Abstract
Objective
To evaluate the human epidermal growth factor receptor 2 (HER2) status in patients with breast cancer using multidetector computed tomography (MDCT)-based handcrafted and deep radiomics features.
Methods
This retrospective study enrolled 339 female patients (primary cohort, n=177; validation cohort, n=162) with pathologically confirmed invasive breast cancer. Handcrafted and deep radiomics features were extracted from the MDCT images during the arterial phase. After the feature selection procedures, handcrafted and deep radiomics signatures and the combined model were built using multivariate logistic regression analysis. Performance was assessed by measures of discrimination, calibration, and clinical usefulness in the primary cohort and validated in the validation cohort.
Results
The handcrafted radiomics signature had a discriminative ability with a C-index of 0.739 [95% confidence interval (95% CI): 0.661−0.818] in the primary cohort and 0.695 (95% CI: 0.609−0.781) in the validation cohort. The deep radiomics signature also had a discriminative ability with a C-index of 0.760 (95% CI: 0.690−0.831) in the primary cohort and 0.777 (95% CI: 0.696−0.857) in the validation cohort. The combined model, which incorporated both the handcrafted and deep radiomics signatures, showed good discriminative ability with a C-index of 0.829 (95% CI: 0.767−0.890) in the primary cohort and 0.809 (95% CI: 0.740−0.879) in the validation cohort.
Conclusions
Handcrafted and deep radiomics features from MDCT images were associated with HER2 status in patients with breast cancer. Thus, these features could provide complementary aid for the radiological evaluation of HER2 status in breast cancer.
Keywords: Breast cancer, human epidermal growth factor receptor 2, multidetector computed tomography, radiomics, deep learning
Introduction
Breast cancer is the most common malignancy in women worldwide and remains the first cause of female cancer-related death (1,2). Breast cancer is a heterogeneous disease with diverse phenotypes that exhibit distinctive biological behaviors (3). Approximately 15%−20% of breast cancers manifest as human epidermal growth factor receptor 2 (HER2) protein overexpression or gene amplification, which is defined as an aggressive subtype associated with metastatic behavior and poor clinical outcomes (4). Nevertheless, the prognosis of the HER2-positive subtype of breast cancer has substantially improved since the development of anti-HER2 targeted therapies. Female patients with HER2-positive breast cancer show good responses and high pathological complete response rates after neoadjuvant chemotherapy with the HER2-blockade agent trastuzumab; further, these patients demonstrate a considerable improvement in both disease-free and overall survival (5-7). Therefore, HER2 status is important for the prognosis of breast cancer and in choosing the optimal individualized treatment strategy for such patients.
HER2-positive cancers tend to accelerate the growth and division of cancer cells, as well as stimulate the cell proliferation and angiogenesis, all of which may cause the tumor heterogeneity (8). In recent years, studies have suggested that the biological characteristics of tumors could be captured using medical images at both genetic and cellular levels (9). Radiomics, the extraction and analysis of quantitative imaging features, enables imaging phenotypes to be correlated with genetic information, which is of great significance for diagnosis, choosing individualized treatment strategies, and predicting the prognosis of tumors (9-13).
Recent studies have tried to use imaging features to assess their associations with the HER2-positive subtype of breast cancer. Although breast radiomics features derived from magnetic resonance imaging (MRI) (14-16) and mammography (MG) (17) are reportedly associated with the HER2-positive subtype, other studies have failed to find a correlation between HER2 status and the features extracted from MRI (18) and positron emission tomography/computed tomography (PET/CT) (19). Accordingly, the current research is still insufficient due to the conflicting relevance; thus, further exploration is required. Multidetector computed tomography (MDCT) also plays an important role in the clinical practice of breast cancer (20), and the 2019 National Comprehensive Cancer Network (NCCN) guidelines of Breast Cancer (version 3. 2019) recommend chest contrast CT examination for patients with breast cancer if pulmonary symptoms are present. Moreover, many patients with breast cancer would undergo MDCT for other reasons as well (e.g., chest pain). The radiomics features within MDCT images may be correlated with the HER2 status in breast cancer, which could provide supplementary assistance with non-invasive imaging evaluation.
Furthermore, conventional radiomics studies generally extracted handcrafted features, which quantify tumor shape, intensity, and texture information based on imaging. However, low-order handcrafted features may be inadequate as they reveal information about medical images from limited aspects; therefore, the tumor heterogeneity may not be fully characterized. Recently, with developments in image recognition and analysis tools, deep learning has drawn increased interest. Deep learning technology, especially convolutional neural network (CNN), is an artificial intelligence algorithm that learns on its own to extract the most predictive features directly from pixel images (21) and has shown remarkable classification and recognition performances in image analysis (22). Compared to handcrafted features, deep features reflect information in medical images from a different perspective and may add further predictive value to HER2 status prediction (23).
To the best of our knowledge, no studies have investigated the correlations between the radiomics features of MDCT images and HER2 status in patients with breast cancer. Therefore, the purpose of this study was to examine if the combination of handcrafted radiomics features and deep learning features based on preoperative MDCT images could evaluate the HER2 status of breast cancer and thus provide complementary aid for the radiological evaluation of HER2 status in patients with breast cancer.
Materials and methods
Patient population
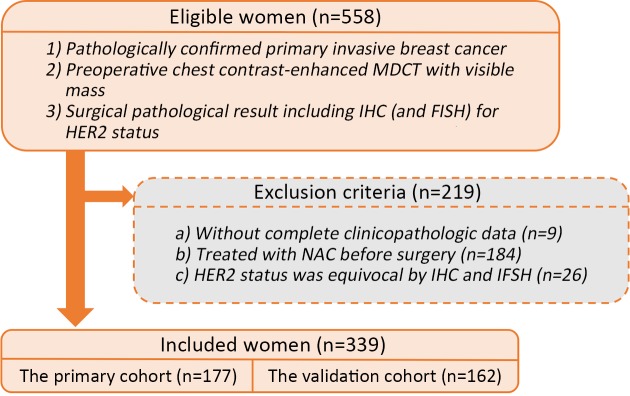
This retrospective study was approved by the Medical Ethics Committee of Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, and the requirement for informed consent was waived. The inclusion criteria were as follows: 1) patients who underwent preoperative contrast-enhanced MDCT between January 2016 and December 2018 with visible breast cancer on the images; 2) histopathological verification of primary invasive breast cancer with surgical resection; and 3) HER2 expression status confirmed by immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH) tests of the surgical specimen. The exclusion criteria were as follows: 1) patients with incomplete clinicopathologic data; 2) patients who were treated with neoadjuvant chemotherapy before surgery; or 3) equivocal HER2 status determined by IHC and FISH tests. The patient recruitment flowchart is presented in Supplementary Figure S1 .
S1. Patient recruitment flowchart. MDCT, multidetector computed tomography; IHC, immunohistochemistry; FISH, fluorescent in situ hybridization; HER2, human epidermal growth factor receptor 2; NAC, neoadjuvant chemotherapy.
In total, 339 female patients who met the criteria were included in this study. Of these, 27 patients had multi-focal or multi-centric lesions, and only the largest tumor lesion of each patient was used for analysis. Finally, the 339 patients were randomly divided into the primary cohort (n=177; age, 50.53±10.49 years old; range, 27−78 years) and the independent validation cohort (n=162; age, 51.88±8.71 years old; range, 35−73 years). The baseline clinical information, including age and tumor location, of recruited patients was collected from the institution archives.
Assessment of HER2 status
The HER2 status of all breast cancer patients included in this study was detected using the surgical specimen without neoadjuvant chemotherapy, and determined using IHC or FISH tests according to the 2013 guideline recommendations of the American Society of Clinical Oncology and College of American Pathologists. The IHC staining intensity of HER2 was graded as 0, 1+, 2+, or 3+. Grades 0 and 1+ were defined as negative, whereas grade 3+ was considered positive. Grade 2+ was equivocal and was further confirmed using FISH, which considered a HER2 gene copy number ≥6 or a HER2/chromosome enumeration probe 17 (CEP17) ratio ≥2.0 as confirmation of HER2 protein overexpression (24).
Radiomics model workflow
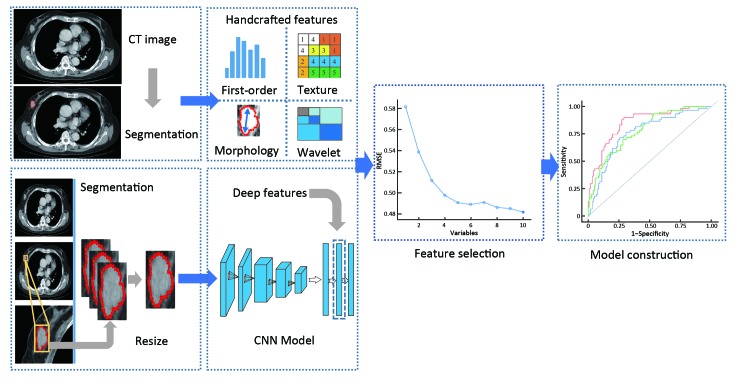
The workflow of radiomics features modelling is illustrated in Figure 1 and includes tumor segmentation and the resized process; handcrafted features and deep features extraction; feature selection; and radiomics signatures and model construction.
1. Flowcharts of radiomics features modelling process. The handcrafted radiomics features were extracted from the manually segmented region of interest. A rectangle containing maximum cross-sectional tumor area was cropped and resized to a fixed size. Deep features were extracted from a generated RGB image after inputting into pre-trained convolutional neural network model. Then both deep and handcrafted features were performed feature selection for radiomics signatures and model building. CT, computed tomography; CNN, convolutional neural network.
MDCT image acquisition and segmentation
Preoperatively, all patients underwent contrast-enhanced chest CT scans, which was performed at different MDCT facilities at Guangdong Provincial People’s Hospital between January 2016 and December 2018. A more detailed description regarding image acquisition and segmentation is provided in Supplementary materials .
Feature extraction
Handcrafted feature extraction
In this study, four categories of handcrafted radiomics features were extracted for analysis: 1) first-order statistics features; 2) size- and shape-based features; 3) texture features; and 4) filter features. The extraction of handcrafted features is described in detail in the Supplementary materials . Handcrafted feature generation was performed via a toolbox developed in-house using MATLAB 2016b (Mathworks, Natick, MA, USA).
Deep feature extraction
Since medical images typically have a limited dataset compared to natural image sets, causing difficulties in training CNN models from scratch, the transfer learning has been proposed to overcome this shortage. Transfer learning is an approach which uses pre-trained models from images of other domains and makes them useful for new datasets (25). Currently, transfer learning is widely implemented in the area of medical deep learning and may alleviate the limitation of small datasets (26).
In this study, transfer learning was performed to extract deep features from two-dimensional MDCT images. Convolutional neural networks fast (CNN-F) (27), as the CNN model used in our study, consisted of five convolutional layers and three fully connected layers, and was pre-trained using the ILSVRC-2012 dataset. The hyperparameters of the pre-trained model were the same as those used by Krizhevsky (28) and are presented in Supplementary materials .
For each patient, the section containing the largest tumor area was selected as the input into the CNN-F. The pre-trained model required three-channel input images (RGB-coded images); however, the medical images in DICOM format were single-channel gray images. Therefore, we first selected and manually segmented the largest tumor area section and cropped the tumor area. Then, the gray values of the segmented region of interest (ROI) were converted into the range (0, 255) using linear transformation (29) and each image was rescaled to 224×224 pixels using bicubic interpolation. Next, three rescaled images were used as the R, G and B channels and stacked into a three-channel image (224×224×3), which met the requirement of the pre-trained CNN-F model input. Finally, the deep features were calculated by forward propagation, and the features of the fully connected layer prior to last fully connected layer were extracted as deep features for subsequent analysis.
The entire deep feature extraction process was performed based on a MATLAB toolbox called MatConvNet (Version 1.0-beta25; http://www.vlfeat.org/matconvnet/).
Feature selection
To construct effective and robust radiomic signatures, a coarse to fine strategy was employed for feature selection. Firstly, depending on the different combinations of the independent segmentation of 100 patients, intra- and inter-class correlation coefficients (ICCs) were used to determine the robust features. Features with ICC values >0.75 were classified as robust features for further analysis. Secondly, univariate analysis (the Mann-Whitney U test) was performed to compare the robust radiomics features between HER2-positive and HER2-negative groups. All features were sorted in ascending order in terms of P values generated from the univariate analysis; of the features, the top 20% were selected. Thirdly, the support vector machine with recursive feature elimination (SVM-RFE) algorithm was used for further feature selection ( 30). SVM-RFE is an efficient feature selection algorithm which ranks the features according to the weight of features based on support vectors. Finally, the number of key features selected for building a radiomics signature was determined by the C-index value using 10-fold cross-validation. This procedure was implemented on both handcrafted feature and deep feature selections in the primary cohort.
Radiomics signature construction
Using the selected key handcrafted features and deep features, the handcrafted- and deep-radiomics signatures for predicting HER2 status were respectively developed using multivariate logistic regression in the primary cohort. We calculated the handcrafted radiomics score (HRad-score) and deep radiomics score (DRad-score) for each patient with a linear combination of the selected features which were respectively weighted by their normalized coefficients. The signatures trained on the primary cohort were applied to the validation cohort for testing in independent cases.
Prediction model development
Multivariate logistic regression was used to select clinical predictive factors (i.e., age, tumor location) for developing the prediction model in the primary cohort. Handcrafted- and deep-radiomics signatures were applied to develop a prediction model in the primary cohort.
Statistical analysis
The differences in clinical characteristics and radiomics scores between the HER2-positive and HER2-negative patients in both the primary and validation cohorts were assessed. The two-sided Mann-Whitney U test was used for continuous variables (i.e., age, Rad-score) and the Chi-square test for categorical variables (i.e., tumor location). The differences in predictive performance between the combined model and the radiomics signatures were tested using the Delong test. Statistical analysis in this study was performed with R software (Version 3.5.2; http://www.R-project.org). The R packages that were used in this study are listed in the Supplementary materials . P-values <0.05 were considered to be statistically significant.
Evaluation of signatures and model performance
To evaluate the radiomics signatures and prediction model performance in this study, we measured the overall performance, discrimination, calibration and clinical usefulness of the model in the primary cohort and then validated it in the validation cohort (31).
Overall performance
Brier score (32) was calculated to assess the overall performance of the radiomics signatures and the prediction model. The Brier score provided a measure of the agreement between the observed binary outcome (i.e., HER2 positive vs. HER2 negative) and the predicted probability of that outcome. The Brier score for a model can range from 0 (perfect model) to 0.25 (non-information model). Generally, a lower Brier score implies better model calibration and discrimination.
Discrimination
The C-index was used to measure the discriminative ability of the radiomics signatures and prediction model. It is equal to the area under the receiver operating characteristic (ROC) curve and varies from 0.5 (no apparent accuracy) to 1.0 (perfect accuracy) (33).
Calibration
Calibration was used to describe the consistency between the actual outcomes and the predictions. The calibration curve was graphically presented as an assessment of calibration, with predictions on the x-axis and the actual outcome on the y-axis. Perfect predictions should be on the 45-degree line. The Hosmer-Lemeshow test was applied to assess the goodness-of-fit of the model, and a high P-value (>0.05) is considered to be reasonable calibration.
Clinical usefulness
To evaluate the clinical utility of the combined model, the decision curve analysis was applied (34). In this study the standardized net benefit (sNB), which ranges from 0 to 1, was used as a function of the risk threshold in the decision curve with visualization. The clinical impact plot was used to visually show the estimated number of patients that had been deemed as high risk for each risk threshold and the true positive cases. An ROC components plot was used to show the constituents of sNB (i.e., the true and false positive rates) (34).
Results
Patients’ clinical characteristics
There was no significant difference in the HER2 status between the primary and validation cohorts (P=0.893). There were also no significant differences in clinical characteristics between the two cohorts (P=0.198−0.893).
The differences in clinical characteristics between the HER2-positive and HER2-negative groups in both the primary and validation cohorts are shown in Table 1 . No significant differences were found in clinical characteristics (age and tumor location) between the HER2-positive and HER2-negative patients in either cohort (P=0.065−0.883).
1. Characteristics of patients in primary and validation cohort.
| Characteristics | Primary cohort | Validation cohort | |||||
| HER2-negative (n=117) | HER2-positive (n=60) | P | HER2-negative (n=105) | HER2-positive (n=57) | P | ||
| HER2, human epidermal growth factor receptor 2; HRad-score, handcrafted radiomics score; DRad-score, deep radiomics score; IQR, interquartile range; P value is calculated from the univariable association analyses between each of the clinicopathological variables and HER2 status. | |||||||
Age (
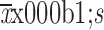 ) (year)
) (year)
|
50.63±9.98 | 50.33±11.48 | 0.864 | 52.64±9.17 | 50.47±7.68 | 0.113 | |
| Tumor location (%) | 0.065 | 0.883 | |||||
| Upper inner quadrant | 42 (23.7) | 14 (7.9) | 31 (19.1) | 20 (12.3) | |||
| Lower inner quadrant | 6 (3.4) | 9 (5.1) | 12 (7.4) | 7 (4.3) | |||
| Lower outer quadrant | 14 (7.9) | 10 (5.6) | 21 (13.0) | 10 (6.2) | |||
| Upper outer quadrant | 55 (31.1) | 27 (15.3) | 41 (25.3) | 20 (12.3) | |||
| HRad-score, median (IQR) | −1.035
(−1.375, −0.590) |
−0.322
(−0.791, 0.095) |
<0.001 | −1.083
(−1.500, −0.581) |
−0.541
(−0.936, 0.0418) |
<0.001 | |
| DRad-score, median (IQR) | −1.012
(−2.041, −0.363) |
−0.143
(−0.691, 0.288) |
<0.001 | −1.308
(−2.056, −0.786) |
−0.278
(−0.777, 0.216) |
<0.001 | |
Feature selection and radiomics signature construction
In total, 5,013 handcrafted and 4,096 deep features were extracted from the ROIs of patients with breast cancer. After feature selection, 7 handcrafted and 7 deep features with preferable predictive value were finally selected in the primary cohort to construct the handcrafted and deep radiomics signatures, respectively. The handcrafted radiomics (HRad)- and deep radiomics (DRad)-scores of each patient were calculated using the formula constructed by the respective selected features (Supplementary materials ).
Evaluation of handcrafted and deep radiomics signatures
The handcrafted radiomics signature demonstrated discriminative ability with a C-index of 0.739 [95% confidence interval (95% CI): 0.661−0.818] in the primary cohort and 0.695 (95% CI: 0.609−0.781) in the validation cohort. The deep radiomics signature demonstrated discriminative ability with a C-index of 0.760 (95% CI: 0.690−0.831) in the primary cohort and 0.777 (95% CI: 0.696−0.857) in the validation cohort. The Brier scores, Hosmer-Lemeshow test results, and sNBs of the handcrafted radiomics signature and deep radiomics signatures are shown inTable 2 .
2. Performance of two radiomics signatures and combined model.
| Performance | Measure | Primary cohort | Validation cohort | |||||
| Handcrafted signature | Deep signature | Combined model | Handcrafted signature | Deep signature | Combined model | |||
| 95% CI, 95% confidence interval; H-L test, Hosmer-Lemeshow test; sNB, standardized net benefit. | ||||||||
| Overall | Brier score | 0.191 | 0.182 | 0.159 | 0.220 | 0.231 | 0.211 | |
| Discrimination | C-index (95% CI) | 0.739 (0.661−0.818) | 0.760 (0.690−0.831) | 0.829 (0.767−0.890) | 0.695 (0.609−0.781) | 0.777 (0.696−0.857) | 0.809 (0.740−0.879) | |
| Calibration | H-L test (P) | 0.674 | 0.294 | 0.887 | 0.416 | 0.161 | 0.528 | |
| Clinical usefulness (T50%) | sNB | 0.100 | 0.150 | 0.333 | 0.070 | 0.263 | 0.360 | |
Performance of combined model
No significant clinical predictors of HER2 status in breast cancer were found using logistic regression analysis, whereas the handcrafted and deep radiomics signatures were identified as independent predictors. Therefore, a combination of the handcrafted and deep radiomics signatures was used to form the prediction model in this study.
The combined model showed better performance for the prediction of HER2 status in breast cancer than either of the two radiomics signatures in both cohorts (Table 2 ). The overall performance of the combined model was improved; the Brier scores decreased from 0.220 and 0.231 to 0.211. The combined model showed good discriminative ability, achieving a C-index of 0.829 (95% CI: 0.767−0.890) in the primary cohort and 0.809 (95% CI: 0.740−0.879) in the validation cohort.
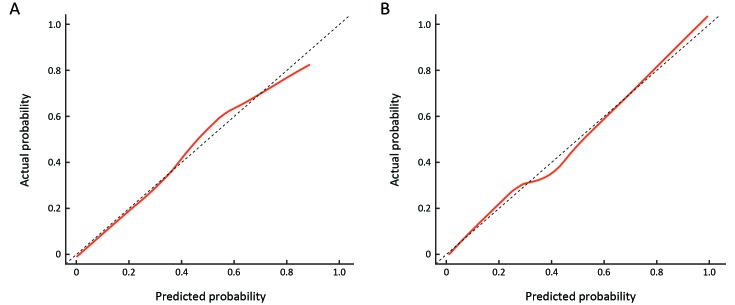
The calibration curve of the combined model for the prediction of HER2 status demonstrated good agreement between the predicted and observed outcomes in both the primary and validation cohorts (Figure 2 ). The Hosmer-Lemeshow test was non-significant in the primary (P=0.887) and validation cohorts (P=0.528), indicating goodness of fitness.
2. Calibration curve of combined model in primary cohort (A) and validation cohort (B). The y-axis represents the actual human epidermal growth factor receptor 2 (HER2)-positive outcome and the x-axis represents the predicted HER2-positive risk. The dotted line represents a perfect prediction. The red solid line represents the predictive performance of the combined model. Perfect prediction should be on the 45° line.
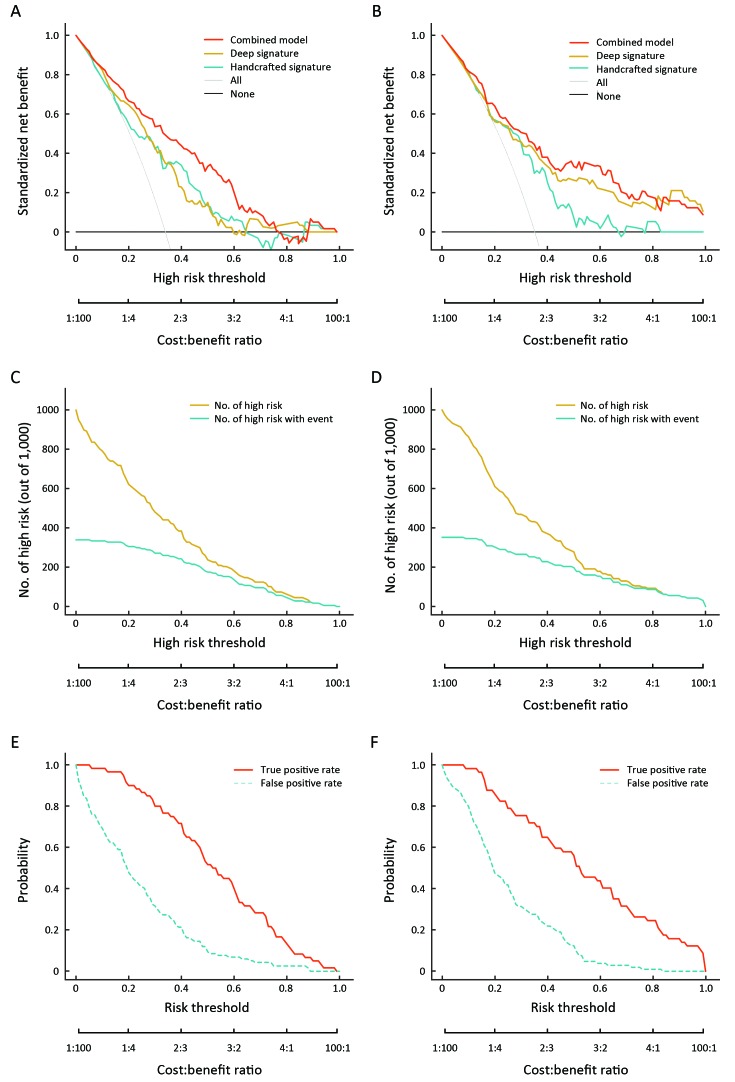
The decision curve analysis for the two radiomics signatures and the combined model in both cohorts is presented in Figure 3A ,B . The decision curve analysis plot showed that the combined model outperformed the two radiomics signatures in the range of thresholds from 0.2 to 0.8. The clinical impact plot showed that, if a 0.5 risk threshold was used, the number of cases identified as high risk of expressing HER2-positive was close to the number of true HER2-positive cases (Figure 3C ,D ). Finally, the true and false positive rates were displayed as functions of the risk threshold in the ROC components plot (Figure 3E ,F ).
3. Evaluation of clinical usefulness. (A,B) Decision curves for the combined model and two radiomics signatures of primary cohort (A) and validation cohort (B). The vertical axis represents standardized net benefit. The two horizontal axes show the correspondence between the risk threshold and the cost-benefit ratio; (C,D) Clinical impact curve for combined model of primary cohort (C) and validation cohort (D). Of 1,000 patients, the solid yellow line represents the number of patients who would be considered high-risk human epidermal growth factor receptor 2 (HER2)-positive for each risk threshold by the combined model. The solid green line indicates the true positive cases with HER2-positive; (E,F) Receiver operating characteristic (ROC) component curve of the combined model for primary cohort (E) and validation cohort (F). The figure shows information similar to that of a ROC curve and also shows the true positive rate (solid red line) and false positive rate (dotted green line) in each risk threshold.
Discussion
In this study, we explored the potential association between the HER2 status of breast cancer and radiomics features extracted from MDCT images. The combined model, which incorporated both handcrafted and deep radiomics features, showed a good performance in evaluating HER2 status; thus, the combined model may be useful as a complementary aid in the radiological evaluation of breast cancer.
HER2-positive status has been shown to be a poor prognostic indicator for breast cancer, which also simultaneously means significant benefits from anti-HER2 targeted therapies (7). Accurately correlating radiological features with the HER2 status in the radiological evaluation of patients with breast cancer is important. Although MDCT is not the primary radiological method for breast cancer evaluation, at Guangdong Provincial People’s Hospital, most patients with breast cancer undergo routine MDCT scans for staging before surgery or neoadjuvant chemotherapy. MDCT is also used for the follow-up of patients with advanced breast cancer. MDCT is advantageous as it has a fast scan time and is capable of multi-planar reconstruction; further, it can be performed when patient are in the supine position (35). It can also simultaneously evaluate the extension of lesions, as well as region lymph nodes, the skin, chest wall and distant metastasis (20).
A previous study by Tamaki et al. found that MDCT features (tumor shape, enhancement pattern and density) are distinct among breast cancer subtypes (36). In recent years, radiomics has appeared as an emerging field that converts medical imaging into quantitative features, which may improve clinical diagnosis, prognosis and prediction, especially in oncology (12,37). In view of the ability of radiomics to noninvasively decode tumor heterogeneity, current studies of other tumors have shown a potential correlation between tumor genotype and CT-based radiomics characteristics (11,38,39). In this study, we examined if radiomics features based on MDCT images reflect HER2 status in patients with breast cancer.
Many previous studies have tried to investigate the relationship between image characteristics and the HER2-enriched subtype in breast cancer; the most commonly used imaging technique was MRI (14,15), followed by MG (17) and PET/CT (19). Chang et al. have demonstrated that breast MRI features that quantify the tumor heterogeneity could be used to indicate HER2 status with an area under the curve (AUC) of 0.8458 (16). Although the AUC was higher than that of our study, it lacked independent validation. Moreover, other studies have found no correlation between the HER2 status and imaging features extracted from MRI (18) and PET/CT (19). As for mammographic features (including tumor size, non-spiculated mass, and calcification) in the study of Nie et al. (40) and MG-based quantitative radiomics features from study of Ma et al. (17) and Zhou et al. (41), above studies reported good performances in differentiating the HER2-enriched subtype, with AUCs of 0.75, 0.78 and 0.787, respectively. The above predictive performances were both slightly poorer than the C-index (which is equal to the AUC) of our study in the validation cohort (C-index: 0.809). Additionally, compared with the generally small HER2-positive dataset in most of the above-mentioned studies, our study enrolled 117 HER2-positive patients (117/339) with surgical pathology for analysis. Therefore, the performance of MDCT radiomics features provides a complementary aid for the radiological evaluation of HER2 status in breast cancer.
In recent years, the integration of multiple markers into one model has been shown to be advantageous for the individualized management of patients and has also been shown to outperform the use of individual markers (12,42). To improve the performance of imaging features in our study, we extracted and incorporated handcrafted and deep radiomics features into the predictive model. CNNs are deep learning models which learn increasingly higher level features from input images through a series of successive linear and nonlinear layers (21,43). Compared to conventional handcrafted radiomics features, high-order deep radiomics features could provide further supplementary information to elevate the performance of the model (44,45). In this study, the model that incorporated both handcrafted and deep radiomics signatures outperformed the individual radiomics signatures in HER2 status evaluation, with a good discriminative ability (C-index=0.829 in the primary cohort; C-index=0.809 in the validation cohort).
As a preliminary study, this study had some limitations. First, we examined a relatively small dataset from a single institution. Second, we only used two-dimensional radiomics features of the largest slice for analysis. Third, MDCT is only a single radiological method; further combinations with radiomics findings from multiple radiological methods including MR and MG are expected to improve the radiological evaluation for HER2 status.
Conclusions
This study indicated that handcrafted and deep radiomics features extracted from MDCT images are associated with HER2 status in breast cancer and may provide complementary, noninvasive assistance in the radiological evaluation of the HER2 status in patients with breast cancer.
Acknowledgements
This work supported by the National Key R&D Program of China (No. 2017YFC1309100), the National Science Fund for Distinguished Young Scholars (No. 81925023), the National Natural Science Foundation of China (No. 81771912, 81701662, 81701782, 81601469, and 81702322) and Science and Technology Planning Project of Guangdong Province (No. 2017B020227012).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Supplementary materials
Image acquisition and segmentation
Acquisition parameters of multidetector computed tomography (MDCT) scanners were listed in Supplementary Table S1 . After all CT images of each patient were retrieved from the picture archiving and communication system (Carestream, Canada) in Guangdong Provincial People’s Hospital, we chose arterial phase for image segmentation and analysis, because this is the phase when the enhancement of tumor lesion showed favorable distinction from adjacent normal glandular tissues.
S1. CT scanning parameters for patients.
| Scanner | Patients No. | Tube voltage (kV) | Tube current (mAs) | Rotation time (s) | Detector collimation (mm) | Field of view (mm) | Matrix | Reconstruction section thickness (mm) | Acquisition time (s) | |
| Pulmonary arterial phase | Arterial phase | |||||||||
| CT, computed tomography; Acquisition time is the time after 80−100 mL of iodinated contrast material (Ultravist 370, Bayer Schering Pharma, Berlin, Germany) was injected into vein at a rate of 4 mL/s with a pump injector (Ulrich CT Plus 150, Ulrich Medical, Ulm, Germany). | ||||||||||
| Philips Ingenuity 64-slice CT (Philips Healthcare, Cleveland, Ohio, USA) | 89 | 120 | 150 | 0.5 | 64×0.625 | 350×350 | 512×512 | 1 | 18 | 35 |
| 256-slice Brilliance iCT (Philips Healthcare, Cleveland, Ohio, USA) | 93 | 120 | 150 | 0.5 | 128×0.625 | 350×350 | 512×512 | 1 | 18 | 35 |
| 64-slice LightSpeed VCT (GE Medical systems, Milwaukee, WI, USA) | 157 | 120 | 150 | 0.4 | 64×0.625 | 350×350 | 512×512 | 1.25 | 18 | 35 |
For handcrafted and deep features extraction, tumor region of interest (ROI) was segmented in the largest cross-sectional area of the CT image using the free open source software ITK-SNAP (Version 3.6.0, http://www.itksnap.org). All ROIs were manually delineated along the tumor outline with exclusion of the air and necrosis area that caused by biopsy. To evaluate the inter-observer and intra-observer reproducibility of the feature extraction, 100 cases were randomly selected for ROI segmentation and feature extraction. The ROI segmentation was performed by two radiologists, Reader 1 and Reader 2, with 8 and 10 years of chest CT interpretation experience, respectively. Reader 1 then repeated the process one month later. Intra- and inter-class correlation coefficients (ICCs) were applied for the evaluation of the intra- and inter- observer agreement of feature extraction. The remaining image segmentation was performed by Reader 1 who was blinded to the patient’s clinicopathological information.
Handcrafted feature extraction
In view of CT images were collected from different CT scanners, we standardized the images to minimize bias and achieve predictive consistency. The ROIs of segmented images were resampled to 1 mm × 1 mm pixels before feature extraction by using bicubic interpolation.
All handcrafted radiomics features were divided into four groups: first order statistics (n=14), size and shape (n=8), textures (n=63), filter features [n=(63+14) ×4×15=4,620]. The process and the radiomics features selected were described in detail below.
Size- and shape-based features
Size- and shape-based features mainly reflects the morphological features of the tumor.
First-order statistics features
First-order statistics features were mainly used to describe the distribution of pixel intensities in CT images.
Texture features
Texture features mainly describe the spatial relationship between pixels. In this study, gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM.), gray level dependence matrix (GLDM), and neighborhood gray tone difference matrix (NGTDM) were used to describe the feature of images.
Filter features
Laplacian of Gaussian filter: A Laplacian of Gaussian spatial band-pass filter (
 ) was utilized to extract image features through adopting different filter parameter σ. The Laplacian of Gaussian filter distribution shows the follow:
) was utilized to extract image features through adopting different filter parameter σ. The Laplacian of Gaussian filter distribution shows the follow:
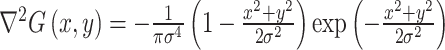 |
Where x, y refers to space coordinates of the pixels, and σ in (1.0, 1.5, 2.0, 2.5).
Wavelet filter: Wavelet features were extracted from decomposed images by different wavelet. Different functions (high-pass or low-pass, represented by H or L) on different scale (X, Y) were represented by a number from 1 to 4 (LL, LH, HL, HH). The following wavelets were used: rbio4.4, rbio3.3, rbio2.6, rbio1.5, bior6.8, bior3.7, bior2.8, bior2.4, bior1.3, coif5, coif2, sym8, sym2, db8, db4.
Deep learning feature extraction
In this study, transfer learning was performed to overcome the challenge of the limited amount of data for deep learning. Convolutional neural network (CNN)-F, as the CNN model used in our study, consists of five convolutional layers and three fully connected layers and is pre-trained on the ILSVRC-2012 dataset. The hyper-parameter of the model is the same as that used by Krizhevsky: weight decay 5 × 10−4, momentum 0.9, initial learning rate 10−2. When the validation error stops reducing, the initial rate drops to 0.1.
A total of 4,096 features was extracted from the last fully-connected layer. These features have a very abstract meaning, and we use a special nomenclature to name them as
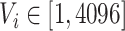 .
.
Statistical analysis
The following R packages were used in the study:
Caret: for feature selection
rms: logistic model building, nomograms, calibration plots, VIF
rmda: decision curve analysis, clinical impact plot
ResourceSelection: Hosmer-Lemeshow goodness of fit test
Calculation of radiomics score
Handcrafted radiomics signature
After feature selection produce, 7 handcrafted features were selected. A handcrafted radiomics signature was constructed with 7 handcrafted features by using the logistic regression model. The handcrafted radiomics score for each patient was computed by follow formula (the P value of coefficients and abbreviation description of 7 handcrafted radiomics features were show in Supplementary Table S2 ):
S2. P value of coefficients in handcrafted radiomics signature.
| Features | Coefficient | P |
| GLSZM, gray level size zone matrix; SALGLE, small area low gray level emphasis; GLCM, gray level co-occurrence matrix; GLRLM, gray level run length matrix; LRHGLE, long run high gray level emphasis; LoG, laplacian of gaussian. | ||
| bior2.8_LH_GLSZM_SALGLE | 8.037 | 0.0430 |
| bior3.7_LL_ GLCM_energy | 1.502 | 0.0009 |
| bior6.8_HL_GLRLM_LRHGLE | −0.006 | 0.0130 |
| coif5_LL_GLCM_energy | −12.186 | 0.0490 |
| db8_HL_GLSZM_SALGLE | −15.893 | 0.0110 |
| LoG_1.5_GLCM_correlation | −0.905 | 0.0440 |
| LoG_1.0_ GLSZM_SALGLE | −2.470 | 0.0140 |
 =
=
 where
where
αX=0.654 + 8.037* bior2.8_LH_GLSZM_SALGLE + 1.502* bior3.7_LL_ GLCM_energy–0.006* bior6.8_HL_GLRLM_LRHGLE – 12.186* coif5_LL_GLCM_energy – 15.893* db8_HL_GLSZM_SALGLE – 0.905* LoG_1.5_GLCM_correlation – 2.47* LoG_1.0_ GLSZM_SALGLE
Deep radiomics signature
Same as the handcrafted radiomics signature, the deep radiomics score was obtained as follow (the P value of coefficients of 7 deep radiomics features were shown in Supplementary Table S3 ):
S3. P value of coefficients in deep radiomics signature.
| Features | Coefficient | P |
| V40 | 2.311 | 0.0070 |
| V60 | 2.531 | 0.0470 |
| V1136 | −1.198 | 0.0320 |
| V1653 | −2.239 | 0.0270 |
| V1781 | −2.256 | 0.0006 |
| V2847 | −7.434 | 0.0020 |
| V4027 | −1.538 | 0.0050 |
 =
=
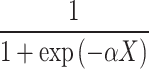 where
where
αX= 2.798 + 2.311*V40 + 2.531*V60 – 1.198*V1136 – 2.239*V1653 – 2.256*V1781 – 7.434*V2847 – 1.538*V4027
Contributor Information
Zaiyi Liu, Email: zyliu@163.com.
Changhong Liang, Email: liangchanghong@gdph.org.cn.
References
- 1.Siegel RL, Miller KD, Jemal A Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553–68. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sørlie T, Eisen MB, et al Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Loibl S, Gianni L HER2-positive breast cancer. Lancet. 2017;389:2415–29. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 5.Prat A, Pineda E, Adamo B, et al Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Prat A, Cheang MC, Galván P, et al Prognostic value of intrinsic subtypes in hormone receptor-positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol. 2016;2:1287–94. doi: 10.1001/jamaoncol.2016.0922. [DOI] [PubMed] [Google Scholar]
- 7.Llombart-Cussac A, Cortés J, Paré L, et al HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18:545–54. doi: 10.1016/s1470-2045(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 8.Mohammed RA, Ellis IO, Mahmmod AM, et al Lymphatic and blood vessels in basal and triple-negative breast cancers: characteristics and prognostic significance. Mod Pathol. 2011;24:774–85. doi: 10.1038/modpathol.2011.4. [DOI] [PubMed] [Google Scholar]
- 9.Aerts HJ, Velazquez ER, Leijenaar RT, et al Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevaert O, Mitchell LA, Achrol AS, et al Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–74. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlo CA, Di Paolo PL, Chaim J, et al Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270:464–71. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YQ, Liang CH, He L, et al Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34:2157–64. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Tian J, Dong D, et al Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23:4259–69. doi: 10.1158/1078-0432.ccr-16-2910. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Zhu Y, Burnside ES, et al Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2(pii):16012. doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutton EJ, Dashevsky BZ, Oh JH, et al Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging. 2016;44:122–9. doi: 10.1002/jmri.25119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang RF, Chen HH, Chang YC, et al Quantification of breast tumor heterogeneity for ER status, HER2 status, and TN molecular subtype evaluation on DCE-MRI. Magn Reson Imaging. 2016;34:809–19. doi: 10.1016/j.mri.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Ma W, Zhao Y, Ji Y, et al Breast cancer molecular subtype prediction by mammographic radiomic features. Acad Radiol. 2019;26:196–201. doi: 10.1016/j.acra.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm LJ Breast MRI radiogenomics: Current status and research implications. J Magn Reson Imaging. 2016;43:1269–78. doi: 10.1002/jmri.25116. [DOI] [PubMed] [Google Scholar]
- 19.Antunovic L, Gallivanone F, Sollini M, et al [18F]FDG PET/CT features for the molecular characterization of primary breast tumors . Eur J Nucl Med Mol Imaging. 2017;44:1945–54. doi: 10.1007/s00259-017-3770-9. [DOI] [PubMed] [Google Scholar]
- 20.Kang DK, Kim MJ, Jung YS, et al Clinical application of multidetector row computed tomography in patient with breast cancer. J Comput Assist Tomogr. 2008;32:583–98. doi: 10.1097/RCT.0b013e31815074ce. [DOI] [PubMed] [Google Scholar]
- 21.LeCun Y, Bottou L, Bengio Y, et al Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998;86:2278–324. doi: 10.1109/9780470544976.ch9>. [DOI] [Google Scholar]
- 22.Cicero M, Bilbily A, Colak E, et al Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol. 2017;52:281–7. doi: 10.1097/RLI.0000000000000341. [DOI] [PubMed] [Google Scholar]
- 23.Anthimopoulos M, Christodoulidis S, Ebner L, et al Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35:1207–16. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond ME, Hicks DG, et al Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–56. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yosinski J, Clune J, Bengio Y, et al How transferable are features in deep neural networks? Eprint Arxiv. 2014:3320–8. [Google Scholar]
- 26.Christodoulidis S, Anthimopoulos M, Ebner L, et al Multisource transfer learning with convolutional neural networks for lung pattern analysis. IEEE J Biomed Health Inform. 2017;21:76–84. doi: 10.1109/JBHI.2016.2636929. [DOI] [PubMed] [Google Scholar]
- 27.Chatfield K, Simonyan K, Vedaldi A, et al. Return of the devil in the details: Delving deep into convolutional nets. Computer Science 2014.
- 28.Krizhevsky A, Sutskever I, Hinton GE Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–05. [Google Scholar]
- 29.Sasikala V, LakshmiPrabha V. A comparative study on the swarm intelligence based feature selection approaches for fake and real fingerprint classification. 2015 International Conference on Soft-Computing & Networks Security (ICSNS) Coimbatore 2015, pp:1-8.
- 30.Guyon I, Weston J, Barnhill S, et al Gene Selection for Cancer Classification using Support Vector Machines. Machine Learning. 2002;46:389–422. doi: 10.1023/a:1012487302797>. [DOI] [Google Scholar]
- 31.Steyerberg EW, Vickers AJ, Cook NR, et al Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology. 2010;21:128–38. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arkes HR, Dawson NV, Speroff T, et al The covariance decomposition of the probability score and its use in evaluating prognostic estimates. Med Decis Making. 1995;15:120–31. doi: 10.1177/0272989X9501500204. [DOI] [PubMed] [Google Scholar]
- 33.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. 1982;143:29-36.
- 34.Kerr KF, Brown MD, Zhu K, et al Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol. 2016;34:2534–40. doi: 10.1200/JCO.2015.65.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrone A, Lo Mele L, Sassi S, et al MDCT of the breast. AJR Am J Roentgenol. 2008;190:1644–51. doi: 10.2214/AJR.07.3145. [DOI] [PubMed] [Google Scholar]
- 36.Tamaki K, Ishida T, Miyashita M, et al Multidetector row helical computed tomography for invasive ductal carcinoma of the breast: correlation between radiological findings and the corresponding biological characteristics of patients. Cancer Sci. 2012;103:67–72. doi: 10.1111/j.1349-7006.2011.02116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambin P, Rios-Velazquez E, Leijenaar R, et al Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mei D, Luo Y, Wang Y, et al CT texture analysis of lung adenocarcinoma: can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging. 2018;18:52. doi: 10.1186/s40644-018-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Dong D, Fang M, et al Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28:2058–67. doi: 10.1007/s00330-017-5146-8. [DOI] [PubMed] [Google Scholar]
- 40.Nie Z, Wang J, Ji XC Microcalcification-associated breast cancer: HER2-enriched molecular subtype is associated with mammographic features. Br J Radiol. 2018:20170942. doi: 10.1259/bjr.20170942. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Tan H, Bai Y, et al Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur J Radiol. 2019;121:108718. doi: 10.1016/j.ejrad.2019.108718. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Ding J, Spuhler K, et al Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2019;49:131–40. doi: 10.1002/jmri.26224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soffer S, Ben-Cohen A, Shimon O, et al Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology. 2019;290:590–606. doi: 10.1148/radiol.2018180547. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Man C, Gong L, et al A deep learning radiomics model for preoperative grading in meningioma. Eur J Radiol. 2019;116:128–34. doi: 10.1016/j.ejrad.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Paul R, Hawkins SH, Hall LO, et al. Combining deep neural network and traditional image features to improve survival prediction accuracy for lung cancer patients from diagnostic CT. 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC) Budapest, 2016, pp:002570-002575.