Abstract
Aims
As of the 28th April 2020, the COVID-19 pandemic has infiltrated over 200 countries and affected over three million confirmed people. We review different biomarkers to evaluate if they are able to predict clinical outcomes and correlate with the severity of COVID-19 disease.
Methods
A systematic review of the literature was carried out to identify relevant articles using six different databases. Keywords to refine the search included ‘COVID-19’, ‘SARS-CoV2’, ‘Biomarkers’, among others. Only studies which reported data on pre-defined outcomes were included.
Key findings
Thirty-four relevant articles were identified which reviewed the following biomarkers: C-reactive protein, serum amyloid A, interleukin-6, lactate dehydrogenase, neutrophil-to-lymphocyte ratio, D-dimer, cardiac troponin, renal biomarkers, lymphocytes and platelet count. Of these, all but two, showed significantly higher levels in patients with severe complications of COVID-19 infection compared to their non-severe counterparts. Lymphocytes and platelet count showed significantly lower levels in severe patients compared to non-severe patients.
Significance
Although research is still in its early stages, the discovery of how different biomarkers behave during the course of the disease could help clinicians in identifying severe disease earlier and subsequently improve prognosis. Nevertheless, we urge for more research across the globe to corroborate these findings.
Keywords: SARS-CoV-2, COVID-19, Biomarkers, Blood tests
1. Introduction
On December 2019, Wuhan City in China, became the epicentre of unexplained cases of pneumonia. On January 2020, Chinese scientists identified this as a novel coronavirus, temporarily labelled as, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,51]. Its name was then changed to coronavirus disease 2019 (COVID-19) by the World Health Organization in February 2020 as the disease spread worldwide [2].
It has been suggested the outbreak has a zoonotic origin, and like other respiratory pathogens, it spreads through human-to-human transmission such as coughing and sneezing [3]. Although limited, research suggests a possibility of transmission even among the asymptomatic [4].
As of the 28th April 2020, over 200 countries have been affected by the COVID-19 disease with over three million confirmed cases leading to over 200,000 deaths. Yet it is believed many remain unreported in certain areas of the world [1].
Infected patients may present with any of the following; fever, high temperature (>37.3 °C), cough, myalgia, sputum production, headache, haemoptysis, diarrhoea, dyspnoea and in some cases, acute respiratory distress syndrome (ARDS), acute cardiac injury or secondary infection [5,54].
Upon examination, these subjective clinical symptoms can be interpreted more confidently with the use of biological markers (biomarkers). These provide objective values throughout the progression of the disease [6]. Henceforth, categorising patients into mild, severe or critical becomes more defined, allowing for earlier interventions [7].
This article aims to explore the role of different biomarkers in the disease pathogenesis of COVID-19 and assess how their levels vary depending on the severity of the disease. By doing so, it gives clinicians a tool to group patients and predict prognosis and mortality. The biomarkers we review include, C-reactive protein (CRP), IL-6, white cell count (WCC), lactate dehydrogenase (LDH), D-dimers, platelet count, cardiac troponin and renal markers.
2. Methods and materials
2.1. Search strategy
A comprehensive literature search was done on PubMed, SCOPUS, Embase, Cochrane database, Google Scholar and Ovid to identify articles discussing biomarkers in this review and its clinical implications on COVID-19 in accordance with Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines. Key words used were ‘C-reactive protein’ ‘COVID-19’, ‘interleukin-6’, ‘lactate dehydrogenase’, ‘SARS’, ‘white cell count’, ‘neutrophil count’, ‘lymphocyte count’, ‘D-dimer’, ‘platelet count’, ‘cardiac troponin’, ‘renal biomarkers’, ‘urea’ ‘creatinine’. The search terms were used as key words and in combination as MeSH terms to maximize the output from literature findings. A staged literature search was done, whereby a separate literature search was performed for each section within this article and all the relevant studies were identified and summarized separately. If a paper reports on multiple biomarker, then the results have been shared between different parts of this review. The relevant articles are cited and referenced within each section separately. No limit has been placed on publication time or language of the article. All the relevant articles were identified and screened by three authors; the results are summarized in narrative manner in each relevant section of this review. A summary table of each section is provided where appropriate.
2.2. Inclusion and exclusion criteria
Studies were included if they have reviewed a correlation between a biomarker and the severity of COVID-19. Exclusion criteria were editorials, consensus documents, commentaries, and studies with no particular definition of the role of biomarkers in COVID-19.
2.3. Data extraction
All articles were screened by two authors and any disagreement was reached by consensus or involvement of third author. Data extracted by two authors and validated by third author.
2.4. Quality assessment
Quality of each publication was evaluated by two independent reviewers. This review addressed key domains: type of biomarker, level of the biomarker, correlation with severity of the disease, survival, number of patients investigated and outcomes.
2.5. Statistical analysis
It was not possible to conduct an appropriate meta-analysis because there were not enough research data among the studies on this subject.
3. Results
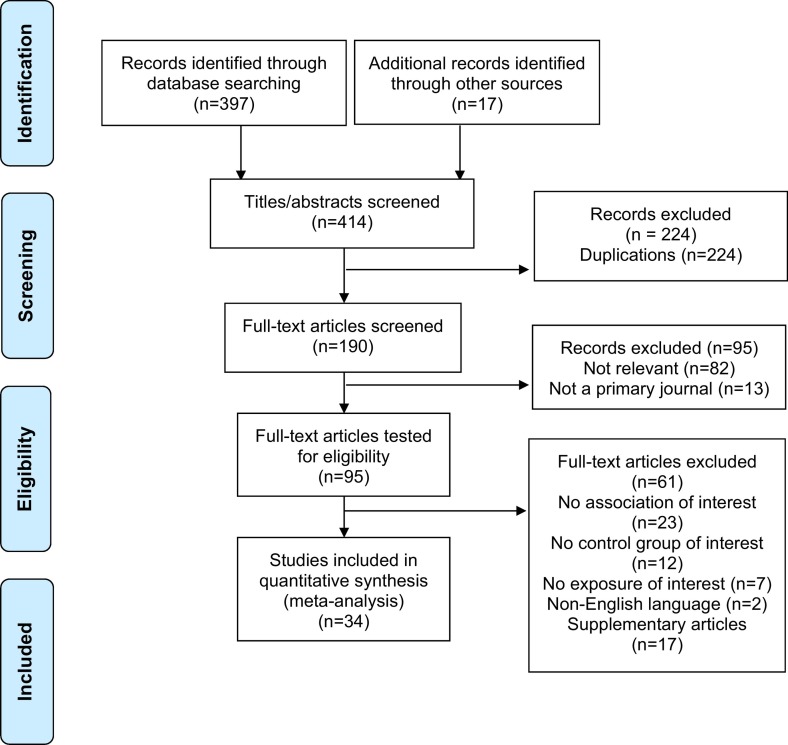
A total of 414 articles were found. After removal of duplicates, 224 articles were used for full-text screening with 34 studies included in our analysis. The complete PRISMA flow chart is reported in Fig. 1 . Biomarkers measured were C-reactive protein (CRP), interleukin-6 (IL-6), White cell count (WCC), lactate dehydrogenase (LDH), D-dimer, platelet count, high-sensitivity troponin I (hs-TnI) and renal markers. Table 1 summarises the study's characteristics.
Fig. 1.
PRISMA flow diagram.
Table 1.
Study characteristics.
| Author | Study design | Country | Cohort size | Biomarkers studied | Comments |
|---|---|---|---|---|---|
| Huang et al. (2020) [5] | Prospective | China | 41 | Lymphocytes, IL-6, D-dimer, platelet count | Compared to non-ICU patients, ICU patients had higher plasma levels of IL-6, D-dimer and platelets. Lymphopenia was more common in ICU patients than non-ICU patients |
| Li et al. 2020 [39] | Retrospective cohort; single centre | China | 132 | CRP, WCC | Critically severe patients had significantly higher CRP and WCC than severe or non-severe patients. |
| Liu et al [9] | Retrospective cohort; single centre | China | 140 | CRP, IL-6 | Significantly more patients in the severe group experienced higher CRP and IL-6 levels vs non-severe. |
| Ji et al. 2020 [10] | Retrospective cohort; single centre | China | 33 | CRP, D-dimer, LC | The preprint study shows CRP, D-dimer significantly increased but LC decreased for patients in direct vs indirect contact to Wuhan – does not assess severity. CRP levels compared by ratio to other blood parameters indicated CRP changes prior to others. |
| Tan et al. 2020 [12] | Retrospective cohort; single centre | China | 27 | CRP, WCC | ROC analysis showed strong association found between CRP levels and progression of disease. Interestingly, absolute values of increased WCC and NLR found in influenza A or B group vs SARS-COV-2. |
| Wang et al. 2020 [40] | Retrospective cohort; single centre | China | 27 | CRP | Greater CRP values correspond with the critical group, as groups were determined based on the diameter of largest lung lesion - CRP levels may indicate lung damage and development of disease. |
| Qin et al. 2020 [8] | Retrospective cohort; single centre | China | 452 | CRP, IL-6 and LDH, WCC | Significantly higher levels of CRP, IL-6, LDH and NC but low LC in severe COVID-19,vs non-severe group. Surveillance may help in early screening of critical illness |
| Wang et al. 2020 [17] | Retrospective cohort; single centre | China | 138 | WCC, NC, LC, LDH, D-dimer, blood urea, creatinine | Higher WCC, specifically higher NC, lower LC and higher NLR in ICU vs non-ICU patients. Prior to the death of critical patients: NC, D-dimer, blood urea, and creatinine levels rose throughout until death, whilst the LC carried on falling. Elevated LDH also correlated with patients in ICU vs non-ICU. |
| Chen N et al. 2020 [15] | Retrospective cohort; single centre | China | 99 | WCC, NC, LC, LDH, CRP, platelet count, D-dimer, IL-6, blood urea, serum creatinine | Levels of LDH, CRP,D-dimer, IL-6 increased over normal ranges in COVID-19- induced pneumonia patients, whereas increases and decreases were seen in WCC (high NC, low LC), blood urea, serum creatinine and platelet count. Absolute LC values decreased across patients; destruction of T lymphocytes may lead to worse outcomes. |
| Tan et al. 2020 [18] | Retrospective cohort; single centre | China | 90 | WCC | Graphical representation of time-lymphocyte% model (TLYM%) using 5 randomly selected individuals to illustrate disease progression until death. TLYM% used for 90 hospitalised patients, it was able to categorise patients consistently. Surveillance may identify critically ill patients earlier. |
| Gong et al. 2020 [7] preprint | Retrospective cohort; single centre | China | 100 | WCC, CRP | Preprint corresponds with other studies that a high WBC, NC and CRP suggests greater severity of disease. |
| Luo et al. 2020 [21] | Retrospective cohort; single centre | China | 35 | LDH | Increased LDH levels are associated with patients in the severe group, indicating LDH may be reflective of severity of disease. |
| Xiong et al. 2020 [23] | Retrospective cohort; single centre | China | 42 | LDH | Correlates LDH values with CT scores but no breakdown is present |
| Mo P et al. 2020 [24] | Retrospective; single centre | China | 155 | LDH, NC, CRP, platelet count | LDH, NC, CRP and platelet count were higher in refractory vs general patients. Refractory patients had more cases of lung abnormalities, suggesting these biomarkers correlate with development of disease |
| Ferrari D et al. (2020) [20] | Retrospective; single centre | Italy | 141 | LDH | LDH measured in COVID-19 positive vs negative patients and higher levels apparent in positive groups. |
| Tang et al. 2020 [26] | Retrospective cohort; single centre | China | 183 | D-dimer | Abnormal coagulation results with markedly elevated D-dimer are common in deaths with COVID-19 |
| Zhou et al. 2020 [32] | Retrospective cohort; multi-centre | China | 191 | D-dimer, hs-TnI, platelet count | D-dimer levels >1 μg/mL can help clinicians in identifying patients with poor prognosis at earlier stage There were significantly higher levels of platelets and hs-TnI in non-survivors compared to survivors |
| Guan et al. 2020 [22] | Retrospective cohort; single centre | China | 1099 | D-dimer, Platelet count, CRP lymphocytes and LDH | Although there were evident differences in lymphocytes (lymphocytopenia), platelet count (rose) and D-dimer (rose) in patients who experienced composite endpoints (ICU admission, invasive mechanical ventilation and death) there was no statistical analysis performed LDH measured in 675. Higher LDH levels present in majority of severe patients vs non-severe. |
| Liu et al. 2020 [41] | Retrospective cohort; single centre | China | 12 | Lymphocytes, Albumin, Neutrophils, CRP, PaO2/FiO2, platelet count, creatinine and LDH | Ct value of virus correlated strongly with CRP, albumin and LDH among others. Murray score for ARDS correlated with the same markers. |
| Liu et al. 2020 [42] | Retrospective cohort; single centre | China | 78 | CRP, Albumin, Platelet count, D-dimer, AST, ALT, Creatinine | Increased CRP and decreased albumin strongly correlated with disease progression |
| Ruan et al. 2020 [43] | Retrospective cohort; single centre | China | 150 | WBC counts, lymphocytes, platelets, albumin, total bilirubin, urea, creatinine, myoglobin, cardiac troponin, CRP and IL-6 | Cardiac troponin, Myoglobin, CRP and IL-6 significantly increased in cases with mortality |
| Yang et al. 2020 [44] | Retrospective cohort; single centre | China | 52 | Haemoglobin, lymphocytes, Platelet count, prothrombin time, bilirubin, creatinine and lactate. | Higher levels of platelets in severe group but lymphocytopenia seen as the most common marker of infection. |
| Young et al. 2020 [45] | Descriptive | Singapore | 18 | Haemoglobin, lymphocytes, platelet count, neutrophils, CRP and LDH | Lower platelets found in severe group, but statistical analysis was not performed |
| Liu et al. 2020 [30] | Retrospective cohort; single centre | China | 383 | Platelet parameters: platelet count, volume, distribution width and larger cell ratio | Thrombocytopenia associated with mortality Patients split on basis of thrombocytopenia not severity |
| Yang et al 2020 [31] | Retrospective cohort; single centre | China | 1476 | Platelet count | Lowest platelet count associated with mortality |
| Shi et al 2020 [33] | Retrospective cohort; single centre | China | 416 | Cardiac troponin, platelet count, creatinine kinase | Greater proportion of cardiac injury, marked as a rise in creatinine kinase and cardiac troponin, is more likely to require non-invasive/invasive ventilation compared to those without |
| Xiang et al. 2020 [35] | Retrospective cohort; single centre | China | 28 | AST, GGT, ALP, albumin, creatinine kinase, CKMB, CRP, creatinine, urea and cystatin C | Serum values of urea, creatinine and cystatin-C significantly increased in severe COVID-19. |
| Cheng et al. 2020 [36] | Prospective cohort; single centre | China | 701 | Creatinine | Raised creatinine levels associated with poor outcome in COVID-19 infection |
| Zhou et al. 2020 [37] | Retrospective cohort; single centre | China | 178 | Urinalysis: proteinuria, haematuria, leucocyturia | Urinalysis on admission can effectively highlight kidney impairment |
| Chen L et al (2020) [46] | Prospective cohort; single centre | China | 29 | IL-6 | Increased expression of IL-2R and IL-6 in serum is expected to predict the severity of COVID-19 |
| Liu et al. (2020) [47] | Retrospective cohort; single centre | China | 80 | IL-6, CRP, LDH, ferritin and D-dimer | Baseline IL-6, CRP, LDH and ferritin was closely related to severity of COVID-19. Elevated IL-6 was significantly related to clinical manifestations of severe type patients |
| Diao et al (2020) [48] | Retrospective cohort; multi-centre | China | 552 COVID; 40 healthy | T-cells, IL-6 | T cells are reduced significantly and negatively correlated to IL-6 in COVID-19 |
| Wu et al (2020) [49] | Retrospective cohort; multi-centre | China | 150 | IL-6, D-dimer, LDH, neutrophil count | ARDS development in COVID-19 is significantly correlated with rise in IL-6, D-dimer, LDH and neutrophil count |
| Zhang et al (2020) [50] | Retrospective; single centre | China | 343 | D-dimer | D-dimer on admission of >2.0 μg/mL could effectively predict in-hospital mortality in patients with COVID-19 and could be an early and helpful marker to improve management |
IL-6 = interleukin-6; ICU = intensive care unit; CRP = C-reactive protein; WCC = white cell count; LC = leucocyte count; ROC = Receiver operating characteristic; NLR = neutrophil:lymphocyte ratio; NC = neutrophil count; LDH = lactate dehydrogenase; hs-TnI = high sensitivity troponin I; AST = aspartate aminotransferase; ALT = alanine aminotransferase; GGT = gamma-glutamyl transpeptidase; ALP = alkaline phosphatase; CKMB = creatine kinase myocardial band.
3.1. C-reactive protein
CRP is a plasma protein produced by the liver and induced by various inflammatory mediators such as IL-6. Despite being non-specific, this acute phase reactant is used clinically as a biomarker for various inflammatory conditions; a rise in CRP levels are associated with an increase in disease severity [7].
The application of CRP in COVID-19 has been highlighted by a retrospective single-centre study in Wuhan, China, where the majority of patients in the severe cohort showed significantly higher levels compared to the non-severe cohort (57.9 mg/L vs 33.2 mg/L, P < 0.001) [8]. A second retrospective cohort study found the likelihood of progressing to severe COVID-19 disease increased in patients with CRP levels >41.8 mg/L [9]. Both studies suggest CRP levels are a strong indicator to reflect the presence and severity of COVID-19 infection.
Furthermore, a study from unpublished observations suggests CRP is one of the first biomarkers within blood plasma that changes to reflect physiological complications; if accepted CRP will be the most effective biomarker to predict the progression of COVID-19 infection. Contrastingly, the same study illustrated some cases of infection which showed changes in serum amyloid A (SAA) instead of evoking significant CRP changes thus requiring further evaluation [10].
Whilst the use of SAA as a biomarker for COVID-19 requires further research, CRP and SAA are commonly used in conjunction to monitor inflammatory diseases. Though SAA is another acute phase reactant, it is responsive to both viral and bacterial infections compared to CRP [11].
Pathologically, computed tomography (CT) scans can identify lung lesions relating to COVID-19. Nonetheless, a study conducted in China revealed CT scores could not differentiate mild cases from severe. However, compared to erythrocyte sedimentation rate (ESR), CRP levels were significantly greater during early periods of severe cases and proved to be a more sensitive biomarker in reflecting disease development [12].
The excellent performance of CRP as a biomarker is reflected in the ‘area under curve’ in the receiver operating analysis of 0.87 (95% CI, 0.10–1.00) where values 83% and 91% represent sensitivity and specificity, respectively. Hence compared to CT scans alone, CRP values are more reliable for earlier identification of case severity [12]. Table 2 summarises the studies used in analysing CRP and COVID-19.
Table 2.
Studies that compare C-reactive protein for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Li H, et al. 2020 [39] | 33.22 ± 32.21 | 66.04 ± 44.89 97.44 ± 58.60 (critically severe) |
P = 0.001 for all | Critically severe patients had significantly higher CRP than severe or non-severe patients. SAA correlated with CRP too consistently, indicating both should be used to reflect severity of disease – but study lacks a control group. |
| Liu et al [9] | >8.0 = 56.1% of patients 0–8.0 = 43.9% of patients |
>8.0 = 93.9% of patients 0–8.0 = 6.1% of patients |
P < 0.001 for all | Significantly more patients in the severe group experienced higher CRP levels vs non-severe. |
| Qin et al. 2020 [8] | 33.2 (8.2–59.7) | 57.9 (20.9–103.2) | P < 0.001 | Higher levels of CRP recorded in the severe group vs non-severe group are suggestive that CRP can be monitored to assess progression of disease. |
| Ji et al. 2020 [10] | 11.89 (9.74–23.36) (Indirect contact) |
5.68 (2.80–13.0) (Direct contact) |
N/A | Stratifies patients by direct and indirect contact to Wuhan – does not assess severity |
| Tan et al. 2020 [12] | N/A | N/A | CRP (R = 0.62; P < .01) ROC = 0.87 (95% CI 0.10–1.00) Cut-off = 20.42 mg/L Sensitivity = 83% Specificity = 91% |
Absolute values not reported – instead performs ROC analysis, showing significant increase in CRP levels prior to changes in CT scores for early periods of severe group. |
| Wang et al. 2020 [40] | 1.52 ± 1.56 (mild) 16.76 ± 18.38 (moderate) |
54.15 ± 1.06 (severe) 105.00 ± 12.73 (critical) |
Mild: moderate P = 0.007 Moderate: severe P = 0.511 Severe: critical P = 0.947 |
Groups were determined based on the diameter of largest lung lesion. Greater CRP values are more prominent in critical group – indicating lung damage. |
CRP = C-reactive protein; ROC = Receiver operating characteristic.
3.2. Interleukin-6
Cytokine release syndrome (CRS) is an over-exaggerated immune response involving an overwhelming release of pro-inflammatory mediators. This mechanism underlies several pathological processes including acute respiratory distress syndrome (ARDS) [13]. Studies investigating the role of cytokines in SARS and MERS have had also found a link between CRS and disease severity [14]. Understanding their role in COVID-19 disease may help facilitate the design of novel immunotherapies.
Studies have revealed that levels of IL-6, the most common type of cytokine released by activated macrophages, rise sharply in severe manifestations of COVID-19 [15]. However, since most studies to date have been observational, it is difficult to extrapolate if the rise is significant enough to cause the manifestations seen in severe forms.
One meta-analysis reviewing six studies show mean IL-6 concentrations were 2.9-fold higher in patients with complicated COVID-19 compared to those with non-complicated disease (n = 1302; 95% CI 1.17–7.19) [16]. In its analysis, the outcomes of the studies include ICU admission, onset of ARDS and mortality. Since the proportionate rise of IL-6 is correlated with disease severity, this study can prove ground-breaking. Although clinicians can use this to identify severity earlier and commence oxygen therapy sooner, the varying outcomes makes it somewhat difficult to ascertain what level of IL-6 corresponds to what negative outcome. Furthermore, many studies recruited participants from the same centre, giving rise to the potential of selection bias. Table 3 summarises the studies used in analysing IL-6 and COVID-19.
Table 3.
Studies that compare IL-6 for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Chen et al (2020) [46] | 34 ± 7 | 72 ± 12 | P < 0.0001 | Increased expression of IL-2R and IL-6 in serum is expected to predict the severity of COVID-19 |
| 23. Liu et al. (2020) [47] | 2.4 (2.1–2.9) | 36.5 (30.8–42) | P < 0.0001 | Severity of COVID-19 could be predicted with baseline IL-6 levels |
| Diao et al (2020) [48] | 51 ± 74 | 186 ± 283 | P < 0.0001 | Significantly higher baseline levels of IL-6 in those requiring ICU compared to those who do not |
| Huang et al (2020) [5] | 5 (0–11.2) | 6.1 (1.8–37.7) | P < 0.0001 | Significantly higher baseline levels of IL-6 in those requiring ICU compared to those who do not |
| Qin et al (2020) [8] | 13.3 (3.9–41.1) | 25.5 (9.5–54.5) | P < 0.0001 | Significantly higher levels of IL-6 in sever and critical COVID-19. Surveillance may help in early screening of critical illness |
| Wu et al (2020) [49] | 6.3 (5.4–7.8) | 7.4 (5.6–10.9) | P < 0.0001 | ARDS development in COVID-19 is related to rise in IL-6 |
IL-6 = interleukin-6; ARDS = Acute Respiratory Distress Syndrome; ICU = intensive care unit.
3.3. White cell count
White blood cells (WBCs), known as leucocytes are a component of blood generated from bone marrow and lymphoid tissue. They are divided into two major groups, granulocytes and agranulocytes. Within the granulocyte group are eosinophils, basophils and neutrophils (NC), whereas lymphocytes (LC) and monocytes are present in agranulocytes. A disproportionate number of these cells may reveal an underlying infection and hence can be measured using blood tests, producing a WCC. However, the reliability of WCC as a biomarker for COVID-19 remains unproven.
A retrospective study found several differences in WCC between severe and non-severe COVID-19 patients [8]. Both groups experienced an increase in leucocytes with the severe group having a significantly greater rise (5.6 vs 4.9 × 109/L; P < 0.001). NCs were predominantly driving this increase as the severe set (4.3 vs 3.2 × 109/L; P < 0.001). Interestingly, the levels of lymphocytes, monocytes, basophils and eosinophils were less, resulting in a greater neutrophil-to-lymphocyte ratio (NLR; 5.5 vs 3.2; P < 0.001). NLR is an infamous biomarker, high in wide-spread inflammatory conditions and can be used to reflect disease severity. However, a larger study is needed to clarify NLR's effectiveness as a biomarker.
Another conducted in China concludes similar findings of high NC and low LC count in severely affected patients, suggesting NLR could be a potential biomarker for early detection of severe COVID-19 [17].
However, other factors may disrupt the accuracy of the WCC results observed. These include glucocorticoid therapy and other underlying viral/bacterial infections [11].
LC is separately addressed in the literature, secondary to NLR. A descriptive study in China reported depleted LC levels in the majority of COVID-19 patients [15].
Another study has found low blood lymphocyte percentage (LYM%) in critically ill patients, suggesting low LC count indicates poor prognosis. However, since the virus can target lymphoid tissue and mechanisms of IL-6, other causes of low LC count must be investigated [18]. Similar to NLR, the clinical benefits of LC count as a biomarker for COVID-19 remains uncertain.
Table 4 summarises the studies used in analysing WCC and COVID-19.
Table 4.
Studies that compare WCC for COVID-19.
| Author | Level in non-severe patient (×109/L) | Level in severe patient (×109/L) | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Qin et al. 2020 [8] | WCC = 4.9 (3.7–6.1) NC = 3.2 (2.1–4.4) LC = 1.0 (0.7–1.3) NLR = 3.2 (1.8–4.9) |
WCC = 5.6 (4.3–8.4) NC = 4.3 (2.9–7.0) LC = 0.8 (0.6–1.1) NLR = 5.5 (3.3–10.0) |
P < 0.001 for all | Retrospective + China + 452 Significantly higher NLR in severe patients. Monitoring may aid in early screening of critical illness. |
| Wang et al. 2020 [17] | WCC = 4.3 (3.3–5.4) NC = 2.7 (1.9–3.9) LC = 0.9 (0.6–1.2) (non-ICU) |
WCC = 6.6 (3.6–9.8) NC = 4.6 (2.6–7.9) LC = 0.8 (0.6–1.1) (ICU) |
P = 0.003 P < 0.001 P = 0.03 |
Retrospective + China + 138 Non-ICU vs ICU patients had drastically lower WCC, checking low LC and high NC may help in early detection of disease progression. |
| Chen N et al. 2020 [15] | WCC = 7.5 (3.5–9.5 normal range) NC = 5.0 (3.3–8.1 normal range) 38% increase LC = 0.9 (1.1–3.2 normal range) 35% decrease |
N/A | Absolute values of 99 patients obtained in patients with pneumonia. Surveillance of NC/LC may reflect severity of lung abnormalities. | |
| Tan et al. 2020 [18] | N/A | P < 0.001 | Only lymphocyte% (LYM%) representation is graphical. TLYM% used for 90 hospitalised patients, it was able to categorise patients consistently. Surveillance may identify critically ill patients earlier. | |
| Gong et al. 2020 [7] preprint | WBC >9.5 × 109/L NC > 7.305 × 109/L |
R = −0.54, P<0.001 R = −0.585, P<0.001 |
The preprint suggests high WBC and NC indicates the higher the likelihood the severity of disease will progress to a critical stage | |
WCC = White cell count; NC = neutrophils; LC = lymphocytes; WBC = white blood cell.
3.4. Lactate dehydrogenase
In glucose metabolism, the enzyme LDH converts pyruvate to lactate. LDH secretion is triggered by necrosis of the cell membrane, hinting to viral infection or lung damage, such as the pneumonia induced by SARS-CoV-2 [19]. There is convincing evidence linking LDH levels to the development of COVID-19 disease [20].
A study found significantly higher levels of LDH in ICU patients than non-ICU patients (248 U/L vs 151 U/L, p=0.002). Since high levels of LDH continued in the ICU patients number of days post-admission (160 U/L vs 218 U/L, p=0.002), LDH may be a predictive biomarker of severe disease. However, the one centre study may be prone to selection bias which could potentially reduce its validity [21].
A multi-centre study involving 1099 patients reported supporting evidence correlating extent of tissue damage and inflammation with increasing levels of LDH [22]. Furthermore, when LDH levels were correlated with CT scans, significantly higher levels reflected the severity of pneumonia [23].
There is increasing confidence in using LDH as a biomarker to measure severity of COVID-19 infection. Another study found that there was a significant rise in LDH levels among refractory COVID-19 patients [24].
Table 5 summarises the studies used in analysing LDH and COVID-19.
Table 5.
Studies that compare LDH for COVID-19.
| Author | Level in non-severe patient (U/L) | Level in severe patient (U/L) | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Luo et al. 2020 [21] | 151 (139–180) | 248 (162–273) | P < 0.002 | Higher LDH levels reported in severe patient's vs non severe. |
| Xiong et al. 2020 [23] | N/A | N/A | R = 0.78, P < 0.001 for correlation between LDH and CT score | LDH values linked to CT scores (used to assess severity)-no absolute values |
| Guan et al. 2020 [22] | ≥250 (205/551) 37.2% of patients | ≥250 (72/144) 58.1% of patients | N/A | Multi-centre + China + 1099 Higher LDH levels present in majority of severe patients vs non-severe. LDH measured in 675. |
| Mo P et al. 2020 [24] | 241 (198–338) (General) |
293 (193–434) (Refractory) |
P = 0.017 | Retrospective + China + 155, refractory vs general |
| Ferrari D et al. 2020 [20] | 276.4 ± 118.3 (COVID-19 negative) |
388.0 ± 154.5 (COVID-19 positive) |
P <0.001 | Retrospective + Italy + 141 LDH measured, COVID-19 positive vs negative |
| Wang et al. 2020 [17] | 212 (171–291) (non-ICU) |
435 (302–596) (ICU) |
P < 0.001 | Retrospective, single-centre + China +138, non-ICU vs ICU |
LDH = lactate dehydrogenase; ICU = intensive care unit.
3.5. D-dimer
D-dimer originate from the lysis of cross-linked fibrin with rising levels indicating the activation of coagulation and fibrinolysis [25]. Early studies have associated COVID-19 with haemostatic abnormalities with one study observing elevated levels of D-dimer, the measure of coagulation, in non-survivors compared to survivors [26].
A retrospective cohort study composed of 191 patients found that D-dimer levels >1.0 μg/mL(p=0.0033) were associated with increased mortality among COVID-19 patients. Furthermore, they found that levels of 2.0 μg/mL or more on admission was the optimum cut-off to predict in-hospital mortality for COVID-19 [27]. Studies have reported that nearly 90% of inpatients with pneumonia had increased coagulation activity marked rising D-dimer levels [28].
Furthermore, Huang et al. found that levels of D-dimer on admission could be used to triage patients into critical care [5]. The researchers found that median D-dimer levels were higher in ICU patients compared to non-ICU patients (2.4 mg/L vs. 0.5 mg/L; p=0.0042). This, along with the previous study, suggests that D-dimer levels can be used as a prognostic marker and help clinicians monitor those who are likely to deteriorate earlier. However, this study confirmed the diagnosis of COVID-19 using lower respiratory tract specimens and did not use paired nasopharyngeal swabs to investigate the viral RNA detection rate between the upper and lower respiratory tract specimens. Secondly, with a cohort size of 41 patients, it is difficult to assess predictors of disease severity and mortality with multivariable-adjusted methods.
Table 6 summarises the studies used to analyse D-dimer and COVID-19.
Table 6.
Studies that compare D-dimer for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Tang et al. (2020) [26] | 0.61 (0.35–1.29) | 2.12 (0.77–5.27) | N/A | Abnormal coagulation results with markedly elevated D-dimer are common in deaths with COVID-19 |
| Zhou et al. (2020) [32] | 0.6 (0.3–1.0) | 5.2 (1.5–21.1) | P < 0.001 | D-dimer levels >1 μg/mL can help clinicians in identifying patients with poor prognosis at earlier stage |
| Guan et al. (2020) [22] | 43.2% with >0.5 mg/L | 59.6% with >0.5 mg/L | N/A | D-dimer levels much higher in those requiring ICU admission and invasive ventilation however statistical analysis not performed |
| Huang et al. (2020) [5] | 0.5 mg/L | 2.4 mg/L | P = 0.0042 | Compared to non-ICU patients, ICU patients had significantly higher levels of D-dimer |
| Zhang et al (2020) [50] |
0.41 mg/L (0.15–0.69) | 4.76 mg/L (2.99–11.9) | P < 0.001 | D-dimer on admission of >2.0 μg/mL could effectively predict in-hospital mortality in patients with COVID-19 and could be an early and helpful marker to improve management |
ICU = intensive care unit.
3.6. Platelet count
As seen with previous coronavirus outbreaks, COVID-19 infection leads to severe haematological changes leading to thrombocytopenia.
Meta-analysis of 1799 patients reveal those with severe COVID-19 infections had significantly lower platelet counts (WMD −31 × 109/L; 95% CI, −35 to −29 × 109/L) [29]. When using mortality as an endpoint, non-survivors evidently had a significantly lower platelet count (WMD, −48 × 109/L; 95% CI, −57 to −39 × 109/L). Using thrombocytopenia as an endpoint also revealed a fivefold greater risk of COVID-19 (OR, 5.13; 95% CI, 1.81–14.58). Despite the varying definitions of disease severity and thrombocytopenia having an influence on results analysis, platelet count could possibly be used clinically to indicate infection severity.
A retrospective study which used Cox proportional hazard regression analysis found that platelet count is an independent risk factor for mortality among COVID-19 patients, where a 50 × 109/L increase is associated with 40% deceased mortality (HR 0.60, 95%CI: 0.43, 0.84) [30]. Here, thrombocytopenia at admission was more likely to occur in non-survivors than in survivors. Although many risk factors were accounted for in this study, the possibility for unmeasured confounder cannot be excluded.
Another study corroborates the previously documented work. The nadir platelet count was significantly associated with mortality – and the lower the nadir, the stronger the association [31]. Again, thrombocytopenia was more likely to occur in non-survivors than survivors. This study is from adequate sample sizes providing statistical power, however, similar to the previous studies, they are all retrospective making the correlation seen difficult to extrapolate from.
Testing the platelet count is a routine part of laboratory tests and the literature suggests it has inherent value in providing more detail on the patient's condition.
Table 7 summarises the studies used in analysing platelet count and COVID-19.
Table 7.
Studies that compare platelet count for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Guan et al. 2020 [22] | 172,000 | 137,500 | N/A | Although there were evident differences in lymphocytes (lymphocytopenia), platelet count (rose) and D-dimer (rose) in patients who experienced composite endpoints (ICU admission, invasive mechanical ventilation and death) there was no statistical analysis performed |
| Huang et al. 2020 [5] | 149,000 | 196,000 | P = 0.45 | Compared to non-ICU patients, ICU patients had higher plasma levels of IL-6, D-dimer and platelets. But this difference was not significant. |
| Liu et al. 2020 [41] | 173,200 ± 55.37 | 143,900 ± 64.81 | P = 0.116 | Ct value of virus correlated strongly with CRP, albumin and LDH among others. Murray score for ARDS correlated with the same markers. |
| Liu et al. 2020 [42] | 186,200 | 139,500 | N/A | Increased CRP and decreased albumin strongly correlated with disease progression |
| Ruan et al. 2020 [43] | 221,000 (78,000) | 173,600 (67,000) | P < 0.001 | Cardiac troponin, Myoglobin, CRP and IL-6 significantly increased in cases with mortality |
| Wang et al. 2020 [17] | 165,000 | 142,000 | P < 0.78 | Higher WCC, specifically higher NC, lower LC and higher NLR in ICU vs non-ICU patients. Prior to the death of critical patients: NC, D-dimer, blood urea, and creatinine levels rose throughout until death, whilst the LC carried on falling. Elevated LDH also correlated with patients in ICU vs non-ICU. |
| Yang et al. 2020 [44] | 191,000 (63,000) | 164,000 (74,000) | N/A | Higher levels of platelets in severe group but lymphocytopenia seen as most common marker of infection Mortality used to indicate severity |
| Young et al. 2020 [45] | 159,000 | 156,000 | N/A | Lower platelets found in severe group, but statistical analysis was not performed. Severity determined by supplemental oxygen requirement |
| Zhou et al. 2020 [32] | 220,000 | 165,500 | P < 0·0001 | D-dimer levels >1 μg/mL can help clinicians in identifying patients with poor prognosis at earlier stage There were significantly higher levels of platelets and hs-TnI in non-survivors compared to survivors Mortality used to determine severity |
| 41. Liu et al. 2020 [30] | 186,000 (160,000–227,000) | 105,000 (92,000–116,000) |
P < 0.001 | Thrombocytopenia associated with mortality Patients split on basis of thrombocytopenia not severity |
| 42. Yang et al. 2020 [31] | 203,000 (155,000–257,000) | 79,000 (43,000–129,000) | P < 0.001 | Lowest platelet count associated with mortality Mortality used to indicate severity Uses nadir platelet count |
IL-6 = interleukin-6; ICU = intensive care unit.
3.7. Cardiac troponin
There is growing evidence of higher mortality rates among those with underlying cardiovascular disease due to COVID-19 infection [22,52,53]. Some have investigated the use of high-sensitivity cardiac troponin I (hs-TnI) as a marker of disease progression and mortality.
A retrospective study performed in China of patients with confirmed COVID-19 based on SARS-CoV-2 RNA detection, revealed a univariable odds ratio for death at 80.1 (95% CI 10.3–620.4, p<0.0001) for hs-TnI [32]. This risk was higher compared to other biomarkers such as D-dimer and lymphocyte count. Another study of 416 hospitalised patients with COVID-19 reported that hs-TnI was elevated in 1 in 5 patients on presentation [33]. These patients were more likely to require invasive (22% vs 4%, p<0.001) or non-invasive (46% vs 4%, p<0.001) ventilation, develop ARDS (59% vs 15%, p<0.001) or acute kidney injury (9% vs 0%, p<0.001).
Early recognition of myocardial injury indicated by elevated hs-TnI aids in appropriate triage to a critical care area and informs the use of inotropes and vasopressors. However, elevated levels are common in hospitalised patients and are likely to be due to non-ischaemic causes of myocardial injury. This may lead to inappropriate use of cardiology consultation and downstream testing and increased risk to cardiac physiology staff.
Table 8 summarises the studies used in analysing hs-TnI and COVID-19.
Table 8.
Studies that compare cardiac troponin levels for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| Zhou et al. (2020) [32] | 3.0 (1.1–5.5) | 22.2 (5.6–83.1) | P < 0.0001 | Significantly higher levels of hs-TnI in non-survivors compared to survivors |
| Shi et al. (2020) [33] | <0.006 μg/L (<0.006–0.009) | 0.19 μg/L (0.08–1.12) | P < 0.0001 | Significantly higher levels of hs-TnI in patients who require mechanical ventilation compared to those who do not |
Hs-TnI = high sensitivity troponin I.
3.8. Renal markers
There is also evidence that chronic kidney disease is associated with severe forms of COVID-19 infection [34].
Studies have demonstrated significantly higher levels of renal biomarkers such as serum urea, creatinine and markers of glomerular filtration rate in severe cases [35]. Since these results stem from the analysis of 28 patients, extrapolation across larger cohorts is more difficult.
A larger study of 701 patients revealed that elevated serum creatinine levels on admission correlated with severity due to significant abnormalities in the coagulation pathway [36]. They also found that these patients were more likely to require mechanical ventilation or be placed in intensive care. Univariate Cox regression analysis found elevated creatinine levels was also associated with in-hospital mortality (HR 2.99, 95% CI: 2.00, 4.47). Proteinuria, haematuria and elevated urea levels had similar, if not larger, hazard ratios.
Interestingly, another study showed a potential role for urinalysis over serum markers of kidney function [37]. Here, abnormalities in the routine urine test on admission correlated strongly with disease severity. They go on to suggest that urinalysis may reveal kidney impairment more readily than evaluation of serum renal biomarkers. However, these tests were only carried out on admission and so patients in earlier stages of the infection had changes in serum levels obscured by compensatory kidney function. Hence renal abnormalities on admission may indicate higher risks of deterioration, ensuring appropriate triaging.
Table 9 summarises the studies used in analysing renal markers and COVID-19.
Table 9.
Studies that compare creatinine levels for COVID-19.
| Author | Level in non-severe patient | Level in severe patient | Confidence interval (CI) range and p value | Comments |
|---|---|---|---|---|
| 45. Xiang et al. 2020 [35] | N/A | N/A | P < 0.001 | Serum values of urea, creatine and cystatin-C significantly increase in severe COVID-19 No values provided – only graphical depiction of results |
| 46. Cheng et al. 2020 [36] | 77 ± 31 | 132 ± 39 | P < 0.001 | Raised creatinine levels associated with poor outcome in COVID-19 infection Patients split on basis of creatine level not severity |
| 47. Zhou et al. 2020 [37] | 65.3 (56.5–74.3) | 71.0 (55.8–89.4) | P = 0.067 | Urinalysis on admission can effectively highlight kidney impairment Patients split on basis of abnormal urinalysis |
4. Discussion
COVID-19 is a rapidly spreading pandemic increasing the burden on medical facilities. Symptoms vary from mild fever to ARDS complicating diagnosis, prognosis, and monitoring. Hence it is vital to ascertain a patient's condition in a timely manner. Biomarkers are quantitative measurements used clinically for many conditions reflecting pathological development. A summary of the biomarkers discussed in this review can be found in Table 10 .
Table 10.
Summary of Changes in Biomarkers Seen in Severe COVID-19 Infection.
| Biomarker | Change in severe COVID-19 infection |
|---|---|
| CRP | Increase |
| SAA | Increase |
| IL-6 | Increase |
| LDH | Increase |
| WCC | NLR increases LC decrease |
| D-dimer | Increase |
| Platelet count | Decrease |
| Cardiac troponin | Increase |
| Renal biomarkers | Urea & creatinine increase |
CRP = C-reactive protein; SAA = serum amyloid A; IL-6 = interleukin 6; LDH = lactate dehydrogenase; WCC = White cell count.
When assessing a patient with COVID-19 infection, biomarkers can be useful to clinicians in starting treatment and close monitoring. Though biomarkers may help improve prognosis and outcomes, their significant variability between patients could affect the findings of the studies.
Compared to other biomarkers, there is hesitancy in using WCC alone as it is influenced by many factors such as glucocorticoid treatment which increases it. Although WCC encompasses many cell types, NCs and LCs are most clinically relevant biomarkers. Multiple studies on COVID-19 have concurred high NLR in severe cases compared to non-severe cases due to high NC and low LC. The use of LC independently has been suggested as a potential biomarker of COVID-19 as patients have consistently low LC, with significant lymphopenia reported in critically ill patients [18,55]. Therefore, further research on WCC accuracy for assessing disease progression is necessary.
Although, most of the studies referenced in this review are single centred studies originating in Wuhan, China, the virus is now a global pandemic thus requiring international studies. As a copious amount of data was collected from the same location, it is possible patients may have been used in more than one study. Furthermore, categorising patients into severe and non-severe sub-groups is dependent on the definitions of severity of COVID-19. Two very different measures of severity used in some studies are ICU admission and onset of ARDS. This questions the heterogeneity across the studies and validity of the findings.
Variability can be accounted to the differences in patient exposure to the virus or intraindividual variability, where differences in laboratory procedures lead to errors. Consequently, the sensitivity and specificity of biomarkers are useful indicators to the effectiveness, though not all studies discussed this. To ascertain the usefulness of the biomarkers listed in this review as indicators of disease progression, and whether they definitively rise in COVID-19 requires further data collection. Statistical analysis using receiver-operator characteristic curves can also provide analysis sensitivity and false-positive rates.
Findings of these studies have discovered changes in biomarker levels and may potentially be useful in creating a therapeutic intervention. For instance, one study has reviewed the use of anticoagulation therapy in patients with coagulopathy or marked rise in D-dimers in the setting of COVID-19 [38]. Low molecular weight heparin was found to be associated with better prognosis in severe cases. To further asses the role of anti-coagulants as a treatment, we encourage large interventional trails to study this.
Many studies in this review were limited due to small sample size and selection bias if conducted at one centre. To improve reliability and reproducibility, more research into the prognostic value of biomarkers is necessary [[51], [52], [53], [54], [55]].
5. Conclusion
In conclusion, the work to date suggests that there is clear evidence of how the levels of biomarkers may change according to severity of COVID-19 infection. This can be used as an adjunct in clinical practice to guide treatment and admission to ICU. By doing so, it may improve prognosis and minimise the mortality rates. However, being in the infant stages of understanding the pathology of this infectious disease, we urge for further research worldwide to better understand the changes noted in this review.
Funding
No funding obtained.
CRediT authorship contribution statement
Muhammed Kermali: Writing - original draft, Writing - review & editing, Formal analysis, Visualization. Raveena Kaur Khalsa: Writing - original draft, Writing - review & editing, Visualization. Kiran Pillai: Writing - original draft, Writing - review & editing, Visualization. Zahra Ismail: Writing - review & editing. Amer Harky: Conceptualization, Project administration, Supervision, Visualization, Writing - review & editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. 2020 Feb 11. Severe Acute Respiratory Syndrome-related Coronavirus: The Species and Its Viruses – A Statement of the Coronavirus Study Group. bioRxiv. 2020.02.07.937862. [Google Scholar]
- 2.Chan J.W.M., Ng C.K., Chan Y.H., Mok T.Y.W., Lee S., Chu S.Y.Y., et al. Short term outcome and risk factors for adverse clinical outcomes in adults with severe acute respiratory syndrome (SARS) Thorax. 2003 Aug;58(8):686–689. doi: 10.1136/thorax.58.8.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth T.F., Kournikakis B., Bastien N., Ho J., Kobasa D., Stadnyk L., et al. Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 2005 May 1;191(9):1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020 [cited 2020 Apr 28]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK554776/. [PubMed]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce J.D., McCabe S., White N., Clancy R.L. Biomarkers: an important clinical assessment tool. Am. J. Nurs. 2012;112(9):52–58. doi: 10.1097/01.NAJ.0000418926.83718.28. Sep. [DOI] [PubMed] [Google Scholar]
- 7.Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., et al. 2020 Feb 27. Correlation Analysis Between Disease Severity and Inflammation-related Parameters in Patients With COVID-19 Pneumonia. medRxiv. 2020.02.25.20025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. (Mar 12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020 Apr 14;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji W., Bishnu G., Cai Z., Shen X. 2020 Mar 13. Analysis Clinical Features of COVID-19 Infection in Secondary Epidemic Area and Report Potential Biomarkers in Evaluation. medRxiv. 2020.03.10.20033613. [Google Scholar]
- 11.Yip T.T.C., Chan J.W.M., Cho W.C.S., Yip T.-T., Wang Z., Kwan T.-L., et al. Protein chip array profiling analysis in patients with severe acute respiratory syndrome identified serum amyloid a protein as a biomarker potentially useful in monitoring the extent of pneumonia. Clin. Chem. 2005 Jan;51(1):47–55. doi: 10.1373/clinchem.2004.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan C., Huang Y., Shi F., Tan K., Ma Q., Chen Y., et al. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J. Med. Virol. 2020 Apr 13 doi: 10.1002/jmv.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahajan S., Decker C.E., Yang Z., Veis D., Mellins E.D., Faccio R. Plcγ2/Tmem178 dependent pathway in myeloid cells modulates the pathogenesis of cytokine storm syndrome. J. Autoimmun. 2019;100:62–74. doi: 10.1016/j.jaut.2019.02.005. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coomes E.A., Haghbayan H. medRxiv; 2020 Apr 3. Interleukin-6 in COVID-19: A Systematic Review and Meta-Analysis. 2020.03.30.20048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 Feb 7;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduction and Targeted Therapy. 2020;5(1):1–3. doi: 10.1038/s41392-020-0148-4. Mar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y., Zhang H., Mu S., Wei W., Jin C., Xue Y., et al. 2020 Mar 27. Lactate Dehydrogenase, a Risk Factor of Severe COVID-19 Patients. medRxiv. 2020.03.24.20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari D., Motta A., Strollo M., Banfi G., Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0398. (Apr 16) Ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Clinical findings of 35 cases with novel coronavirus pneumonia outside of Wuhan. 2020 Apr 17. https://www.researchsquare.com/article/rs-22554/v1 [cited 2020 Apr 29]; Available from.
- 22.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. (Feb 28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y., Sun D., Liu Y., Fan Y., Zhao L., Li X., et al. Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Investig. Radiol. 2020;55(6):332–339. doi: 10.1097/RLI.0000000000000674. (Mar 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect. Dis. 2020 Mar 16 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Long Y., Xiao H., Yang J., Toulon P., Zhang Z. Use of D-dimer in oral anticoagulation therapy. Int. J. Lab. Hematol. 2018;40(5):503–507. doi: 10.1111/ijlh.12864. [DOI] [PubMed] [Google Scholar]
- 26.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Yan X., Fan Q., Liu H., Liu X., Liu Z., et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J. Thromb. Haemost. 2020 Apr 19 doi: 10.1111/jth.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milbrandt E.B., Reade M.C., Lee M., Shook S.L., Angus D.C., Kong L., et al. Prevalence and significance of coagulation abnormalities in community-acquired pneumonia. Mol. Med. 2009 Dec;15(11−12):438–445. doi: 10.2119/molmed.2009.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin. Chim. Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. Mar 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Sun W., Guo Y., Chen L., Zhang L., Zhao S., et al. Association between platelet parameters and mortality in coronavirus disease 2019: retrospective cohort study. Platelets. 2020 Apr 16;0(0):1–7. doi: 10.1080/09537104.2020.1754383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Yang Q, Wang Y, Wu Y, Xu J, Yu Y, et al. Thrombocytopenia and its association with mortality in patients with COVID-19. J. Thromb. Haemost. [Internet]. [cited 2020 Apr 30];n/a(n/a). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jth.14848. [DOI] [PMC free article] [PubMed]
- 32.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020 Mar 25 doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henry B.M., Lippi G. Chronic kidney disease is associated with severe coronavirus disease 2019 (COVID-19) infection. Int Urol Nephrol [Internet] 2020 Mar 28 doi: 10.1007/s11255-020-02451-9. [cited 2020 Apr 30]; Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang J., Wen J., Yuan X., Xiong S., Zhou X., Liu C., et al. Potential biochemical markers to identify severe cases among COVID-19 patients. medRxiv. 2020 Mar 23;2020.03.19.20034447. [Google Scholar]
- 36.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020 May 1;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H., Zhang Z., Fan H., Li J., Li M., Dong Y., et al. 2020 Apr 6. Urinalysis, but Not Blood Biochemistry, Detects the Early Renal-impairment in Patients With COVID-19. medRxiv. 2020.04.03.20051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020;18(5):1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Xiang X., Ren H., Xu L., Zhao L., Chen X., et al. SAA is a biomarker to distinguish the severity and prognosis of coronavirus disease 2019 (COVID-19) J Infect [Internet] 2020 Apr 8 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7141628/ [cited 2020 Apr 29]; Available from: Ahead of print. [Google Scholar]
- 40.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020 Mar 31 doi: 10.1016/j.medmal.2020.03.007. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7146693/ [Internet]. [cited 2020 Apr 29]; Available from. S0399-077X(20)30086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W., Tao Z.-W., Wang L., Yuan M.-L., Liu K., Zhou L., et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin. Med. J. 2020;133(9):1032–1038. doi: 10.1097/CM9.0000000000000775. May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 Mar 3;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020 Feb 24;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. (Mar 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L., Liu H.G., Liu W., Liu J., Liu K., Shang J., et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020 Feb 6;43(0) doi: 10.3760/cma.j.issn.1001-0939.2020.0005. [DOI] [PubMed] [Google Scholar]
- 47.Liu T., Zhang J., Yang Y., Ma H., Li Z., Zhang J., et al. The potential role of IL-6 in monitoring severe case of coronavirus disease 2019. medRxiv. 2020 doi: 10.15252/emmm.202012421. Mar 10;2020.03.01.20029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. medRxiv; 2020 Feb 20. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) 2020.02.18.20024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 Mar 13 doi: 10.1001/jamainternmed.2020.0994. Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z, et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. Journal of Thrombosis and Haemostasis [Internet]. [cited 2020 Apr 25];n/a(n/a). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jth.14859. [DOI] [PMC free article] [PubMed]
- 51.Abuelgasim E., Saw L.J., Shirke M., Zeinah M., Harky A. COVID-19: Unique public health issues facing Black, Asian and minority ethnic communities [published online ahead of print, 2020 May 8] Curr. Probl. Cardiol. 2020:100621. doi: 10.1016/j.cpcardiol.2020.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan I.H., Zahra S.A., Zaim S., Harky A. At the heart of COVID-19. J. Card. Surg. 2020 May 5 doi: 10.1111/jocs.14596. [Epub ahead of print] Review. PubMed PMID: 32369872. [DOI] [PubMed] [Google Scholar]
- 53.Hashkhusha T.R., Chan J.S.K., Harky A. ACE inhibitors and COVID-19: We don't know yet. J. Card. Surg. 2020 Apr 27 doi: 10.1111/jocs.14582. [Epub ahead of print] PubMed PMID: 32340070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and Multi-Organ Response [published online ahead of print, 2020 Apr 28] Curr. Probl. Cardiol. 2020:100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savarimuthu S., BinSaeid J., Harky A. The role of ECMO in COVID-19: Can it provide rescue therapy in those who are critically ill? J. Card. Surg. 2020 doi: 10.1111/jocs.14635. (n.d.) 0–0. [DOI] [PMC free article] [PubMed] [Google Scholar]