct fertilization of the ovules influencing not just the pollen tube growth but also the correct fusion of the two carpels that permits to form the septum and transmitting tract. Moreover, in the bee1 bee3 stk ces-4 quadruple mutant we detected the overexpression of three genes encoding for GDSL esterase/lipase involved in lipid catabolism (Table 1 and Figure 6). The overexpression of a GDSL named, CUTICLE DESTRUCTING FACTOR 1 (CDEF1), causes abnormal organ fusion in leaves, stems and flowers (Takahashi et al., 2010). This gene codes for a protein which is considered a cutinase (Takahashi et al., 2010). It is possible that like for the CDEF1 overexpression, the excess of cutinase in the quadruple mutant respect to wild type caused abnormal organ fusion.
STK and CES Influence Programmed Cell Death in Transmitting Tract
Cell death and tissue degeneration are required for pollen tube growth in the transmitting tract of Arabidopsis (Crawford et al., 2007). The phenotype described for combined ces and bee mutants showed that they fail to form the ECM and to accomplish PCD within the transmitting tract (Crawford and Yanofsky, 2011). Interestingly, STK has already been shown to control PCD in transmitting tract with NTT (Herrera-Ubaldo et al., 2019) but also in the receptive synergid cell through regulation of its direct target genes, VDD and VAL, two Reproductive Meristem (REM) transcription factors (Matias-Hernandez et al., 2010; Mendes et al., 2016).
As we show in the Table 1, in the quadruple mutant bee1 bee3 stk ces-4 several genes involved in cell death are downregulated. The ATWSCP gene encodes for a Kunitz-type protease inhibitor localized preferentially in the transmitting tract and septum, which is involved in ECM and PCD but also in the exit of pollen tubes from the septum to reach the ovules during the fertilization phase (Bektas et al., 2012; Boex-Fontvieille et al., 2015). Also, ACD6 is downregulated (Table 1 and Figure 6), this gene encodes for a protein that promotes cell death in Arabidopsis (Rate et al., 2007). Furthermore, three Arabinogalactan genes (AGPs) resulted downregulated in the quadruple mutant (Table 1 and Figure 6). The AGPs have been associated with ECM composition of the transmitting tract in different plant species (Hoggart and Clarke, 1984; Webb and Williams, 1988; Gane et al., 1995) and they have been shown to play a role in the regulation of PCD during pollen tube growth (Coimbra et al., 2009).
Does Auxin-Brassinosteroid Crosstalk Influence Transmitting Tract Development?
It was recently suggested that the auxin pathway plays an important role in the specification and formation of the tissues along the pistil axis (Larsson et al., 2013; Robert et al., 2015). However, the role of auxin signaling in transmitting tract development is still not fully understood. ARF6 and ARF8 regulate CES expression in the medial domain of the pistil during transmitting tract development (Crawford and Yanofsky, 2011). The double mutant arf6 arf8 displayed reduced alcian blue staining, that detects acidic polysaccharides, the major components of the ECM (Crawford and Yanofsky, 2011). According to our RNAseq data, four Aux/IAAs and two SAURs genes were downregulated in the quadruple mutant in respect to wild type (Table 1 and Figure 6). Aux/IAA proteins and SAURs might be activated in presence of brassinosteroids since evidences of a crosstalk between auxin and brassinosteroid have already been described (Nakamura et al., 2006). Moreover, SAUR9 expression is induced by a combination of auxin and brassinosteroids (van Mourik et al., 2017). Probably a STK-CES-BEE1-BEE3 complex is able to connect the auxin-brassinosteroid crosstalk in transmitting tract tissue, however, the precise role is of this crosstalk in the regulation of transmitting tract development remains unclear.
Finally, from the analysis of the enriched binding sites an interestingly candidate was found to be a possible direct target of the complex composed by the MADS and bHLH transcription factors described in this work. The AT3G49270 gene (upregulated, see Supplementary Table S5) has both CArG and G boxes binding sites. It encodes for an extensin-like protein that has been suggested to have a possible role in postpollination responses such as cell wall protein modification or degradation associated with pistil receptivity (Boavida et al., 2011).
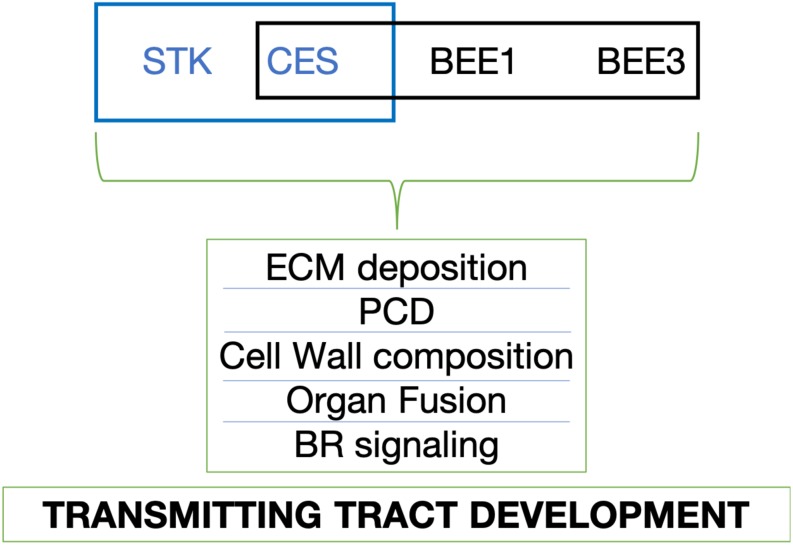
In summary, the data presented here suggest that a MADS-box-bHLH protein complex mastered by the physical interaction of STK-CES, plays a role in the regulation of genes involved in the PCD, ECM and cell wall biosynthesis pathways and by that controlling correct carpel fusion and transmitting tract development to guarantee efficient ovule fertilization (Figure 7).
FIGURE 7.
Proposed model of transmitting tract development controlled by STK, CES, BEE1, and BEE3. The blue box symbolizes the interaction (STK-CES) that we found in this research project; The black box represents the interaction (CES-BEE1-BEE3) that has been previously described (Crawford and Yanofsky, 2011; Poppenberger et al., 2011). ECM, extracellular matrix; PCD, programmed cell death; BR, brassinosteroid.
Data Availability Statement
The datasets generated for this study can be found in the Gene Expression Omnibus (GEO) database under the accession number GSE135559 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135559).
Author Contributions
MD and IR-V designed, performed, analyzed experiments, and wrote the manuscript. EZ, PB, AG, FC, EC, and VG performed experiments. MC performed bio-informatics analyses. MM performed aniline blue staining, analyzed all the results and, contributed writing the manuscript. LC and MK designed experiments, research strategies, and contributed to the writing of the manuscript. All the authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. J. Chory and B. Poppenberger for seed sharing. Prof. J. Martínez for assistance in the genotyping of the bee mutants. We are grateful to I. Latorre for help with in situ hybridizations and general laboratory assistance. We also thank Domenico Loperfido for help with quadruple mutant genotyping and for the collection of the material for RNAseq experiment.
Funding. MD and LC were funded by the SEXSEED project-Horizon2020-MSCA RISE 2016 (690946). IR-V was funded by an EMBO Post-doctoral fellowship and by Fondo per gli Investimenti della Ricerca di Base-Futuro in Ricerca. EZ was funded by EVOCODE. VG was supported by Ministero dell’Istruzione, dell’Universitá e della Ricerca MIUR, SIR2014 MADSMEC, Proposal number RBSI14BTZR. The Ph.D. fellowship of FC was supported by the Doctorate School in Molecular and Cellular Biology, Universitá degli Studi di Milano. MM was funded by SEXSEED and Foundation for Science and technology (FCT), Portugal (SFRH/BPD/99936).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00526/full#supplementary-material
References
- Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43 534–552. 10.1016/j.plipres.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N. (2003). “T-DNA mutagenesis in Arabidopsis BT - plant functional genomics,” in Plant Molecular Biology, ed. Grotewold E. (Totowa, NJ: Humana Press; ), 177–187. [Google Scholar]
- Aoki K., Ogata Y., Shibata D. (2007). Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 48 381–390. 10.1093/pcp/pcm013 [DOI] [PubMed] [Google Scholar]
- Aspeborg H., Coutinho P. M., Wang Y., Brumer H., Henrissat B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. 10.1186/1471-2148-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzà V., Roig-Villanova I., Di Marzo M., Masiero S., Colombo L. (2016). Seed abscission and fruit dehiscence required for seed dispersal rely on similar genetic networks. Development 143 3372–3381. 10.1242/dev.135202 [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J. S., Lister C., Lippman Z., Martienssen R. A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. 10.1038/nature02269 [DOI] [PubMed] [Google Scholar]
- Bektas I., Fellenberg C., Paulsen H. (2012). Water-soluble chlorophyll protein (WSCP) of Arabidopsis is expressed in the gynoecium and developing silique. Planta 236 251–259. 10.1007/s00425-012-1609-y [DOI] [PubMed] [Google Scholar]
- Boavida L. C., Borges F., Becker J. D., Feijó J. A. (2011). Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 155 2066–2080. 10.1104/pp.110.169813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boex-Fontvieille E., Rustgi S., von Wettstein D., Reinbothe S., Reinbothe C. (2015). Water-soluble chlorophyll protein is involved in herbivore resistance activation during greening of Arabidopsis thaliana. J. Exp. Bot. 66 6119–6135. 10.1073/pnas.1507714112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Baum S. F., Eshed Y., Putterill J., Alvarez J. (1999). “Molecular genetics of gynoecium development in Arabidopsis,” in Current Topics in Developmental Biology, eds Pedersen R. A., Schatten G. P. (Cambridge, MA: Academic Press; ), 155–205. 10.1016/s0070-2153(08)60316-6 [DOI] [PubMed] [Google Scholar]
- Chae K., Kieslich C. A., Morikis D., Kim S. C., Lord E. M. (2009). A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21 3902–3914. 10.1105/tpc.109.070854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski I., Wanke D., Birkenbihl R. P., Somssich I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68 81–92. 10.1007/s11103-008-9353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S., Almeida J., Junqueira V., Costa M. L., Pereira L. G. (2007). Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. J. Exp. Bot. 58 4027–4035. 10.1093/jxb/erm259 [DOI] [PubMed] [Google Scholar]
- Coimbra S., Costa M., Jones B., Mendes M. A., Pereira L. G. (2009). Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J. Exp. Bot. 60 3133–3142. 10.1093/jxb/erp148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B. C. W., Ditta G., Yanofsky M. F. (2007). The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 17 1101–1108. 10.1016/j.cub.2007.05.079 [DOI] [PubMed] [Google Scholar]
- Crawford B. C. W., Yanofsky M. F. (2011). HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 138 2999–3009. 10.1242/dev.067793 [DOI] [PubMed] [Google Scholar]
- de Folter S., Angenent G. C. (2006). trans meets cis in MADS science. Trends Plant Sci. 11 224–231. 10.1016/j.tplants.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Di Marzo M., Herrera-Ubaldo H., Caporali E., Novák O., Strnad M., Balanzà V., et al. (2020). SEEDSTICK controls Arabidopsis fruit size by regulating cytokinin levels and FRUITFULL. Cell Rep. 30 2846–2857.e3. 10.1016/j.celrep.2020.01.101 [DOI] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D., Erreni S., Pajoro A., Caporali E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23 2850–2863. 10.1105/tpc.111.087007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18 5370–5379. 10.1093/emboj/18.19.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M., Unterholzner S. J., Rathnayake A. I., Castellanos M., Khan M., Kugler K. G., et al. (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. U.S.A. 113 E5982–E5991. 10.1073/pnas.1611477113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O., Castro-Mondragon J. A., Khan A., van der Lee R., Zhang X., Richmond P. A., et al. (2019). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48 D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D. M., Nemhauser J., Muramitsu T., Maloof J. N., Alonso J., Ecker J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M., Tasaka Y., Tanaka Y., Ashikari T., Kusumi T., Murata N. (1998). Characterization of Δ9 acyl-lipid desaturase homologues from Arabidopsis thaliana. Plant Cell Physiol. 39 247–253. 10.1093/oxfordjournals.pcp.a029364 [DOI] [PubMed] [Google Scholar]
- Gane A. M., Clarke A. E., Bacic A. (1995). Localisation and expression of arabinogalactan-proteins in the ovaries of Nicotiana alata Link and Otto. Sex. Plant Reprod. 8 278–282. 10.1007/BF00229384 [DOI] [Google Scholar]
- Gao M., Showalter A. M. (1999). Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 19 321–331. 10.1046/j.1365-313X.1999.00544.x [DOI] [PubMed] [Google Scholar]
- Guan Y., Nothnagel E. A. (2004). Binding of arabinogalactan proteins by yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol. 135 1346–1366. 10.1104/pp.104.039370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann I., Pidkowich M. S., Girke T., Shanklin J. (2004). Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. U.S.A. 101 10266–10271. 10.1073/pnas.0402200101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Ubaldo H., de Folter S. (2018). Exploring cell wall composition and modifications during the development of the gynoecium medial domain in Arabidopsis. Front. Plant Sci. 9:454. 10.3389/fpls.2018.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Ubaldo H., Lozano-Sotomayor P., Ezquer I., Di Marzo M., Chávez Montes R. A., Gómez-Felipe A., et al. (2019). New roles of NO TRANSMITTING TRACT and SEEDSTICK during medial domain development in Arabidopsis fruits. Development 146:dev172395. 10.1242/dev.172395 [DOI] [PubMed] [Google Scholar]
- Hoggart R. M., Clarke A. E. (1984). Arabinogalactans are common components of angiosperm styles. Phytochemistry 23 1571–1573. [Google Scholar]
- Johnson M. A., Lord E. (2006). “Extracellular guidance cues and intracellular signaling pathways that direct pollen tube growth,” in The Pollen Tube: A Cellular and Molecular Perspective, ed. Malhó R. (Berlin: Springer; ), 223–242. 10.1007/7089_051 [DOI] [Google Scholar]
- Johnson M. A., Preuss D. (2002). Plotting a course: multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. 10.1016/s1534-5807(02)00130-2 [DOI] [PubMed] [Google Scholar]
- Khan M., Rozhon W., Unterholzner S. J., Chen T., Eremina M., Wurzinger B., et al. (2014). Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 5:4687. 10.1038/ncomms5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T., Zhang K., Dong J., Lord E. M. (2006). Exogenous free ubiquitin enhances lily pollen tube adhesion to an in vitro stylar matrix and may facilitate endocytosis of SCA. Plant Physiol. 142 1397–1411. 10.1104/pp.106.086801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Wang Z. Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61 681–704. 10.1146/annurev.arplant.043008.092057 [DOI] [PubMed] [Google Scholar]
- Knauss S., Rohrmeier T., Lehle L. (2003). The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem. 278 23936–23943. 10.1074/jbc.M212585200 [DOI] [PubMed] [Google Scholar]
- Kunst L., Samuels A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42 51–80. 10.1016/s0163-7827(02)00045-0 [DOI] [PubMed] [Google Scholar]
- Langmead B., Steven S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923.Fast [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E., Franks R. G., Sundberg E. (2013). Auxin and the Arabidopsis thaliana gynoecium. J. Exp. Bot. 64 2619–2627. 10.1093/jxb/ert099 [DOI] [PubMed] [Google Scholar]
- Lavy M., Estelle M. (2016). Mechanisms of auxin signaling. Development 143 3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon K. A., Stéphane R., Peter H. K., Lord E. M. (1998). The structure of the transmitting tissue of Arabidopsis thaliana (L.) and the path of pollen tube growth. Sex. Plant Reprod. 11 49–59. [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bionformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42 490–495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord E. M., Russell S. D. (2002). The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18 81–105. 10.1146/annurev.cellbio.18.012502.083438 [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L., Battaglia R., Galbiati F., Rubes M., Eichenberger C., Grossniklaus U., et al. (2010). VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22 1702–1715. 10.1105/tpc.109.068627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B. A., Guilfoyle T. (1987). Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol. Biol. 9 611–623. 10.1007/BF00020537 [DOI] [PubMed] [Google Scholar]
- Mendes M. A., Guerra R. F., Castelnovo B., Silva-Velazquez Y., Morandini P., Manrique S., et al. (2016). Live and let die: a REM complex promotes fertilization through synergid cell death in Arabidopsis. Development 143 2780–2790. 10.1242/dev.134916 [DOI] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (2007). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. 10.2307/3870827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z., Do C. T., Rihouey C., Morin H., Lerouge P., Jouanin L. (2006). Purification, functional characterization, cloning, and identification of mutants of a seed-specific arabinan hydrolase in Arabidopsis. J. Exp. Bot. 57 2339–2351. 10.1093/jxb/erj205 [DOI] [PubMed] [Google Scholar]
- Mizzotti C., Ezquer I., Paolo D., Rueda-Romero P., Guerra R. F., Battaglia R., et al. (2014). SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLoS Genet. 10:e1004856. 10.1371/journal.pgen.1004856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995). Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83 1125–1136. 10.1016/0092-8674(95)90139-6 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Nakajima N., Goda H., Shimada Y., Hayashi K. I., Nozaki H., et al. (2006). Arabidopsis Aux/IAA genes are involved in brassinosteroid-mediated growth responses in a manner dependent on organ type. Plant J. 45 193–205. 10.1111/j.1365-313X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Nawrath C. (2006). Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9 281–287. 10.1016/j.pbi.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Nieuwland J., Feron R., Huisman B. A. H., Fasolino A., Hilbers C. W., Derksen J., et al. (2005). Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17 2009–2019. 10.1105/tpc.105.032094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D., Shi J. X., Bocobza S., Franke R. B., Schreiber L., Aharoni A. (2010). The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Mol. Plant 3 563–575. 10.1093/mp/ssp103 [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E., Wisman E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Poppenberger B., Rozhon W., Khan M., Husar S., Adam G., Luschnig C., et al. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30 1149–1161. 10.1038/emboj.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H. (2000). Phytochrome-interacting factors. Semin. Cell Dev. Biol. 11 457–466. [DOI] [PubMed] [Google Scholar]
- Rate D. N., Cuenca J. V., Bowman G. R., Guttman D. S., Greenberg J. T. (2007). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708. 10.2307/3871047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde J. I., Zuñiga-Mayo V. M., Chávez Montes R. A., Marsch-Martínez N., de Folter S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18 644–655. 10.1016/j.tplants.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Ripoll J. J., Zhu M., Brocke S., Hon C. T., Yanofsky M. F., Boudaoud A., et al. (2019). Growth dynamics of the Arabidopsis fruit is mediated by cell expansion. Proc. Natl. Acad. Sci. U.S.A. 116 25333–25342. 10.1073/pnas.1914096116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H. S., Crhak Khaitova L., Mroue S., Benková E. (2015). The importance of localized auxin production for morphogenesis of reproductive organs and embryos in Arabidopsis. J. Exp. Bot. 66 5029–5042. 10.1093/jxb/erv256 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A. H. K., Yanofsky M. F. (2006). Fruit Development in Arabidopsis. Arabidopsis Book 4:e0075. 10.1199/tab.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., et al. (2007). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. 10.2307/3870872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P., Schorderet M., Ryser U., Buchala A., Kolattukudy P., Metraux J.-P., et al. (2007). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12 721–738. 10.2307/3870997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Shimada T., Kondo M., Tamai A., Mori M., Nishimura M., et al. (2010). Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol. 51 123–131. 10.1093/pcp/pcp173 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. 10.1105/tpc.013839.et [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C. W., Dwyer K. G., Nasrallah M. E., Nasrallah J. B. (2005). Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 138 977–989. 10.1104/pp.105.060558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbant A., Fournet F., Lequart M., Zabijak L., Pageau K., Bouton S., et al. (2016). PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J. Exp. Bot. 67 2177–2190. 10.1093/jxb/erw025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mourik H., van Dijk A. D. J., Stortenbeker N., Angenent G. C., Bemer M. (2017). Divergent regulation of Arabidopsis SAUR genes: a focus on the SAUR10-clade. BMC Plant Biol. 17:245. 10.1186/s12870-017-1210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Nemhauser J. L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21 177–201. 10.1146/annurev.cellbio.21.090704.151241 [DOI] [PubMed] [Google Scholar]
- Verwoerd T. C., Dekker B. M., Hoekema A. (1989). A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acid Res. 17:2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M. C., Williams E. G. (1988). The pollen tube pathway in the pistil of Lycopersicon peruvianum. Ann. Bot. 61 415–423. 10.1093/oxfordjournals.aob.a087573 [DOI] [Google Scholar]
- Wrzaczek M., Brosche M., Kollist H., Kangasjarvi J. (2009). Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc. Natl. Acad. Sci. U.S.A. 106 5412–5417. 10.1073/pnas.0808980106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M., Vainonen J. P., Stael S., Tsiatsiani L., Help-Rinta-Rahko H., Gauthier A., et al. (2015). GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34 55–66. 10.15252/embj.201488582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K., Bacchetto R. G., Lockhart K. M., Friesen L. J., Potts D. A., Covello P. S., et al. (2003). Expression of the Arabidopsis ADS1 gene in Brassica juncea results in a decreased level of total saturated fatty acids. Plant Biotechnol. J. 1 221–229. 10.1046/j.1467-7652.2003.00021.x [DOI] [PubMed] [Google Scholar]
- Yeats T. H., Rose J. K. C. (2013). The formation and function of plant cuticles. Plant Physiol. 163 5–20. 10.1104/pp.113.222737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., Pavesi G. (2009). Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37 247–252. 10.1093/nar/gkp464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Gene Expression Omnibus (GEO) database under the accession number GSE135559 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135559).