Summary
The ongoing outbreak of the novel coronavirus pneumonia COVID-19 has caused great number of cases and deaths, but our understanding about the pathogen SARS-CoV-2 remains largely unclear. The attachment of the virus with the cell-surface receptor and a cofactor is the first step for the infection. Here, bioinformatics approaches combining human-virus protein interaction prediction and protein docking based on crystal structures have revealed the high affinity between human dipeptidylpeptidase 4 (DPP4) and the spike (S) receptor-binding domain of SARS-CoV-2. Intriguingly, the crucial binding residues of DPP4 are identical to those that are bound to the MERS-CoV-S. Moreover, E484 insertion and adjacent substitutions should be most essential for this DPP4-binding ability acquirement of SARS-CoV-2-S compared with SARS-CoV-S. This potential utilization of DPP4 as a binding target for SARS-CoV-2 may offer novel insight into the viral pathogenesis and help the surveillance and therapeutics strategy for meeting the challenge of COVID-19.
Subject Areas: Virology, Molecular Structure, Crystallography
Graphical Abstract
Highlights
-
•
SARS-CoV-2 spike receptor-binding domain has a potentially high affinity to DPP4
-
•
SARS-CoV-2-S/DPP4 binding shares key DPP4 residues with that of MERS-CoV-S/DPP4
-
•
E484 and adjacent mutations are critical for the DPP4-binding ability of SARS-CoV-2-S
Virology; Molecular Structure; Crystallography
Introduction
The ongoing outbreak of the novel coronavirus disease (COVID-19) that was initially reported at Wuhan city, China, in December 2019 has rapidly been characterized as a pandemic, transmitting to almost all the countries or territories in the world. As of March 27, 2020, confirmed cases have exceeded 500,000, including 23,335 deaths globally (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports). As an emerging disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, previously named 2019-nCoV), COVID-19 has since become a global concern with the very-high level of risk. The pathogen SARS-CoV-2 has been shown to be a member of Betacoronavirus closely related to the SARS-CoV and the strain RaTG13 of bat origin (Zhou et al., 2020).
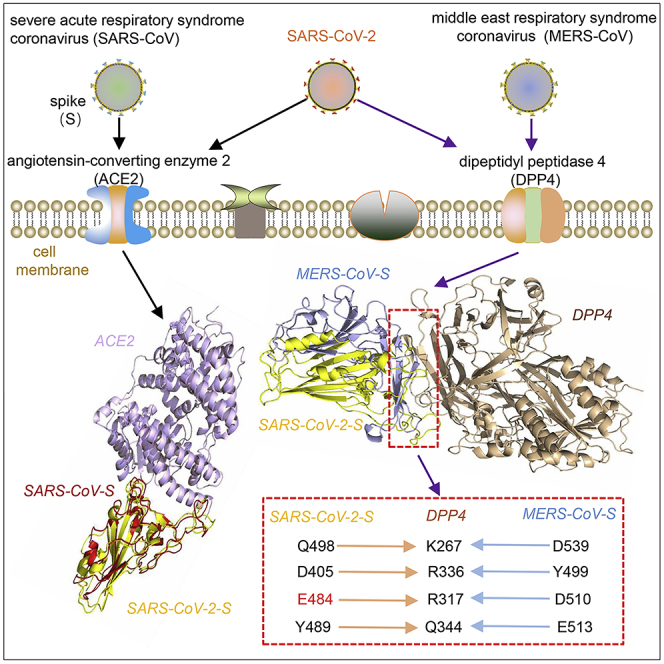
The coronaviruses are enveloped ranking among the dangerous zoonotically emerging pathogens (Song et al., 2019). There were two more severe Betacoronavirus disease outbreaks in humans since this century. They were SARS in 2003 and the Middle East respiratory syndrome (MERS) that started in 2012 (Song et al., 2019). Both SARS-CoV and MERS-CoV were zoonotic agents transmitted from bats to intermediate hosts such as palm civets or dromedary camels, and finally to humans (Cui et al., 2019, Song et al., 2019). The coronavirus is a type of positive-stranded RNA virus with the spike (S) glycoprotein on the virion envelope mediating receptor recognition through the receptor-binding domain (RBD) and membrane fusion following the proteolytic cleavage into S1 and S2 subunits (Cui et al., 2019). The receptors are predominant determinants for the host tropism and pathogenicity of the viruses (Cui et al., 2019). Coronaviruses have evolved sophisticated receptor recognition patterns. Spikes from related coronaviruses can recognize distinct receptors, whereas spikes of distant coronaviruses can employ the same entry receptor. For instance, SARS-CoV uses angiotensin-converting enzyme 2 (ACE2) as its main receptor and MERS-CoV engages the transmembrane dipeptidylpeptidase 4 (DPP4, also known as CD26) as the primary receptor (Cui et al., 2019, Song et al., 2019, Wang et al., 2013). A few reports have proposed that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for its cellular entry (Hoffmann et al., 2020, Letko et al., 2020, Zhou et al., 2020), although four of five key residues within the RBD of SARS-CoV-2 are mutated when compared with that of SARS-CoV (Zhou et al., 2020). ACE2 is also shared as a receptor of another human coronavirus, NL63, which belongs to the Alphacoronavirus genus (Chan et al., 2016, Cui et al., 2019, Song et al., 2019). In addition, the natural hosts of SARS-CoV-2 have been suggested to be bats and pangolins resulting from analyses of the viral genome characteristics (Lam et al., 2020, Zhang et al., 2020, Zhou et al., 2020), but an intermediate animal that directly transmits SARS-CoV-2 to human has not been confirmed.
Intriguingly, the viral cell entry always requires multiple transmembrane proteins in the target cell apart from the primary receptor (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). The coronavirus spikes are able to recognize a broad range of cell-surface molecules in addition to the designated receptors for entry. These molecules are called coreceptors or attachment factors, which have been reported to play critical roles in the viral propagation (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). The cellular susceptibility to virus infection may correlate with these proteins' interactions, which helps ensure the triggering of viral entry at an appropriate place (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). A receptor for one kind of coronavirus can be utilized for a cofactor of the infection of another or even a distant coronavirus. For example, the human coronavirus OC43 (hCoV-OC43) receptor, O-acetylated sialic acid, has been suggested as an attachment factor in the binding of hCoV-HKU1 to the cell surface (Chu et al., 2018, Cui et al., 2019). The carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) and glucose-regulated protein GRP78 are identified as attachment factors that could enhance the attachment in nonpermissive cells and entry in susceptible cells for MERS-CoV (Chan et al., 2016, Chu et al., 2018). GRP78 can simultaneously help the cell entry of the bat coronavirus HKU9 except for MERS-CoV (Chu et al., 2018). Like CEACAM5, CEACAM1 is a member of the same family, which serves as the receptor of the animal coronavirus, mouse hepatitis virus (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). Moreover, the tetraspanin CD9 was identified as a host cell-surface factor that augments MERS-CoV entry by scaffolding the receptor DPP4 and the protease TMPRSS2 (Earnest et al., 2017). The latter can act as a priming factor in the early infection of SARS-CoV, MERS-CoV, or SARS-CoV-2 (Chan et al., 2016, Chu et al., 2018, Hoffmann et al., 2020). In the case of SARS-CoV, dendritic cell-specific inter-cellular adhesion molecule-3-grabbing nonintegrin (DCSIGN) and DCSIGN-related facilitate virus entry by interacting with S protein (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). It remains unclear whether coronaviruses bind to these attachment factors through RBDs of the viral spikes. Little attention is given to the cell-surface co-factors in facilitating the attachment and entry of SARS-CoV-2 currently. However, study on this aspect may help understand the viral pathogenesis, which remains largely unknown.
Results
The Prediction of Interaction between Human Proteins and the SARS-CoV-2 Spike
The interaction of the receptor-binding domain (RBD) of SARS-CoV-2 spike (S) protein and ACE2, as well as the monomer proteins of DPP4, ACE2, and coronavirus spike proteins, have been resolved by crystallography (Lan et al., 2020, Walls et al., 2020). An integrative human-virus protein interaction inference based on protein crystal structures was used to predict human proteins probably interacting with SARS-CoV-2 spike protein. In the prediction, interolog mapping, domain-domain interaction, and machine learning were combined to evaluate the score for each protein-protein interaction (Schleker et al., 2012, von Mering et al., 2005, Yang et al., 2020, Zhou et al., 2014). The result showed that both ACE2 and DPP4 ranked in the top five human proteins (Table 1). It is known that ACE2 has been proposed to be a receptor of SARS-CoV-2. As DPP4 obtained the highest integration score (Table 1), and is also associated with a Betacoronavirus by acting as the receptor of MERS-CoV, we have focused on it next.
Table 1.
The Top Five Predicted Human Proteins Interacting with SARS-CoV-2 S by Three Methods
| UniProt ID | Human Protein | Virus Protein | IM | DDI | Integration |
|---|---|---|---|---|---|
| P27487 | DPP4 | SARS-CoV-2 S | 0.8413 | 0.5357 | 0.9232 |
| Q12884 | FAP | SARS-CoV-2 S | 0.8413 | 0.5062 | 0.9183 |
| Q8N608 | DPP10 | SARS-CoV-2 S | 0.8413 | 0.5062 | 0.9183 |
| P42658 | DPP6 | SARS-CoV-2 S | 0.8413 | 0.5062 | 0.9183 |
| Q9BYF1 | ACE2 | SARS-CoV-2 S | 0.4628 | 0.5459 | 0.7456 |
Machine learning (ML) did not predict the interactions (value = 0). IM, interolog mapping; DDI, domain-domain interaction; DPP, dipeptidylpeptidase; ACE2, angiotensin-converting enzyme 2; FAP, fibroblast activation protein.
The Affinity of Three Betacoronavirus Spikes and DPP4 or ACE2
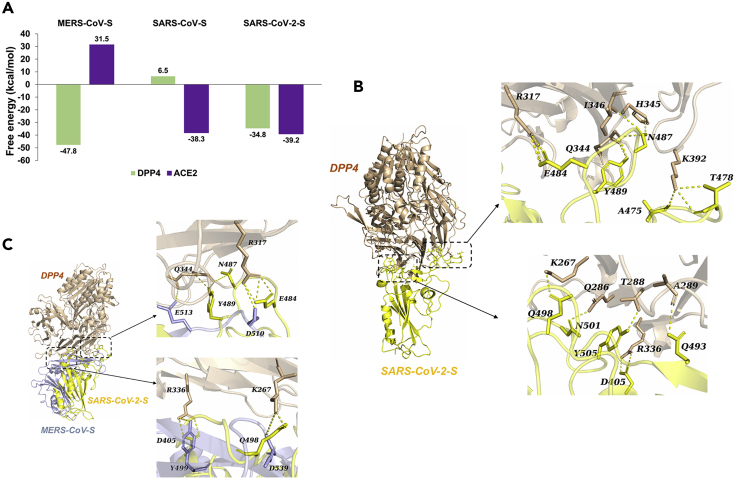
By using a set of methods combining mechanics energies with the generalized Born and surface area continuum solvation and molecular dynamics simulation (Genheden and Ryde, 2015, Maier et al., 2015, Miller et al., 2012, Miyamoto and Kollman, 1992, Salomon-Ferrer et al., 2013, Toukmaji et al., 2000), the binding free energies for the RBDs of coronavirus spike (CoV-S) proteins to DPP4 or ACE2 were calculated (Figure 1A). According to the free energy values (far less than zero), the bindings of SARS-CoV-S/ACE2 and MERS-CoV/DPP4 are consistent with what has been reported. There were no potential interactions of SARS-CoV-S/DPP4 and MERS-CoV/ACE2 (free energy values > 0 kcal/mol). Among the three coronaviruses, only SARS-CoV-2-S could theoretically bind to both ACE2 and DPP4. In terms of the binding of DPP4 with SARS-CoV-2-S, the affinity (−34.8 kcal/mol) was lower than that of DPP4 with MERS-CoV-S (−47.8 kcal/mol), ACE2 with SARS-CoV-2-S (−39.2 kcal/mol), as well as ACE2 with SARS-CoV-S (−38.3 kcal/mol) (Figure 1A).
Figure 1.
The Analyses in the Binding of the Viral Spike Proteins and DPP4 or ACE2
(A) Free energy for the binding between each of three CoV spike (S) proteins and DPP4 or ACE2. Note that all the complexes were downloaded or predicted through ZDOCK based on protein crystal structures.
(B) The structure of predicted interaction complex of SARS-CoV-2-S RBD (yellow) and DPP4 (wheat). The detailed binding interface is magnified separately as indicated.
(C) The structural model of SARS-CoV-2-S RBD bound with DPP4 is superimposed with the crystal structure of MERS-CoV-S RBD. The contact residues of DPP4 (wheat) bound with either MERS-CoV-S RBD (light blue) or SARS-CoV-2-S RBD (yellow) are displayed in the right two panels.
Analysis of the Interaction Interfaces between DPP4 and RBD of SARS-CoV-2-S or MERS-CoV-S
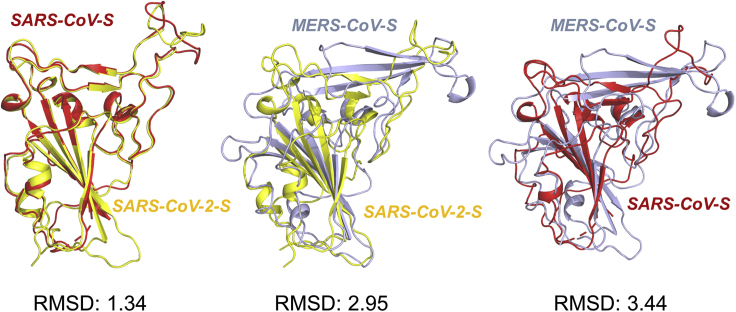
By employing protein docking strategy, we investigated the potential interaction mode between SARS-CoV-2-S RBD and DPP4. The atomic interaction details of the binding interface showed that almost all the contacting residues of DPP4 with SARS-CoV-2-S RBD were consistent with those for binding with MERS-CoV-S RBD (Table S1) (Song et al., 2014). These residues include DPP4 residue K267 with SARS-CoV-2-S RBD Q498, DPP4 R336 with RBD D405, R317 with RBD E484, DPP4 Q344 with RBD Y489/N487, DPP4 Q286 with N501, and DPP4 T288 with Y505 (Figure 1B and Table S1). In addition, DPP4 residues L294 and I295 can contribute van der Waals' forces to the binding (Table S1), not forming hydrogen bonds as in the binding to MERS-CoV-S (Song et al., 2014). Although the homology between SARS-CoV-2-S and MERS-CoV-S RBDs is low (19.1%), they shared identical binding residues of DPP4 at the interfaces (Figure 1C). According to the known structural information of DPP4 interacting with MERS-CoV-S RBD, the residue pairs (DPP4 residue K267, R336, R317, and Q344 with MERS-CoV-S RBD D539, Y499, D510, and E513, respectively) contributed significantly to the interaction (Table S1) (Song et al., 2014). All the four DPP4 residues were also observed to be involved in the binding interface of DPP4/SARS-CoV-2 RBD, contributing to free energy by forming hydrogen bonds. Thus, these four DPP4 residues were regarded as the crucial residues of DPP4 interacting with SARS-CoV-2-S RBD. In one-to-another comparison of the three three CoV-S RBD structures, SARS-CoV-2-S RBD and SARS-CoV-S RBD also showed the lowest root-mean-square deviation (RMSD) value (1.34Å), which is in line with their high identity (Figure 2). However, SARS-CoV-2-S RBD is more structurally similar to MERS-CoV RBD than SARS-CoV RBD to MERS-CoV RBD, with 2.95 Å and 3.44 Å RMSD values, respectively (Figure 2). This may help explain why SARS-CoV-2-S RBD can bind to the MERS-CoV receptor of DPP4, but SARS-CoV-S RBD cannot.
Figure 2.
Comparison of the RBD Structures of Every Two Among Three Viruses
RMSD (root-mean-square deviation) value (Å) can be used for the estimation of the structural similarity of every two proteins. The smaller the RMSD, higher the similarity between both structures.
The Analysis of Key Residues for DPP4-Binding Ability Acquirement of SARS-CoV-2-S
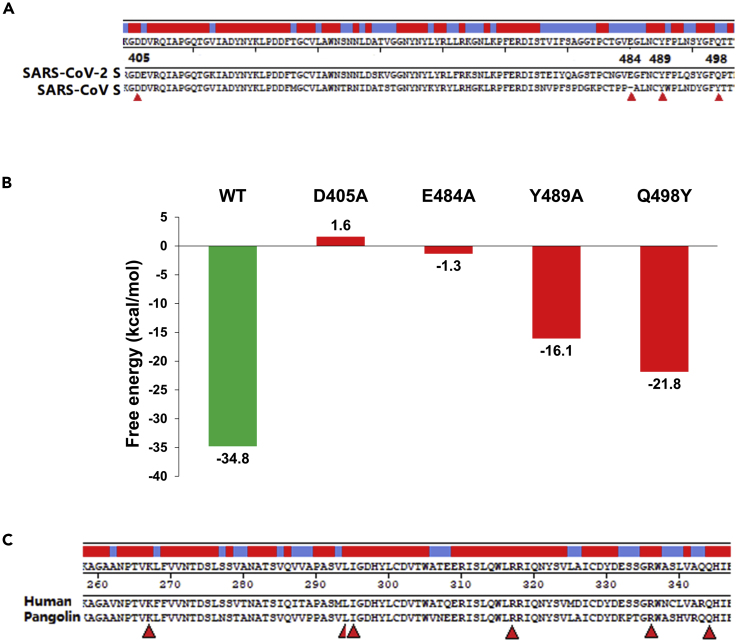
According to the aforementioned results (Figures 1A and 2), SARS-CoV-S does not bind with DPP4. We compared the sequences at the key residues of SARS-CoV-2-S RBD equivalent to that of SARS-CoV-S RBD. Two out of four substitutions or insertion happened in SARS-CoV-2-S RBD (Figure 3A). In particular, at E484 (potentially binding with DPP4 R317) and adjacent residues, there was an insertion and several substitutions. The substitution of Q498Y also made the potential binding between SARS-CoV-2-S RBD with DPP4 K267. Furthermore, the virtual mutation of even one of these residues in SARS-CoV-2-S RBD resulted in considerable reduction of affinity, especially for the substitutions of D405A and E484A (Figure 3B). Actually, there is no mutation at D405 and Y489/N487 compared with the equivalent residues of SARS-CoV-S RBD (Figure 3A). Thus, E484 should be the most important residue for the binding ability. We also checked the amino acids in the spike RBD of the bat strain RaTG13 (Zhou et al., 2020): an insertion of T (not E) and a few different substitutions existed at the sites equivalent to E484 and adjacent residues of SARS-CoV-2 and there were no mutations in the remaining three key residues (Figure S1). In addition, as the pangolin has been suspected as a zoonotic host of SARS-CoV-2, we compared the sequences of human and pangolin DPP4 (Figure 3C). The result showed that they shared all key residues ready to bind to both SARS-CoV-2 and MERS-CoV spikes (Song et al., 2014). These key residues presented in human DPP4 are also conserved in other animal species, including macaque, pig, camel, horse, sheep, bovine, rabbit, and bat (Song et al., 2014).
Figure 3.
Comparison and Mutation of the Key Residues for the Binding of SARS-CoV-2-S RBD and DPP4
(A) Sequence comparison of the key residues between SARS-CoV-2-S and SARS-CoV-S RBDs. There are an insertion and adjacent substitutions at E484, and a Q498Y substitution in SARS-CoV-2-S RBD.
(B) Effect of mutation at the key residues on the binding free energy. Virtual mutations of single key residues could impact the free energy, especially for D405A and E484A. WT, wild-type.
(C) Comparison of the key residues between human and pangolin DPP4 protein sequences. Both shared the completely identical key residues including K267, L294, I295, R317, R336, and Q344.
Discussion
Coronaviruses commonly recognize a wide range of cell-surface targets in addition to the receptors (Chan et al., 2016, Chu et al., 2018, Cui et al., 2019). For MERS-CoV, when the receptor DPP4 and the protease TMPRSS2 are sparse on the cell surface, CD9 can play an important role in rendering the early, efficient entry (Earnest et al., 2017). Before the fusion occurs, sufficient proteolytic cleavage is likely to form an activated cluster in regions of the cell membrane with a relatively high local concentration of these entry factors (Cui et al., 2019, Earnest et al., 2017). This sort of characteristics may give the coronaviruses a physiological advantage in establishing efficient infections in animal or human hosts, thus contributing to their high pathogenicity. With the ongoing pandemic of COVID-19, the causes for a lot of clinical features remain largely to be investigated. Experiments in an animal model have showed that the severity of COVID-19 seems intermediate between those of SARS and MERS (Rockx et al., 2020). Also, the receptor ACE2 is suboptimal for the infection of SARS-CoV-2 in binding to the spike (Procko, 2020). The immunological characterization study showed the infection of SARS-CoV-2 in immune cells, which is especially not correlated with the expression of ACE2 (Wei et al., 2020). Therefore, it seems that ACE2 should have employed some helpers in the swift infection and pathogenesis of SARS-CoV-2.
In this study, according to the binding free energy, the affinity of ACE2/SARS-CoV-2-S is stronger than that of DPP4/SARS-CoV-2-S (Figure 1A), suggesting the priority of the binding of ACE2 to SARS-CoV-2-S and the leading role of ACE2 as a receptor. The results demonstrated that both SARS-CoV-2-S and MERS-CoV-S could bind to DPP4, sharing the identical key binding residues of DPP4 at the interfaces (Figure 1). The results also suggested that the insertion and substitutions at E484 and adjacent residues are essential for the entirely different ability in binding to DPP4 between SARS-CoV-2 and SARS-CoV spikes, for adjacent residues may also contribute to the structure change. It seems that the binding of SARS-CoV-2-S and DPP4 is unique. However, these kinds of mutations can naturally occur as in the bat strain RaTG13 (Figure S1), although the residues may be different. The only animal-origin Betacoronavirus that possesses this potential binding ability currently is the one isolated from pangolins, containing these identical key residues in the spike RBD sequences (Lam et al., 2020). In addition, the models evaluated the binding potential, interface residues, and structures that were consistent with those known virus-host interactions, including SARS-CoV-S/ACE2, and MERS-CoV-S/DPP4, verifying the feasibility of our methodology.
DPP4 is the MERS-CoV receptor, thus possessing advantages in mediating the infection of a coronavirus. It was shown that DPP4 was not able to mediate the SARS-CoV-2 entry independently in nonpermissive cells such as HeLa and BHK2 (Hoffmann et al., 2020, Letko et al., 2020, Zhou et al., 2020). However, its role in the attachment and/or entry during SARS-CoV-2 infection into permissive cells remains unknown. In the present approach, we have revealed that DPP4 can act as a candidate binding target of the SARS-CoV-2-S RBD. DPP4 is a serine protease that is abundantly distributed in human tissues, playing roles as a multi-functional protein (Aliyari Serej et al., 2017). Apart from the lower respiratory tract, kidney, liver, small intestine, and prostate, DPP4 presents in the placenta, fibroblasts of lung and wounded skin, muscle, as well as central nervous systems (Aliyari Serej et al., 2017, Cheng et al., 2019, Cui et al., 2019, Song et al., 2019). Moreover, DPP4 is widely expressed on activated immune cells including CD4(+) and CD8(+) T cells, B cells, natural killer cells, dendritic cells, and macrophages (Shao et al., 2020, Song et al., 2019). It is capable of modulating the production of cytokines, chemokines, and peptide hormones, thus being involved in various immune or inflammatory diseases (Shao et al., 2020). But for all, we will have to know the interaction between DPP4 and the primary receptor ACE2, and how DPP4 may help the entry of SARS-CoV-2 into host cells.
The serine protease inhibitor, camostat mesylate, has been confirmed to efficiently suppress the infection of SARS-CoV-2 for the purpose of TMPRSS2 inhibition (Hoffmann et al., 2020). As DPP4 is also a serine protease, this would be reasonably suitable for the inhibition of SARS-CoV-2 infection even if DPP4 is a cell-surface target of the viral spike. Inhibitors against DPP4 could be developed for the COVID-19 therapeutics. The conservative property of DPP4 among mammals might allow the SARS-CoV-2 to maintain in animal reservoirs and to confer recombination events. Thus, it would be necessary to consider as wide as animals in the surveillance of SARS-CoV-2.
In conclusion, bioinformatics approaches based on the protein crystal structures have predicted that the MERS-CoV receptor DDP4 might act as a candidate-binding target or coreceptor of SARS-CoV-2. Intriguingly, the crucial binding residues in DPP4 are identical with those for DPP4 as bound to MERS-CoV-S RBD. The mutations at E484 and adjacent residues might be mostly important to contribute to the acquirement of this binding ability. This model provides new insight into the interaction between SARS-CoV-2 and the host. In-depth study around this aspect may provide better understanding on the host tropism and pathogenicity of SARS-CoV-2 and help the surveillance and therapeutics strategy for meeting the challenge of the viral infection.
Limitations of the Study
By employing bioinformatics approaches based on protein crystal structures, our study revealed that SARS-CoV-2 may utilize DPP4, the MERS-CoV receptor, as its binding target. The methodology resolved all the protein complexes between virus spikes and human receptors with fundamentally consistent interfaces to those reported, demonstrating the feasibility of using the methods. However, the limitation of the methodology is that it is not able to predict how two human proteins compete for the binding site in structural space, as both ACE2 and DPP4 can bind at the same RBD of SARS-CoV-2. According to the result of free energy calculation, the affinity of ACE2/SARS-CoV-2-S is stronger than that of DPP4/SARS-CoV-2-S; this may result in the priority of the binding of ACE2 to SARS-CoV-2-S. However, the expression amount of these two human proteins may be different in different cell types or patients and result in an unknown binding mode. The questions of this aspect should be answered by experimental investigations.
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jianhong Lu (jianhlu@csu.edu.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets/code generated or analyzed during this study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFC1200204 and 2017YFC1200205) and the National Natural Science Foundation of China (31670171 and 81974427). We also thank all researchers submitting the viral sequences and protein crystal structures to the public databases.
Author Contributions
Y.L. and Z.Z. performed the protein docking on crystal structural analyses and revised the manuscript. L.Y., X.L., and Y.X. did the prediction of human-virus proteins and function annotation. S.L., S.X., and P.C. analyzed the sequences. J.L. designed the study and wrote and revised the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: June 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101160.
Supplemental Information
References
- Aliyari Serej Z., Ebrahimi Kalan A., Mehdipour A., Nozad Charoudeh H. Regulation and roles of CD26/DPPIV in hematopoiesis and diseases. Biomed. Pharmacother. 2017;91:88–94. doi: 10.1016/j.biopha.2017.04.074. [DOI] [PubMed] [Google Scholar]
- Chan C.M., Chu H., Wang Y., Wong B.H., Zhao X., Zhou J., Yang D., Leung S.P., Chan J.F., Yeung M.L. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of middle east respiratory syndrome coronavirus. J. Virol. 2016;90:9114–9127. doi: 10.1128/JVI.01133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Yuan G., He J., Shao Y., Zhang J., Guo X. Dysregulation of DPP4 is associated with the AMPK/JAK2/STAT3 pathway in adipocytes under insulin resistance status and liraglutide intervention. Diabetes Metab. Syndr. Obes. 2019;12:2635–2644. doi: 10.2147/DMSO.S229838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan C.M., Zhang X., Wang Y., Yuan S., Zhou J., Au-Yeung R.K., Sze K.H., Yang D., Shuai H. Middle east respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018;293:11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Hantak M.P., Li K., McCray P.B., Jr., Perlman S., Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS. Pathog. 2017;13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H., Tong Y., Ni X., Liao Y., Wei W., Cheung W.Y., Li W., Li L. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., Jensen L.J., Snel B., Hooper S.D., Krupp M., Foglierini M., Jouffre N., Huynen M.A., Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.R., Mcgee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- Procko E. 2020. The Sequence of Human ACE2 Is Suboptimal for Binding the S Spike Protein of SARS Coronavirus 2.https://www.biorxiv.org/content/10.1101/2020.03.16.994236v1 [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Meulder D.D., Amerongen G.V., Brand J.V.D., Okba N.M.A., Schipper D. Comparative pathogenesis of COVID-19, MERS and SARS in a non-human primate model. Science. 2020 doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- Schleker S., Garcia-Garcia J., Klein-Seetharaman J., Oliva B. Prediction and comparison of Salmonella-human and Salmonella-Arabidopsis interactomes. Chem. Biodivers. 2012;9:991–1018. doi: 10.1002/cbdv.201100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S., Xu Q., Yu X., Pan R., Chen Y. Dipeptidyl peptidase 4 inhibitors and their potential immune modulatory functions. Pharmacol.Ther. 2020;209:107503. doi: 10.1016/j.pharmthera.2020.107503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang Y., Wang N., Wang D., Guo J., Fu L., Shi X. Identification of residues on human receptor DPP4 critical for MERS-CoV binding and entry. Virology. 2014;471-473:49–53. doi: 10.1016/j.virol.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukmaji A., Sagui C., Board J., Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J. Chem. Phys. 2000;113:10913–10927. [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell. Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, L., Ming, S., Zou, B., Wu, Y., Hong, Z., Li, Z., Zheng, X., Huang, M., Luo, L., Liang, J., et al. (2020). Viral invasion and type I interferon response characterize the immunophenotypes during Covid-19 infection. https://ssrn.com/abstract=3555695.
- Yang X., Yang S., Li Q., Wuchty S., Zhang Z. Prediction of human-virus protein-protein interactions through a sequence embedding-based machine learning method. Comput. Struct. Biotechnol. J. 2020;18:153–161. doi: 10.1016/j.csbj.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Gao S., Nguyen N., Fan M., Jin J., Liu B., Zhao L., Xiong G., Tan M., Li S. Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rv protein-protein interactions. Biol. Direct. 2014;9:5. doi: 10.1186/1745-6150-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets/code generated or analyzed during this study.