Standfirst
Cell lines are valuable tools for developing treatments that can minimize disparities in prostate cancer. Nevertheless, limitations in their usage, primarily from the inadequate representation of ethnically diverse cell lines, continue to hinder the advance of drug leads that equally benefit men from both African and European ancestry.
The dawn of technical success in culturing human cancer cell lines in the 1950s was originally believed to hold the key for developing cures. The availability of cultured cell lines, such as HeLa, created a cataclysmic surge in Biomedical Research and led to advances in disease elucidation and screening for effective therapeutic agents. At first, the model for anticancer drug development using cell lines was achieved through lock and key approaches whereby one cancer cell line, for example, prostate, was considered representative of all prostate cancers across different races, geographic locale and gender. Soon, this model proved futile as drug companies reported that less than fifty percent of drugs developed from these early cell lines actually work in less than half the population. It became evident that a personalized approach was required, and this necessitated a more in-depth understanding of the molecular diversity of cancer cell biology.
Despite the high failure rates of drug leads in clinical trials, high-throughput in vitro screening assays using cell lines have enabled identification of more drug leads than previous in vivo models, which were mostly performed in mice. Most chemotherapy drugs currently in use have been screened against a variety of established cell lines to define chemotherapy as a primary pillar of cancer treatment, but the use of cell lines is not without challenges. Indeed, cancer cell lines do not faultlessly mimic their tumours of origin and they lack the native tumour micro-environment. While improved cell culture models, like 3D cell culture technologies, are currently being aggressively pursued to better mimic the in vivo cell environment, numerous challenges remain, such as genetic changes that occur during culture, as well as the inability of the cell lines to retain the heterogeneity of the primary tumour. Despite these weaknesses, cancer cell lines still provide valuable models for drug development, particularly in conjunction with 3D culture techniques. We argue that the potentially improved success rate of identifying useful therapeutics with the 3D cell culture models should also take into consideration the ethnic and gender diversity of cell culture model systems to avoid failures of cancer drugs used to treat a diverse human population.
A cancer of noteworthy concern and one with the greatest disparities is prostate, with an estimated 1,600,000 cases and 336,000 deaths annually worldwide1. A decline in prostate cancer (PCa) mortality rates has been reported, but this decline is most evident among Americans than men from Africa and the Caribbean, which have the highest PCa rates and continue to experience elevated PCa incidence and mortality outcomes2, 3. Furthermore, African American men remain two and a half times more likely to die from the disease than Caucasian American men. This leads us to the question, why?
Prostate cancer is one of the most heterogeneously expressed cancers and is believed to be among the more readily cured if identified at early stage. Men of African Ancestry (MAA) are most often diagnosed with advanced stage. Yet, it is reported that advanced PCas among Men of European Ancestry (MEA) respond more readily to the current chemotherapies than the same grade tumours in MAA4. Therefore, MAA have worse outcomes for PCa treatment stage-for-stage and grade-for-grade compared to their European counterparts.
Despite the increased mortality among MAA, of the thirty-two listed human prostate cell lines available from major suppliers, ATCC, Sigma Aldrich and ECACC, 97% are European in origin where ethnicity is known. This is a striking example of disparity in available human cellular models for cancer research. Cancer in vivo models that are primarily patient derived xenografts, PDX, are typically developed from established cancer cell lines, which at this point also means that the majority of these models are also more representative of the European populous. Furthermore, more than 90% of patients enrolled in clinical trials are Caucasians. Taking all of these factors into consideration, MAA are rarely to not represented at all stages of preclinical or clinical anticancer drug development.
The archetypical and stable nature of tumour cell lines has rendered them acceptable models of the biological make-up of their cancers of origin and drug response. The reality is that almost every anticancer drug used today has been tested at some level in these models. To improve the identification of lead drug molecules for clinical trials, having representation of tumour diversity is crucial and clearly not fully represented by one or a few cell lines. To combat this deficiency, the NCI attempted a “disease oriented” drug screening approach by developing a panel of 60 human cancer cell lines derived from nine cancers, resulting in roughly six to seven cell lines representative of each cancer5. The intention was to identify a small selection of drugs from a large pool that were all screened against the panel with sufficient discrimination, but the model failed. Importantly, the number of cell lines per disease was insufficient to fully represent the myriad of tumour heterogeneity. This model was later improved upon by the Bodmer laboratory, which generated one of the largest tissue specific panels comprising 120 colorectal cancer cell lines, thereby providing an ideal example of using available cancer cell lines as research tools. The panel was able to reflect a wide array of subtypes typical to colon cancer at the levels of mRNA expression6 and relevant mutations, APC, TP53, CTNNB1, BRAF, PIK3CA and FBXW77. A relationship between 5-fluorouracil sensitivity and mismatch repair status in a subset of this panel was established. Indeed, this was a win as researchers were able to identify a model that capitalized on enhancing the selection process for drug leads using cancer cell lines. The drawback, however, is that representative ethnic background of these cell lines was still lacking. The use of cancer cell line panels is becoming more widespread and around 16 panels representing different tissues are available for purchase from ATCC, but when a detailed analysis was conducted on the origin of these human cancer cell lines, as shown in Figure 1, only two of the 16 panels had representation by men and women of African ancestry. Of these, the breast cancer panel had 50% representation out of four cell lines and the lymphoma panel had 33%.
Figure 1.
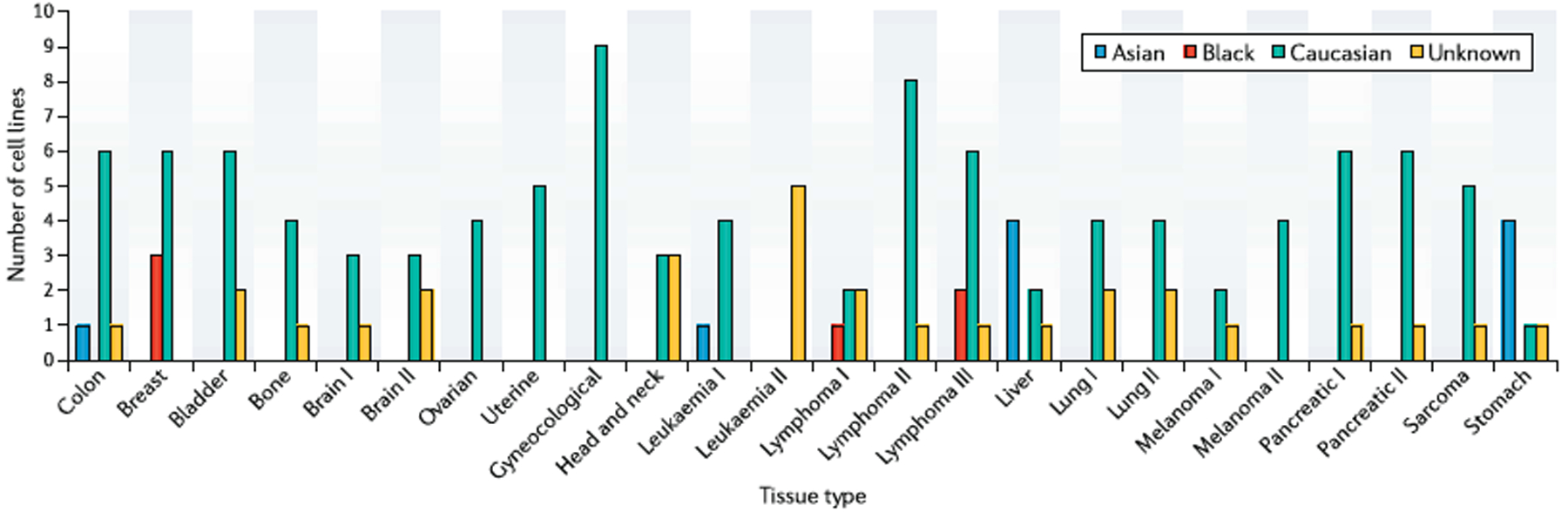
The ethnic representation of panels of cancer cell lines according to tissue specificity on the ATCC website. In some instances, for example, Brain I and Brain II represent different mutational changes within each tissue.
Identifying ways to fully exploit the efficacy of cancer cell lines should not be done in isolation of increasing the representation of varied ethnic groups among the cell lines employed. From the library of cancer cell line panels on ATCC’s website, none is listed for prostate cancer. We know that anticancer drugs respond differently to cancers from different tissue types and to similar cancers from ethnically diverse backgrounds. A novel panel of three PCa cell lines each from MAA and MEA identified the potential role of miRNAs in PCa aggression among MAA8. Although a wider representation of PCa cell lines is required to ascertain frequency of biomarker associations to PCa, the value of this approach is evident. The first two established MAA PCa cell lines, MDA PCa 2a and MDA PCa 2b, are from the same patient and developed more than two decades ago. An online search revealed just a little over 100 publications using these lines. Meanwhile, only 2 citations were noted for the prostate cell lines, RC-77N/E (normal) and RC77T/E (cancer), which were more recently developed from a single MAA patient.
Irrespective of the inherent value of the MAA cell lines9, it seems that the issues to be addressed are two-fold: 1) to develop more MAA PCa cell lines, which of course is paramount to advance the development of drugs to treat PCa in MAA, and 2) to also create a research strategy that encourages their application. In the current research model, demographic data are not required for cell lines to be included in library screening experiments and also not a requirement for genomic sequencing efforts from cell lines or primary tissues. This not only encourages the cycle of a lack of equal representation of PCa tools for both MAA and MEA, but it also limits our full understanding of the disease in both ethnic groups. Developing new PCa cell lines remains a challenge and to achieve the large number of cell lines required to fully represent PCa tumours in MAA requires a strategic and systematic approach. Regions of the world with the highest reported incidence and mortality rates of PCa, such as parts of the Caribbean and Africa, currently lack expertise to effectively develop such cell lines. Collaborative efforts between established laboratories, like the Bodmer laboratory, and institutions within these regions should be established. As an incentive to combat this challenge, the ACRJ Cell Culture lab at The University of the West Indies, Mona is embarking on developing novel PCa cell lines with support from Fox Chase Cancer Center in Philadelphia. It is vital to incorporate more innovative and standardized protocols in these efforts to overcome the hurdles involved in creating the number of PCa cell lines necessary to fully represent the heterogeneous PCa disease within these ethnic groups. The inauguration of an international consortium of researchers could bring together the necessary resources, expertise, and tissue sourcing to ultimately establish a robust panel of PCa cell lines that is truly representative of the genetically diverse human populations suffering from this deadly disease. A more representative panel of PCa cell lines could then serve as a valuable resource to develop targeted therapies that improve outcomes in both MAA and MEA populations, as well as create a paradigm for establishing cell line resources in other cancers that more faithfully represent patient diversity in terms of ethnicity and gender.
References
- 1.Torre LA et al. Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Chokunonga E, Windridge P, Sasieni P, Borok M & Parkin DM Black-white differences in cancer risk in Harare, Zimbabwe, during 1991–2010. Int J Cancer 138, 1416–21 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Brown CR et al. Social determinants of prostate cancer in the Caribbean: a systematic review and meta-analysis. BMC public health 18, 900–900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastina I et al. Cytochrome 450 1B1 (CYP1B1) polymorphisms associated with response to docetaxel in Castration-Resistant Prostate Cancer (CRPC) patients. BMC Cancer 10, 511 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monks A et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 83, 757–66 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Wilding JL, McGowan S, Liu Y & Bodmer WF Replication error deficient and proficient colorectal cancer gene expression differences caused by 3’UTR polyT sequence deletions. Proc Natl Acad Sci U S A 107, 21058–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y & Bodmer WF Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci U S A 103, 976–81 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theodore SC, Rhim JS, Turner T & Yates C MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis 20, S1–96–100 (2010). [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao XY et al. Two mutations identified in the androgen receptor of the new human prostate cancer cell line MDA PCa 2a. J Urol 162, 2192–9 (1999). [DOI] [PubMed] [Google Scholar]


