Abstract
More than 40 different neurological diseases are caused by microsatellite repeat expansions. Since the discovery of RAN translation by Zu et al. in 2011, eight expansion disorders have been identified as RAN-positive diseases. RAN proteins are translated from different types of nucleotide repeat expansions and can be produced from both sense and antisense transcripts. In some diseases, RAN proteins have been shown to accumulate in affected brain regions. Here we review the pathological and molecular aspects associated with RAN protein accumulation for each particular disorder, the correlation between disease pathology and the available in vivo models and the common aspects shared by some of the newly discovered RAN proteins.
Introduction
More than 40 neurological diseases are caused by microsatellites repeat expansions, including Huntington disease (HD), fragile X syndrome (FXS), myotonic dystrophy types 1 and 2 (DM1, DM2), C9orf72 amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD), spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7, 8, 10, 12, 17, 31 and 36 and Fuchs endothelial corneal dystrophy (FECD)1–4
Microsatellites repeats are tandem stretches of 2–10 nucleotides in the DNA. These repeats, which are normally found within the genome, are often polymorphic in length. At some genetic loci microsatellite repeats become genetically unstable5,6 and result in disease when the sequence length exceeds a certain threshold. This size threshold differs for each repeat- harboring gene. In HD, which is caused by a CAG•CTG expansion mutation within the huntingtin (HTT) gene, disease causing mutations typically range from 40–70 CAG repeats, whereas individuals with DM1 harbor hundreds to thousand CTG•CAG repeats in the DMPK gene7,8.
Disease-causing repeat expansion mutations can be found in exonic, intronic as well as 5’ or 3’ untranslated regions (UTRs). The location of various expansion mutations within their respective genes has been historically used to group specific expansion diseases into one of the following mechanistic categories: 1)Protein Gain of function (GOF), exemplified by HD and other polyglutamine expansion diseases in which the expansion mutation is contained within a ORF and translated into polyGln expanded motifs; 2)RNA GOF, for mutations located in the non-coding regions of their corresponding genes. One clear example occurs in DM1, where CUG expanded DMPK transcripts accumulate as intranuclear RNA foci and sequester RNA binding proteins belonging to the muscleblind-like (MBNL) family, which prevents them from performing their function; 3) FXS provides a protein loss of function (LOF) example. FXS is caused by expansions of >200 repeats of a CGG located in the promoter and 5’-UTR of the FMR1 gene. The expanded CGG region undergoes methylation, leading to transcriptional silencing of the FMR1 gene and therefore, the absence or loss of function of the FMR1 protein9.
A “Pathological mystery “
Although significant data supports the contribution of the expanded RNA and expanded polyglutamine proteins in specific dominantly inherited repeat expansion disorders, the molecular mechanisms underlying these diseases is still unclear. Several observations suggest a lack of correlation between the pathological findings and the expression of the corresponding expanded polyglutamine protein or RNA, suggesting that some pieces of the “pathogenic mechanism” puzzle remain to be uncovered.
Some examples come from the CAG/polyGln expansion disorders, from cases in which the established causative mutant protein accumulates in brain regions not typically associated with neurodegeneration. For example, white matter alterations reported in HD or SCA3 can be detected at very early stages of the disease, but these changes appear to occur in the absence or with minimal accumulation of Huntingtin or ataxin-3 polyGln aggregates. These polyGln aggregates preferentially accumulate inside nuclei of specific types of neurons7,10–13. One clear example is the HD cerebellum, where recent reports describe white matter alterations7,14,15 that are mainly negative for polyglutamine16, and still haven’t been sufficiently explored despite signs of possible cerebellar dysfunction in HD. Another example is the accumulation of SCA7 polyGln aggregates in the cerebral cortex17 a brain region typically not considered to be a primary site of neuropathology. Additionally, several studies propose that soluble or oligomeric polyglutamine proteins are toxic and the aggregates themselves may be neuroprotective18. Taken together, these observations suggest that in addition to polyGln other molecular factors contribute to disease.
An additional pathological puzzle is illustrated by fragile X tremor ataxia syndrome (FXTAS). After the disease was first recognized in elderly men who carry CGG premutation alleles in FMR1, large ubiquitin positive inclusions which colocalize with CGG expansion RNAs were observed in neurons and astrocytes throughout the brain19–24. While the number of inclusions correlated with the size of the CGG repeat expansion21,22, they did not contain the CGG-expanded mutant protein FMRP and in a FXTAs mouse model, the ubiquitin aggregates contained a very limited amount of FMR1 mRNA25. These data suggested that additional molecular components contribute to these FXTAS inclusions.
Changing perspectives: bidirectional transcription and RAN translation
The additional discovery that bidirectional transcription occurs at the DM126 and SCA827 loci offered a different view of how expansion mutations could cause disease. For DM1, small 21nt RNAs were reported26. For SCA8, CUG and CAG expansion transcripts are expressed27,28. Bidirectional transcription is now recognized as common across many microsatellite expansion29–31 raising the possibility that sense and antisense transcripts and resulting proteins contribute to disease.
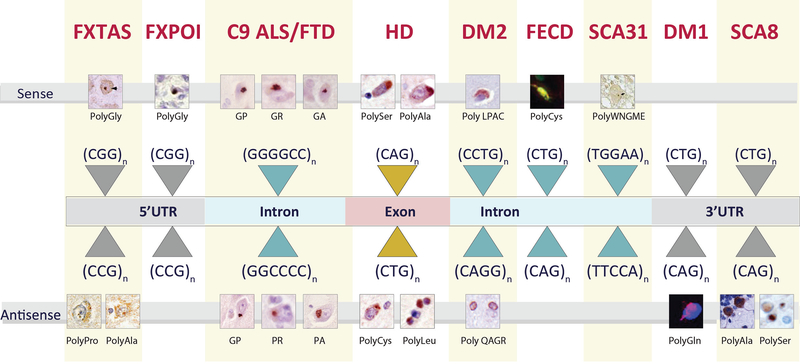
The discovery of repeat associated non-AUG (RAN) translation in 201132 added another level of complexity to possible molecular mechanisms underlying repeat diseases. Several types of microsatellite expansion mutations have been shown to undergo RAN translation. Because RAN translation can occur in all three reading frames across both sense and antisense expanded transcripts, a cocktail of up to six proteins may result from a single mutation. RAN translation was initially reported in SCA8 and DM132 and has become a fast-growing field with nine different RAN diseases reported to date. The current list includes SCA8 (CTG•CAG expansion), DM1 (CTG•CAG), C9orf72 ALS/FTD (GGGGCC•CCCGG), fragile X tremor ataxia syndrome (FXTAS) and fragile X premature ovarian insufficiency (FXPOI) (CGG•CCG), HD (CAG•CTG), SCA31 (TGGAA•TTCCA), DM2 (CCTG•CAGG) and FECD (CTG•CAG)16,30,32–39. Here we review the pathological features of RAN protein accumulation across these diseases.
RAN proteins accumulation and disease pathology
SCA8
Spinocerebellar ataxia type 8 (SCA8) is a dominantly inherited progressive ataxia caused by a CTG•CAG expansion in the overlapping ATXN8OS and ATXN8 genes. SCA8 symptoms include limb and gait ataxia, dysarthria and nystagmus40. Although SCA8 is generally an adult-onset disease, infantile and juvenile forms have been reported41.
The CUG-expanded ATNX8OS transcripts lead to the formation of RNA foci and the sequestration of MBNL proteins42. The initial observation of SCA8 CUG expansion transcripts43 was followed by the discoveries of a CAG expansion transcript that encodes an ATG-initiated polyGln protein27. Later, polyAla32 and polySer33 RAN proteins, expressed from ATXN8 CAG expansion transcripts, were found.
The RAN polyAla protein (translated from the GCA frame) was detected in the soma and dendrites of Purkinje cells from SCA8 human postmortem tissue and SCA8 BAC mice32. In contrast, SCA8 RAN polySer protein (from the AGC frame) accumulates in the cerebellum of SCA8 patients and SCA8 transgenic mice33 (Fig. 1). Curiously, the ATG-initiated polyGln protein and RAN polySer protein show strikingly different patterns of accumulation, with intranuclear polyGln aggregates appearing primarily in Purkinje cells, whereas the RAN polySer accumulates primarily subcortical, and deep white matter regions of the cerebellum (Table 1). SCA8 polyGln and polySer aggregates are also detected in the frontal cortex, hippocampus and brain stem, in similar regions, but do not colocalize33.
Fig. 1.
RAN proteins identified in patient tissue
Table 1.
Summary of RAN proteins identified in neurologic disease.
| Transcript | RAN protein | Protein accumulation in vivo |
|---|---|---|
| C9 ALS/FTD | ||
| Sense | GP | All C9 RNA protein are found as cytoplasmic or perinuclearaggregates in different brain regions of C9 ALS/FTD postmortem tissue, including frontal and motor cortex, hippocampus, dentate gyrus granular layer, cerebellar molecular layer and spinal cord of. In a C9 BAC transgenic mouse model with motor and neurodegenerative phenotype, PolyGlyAla accumulation increase with mouse age and phenotype. Individual C9 RAN proteins induce toxicity both in vitro and in vivo. Some of the C9-RAN mediated toxic mechanisms include alterations of the nucleocytoplasmic transport, ER stress, oxidative stress, stress granule formation, deficits in protein translation or DNA damage (26, 27,29,31,58,59,61–68,70–78,80–82). |
| GR | ||
| GA | ||
| Antisense | GP | |
| PR | ||
| PA | ||
| FXTAS | ||
| Sense | PolyGly | PolyGly protein accumulates as nuclear and perinuclear aggregates in the frontal cortex and hippocampus of FXTAS patients, frequently colocalizimg with ubiquitin. A specific signal for polyGly protein is detected by Western blotting in lysates from FXTAS cerebellum. Colocalization between polyGly and ubiquitin is also detected in a FXTAS Drosophila model carrying 90 CGG repeats (32). |
| Antisense | PolyPro | Specific PolyPro intranuclear aggregates detected t in cortex, hippocampus, cerebellum, midbrain and pons from FXTAS brain tissue. These aggregates are mainly found un neurons and colocalize with Ubiquitin (90). |
| PolyAla | Show a perinuclear and intranuclear accumulation in the neurons of FXTAS hippocampus and cortex, being also found in midbrain and pons. Intranuclear PolyAla inclusions are Ubiquitin positive (90). | |
| FXPOI | ||
| Sense | PolyGly | FXPOI RAN PolyGly accumulates in the ovarian stromal cells of a FXPOI patient and FXTAS mice. Mice show and increase number of primordial follicules at 40 weeks and polyGly aggregates in the pituitary gland. PolyGly accumulates as intranuclear inclusions that are ubiquitin positive (104). |
| HD | ||
| Sense | polySer | All four proteins accumulate in grey matter regions and more abundantly in white matter regions of the striatum, frontal cortex and cerebellum of HD postmortem brain tissue. RAN positive show apoptotic and neuroinflammatory markers. HD-RAN proteins primarily show cytoplasmic, nuclear or perinuclear localization in neurons. In glial cells, the proteins predominantly show nuclear localization. Although both soluble and aggregated accumulation patters are frequently detected, polySer proteins accumulate as dotlike aggregates more frequently than polyAla, polyCys and polyLeu (33). |
| polyAla | ||
| Antisense | PolyCys | |
| polyLeu | ||
| SCA8 | ||
| Sense | PolySer | Detected in the cerebellum of SCA8 patients and in the cortex, hippocampus, brain stem and cerebellum of SCA8 BAC transfenic mice. In mice, polySer accumulates as dot-like agreggates in an age-dependent manner that correlates with the mouse motor dysfunction phenotype. PolySer preferentially accumulates in white matter regions that show demyelination and axonal degeneration (30). |
| Antisense | PolyAla | Accumulates in the cerebellum SCA8 mice and in the cerebellum of SCA8 patients. RAN SCA8 PolyAla is detected in the soma and dendrites of Purkinje cells (28). |
| DM1 | ||
| Antisense | PolyGIn | Accumulates as nuclear aggregates in DM1 cardiac myocytes and leukocytes. PolyGIn inclusions are also found in myoblasts and skeletal muscle at low frequency. DM1 mice carrying 55CTG repeats show polyGIn staining in leukocytes in cardiac tissue (28). |
| DM2 | ||
| Sense | LPAC | Primarily accumulates as small cytoplasmic or perinuclear aggregates in neurons, but also occasionally in glial cells, in brain regions showing abundant macrophages. Cortex, hippocampus and striatum are positive for LPAC acumulation (34). |
| Antisense | QAGR | Show nuclear accumulation that forms small agregates around the nuclear membrane. Preferentially accumulates in white matter regions displaying rarefaction of the fibers. QAGR is detected in DM2 cortex, hippocampus and striatum (34). |
| SCA31 | ||
| Sense | WNGME | Detected in the eye imaginal discs of a SCA31 Drosophila model and in the cerebellum of SCA31 patients, where it accumulates as dot-like granules in the cell bodies and dendrites of Purkinje cells. In flies, WNGME levels correlate with increased eye degeneration (35). |
| FECD | ||
| Sense | PolyCys | Detected as nuclear immunostaining in the comeal endothelium of FECD patients carrying ≥ 55 repeats (36). |
| Antisense | PolyGIn | Detected by Western blot in fibroblast derived from FECD patients that are homozygous for the repeat expansion (36). |
In SCA8 BAC transgenic mice, polySer aggregates increase with age and disease progression, initially accumulating in the brainstem at two months of age, which is consistent with the early motor defects observed in these mice27,33. In both SCA8 mice and human post mortem tissue, SCA8 polySer preferentially accumulates in white matter regions that also show signs of axonal breakage and demyelination. In contrast, no signs of axonal loss or demyelination were found in brain regions negative for polySer accumulation33.
Taken together, these data support possible roles for polyGln, RAN polyAla and RAN polySer proteins in SCA8 pathology. The location of polyGln and RAN polyAla suggest a role in Purkinje cell loss32 and the accumulation of RAN polySer in the white matter regions suggest its involvement in demyelination32,33,44,45. Additionally, the CUG transcripts form RNA foci in Purkinje cells. It is also possible that RAN proteins expressed from CUG expansion transcripts will be found in the future and may contribute to disease.
C9orf72 ALS/FTD
C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia (C9-ALS/FTD) is a dominantly inherited disorder caused by a GGGGCC hexanucleotide repeat expansion in the first intron of the C9orf72 gene. This mutation is the most common known genetic cause of both familial and sporadic ALS and FTD46–48.
C9orf72 ALS, is characterized by the degeneration of upper and lower motor neurons, which leads to muscle weakness and paralysis, respiratory failure and death, usually within 2–5 years49. For FTD, marked neurodegeneration occurs in the frontal and anterior temporal lobes, leading to speech difficulties and cognitive and behavioral abnormalities, such as loss of empathy, abrupt mood changes, disinhibition and behavioral changes50,51.
The C9orf72 expansion mutation is bidirectional transcribed and both sense (GGGGCC) and antisense (GGCCCC) RNA foci accumulate throughout the brain and spinal cord52. The expanded transcripts also undergo RAN translation, generating C9-polyGlyPro (GP), C9-polyGlyArg (GR) and C9-polyGlyAla (GA) RAN proteins from the sense strand; and C9-polyGlyPro (GP), C9-polyProArg (PR) and C9-polyProAla (PA) proteins from the antisense transcript30,31,53.
Although the 2011 discovery of the GGGGCC mutation that connects ALS/FTD with the microsatellite repeat disorders is quite recent46,47, there has been an intense research focus on understanding the pathogenic mechanisms. Data supporting C9orf72 protein LOF, sense and antisense RNA GOF, and RAN protein toxicity are all being hotly pursued. Here we focus on the role of C9 RAN proteins in disease.
RAN proteins are found in multiple regions in postmortem CNS tissue, including frontal and motor cortex, hippocampus, cerebellum and spinal cord. RAN proteins typically accumulate as cytoplasmic or perinuclear cytoplasmic aggregates primarily in neurons. While RAN protein aggregates have been observed in motor neurons, they are relatively rare in these ALS vulnerable cells yet abundant in other brain regions that are not typically thought to be affected in ALS patients including the cerebellum. These data raise questions about whether or not RAN proteins are a primary driver of disease54–57 It is also possible that cells which are most vulnerable in C9 ALS/FTD patients, including motor neurons, are highly sensitive to RAN proteins and die before aggregates are visible or that cells that had RAN protein aggregates had already died, and are no longer detectable in most postmortem cases. RAN positive immunostaining shows variable density, with some brain areas showing intense and clustered accumulation of C9-RAN proteins, while other regions show a more scattered pattern of RAN protein aggregates. C9-RAN protein aggregates colocalize with p62 and are TDP-43 negative30,31,34,35,57,58. While TDP43 aggregates do not co-localize with C9 RAN proteins, a hallmark feature of C9 and other forms of ALS are cytoplasmic TDP43 aggregates. A recent Drosophila study links RAN protein accumulation, karyopherin-α pathology and TDP-43 accumulation and mislocalization59.
Additional experimental models support a toxic role for individual C9 RAN proteins in the absence of RNA GOF effects: PR and GR have been shown to be highly toxic in multiple model systems including cultured cells60–65, zebrafish66, Drosophila60,67–73 and mice74, where the can affect development, motor performance and cellular function. GA and PA proteins have also been reported to be toxic in a variety of in vitro and in vivo models66,67,75–79 and GA accumulation have been correlated with neurodegeneration across both C9 ALS and FTD pathology80.
The cellular pathways affected by RAN protein overexpression are numerous and include ER stress74,76, oxidative stress and DNA damage81, protein translation abnormalities73, nuclear transport deficits69,70,82–84 and stress granule formation74. Nevertheless, it is less clear if RAN proteins play a major role in disease when expressed at endogenous levels. To address this issue, number of BAC transgenic mouse models have been developed using the endogenous human promoters85–88. Surprisingly, two of these models showed molecular phenotypes including the accumulation of RAN proteins but did not develop phenotypic manifestations of the disease. Two models developed molecular and disease phenotypes. The Zhu model showed mild behavioral abnormalities and subtle neuronal loss in the hippocampus. The Liu et al., model developed classic features of ALS and FTD including weight loss, paralysis, motor neuron loss, cortical and hippocampal degeneration, muscle denervation, anxiety-like behavior and decrease survival. RAN protein aggregates are found in regions showing neurodegeneration and increase with disease onset and phenotype.
FXTAS and FXPOI
Fragile X tremor ataxia syndrome (FXTAS) is a late-onset neurodegenerative disorder caused by a 55–200 long CGG repeat in the 5’UTR of the FMR1 gene on the X chromosome. The disease primarily affects older men who develop progressively worsening tremor, gait ataxia, parkinsonism and dementia20,89–92. A pathological hallmark of FXTAS is the accumulation of eosinophilic nuclear inclusions that are ubiquitin positive in both neurons and astrocytes21,22. In contrast to Fragile X syndrome patients (FXS), in which expansion mutation (>200 repeats) shut down gene expression, FMR1 transcript levels are 2–8 times higher in FXTAS patients than in control individuals93,94. FXTAS CGG-expansion transcripts RNAs form foci that have been reported to sequester RBPs including Purα95 hnRNP A2/B196 and Sam6897 causing splicing deficits in FXTAS brain samples and neurodegeneration in Drosophila models96–98
FMR1 CGG expansion RNAs undergo RAN translation in a length-dependent manner, producing polyGly and polyAla proteins in the GGC and GCG reading frames, respectively36. PolyGly accumulation was detected in FXTAS fly and mouse models, and in the frontal cortex, cerebellum and hippocampus of postmortem FXTAS brains36. Homopolymeric RAN proteins are also expressed from antisense CCG expansion RNAs. The accumulation of antisense polypro (CCG), polyArg (CGC) and polyAla (GCC) have been detected in cell culture99. PolyPro accumulates in hippocampus, cortex, midbrain and pons while polyAla is detected in hippocampus, cortex, and midbrain99 (Fig. 1, Table 1).
Interestingly, polyGly, polyPro and polyAla RAN proteins all preferentially accumulate as neuronal perinuclear or intranuclear inclusions, which are also ubiquitin positive, a previously established pathological marker of FXTAs21,22,100, suggesting that RAN proteins contribute to disease. Further evidence supporting a role for polyGly in disease comes from a mouse model study. Sellier et al., compared FXTAS mice with or without a close cognate initiation codon required for polyGly expression101. The first model, expressing both CGG-expanded RNA and polyGly aggregates showed brain inflammation, Purkinje cell loss, motor impairment and decreased survival. In contrast, no significant pathology was observed in mice expressing CGG expansion transcripts in the absence of polyGly protein. The expression of the FMR1 polyGly protein also causes death in neuronal cell cultures101. The polyGly repeat motif was shown to be necessary for protein aggregation and the C-terminal region is required to drive cell toxicity, possibly through interactions with LAP2b and disruptions in nuclear lamina101.
Fragile X premature ovarian insufficiency (FXPOI), like FXTAS, is caused by expansions of 55–200 CGGs in the 5’UTR of the FMR1 gene. FXPOI causes early (≤40 years old) ovarian dysfunction in women carrying the repeat expansion, who are also at risk of developing FXTAS later in life89,102,103. For many years it has been known that CGG-expanded FMR1 transcripts are upregulated in FXPOI patients and ubiquitin-positive intranuclear inclusions accumulate in the ovarian stromal cells of FXPOI patients104. A recent study identified RAN polyGly aggregates in ovarian stromal cells of a single FXPOI patient, which are also positive for ubiquitin105.
PolyGly/ubiquitin aggregates are also found in a FXTAS knock-in mice carrying 98 CGG repeats. Animal studies show polyGly aggregates increase with age and are abundant in 40 week old mice. Older animals also show a higher number of primordial follicles (1.5 fold) compared to wild type animals105. Additionally, RAN polyGly aggregates are found in the pituitary gland of FXTAS mice but not control mice105, suggesting that polyGly might contribute to ovarian failure by affecting the hypothalamus-pituitary-adrenal axis.
Taken together, these studies show RAN proteins accumulate in patient samples and in animal models. While there is substantial evidence that polyGly is toxic, additional work will be required to understand the contributions that polyGly and other known and yet to be detected RAN proteins play in FXTAS and FXPOI.
HD
Huntington disease (HD) is a dominantly inherited disorder caused by a CAG•CTG expansion mutation in the first exon of the HTT gene2,7. The disease is characterized by severe motor, cognitive and psychiatric alterations that normally presents adult-onset, but can also manifest early in life with more severe and faster progressing juvenile onset cases. HD is fully penetrant at 40 CAG•CTG repeats, and there is a clear correlation between longer repeat expansion and earlier onset and more severe forms of the disease.
The HD expansion mutation results in an abnormally expanded polyGln tract in the huntingtin (HTT) protein. Since the discovery of the causative gene in 1993106, most research has focused on understanding the toxic role of the mutant, polyGln-expanded, ATG-initiated HTT protein7,107. RNA-mediated toxicity has also been reported to play a role in HD through the expression of CUG-expanded antisense transcripts108 and the generation of CAG microRNA species109. However, critical aspects of HD, such as the differential vulnerability of specific brain regions and the distinct and more severe pathology in juvenile onset cases are not fully understood.
The possibility of RAN translation occurring in HD has been explored by different groups with polyAla and polySer expressed from HTT minigenes in cultured cells32,110. In 2015, Banez-Coronel et al.16 demonstrated that RAN translation can also occur across relatively small repeat expansions located in protein coding regions and also in vivo. The HD CAG/CTG expansion expressed four novel homopolymeric RAN proteins from both sense and antisense transcripts. Banez et al., showed polyAla, polySer, polyLeu and polyCys RAN proteins accumulate in several brain regions including caudate/putamen, cortical white matter and cerebellum (Fig. 1, Table 1), with RAN protein signal dramatically higher in the cerebellum of juvenile-onset cases showing severe cerebellar atrophy.
HD RAN protein accumulation is variable, and can be detected by increased nuclear or cytoplasmic staining, with soluble or aggregated patterns. Although HD-RAN proteins are detected in neurons, they are more frequently found in glial cells. Strikingly, prominent RAN protein staining was observed in the white matter bundles of the striatum, Bergman glia, white matter regions of the cerebellum and white matter regions around the dentate nuclei, regions in which polyGln aggregates are absent or minimal. These data suggest RAN proteins contribute to the white matter abnormalities reported in HD111–115.
Another important link between HD-RAN accumulation and disease pathology comes from the frequent colocalization between RAN proteins and active Caspase3. These data suggest that RAN positive cells are damaged or undergoing cell death. HD-RAN proteins are more frequently found in brain regions showing atrophy, astrogliosis and microglial activation, and areas showing severe atrophy16. All four identified HD-RAN proteins are toxic to neural cells in vitro, independent of RNA-mediated effects16.
Taken together, these findings demonstrate that sense and antisense RAN proteins accumulate in HD brains and correlate with sites of degenerative changes.
DM2
Myotonic Dystrophy type 2 is caused by an intronic CCTG expansion in the CNBP gene116. This multisystemic disease, which causes muscle weakness and myotonia, also affects the heart, the eye, the endocrine system and the brain. CNS features include executive function deficits and white matter abnormalities117–122. DM2 CCUG-expanded transcripts form nuclear RNA foci which sequester the muscleblind (MBNL) family of RNA binding proteins, causing abnormalities in RNA localization and processing1,123,124. For many years, RNA GOF effects have been considered to be the major driver of disease in DM2. However, a recent study by Zu et al.38 showed the DM2 tetranucleotide expansion is bidirectionally transcribed and produces both, sense (CCUG) and antisense (CAGG), expansion transcripts. These transcripts, in turn, undergo RAN translation, generating tetrapeptide leucine-proline-alanine-cysteine (LPAC) and glutamine-alanine-glycine-arginine (QAGR) repeat expansion proteins without an AUG-initiation codon. These RAN proteins accumulate in the cortex, hippocampus and striatum of DM2 patient brains with specific patterns (Fig. 1): LPAC accumulates as small cytoplasmic punctate aggregates that can be perinuclear or located in the cell processes, while QAGR immunostaining is primarily nuclear, with small aggregates at or in close proximity to the nuclear membrane (Table 1).
LPAC is consistently detected in grey matter regions of the brain. LPAC aggregates are found primarily in neurons, and occasionally in glia. There is considerable variability in the frequency of LPAC positive cells within the brain, even within the same brain regions. Interestingly, LPAC positive regions show abundant macrophage staining, suggesting that inflammation can be a trigger that favors RAN translation or that RAN LPAC proteins trigger inflammatory processes. In contrast, QAGR primarily accumulates in the white matter regions of the brain and QAGR positive white matter regions show rarefaction of fibers, suggesting a role for QAGR in axonal loss and increased water intercalation38 (Table 1). Additionally, both LPAC and QAGR are toxic to cultured cells in the absence of RNA-mediated effects, supporting a pathogenic role of these RAN proteins. Zu et al. also showed that MBNL overexpression decreases RAN protein levels by sequestering CCUG expansion transcripts within the nuclei. When CCUG repeats are not sequestered they can be exported into the cytoplasm where they undergo RAN translation.
DM1
Myotonic dystrophy type 1 is a dominantly inherited neuromuscular disorder with multisystemic features including myotonia, progressive muscle weakness and wasting, cataracts, cardiac effects, testicular atrophy and CNS abnormalities119. The disease is caused by a CAG•CTG expansion in the 3’UTR of the DMPK gene. Healthy individuals have 5–38 repeats while DM1 patients carry large expansions containing hundreds or thousands of repeats8.
DM1 is generally considered an RNA GOF disease based on a wealth of genetic and biochemical data and on the location of the causative expansion in a non-coding region125,126. CUG expansion transcripts expressed from the DM1 locus form nuclear RNA foci which sequester the RBP MBNL1 and cause alternative splicing abnormalities127,128.
The detection of a polyGln RAN protein in DM1 raises the possibility that RAN proteins contribute to this disease32. DM1 polyGln nuclear aggregates have been detected in cardiac myocytes of DM1 mice, as well as in skeletal muscle and peripheral leucocytes in DM1 human samples32 (Fig. 1, Table 1). The colocalization of DM1 PolyGln with caspase 8 in human leukocytes suggests polyGln RAN proteins can be toxic. While RAN proteins expressed in the polyGln frame are found in several DM1 mouse and human tissues, additional work and better antibody tools are needed to understand when and where DM1 RAN proteins accumulate, if proteins from multiple frames are expressed and if they accumulate in the brain, a common theme in other repeat expansion disorders.
SCA31
Spinocerebellar ataxia type 31 (SCA31) is an adult-onset autosomal dominant neurodegenerative disorder that shows progressive cerebellar ataxia and Purkinje cell degeneration129. SCA31 is associated with a TGGAA•TAAAA pentanucleotide repeat expansion. The mutation is located in an intronic region shared by the genes brain expressed, associated with Nedd4 (BEAN1) from one DNA strand, and thymidine kinase 2 (TK2) from the opposite strand130,131. In support of an RNA GOF model, Nimi et al., show that UGGAA expansion transcripts form nuclear RNA foci in Purkinje cells132. Additionally, Ishiguro et al.37 demonstrated the accumulation of a tryptophan-asparagine-glycine-methionine-glutamic acid (WNGME) pentapeptide repeat protein in vivo. This repeat is distinct from other expansion mutations in that an ATG codon, which may be used for translation initiation is embedded throughout the repeat tract.
Both RNA foci and WNGME RAN proteins accumulate in an SCA31 fly model and induce severe neurodegeneration and a shortened life span. SCA31 RAN pentapeptides accumulate as granular structures in the cell body and dendrites of Purkinje cells in the cerebellum of SCA31 patients, but not in control cases (Fig. 1, Table 1). Interestingly, the RBPs TDP-43, FUS and hnRNPA2B1 bind to the UGGAA RNA expansion acting as chaperones and reducing RNA foci, RAN protein levels and toxicity in the fly eye37. Although new studies are necessary to further unravel the pathogenic mechanisms in SCA31, these results suggest that RNA GOF, RAN protein GOF or both mechanisms are involved in SCA31 onset and progression.
FECD
The most recent addition to the list of RAN protein disorders is Fuchs Endothelial Corneal Distrophy (FECD). FECD is an inherited degenerative disease that severely affects the corneal endothelium and results in corneal edema and, in severe cases, vision loss133,134. There are several genes associated with FECD, but the most specific genetic association known is a CTG•CAG expansion in the third intron of TCF4135.
Similar to DM1 and SCA8 disorders, CUG expansion transcripts expressed from TCF4 form nuclear RNA foci that colocalize with MBNL1, which is believed to underlie the aberrant RNA splicing found in the cornea of FECD patients136. Additional experiments by Soragni et al.39 showed that overexpression CUG expansion transcripts from FECD minigenes, generates RAN polyCys proteins in cultured cells, which cause toxicity and oxidative stress in immortalized corneal endothelial cells. PolyCys is also detected in in the corneal endothelium of patients with FECD39 (Fig. 1, Table 1). Additionally, this group detected a polyGln protein in FECD fibroblasts. Similar to other expansion diseases, further studies are needed to understand the contributions that RNA GOF and RAN proteins have in FECD.
Common aspects across RAN protein diseases
Although each RAN positive disorder has its own set of distinct clinical features and pathology, individual RAN proteins, even if they harbor the same repeat motif, should be considered distinct because they often have unique flanking sequences and tissue accumulation patterns. Nevertheless, there are several common themes underlying RAN translation and RAN-mediated pathology that provide insight.
Repeat length dependence.
RAN translation is often repeat length dependent, with a length threshold required for RAN protein production and increased RAN proteins accumulation with longer repeat tracts. Length-dependent RAN translation has been observed using a variety of minigene constructs for SCA832, HD109, FXTAS36 and C9orf72 ALS/FTD30.
In FXTAS cell culture experiments, RAN polyAla proteins are detected from constructs expressing 88 CGG repeats but not from constructs with 30 repeats36. Similarly, for HD, transfection experiments using HTT exon1 minigenes showed RAN polyAla proteins were detected in cells expressing ≥50 CAG repeats and RAN polySer detected with 35 CAG repeats. The polySer aggregates showed progressively increasing aggregation with longer repeat lengths16. The correlation of higher RAN protein levels and increased RAN protein aggregates may contribute to the repeat-length dependent genetic anticipation that is found in most of the repeat expansion disorders.
White matter accumulation.
The prominent RAN protein accumulation in damaged white matter brain regions in HD, DM2 and SCA8 cases16,33,38 supports the hypothesis that RAN proteins play a role in the white matter alterations that can occur very early in each of these diseases. The mechanisms underlying these white matter abnormalities have historically been considered a consequence of neuronal death process since the expected toxic CUG RNA or polyGln proteins do not accumulate in these regions. The findings of astroglyosis, demyelinization, microglial and/or caspase activation in white matter regions with abundant RAN protein accumulation offer an exciting new pathogenic hypothesis that needs further study. It is possible that RAN white matter accumulation will be found in other known and yet to be identified RAN protein diseases137–139.
Variable RAN protein accumulation.
Immunohistochemistry studies have shown that RAN protein aggregates frequently show variable densities. For example, some brain areas show clustered accumulation and high-dense staining while other areas within the same brain region (e.g. the hippocampus, cerebellum of frontal cortex) show less frequent or no RAN positive cells30,33,109. These data suggest localized molecular or environmental triggers can activate RAN translation. The observation of RAN translation clusters in regions of infarction (DM2)38 suggest that external factors such as hypoxia, inflammatory molecules, free radicals, specific metabolites or energy deficits might promote RAN translation140–142. Another possibility is that RAN proteins or factors required for RAN translation can propagate from cell to cell, facilitating RAN accumulation in neighboring cells. This hypothesis has been validated for C9orf72 GA, GP and PA RAN proteins in cultured cells143,144.
A clear understanding of the mechanisms underlying variable or patchy RAN protein accumulation may provide valuable knowledge about disease progression and regional susceptibility, a possibility suggested by Zu et al30.
Prospecting for proteins in other diseases
RAN translation is a rapid moving field, with nine different repeat disorders identified as RAN positive diseases since the discovery of RAN translation by Zu et al. in 201132. The demonstration that RAN translation can occur from repeats located in both coding and non-coding regions suggests RAN proteins may be found in additional polyGln expansion disorders. Interestingly, recent reports show ATXN8OS expansions and FXTAS mutations, as well as FXTAS-polyGly proteins are found in autopsy brains from patients with progressive supranuclear palsy (PSP) and Parkinsonism145,146. These findings highlight that known or yet to be discovered repeat expansion mutations may contribute to the symptomatology of additional neurological disorders.
Better detection tools are badly needed to identify novel RAN-translated proteins and to characterize diseases already known to express one or more RAN proteins36,39,101. Reduced protein solubility, low protein levels, age- or stress-dependent RAN protein expression140–142 all present technical challenges for protein detection in patient samples. Additionally, with the exception of polyGln antibodies, it has been difficult to generate specific antibodies to detect homopolymeric repeat tracts, which would allow screening for RAN proteins across diseases with the same repeat expansion mutation (e.g. the CAG•CTG SCAs). It is also likely that with the recent advances in DNA including long sequencing technology147, novel repeat expansion diseases will be identified.
Blocking RAN proteins as a therapeutic strategy
The discovery of RAN proteins in an increasing number of expansion disorders and the various lines of evidence that support their pathogenic role highlights the need to develop strategies to block RAN translation. This would allow scientist to test and better understand the contribution of RNA GOF vs. RAN protein toxicity and may lead to the development of therapeutic strategies. The use of antisense oligonucleotides (ASOs) to degrade expansion RNAs has received a lot of attention. While ASOs provide a platform to degrade multiple types of disease-causing expansion transcripts and therefore would reduce RAN protein levels expressed from those transcripts at the same time, ASO-based therapeutic efforts have focused on targeting the expanded transcript that was initially discovered and not the other strand148–154.
The recurrent observations that bidirectional transcription and RAN translation occur across both strands of the repeat expansion mutation suggests that targeting both sense and antisense transcripts may be required for some of these disorders. It will also be important to consider the possibility that downregulation of sense transcripts may lead to the upregulation of antisense transcripts in some of these diseases155,156.
An interesting alternative therapeutic approach would be to block RAN translation. This could be achieved using strategies to prevent expansions RNAs from leaving the nucleus including the overexpression of RBPs such as MBNL38 or TDP-4337 or by reducing levels of factors that favor RAN translation such as the eukaryotic initiation factor 3F (eIF3F)33.
Additional therapeutic approaches can be aimed at preventing the toxicity of specific RAN proteins. Efforts aligned with this strategy include the generation of drugs that prevent RAN protein function and/or interactions, targeting RAN proteins for clearance through ubiquitination, or the use of antibodies to specifically target individual or sets of RAN proteins. For example, anti-RAN protein antibodies have been shown to prevent protein propagation in cell culture143.
Conclusions
RAN translation has now been reported in nine diseases caused by repeat expansion mutations. Different microsatellite motifs, including tri-, tetra-, penta- and hexanucleotide repeat expansions are permissive for RAN translation, suggesting RAN translation may be a common process across most microsatellite expansion disorders. The development of better tools to detect RAN products is likely to identify new RAN-positive diseases as well as novel RAN protein motifs. Because the production of both sense and antisense RAN proteins is a common feature found in many of these disorders, developing therapeutic strategies to target both sense and antisense RAN proteins will be important. Finally, the development of new in vivo models in that allow both sense and antisense expression will help the community to test and better understand the promise and limitations of various therapeutic strategies.
Acknowledgments
We thank the National Institutes of Health (RO1 NS098819, R37NS040389 and PO1-NS058901), Target ALS, the ALS Association, Packard Center, National Ataxia Foundation, Myotonic Dystrophy Foundation and Muscular Dystrophy Association for support.
References
- 1.Ranum LP & Cooper TA RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29, 259–277, doi: 10.1146/annurev.neuro.29.051605.113014 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Orr HT & Zoghbi HY Trinucleotide repeat disorders. Annu Rev Neurosci 30, 575–621, doi: 10.1146/annurev.neuro.29.051605.113042 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Wieben ED et al. A Common Trinucleotide Repeat Expansion within the Transcription Factor 4 (TCF4, E2–2) Gene Predicts Fuchs Corneal Dystrophy. Plos One 7, doi:ARTN e49083 10.1371/journal.pone.0049083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.paulson HL Repeat expansion diseases. Vol. 147 105–123 (Elsevier, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson DL, Orr HT & Warren ST The unstable repeats--three evolving faces of neurological disease. Neuron 77, 825–843, doi: 10.1016/j.neuron.2013.02.022 S0896–6273(13)00180–3 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson CE, Nichol Edamura K & Cleary JD Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 6, 729–742, doi: 10.1038/nrg1689 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bates G, Tabrizi SJ & Jones L (Oxford University Press, New York, 2014). [Google Scholar]
- 8.Zerylnick C, Torroni A, Sherman SL & Warren ST Normal variation at the myotonic dystrophy locus in global human populations. Am J Hum Genet 56, 123–130 (1995). [PMC free article] [PubMed] [Google Scholar]
- 9.Willemsen R, Levenga J & Oostra BA CGG repeat in the FMR1 gene: size matters. Clin Genet 80, 214–225, doi: 10.1111/j.1399-0004.2011.01723.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson HL The spinocerebellar ataxias. J Neuroophthalmol 29, 227–237, doi: 10.1097/WNO0b013e3181b416de (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulson HL et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Schmidt T et al. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol 8, 669–679 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz E et al. Intranuclear inclusions, neuronal loss and CAG mosaicism in two patients with Machado-Joseph disease. J Neurol Sci 200, 19–25 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Rub U et al. Degeneration of the cerebellum in Huntington’s disease (HD): possible relevance for the clinical picture and potential gateway to pathological mechanisms of the disease process. Brain Pathol 23, 165–177, doi: 10.1111/j.1750-3639.2012.00629.x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees EM et al. Cerebellar Abnormalities in Huntington’s Disease: A Role in Motor and Psychiatric Impairment? Movement Disord 29, 1648–1654, doi: 10.1002/mds.25984 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Banez-Coronel M et al. RAN Translation in Huntington Disease. Neuron 88, 667–677, doi: 10.1016/j.neuron.2015.10.038 S0896–6273(15)00932–0 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmberg M et al. Spinocerebellar ataxia type 7 (SCA7): a neurodegenerative disorder with neuronal intranuclear inclusions. Hum Mol Genet 7, 913–918 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Michalik A & Van Broeckhoven C Pathogenesis of polyglutamine disorders: aggregation revisited. Hum Mol Genet 12 Spec No 2, R173–186, doi: 10.1093/hmg/ddg295 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Brouwer JR, Willemsen R & Oostra BA The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am J Med Genet B Neuropsychiatr Genet 150B, 782–798, doi: 10.1002/ajmg.b.30910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquemont S, Hagerman RJ, Hagerman PJ & Leehey MA Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol 6, 45–55, doi: 10.1016/S1474-4422(06)70676-7 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Greco CM et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain 125, 1760–1771 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Greco CM et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain 129, 243–255, doi: 10.1093/brain/awh683 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Greco CM et al. Testicular and pituitary inclusion formation in fragile X associated tremor/ataxia syndrome. J Urol 177, 1434–1437, doi: 10.1016/j.juro.2006.11.097 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Iwahashi CK et al. Protein composition of the intranuclear inclusions of FXTAS. Brain 129, 256–271, doi: 10.1093/brain/awh650 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Hagerman PJ Current Gaps in Understanding the Molecular Basis of FXTAS. Tremor Other Hyperkinet Mov (N Y) 2, doi: 10.7916/D80C4TH0 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho DH et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20, 483–489 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Moseley ML et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38, 758–769, doi:ng1827 [pii] 10.1038/ng1827 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Ranum LP et al. Double the trouble: bidirectional expression of the SCA8 CAG/CTG expansion mutation - evidence for RNA and protein gain of function effects. Rinsho Shinkeigaku 50, 982–983, doi:JST.JSTAGE/clinicalneurol/50.982 [pii] (2010). [DOI] [PubMed] [Google Scholar]
- 29.Batra R, Charizanis K & Swanson MS Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet 19, R77–82, doi: 10.1093/hmg/ddq132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zu T et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A 110, E4968–4977, doi: 10.1073/pnas.1315438110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendron TF et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126, 829–844, doi: 10.1007/s00401-013-1192-8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zu T et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 108, 260–265, doi: 10.1073/pnas.1013343108 1013343108 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayhan F et al. SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by eIF3F. EMBO J 37, doi: 10.15252/embj.201899023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ash PE et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646, doi: 10.1016/j.neuron.2013.02.004 S0896–6273(13)00135–9 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori K et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338, doi: 10.1126/science.1232927 science.1232927 [pii] (2013). [DOI] [PubMed] [Google Scholar]
- 36.Todd PK et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455, doi: 10.1016/j.neuron.2013.03.026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishiguro T et al. Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31. Neuron 94, 108–124 e107, doi: 10.1016/j.neuron.2017.02.046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zu T et al. RAN Translation Regulated by Muscleblind Proteins in Myotonic Dystrophy Type 2. Neuron 95, 1292–1305 e1295, doi: 10.1016/j.neuron.2017.08.039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soragni E et al. Repeat-Associated Non-ATG (RAN) Translation in Fuchs’ Endothelial Corneal Dystrophy. Invest Ophthalmol Vis Sci 59, 1888–1896, doi: 10.1167/iovs.17-23265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Day JW, Schut LJ, Moseley ML, Durand AC & Ranum LP Spinocerebellar ataxia type 8: clinical features in a large family. Neurology 55, 649–657 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Felling RJ & Barron TF Early onset of ataxia in a child with a pathogenic SCA8 allele. Pediatr Neurol 33, 136–138, doi: 10.1016/j.pediatrneurol.2005.02.010 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Daughters RS et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 5, e1000600, doi: 10.1371/journal.pgen.1000600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koob MD et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 21, 379–384, doi: 10.1038/7710 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Kumar N & Miller GM White matter hyperintense lesions in genetically proven spinocerebellar ataxia 8. Clin Neurol Neurosurg 110, 65–68, doi: 10.1016/j.clineuro.2007.08.014 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Kim JS, Son TO, Youn J, Ki CS & Cho JW Non-Ataxic Phenotypes of SCA8 Mimicking Amyotrophic Lateral Sclerosis and Parkinson Disease. J Clin Neurol 9, 274–279, doi: 10.3988/jcn.2013.9.4.274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeJesus-Hernandez M et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256, doi: 10.1016/j.neuron.2011.09.011 S0896–6273(11)00828–2 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renton AE et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268, doi: 10.1016/j.neuron.2011.09.010 S0896–6273(11)00797–5 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majounie E et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 11, 323–330, doi: 10.1016/S1474-4422(12)70043-1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowland LP & Shneider NA Amyotrophic lateral sclerosis. N Engl J Med 344, 1688–1700, doi: 10.1056/NEJM200105313442207 (2001). [DOI] [PubMed] [Google Scholar]
- 50.McKhann GM et al. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol 58, 1803–1809, doi:nsa10000 [pii] (2001). [DOI] [PubMed] [Google Scholar]
- 51.Bang J, Spina S & Miller BL Frontotemporal dementia. Lancet 386, 1672–1682, doi: 10.1016/S0140-6736(15)00461-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizielinska S et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol 126, 845–857, doi: 10.1007/s00401-013-1200-z (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori K et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126, 881–893, doi: 10.1007/s00401-013-1189-3 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Gomez-Deza J et al. Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathol Com 3, doi:UNSP 38 10.1186/s40478-015-0218-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackenzie IRA et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathologica 130, 845–861, doi: 10.1007/s00401-015-1476-2 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Davidson YS et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Com 2, doi:Artn 70 10.1186/2051-5960-2-70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackenzie IR et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126, 859–879, doi: 10.1007/s00401-013-1181-y (2013). [DOI] [PubMed] [Google Scholar]
- 58.Mackenzie IR & Neumann M Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J Neurochem 138 Suppl 1, 54–70, doi: 10.1111/jnc.13588 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Solomon DA et al. A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-alpha mediates C9orf72-related neurodegeneration. Brain 141, 2908–2924, doi: 10.1093/brain/awy241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen X et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225, doi: 10.1016/j.neuron.2014.12.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamakawa M et al. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24, 1630–1645, doi: 10.1093/hmg/ddu576 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Tao Z et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24, 2426–2441, doi: 10.1093/hmg/ddv005 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Kanekura K et al. Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet 25, 1803–1813, doi: 10.1093/hmg/ddw052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moens TG, Partridge L & Isaacs AM Genetic models of C9orf72: what is toxic? Curr Opin Genet Dev 44, 92–101, doi: 10.1016/j.gde.2017.01.006 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Kwon I et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145, doi: 10.1126/science.1254917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swaminathan A et al. Expression of C9orf72-related dipeptides impairs motor function in a vertebrate model. Hum Mol Genet 27, 1754–1762, doi: 10.1093/hmg/ddy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizielinska S et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194, doi: 10.1126/science.1256800 science.1256800 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee KH et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 167, 774–788 e717, doi: 10.1016/j.cell.2016.10.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freibaum BD et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133, doi: 10.1038/nature14974 nature14974 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boeynaems S et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep 6, 20877, doi: 10.1038/srep20877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang D et al. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol 130, 525–535, doi: 10.1007/s00401-015-1448-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon I et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145, doi: 10.1126/science.1254917 science.1254917 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moens TGN,T; Wilson KM; Atilano ML; Birsa N; Gittings LM; Holbling BV; Dyson MC; Thoeng A; Neeves J; Glaria I; Yu L.; Bussmann J,; Storkebaum E; Pardo M; Choudhary JS; Fratta P; Partridge L; Isaacs AM (Acta Neuropathol, 2019). [DOI] [PMC free article] [PubMed]
- 74.Zhang YJ et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat Med 24, 1136–1142, doi: 10.1038/s41591-018-0071-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohki Y et al. Glycine-alanine dipeptide repeat protein contributes to toxicity in a zebrafish model of C9orf72 associated neurodegeneration. Mol Neurodegener 12, 6, doi: 10.1186/s13024-016-0146-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang YJ et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128, 505–524, doi: 10.1007/s00401-014-1336-5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.May S et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128, 485–503, doi: 10.1007/s00401-014-1329-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang YJ, Jeng US, Chiang YL, Hwang IS & Chen YR The Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-Cell Transmission Properties. J Biol Chem 291, 4903–4911, doi: 10.1074/jbc.M115.694273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schludi MH et al. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathologica 134, 241–254, doi: 10.1007/s00401-017-1711-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakae N et al. Poly-GR dipeptide repeat polymers correlate with neurodegeneration and Clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol Commun 6, 63, doi: 10.1186/s40478-018-0564-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopez-Gonzalez R et al. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron 92, 383–391, doi: 10.1016/j.neuron.2016.09.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jovicic A et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18, 1226–1229, doi: 10.1038/nn.4085 nn.4085 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang K et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61, doi: 10.1038/nature14973 nature14973 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boeynaems S, Bogaert E, Van Damme P & Van Den Bosch L Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol 132, 159–173, doi: 10.1007/s00401-016-1586-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peters OM et al. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron 88, 902–909 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Rourke JG et al. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron 88, 892–901, doi: 10.1016/j.neuron.2015.10.027 S0896–6273(15)00921–6 [pii] (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang J et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron 90, 535–550, doi: 10.1016/j.neuron.2016.04.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Y et al. C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron 90, 521–534, doi: 10.1016/j.neuron.2016.04.005 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Hagerman RJ et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57, 127–130 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Hagerman RJ & Hagerman P Fragile X-associated tremor/ataxia syndrome -features, mechanisms and management. Nat Rev Neurol 12, 403–412, doi: 10.1038/nrneurol.2016.82 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Jacquemont S et al. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am J Hum Genet 72, 869–878, doi: 10.1086/374321 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leehey MA et al. The fragile X premutation presenting as essential tremor. Arch Neurol 60, 117–121 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Tassone F et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet 66, 6–15, doi: 10.1086/302720 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kenneson A, Zhang F, Hagedorn CH & Warren ST Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet 10, 1449–1454 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Jin P et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55, 556–564, doi: 10.1016/j.neuron.2007.07.020 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sofola OA et al. RNA-Binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55, 565–571, doi: 10.1016/j.neuron.2007.07.021 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sellier C et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29, 1248–1261, doi: 10.1038/emboj.2010.21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qurashi A, Li WD, Zhou JY, Peng JM & Jin P Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats. Plos Genetics 7, doi:ARTN e1002102 10.1371/journal.pgen.1002102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krans A, Kearse MG & Todd PK Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome. Ann Neurol 80, 871–881, doi: 10.1002/ana.24800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ariza J et al. A Majority of FXTAS Cases Present with Intranuclear Inclusions Within Purkinje Cells. Cerebellum 15, 546–551, doi: 10.1007/s12311-016-0776-y (2016). [DOI] [PubMed] [Google Scholar]
- 101.Sellier C et al. Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93, 331–347, doi: 10.1016/j.neuron.2016.12.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allingham-Hawkins SJ et al. Fragile X premutation is a significant risk factor for premature ovarian failure: The international collaborative POF in fragile X study - Preliminary data. American Journal of Medical Genetics 83, 322–325, doi:Doi (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nolin SL et al. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. American Journal of Human Genetics 72, 454–464, doi:Doi 10.1086/367713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang MC et al. Ovarian histopathological and ubiquitin-immunophenotypic features in fragile X-associated primary ovarian insufficiency: a study of five cases and selected controls. Histopathology 59, 1018–1023, doi: 10.1111/j.1365-2559.2011.03959.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buijsen RAM et al. Presence of inclusions positive for polyglycine containing protein, FMRpolyG, indicates that repeat-associated non-AUG translation plays a role in fragile X-associated primary ovarian insufficiency. Hum Reprod 31, 158–168, doi: 10.1093/humrep/dev280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 72, 971–983 (1993). [DOI] [PubMed] [Google Scholar]
- 107.La Spada AR & Taylor JP Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 11, 247–258, doi: 10.1038/nrg2748 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chung DW, Rudnicki DD, Yu L & Margolis RL A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet 20, 3467–3477, doi:ddr263 [pii] 10.1093/hmg/ddr263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Banez-Coronel M et al. A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet 8, e1002481, doi: 10.1371/journal.pgen.1002481 PGENETICS-D-11–00806 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun X et al. Nuclear retention of full-length HTT RNA is mediated by splicing factors MBNL1 and U2AF65. Sci Rep 5, 12521, doi: 10.1038/srep12521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fennema-Notestine C et al. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology 63, 989–995, doi:63/6/989 [pii] (2004). [DOI] [PubMed] [Google Scholar]
- 112.Jernigan TL & Fennema-Notestine C White matter mapping is needed. Neurobiol Aging 25, 37–39, doi:S0197458003001684 [pii] (2004). [DOI] [PubMed] [Google Scholar]
- 113.Rosas HD et al. Diffusion tensor imaging in presymptomatic and early Huntington’s disease: Selective white matter pathology and its relationship to clinical measures. Mov Disord 21, 1317–1325, doi: 10.1002/mds.20979 (2006). [DOI] [PubMed] [Google Scholar]
- 114.van den Bogaard SJ et al. Magnetization transfer imaging in premanifest and manifest huntington disease: a 2-year follow-up. AJNR Am J Neuroradiol 34, 317–322, doi: 10.3174/ajnr.A3303 ajnr.A3303 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Aylward EH et al. Regional atrophy associated with cognitive and motor function in prodromal Huntington disease. J Huntingtons Dis 2, 477–489, doi: 10.3233/JHD-130076 203244317VK08753 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liquori CL et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293, 864–867, doi: 10.1126/science.1062125 293/5531/864 [pii] (2001). [DOI] [PubMed] [Google Scholar]
- 117.Meola G & Moxley RT 3rd. Myotonic dystrophy type 2 and related myotonic disorders. J Neurol 251, 1173–1182, doi: 10.1007/s00415-004-0590-1 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Meola G Myotonic dystrophies as a brain disorder. Neurol Sci 31, 863–864, doi: 10.1007/s10072-010-0414-2 (2010). [DOI] [PubMed] [Google Scholar]
- 119.Machuca-Tzili L, Brook D & Hilton-Jones D Clinical and molecular aspects of the myotonic dystrophies: a review. Muscle Nerve 32, 1–18, doi: 10.1002/mus.20301 (2005). [DOI] [PubMed] [Google Scholar]
- 120.Minnerop M et al. The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain 134, 3530–3546, doi: 10.1093/brain/awr299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Charizanis K et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450, doi: 10.1016/j.neuron.2012.05.029 S0896–6273(12)00525–9 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Day JW & Ranum LP RNA pathogenesis of the myotonic dystrophies. Neuromuscul Disord 15, 5–16, doi: 10.1016/j.nmd.2004.09.012 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Mankodi A et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet 10, 2165–2170 (2001). [DOI] [PubMed] [Google Scholar]
- 124.Sznajder LJ et al. Intron retention induced by microsatellite expansions as a disease biomarker. Proc Natl Acad Sci U S A 115, 4234–4239, doi: 10.1073/pnas.1716617115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee JE & Cooper TA Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans 37, 1281–1286, doi:BST0371281 [pii] 10.1042/BST0371281 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O’Rourke JR & Swanson MS Mechanisms of RNA-mediated Disease. Journal of Biological Chemistry 284, 7419–7423, doi: 10.1074/jbc.R800025200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Osborne RJ et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet 18, 1471–1481, doi: 10.1093/hmg/ddp058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yuan Y et al. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res 35, 5474–5486, doi:gkm601 [pii] 10.1093/nar/gkm601 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoshida K et al. Distinctive features of degenerating Purkinje cells in spinocerebellar ataxia type 31. Neuropathology 34, 261–267, doi: 10.1111/neup.12090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ishikawa K et al. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5’ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am J Hum Genet 77, 280–296, doi: 10.1086/432518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sato N et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet 85, 544–557, doi: 10.1016/j.ajhg.2009.09.019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Niimi Y et al. Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA)n in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis. Neuropathology 33, 600–611, doi: 10.1111/neup.12032 (2013). [DOI] [PubMed] [Google Scholar]
- 133.Baratz KH et al. E2–2 protein and Fuchs’s corneal dystrophy. N Engl J Med 363, 1016–1024, doi: 10.1056/NEJMoa1007064 (2010). [DOI] [PubMed] [Google Scholar]
- 134.Zhu AY, Eberhart CG & Jun AS Fuchs endothelial corneal dystrophy: a neurodegenerative disorder? JAMA Ophthalmol 132, 377–378, doi: 10.1001/jamaophthalmol.2013.7993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wieben ED et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2–2) gene predicts Fuchs corneal dystrophy. PLoS One 7, e49083, doi: 10.1371/journal.pone.0049083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Du J et al. RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy. J Biol Chem 290, 5979–5990, doi: 10.1074/jbc.M114.621607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garden GA & La Spada AR Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum 7, 138–149, doi: 10.1007/s12311-008-0027-y (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kang JS et al. White matter damage is related to ataxia severity in SCA3. J Neurol 261, 291–299, doi: 10.1007/s00415-013-7186-6 (2014). [DOI] [PubMed] [Google Scholar]
- 139.Hernandez-Castillo CR et al. Extensive White Matter Alterations and Its Correlations with Ataxia Severity in SCA 2 Patients. PLoS One 10, e0135449, doi: 10.1371/journal.pone.0135449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheng WW et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2 alpha phosphorylation. Nature Communications 9, doi:ARTN 51 10.1038/s41467-017-02495-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Green KM et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nature Communications 8, doi:ARTN 2005 10.1038/s41467-017-02200-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Westergard T et al. Repeat-associated non-AUG translation in C9orf72 ALF/FTD is driven by neuronal excitation and stress. EMBO Mol Med e9423, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou Q et al. Antibodies inhibit transmission and aggregation of C9orf72 poly-GA dipeptide repeat proteins. EMBO Mol Med 9, 687–702, doi: 10.15252/emmm.201607054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Westergard T et al. Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72-ALS/FTD. Cell Reports 17, 645–652, doi: 10.1016/j.celrep.2016.09.032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Samukawa M et al. PSP-Phenotype in SCA8: Case Report and Systemic Review. Cerebellum, doi: 10.1007/s12311-018-0955-0 (2018). [DOI] [PubMed] [Google Scholar]
- 146.Paucar M, Nennesmo I & Svenningsson P Pathological Study of a FMR1 Premutation Carrier With Progressive Supranuclear Palsy. Front Genet 9, 317, doi: 10.3389/fgene.2018.00317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ebbert MTW et al. Long-read sequencing across the C9orf72 ‘GGGGCC’ repeat expansion: implications for clinical use and genetic discovery efforts in human disease. Molecular Neurodegeneration 13, doi:ARTN 46 10.1186/s13024-018-0274-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Denovan-Wright EM & Davidson BL RNAi: a potential therapy for the dominantly inherited nucleotide repeat diseases. Gene Ther 13, 525–531, doi: 10.1038/sj.gt.3302664 (2006). [DOI] [PubMed] [Google Scholar]
- 149.Kordasiewicz HB et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044, doi: 10.1016/j.neuron.2012.05.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wheeler TM et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488, 111–115, doi: 10.1038/nature11362 nature11362 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lagier-Tourenne C et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 110, E4530–4539, doi: 10.1073/pnas.1318835110 1318835110 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rodriguez-Lebron E et al. Silencing mutant ATXN3 expression resolves molecular phenotypes in SCA3 transgenic mice. Mol Ther 21, 1909–1918, doi: 10.1038/mt.2013.152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Keiser MS, Boudreau RL & Davidson BL Broad Therapeutic Benefit After RNAi Expression Vector Delivery to Deep Cerebellar Nuclei: Implications for Spinocerebellar Ataxia Type 1 Therapy. Mol Ther 22, 588–595, doi: 10.1038/mt.2013.279 mt2013279 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rue L et al. Targeting CAG repeat RNAs reduces Huntington’s disease phenotype independently of huntingtin levels. J Clin Invest 126, 4319–4330, doi: 10.1172/JCI83185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Modarresi F et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature Biotechnology 30, 453–+, doi: 10.1038/nbt.2158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sopher BL et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron 70, 1071–1084, doi:S0896–6273(11)00445–4 [pii] 10.1016/j.neuron.2011.05.027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]