Highlights
-
•
CD3+, CD4+ and CD8+ T lymphocytes were decreased in COVID-19-infected patients.
-
•
The populations of T cells are comparable between moderate and severe patients.
-
•
T cells play an important role in regulating the development of COVID-19.
Keywords: COVID-19, Lymphocyte subsets, CD3+ T cells, CD4+ T cells, CD8+ T cells
Abstract
Background
We observe changes of the main lymphocyte subsets (CD16+CD56、CD19、CD3、CD4、and CD8) in COVID-19-infected patients and explore whether the changes are associated with disease severity.
Methods
One-hundred and fifty-four cases of COVID-19-infected patients were selected and divided into 3 groups (moderate group, severe group and critical group). The flow cytometry assay was performed to examine the numbers of lymphocyte subsets.
Results
CD3+, CD4+ and CD8 + T lymphocyte subsets were decreased in COVID-19-infected patients. Compared with the moderate group and the sever group, CD3+, CD4+ and CD8+ T cells in the critical group decreased greatly (P < 0.001, P = 0.005 or P = 0.001).
Conclusions
Reduced CD3+, CD4+, CD8+ T lymphocyte counts may reflect the severity of the COVID-19. Monitoring T cell changes has important implications for the diagnosis and treatment of severe patients who may become critically ill.
1. Introduction
The COVID-19 is highly contagious and its pathologic mechanism has attracted a lot of attention [1], [2], [3]. Most patients eventually made a recovery after careful treatment, but, some had developed into severe and even critical illness [4]. To date, the detailed mechanisms underlying severe respiratory syndrome caused by COVID-19 coronavirus remain unclear [5]. What’s worse, there is no vaccine or effective treatment to rescue severe patients who might be turned into patients with critical illness [3]. Therefore, to clarify the pathological differences between moderate, severe, and critical patients is urgent.
Cytokine storm, which is caused by excessive proinflammatory cytokine responses, is associated with a wide variety of infectious and noninfectious diseases [6], [7]. It is suggested that a cytokine storm plays an important role in the aggravation of the COVID-19 [8]. One important immune cell type that can be activated by virus and initiate a cytokine storm is T cells. After activation by antigen-presenting cells, T cells undergo proliferation and differentiation into specific effector T cells to clear pathogens through producing a series of pro-inflammatory cytokines including IFN-γ, TNF-a, IL-6 [6], [7]. However, whether T cells are involved in the progression of the COVID-19 is unknown.
2. Subjects & methods
2.1. Study population
The study covered 154 cases of novel coronavirus patients from the Renmin Hospital of Wuhan University between 22 January 2020 to 25 February 2020. The patients were divided into 3 groups according to the Diagnosis and Treatment of Novel Coronavirus Patients (the Fifth Pilot Ed.), moderate group (total 49, male 26 and female 23), sever group (total 61, male 34 and female 27), critical group (total 44, male 24 and female 20). The average age of the moderate group, the sever group, and the critical group is 63 ± 13, 63 ± 14, and 65 ± 15 y, respectively. There is no statistical significance between gender and age among these 3 groups (P > 0.05).
All the experiments were performed at the Renmin Hospital of Wuhan University and the research was approved by the Ethics Committee of the Renmin Hospital of Wuhan University. All data were collected anonymously and recorded following the international standards for the protection of privacy and personal information.
2.2. Equipment and reagents
Flow cytometric analysis in our study was performed using a Flow Cytometer BNII (Becton Dickinson). Multitest™ 6-colour TBNK reagents were obtained from BD Biosciences.
2.3. Methods
Fasting blood were collected in heparin on all the patients involved was collected in early morning, all the tests were performed within 2 h. The expression of CD16+CD56、CD19、CD3、CD4、CD8 were analyzed by a Flow Cytometer BNII and the data were analyzed using FACS Diva software (BD Biosciences).
2.4. Statistical analysis
Statistical analysis was performed using SPSS25.0, the non-normal distribution data were analyzed using Kruskal-Wallis test, medium values (P25-P75) were used to present the analysis data. The enumeration data were analyzed using χ2 test or Fisher’s test. P < 0.05 was considered statistically significant.
3. Results
3.1. Baseline characteristics
We collected data on demographic characteristics, comorbidities (diabetes mellitus, hypertension, cardiovascular, and cerebrovascular diseases, chronic liver disease, respiratory system disease and malignant tumor), and the outcome observed. A total of 154 patients hospitalized with COVID-19 were randomly selected, of whom 91 (59.09%) had at least one comorbidity (Table 1 ). It seems that patients with diabetes or cardiovascular and cerebrovascular diseases were more likely to develop into critically ill patients. While in patients with hypertension, chronic liver disease, and malignant tumor, there was no statistical difference among the moderate group, severe group and critical group. Surprisingly, patients with respiratory system disease also showed no effect on the severity of COVID-19. We also found that the patients in critical groups showed definite higher mortality than that in the other 2 groups. And there were no differences in the outcomes between the moderate group and the severe group. Differences in peripheral blood lymphocyte subsets among three groups of patients with COVID-19.
Table 1.
Demographics and baseline characteristics of patients infected with COVID-19.
| All patient (n = 154) | moderate group (n = 49) | severe group (n = 61) | critical group (n = 44) | p value | |
|---|---|---|---|---|---|
| Age (y) | 64 ± 14 | 63 ± 13 | 63 ± 14 | 65 ± 15 | NS |
| Gender (male/Female) | 84/70 | 26/23 | 34/27 | 24/20 | NS |
| Chronic medical illness | |||||
| Any comorbidity | 91 (59.09%) | 28 (57.14%) | 31 (50.82%) | 32 (72.73%) | NS |
| Diabetes | 19 (12.34%) | 5 (10.20%) | 4 (6.56%) | 10 (22.73%) | 0.039 |
| Hypertension | 48 (31.17%) | 13 (26.53%) | 16 (26.23%) | 19 (43.18%) | NS |
| Cardiovascular and cerebrovascular diseases | 30 (19.48%) | 13 (26.53%) | 2 (3.28%) | 15 (34.09%) | <0.001 |
| Chronic liver disease | 5 (3.25%) | 2 (4.08%) | 2 (3.28%) | 1 (2.27%) | NS |
| Respiratory systemdisease | 15 (9.74%) | 2 (4.08%) | 6 (9.84%) | 7 (15.91%) | NS |
| Malignant tumor | 3 (1.95%) | 0 (0.00%) | 2 (3.28%) | 1 (2.27%) | NS |
| Clinical outcome | |||||
| Remained in hospital | 41 (26.62%) | 16 (32.65%) | 13 (21.31%) | 12 (27.27%) | NS |
| Transferred | 10 (6.49%) | 6 (12.24%) | 4 (6.56%) | 0 (0.00%) | NS |
| Discharged | 76 (49.35%) | 24 (48.98%) | 42 (68.85%) | 10 (22.73%) | <0.001 |
| Died | 24 (15.58%) | 2 (4.08%) | 2 (3.28%) | 20 (45.45%) | <0.001 |
Note: Three patients lost case information and data are mean ± SD, n/n, or n (%). p values comparing moderate group, severe group and critical group are from χ2 test or Fisher’s exact test, or Variance analysis, P < 0.05 was defined as statistically significant.
We examined the distribution of peripheral blood lymphocytes in patients infected with COVID-19 and correlations between the COVID-19 and the lymphocyte subsets. As shown in the following Table 2 , the lymphocytes were decreased in most patients (72.73%). Compared to the moderate group (63.27%) and the severe group (73.77%), the lymphocytes numbers decreased significantly in the critical group (81.82%).
Table 2.
The comparison of lymphocyte subsets of patients infected with COVID-19 between different groups.
| All patient (n=154) |
moderate group (n=49) |
severe group (n=61) |
critical group (n=44) |
p value | |
|---|---|---|---|---|---|
| CD16+56 count, per ul |
109 (64.75~170) |
118 (76.5~184) |
99 (54.5~160) |
95 (61.25~181.75) |
NS |
| <84 | 62/154(40.26%) | 14/49(28.57%) | 27/61(44.26%) | 21/44(47.73%) | NS |
| ≥84 | 92/154(59.74%) | 35/49(71.43%) | 34/61(55.74%) | 23/44(52.27%) | - |
| CD19 count, per ul |
119.5 (77.75~178.75) |
114 (62.5~197) |
131 (95.5~194.5) |
106.5 (70~165) |
NS |
| <80 | 39/154(25.32%) | 14/49(28.57%) | 10/61(16.39%) | 15/44(34.09%) | NS |
| ≥80 | 115/154(74.68%) | 35/49(71.43%) | 51/61(83.61%) | 29/44(65.91) | - |
| CD3 count, per ul |
437.5 (308.75~648.25) |
509 (336.5~873.5) |
517 (376~651.5) |
341.5 (226.75~440.5) |
<0.001 |
| <723 | 120/154(77.92%) | 32/49(65.31%) | 50/61(81.97%) | 38/44(86.36%) | 0.031 |
| ≥723 | 34/154(22.08%) | 17/49(34.69%) | 11/61(18.03%) | 6/44(13.64%) | - |
| CD4 count, per ul |
276 (178~402.25) |
290 (172.5~506) |
292 (237.5~399.5) |
192.5 (125.25~335.5) |
0.005 |
| <404 | 117/154(75.97%) | 30/49(61.22%) | 47/61(77.05%) | 40/44(90.91%) | 0.004 |
| ≥404 | 37/154(24.03%) | 19/49(38.78%) | 14/61(22.95%) | 4/44(9.09%) | - |
| CD8 count, per ul |
146.5 (89.75~257.25) |
161 (101~302.5) |
200 (93~261) |
98.5 (54.5~158.75) |
0.001 |
| <220 | 105/154(68.18%) | 31/49(63.27%) | 37/61(60.66%) | 37/44(84.09%) | 0.026 |
| ≥220 | 49/154(31.82%) | 18/49(36.73%) | 24/61(39.34%) | 7/44(15.91%) | - |
| CD4 count /CD8 count |
1.76 (1.18~2.77) |
1.89 (1.25~2.64) |
1.75 (1.09~2.75) |
1.74 (1.19~3.33) |
NS |
| <0.9 | 24/154(15.58%) | 9/49(18.37%) | 10/61(16.39%) | 5/44(11.36%) | NS |
| 0.9-2.0 | 63/154(40.91%) | 18/49(36.73%) | 24/61(39.34%) | 21/44(47.73%) | - |
| ≥2.0 | 67/154(43.51%) | 22/49(44.90%) | 27/61(44.26%) | 18/44(40.91%) | - |
| Lymphocyte count, ×109 per L | 0.835 (0.608~1.13) |
0.96 (0.635~1.35) |
0.91 (0.67~1.12) |
0.75 (0.533~0.888) |
0.012 |
| <1.1 | 112/154(72.73%) | 31(63.27%) | 45(73.77%) | 36(81.82%) | 0.13 |
| 1.1-3.2 | 42/154(27.27%) | 18(36.73%) | 16(26.23%) | 8(18.18%) | - |
Note: Data are median (IQR) or n/N (%), where N is the total number of patients with available data. p values comparing moderate group, severe group and critical group are from χ2, Fisher’s exact test, or Kruskal-Wallis test, P<0.05 was defined as statistically significant.
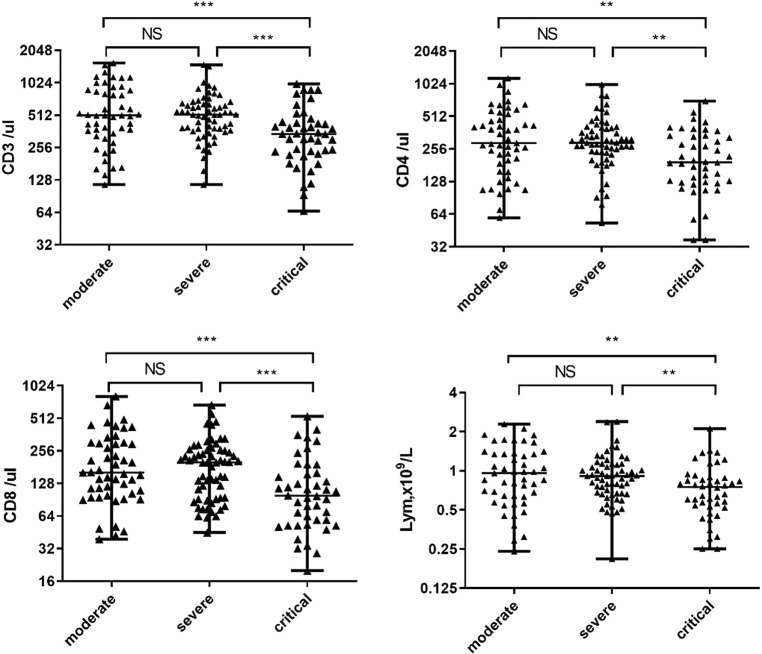
In our study (Fig. 1 ), the absolute value of CD3+ T cells was below the normal range in 120 (77.92%) patients. Compared to the severe group, the median absolute value of CD3+ T cells was significantly lowered (P < 0.001) in the critical group, whereas no significant difference was found between the moderate group and the severe group. The absolute value of CD4+ T cells was below the normal range in 117 (75.97%) patients. Almost all the patients in the critical group showed low CD4+ T cell counts (40/total 44, 90.91%). Moreover, a significantly lower median absolute value of CD4+ T cells was observed in the critical group compared to the severe group (P = 0.005), but not in the moderate group versus the severe group. The absolute value of CD8+ T cells was below the normal rage in 2/3 patients (105/total 154). Similar to CD4+ T cells, compared to the severe group, the median absolute value of CD8+ T cells was significantly lower in the critical group (P = 0.005), while no significant differences were found between the moderate group and the severe group. Meanwhile, the CD4+/CD8+ T cell ratio was below the normal range in 24 patients (15.58%) and above the normal range in 67 patients (43.51%). No statistical significant difference was observed among the three study groups in the two different disorders.
Fig. 1.
Absolute values of CD3+, CD4+, and CD8+ lymphocytes, in patients with moderate, severe or critical COVID-19 infection. The longer horizontal line indicates the median value for each group. *** indicates P < 0.01, ** indicates P < 0.05.
We also observed a reduction concerning the normal range of NK cells (CD16+CD56) in 62 patients (40.26%) and B cells (CD19+) in 39 patients (25.32%), but no statistical differences were found in absolute values of NK cells or B cells among the 3 groups.
4. Discussion
Whether T cells are involved in the progression of the COVID-19 is not clear. In this study, we found there was a strong correlation between the severity of COVID-19 and the CD3+, CD4+ and CD8+ T lymphocytes. we have shown that the population of CD3+, CD4+ and CD8+ Lymphocyte subsets is decreased when patients went from severe to critical whereas the populations of T cells are comparable between moderate and severe patients. However, there was no significant changes in the number of NK cells and B cells between these groups. Based on the data, our study has strengthened the significance of T cells in the clearance of the COVID-19 coronavirus.
Of the 154 patients with COVID-19 recruited, the median age was 63.90 y and 84 were male. In the cohort, we observed that 59% of patients had at least one underlying disorder including diabetes, hypertension, cardiovascular and cerebrovascular diseases, and chronic liver disease, respiratory system disease and malignant tumor. In accordance with previous reports, we observed that patients with diabetes or cardiovascular and cerebrovascular diseases were more likely to develop into critically ill patients [1], [11]. However, there was no statistical difference in disease severity in patients with hypertension, chronic liver disease, malignant tumor, or even respiratory system disease, suggesting that different underlying diseases contribute differently to disease progression of COVID-19.
CD3+ T cells are mainly composed of CD4+ T cells and CD8+ T cells. CD4+ helper T cells have a crucial role in adaptive immune responses [9]. Upon antigen presentation, naïve CD4+ T cells can differentiate into distinct subsets [10]. Among them, Th1 cells, which are induced by IL-12 and produce large quantities of IFN-γ, are involved in enhancing the clearance of certain intracellular pathogens, including viruses [10], [11]. Besides, CD8+ T cells restricted by class I major histocompatibility complex molecules are important in establishing immunity to influenza virus because they recognize internal viral proteins that are conserved between multiple viral strains [12]. Both CD4+T cells and CD8+ T cells are critical in defending against influenza viruses [13], [14], [15]. Since the sequence of the new coronavirus is highly homologous to SARS and MERS virus [16], [17], both of which are severe respiration viruses, probably that CD4+T cells and CD8+ T cells are also responsible for controlling the new coronavirus. Here, we have shown that the reduction of those populations is associated with the enhancement of the severity of patients with the COVID-19. Although further studies are needed to examine the detailed mechanisms of how T cell controlling the COVID-19 coronavirus, our findings broaden our horizons in understanding the pathogenesis of the COVID-19.
Complications or ultimately death caused by COVID-19 are often associated with cytokine storms [18]. It was reported that people with weakened immune systems due to pre-existing diseases, such as cancer, diabetes, liver or kidney disease, are more likely to get infected by the new coronavirus or even more easily turned into severe or critical illness [4], [19]. Since depressed immunity is manifested by decreased immune cell populations including T cells, patients can easily cause the onset of cytokine storm when encountering massive virus invasion. Nowadays, it is common to use glucocorticoid [20] or specific neutralization antibodies to fight against cytokine storms [21]. The former causes a lot of complications while the latter is costly [22]. It has been proposed that immuno-modulatory therapy may prevent cytokine storms [14]. Here, we proposed that enhancing T cell populations or improving their functions through medical features or diet may deter the disease from getting worse. Our results further support the concept that boosting your immunity is effective in dealing with viruses [23], [24].
5. Conclusion
Taken together, we suggested that T cells may play an important role in regulating the development of COVID-19. Enhancing T cell populations may be effective in preventing severe patients from getting worse.
CRediT authorship contribution statement
Rui Liu: Conceptualization, Writing - original draft. Ying Wang: Conceptualization, Writing - original draft. Jie Li: Data curation. Huan Han: Data curation. Zunen Xia: Data curation. Fang Liu: Investigation. Kailang Wu: Investigation. Lan Yang: Investigation. Xinghui Liu: Supervision, Writing - review & editing. Chengliang Zhu: Supervision, Writing - review & editing.
Acknowledgement
This work was supported by The National Natural Science Foundation of China, the National Mega Project on Major Infectious Disease Prevention, the Open Research Program of the State Key Laboratory of Virology of China, the Outstanding Leaders Training Program of Pudong Health Bureau of Shanghai, the Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai and Postdoctoral Sustentation Fund of Shanghai Gongli Hospital, Secondary Military Medical University.
References
- 1.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J. Med.Virol. 2020 doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hageman J.R. The Coronavirus Disease COVID-19. Pediatr. Ann. 2019;49(2020):e99–e100. doi: 10.3928/19382359-20200219-01. [DOI] [PubMed] [Google Scholar]
- 4.C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet 395 (2020) 497-506. doi:10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed]
- 5.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., Guo L., Zhang G., Li H., Xu Y., Chen M., Gao Z., Wang J., Ren L., Li M. Genomic diversity of SARS-CoV-2 in Coronavirus Disease 2019 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39:517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 7.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/mmbr.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H., Liu F. Regulation, Communication, and Functional Roles of Adipose Tissue-Resident CD4+ T Cells in the Control of Metabolic Homeostasis. Front. Immuno. 2018;9 doi: 10.3389/fimmu.2018.01961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J., Paul W.E. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macatonia S.E., Hosken N.A., Litton M., Vieira P., Hsieh C.S., Culpepper J.A., Wysocka M., Trinchieri G., Murphy K.M., O'Garra A. Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J. Immunol. 1995;154:5071–5079. [PubMed] [Google Scholar]
- 12.Jansen J.M., Gerlach T., Elbahesh H., Rimmelzwaan G.F., Saletti G. Influenza virus-specific CD4+ and CD8+ T cell-mediated immunity induced by infection and vaccination. J. Clin. Virol. 2019;119:44–52. doi: 10.1016/j.jcv.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Dunning J., Blankley S., Hoang L.T., Cox M., Graham C.M., James P.L., Bloom C.I., Chaussabel D., Banchereau J., Brett S.J., Moffatt M.F., O'Garra A., Openshaw P.J.M. Progression of whole-blood transcriptional signatures from interferon-induced to neutrophil-associated patterns in severe influenza. Nat. Immunol. 2018;19:625–635. doi: 10.1038/s41590-018-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q., Zhou Y.H., Yang Z.Q. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol. Immunol. 2016;13:3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y., Hsu A.C., Pang Z., Pan H., Zuo X., Wang G., Zheng J., Wang F. Role of the Innate Cytokine Storm Induced by the Influenza A Virus. Viral Immunol. 2019;32:244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 16.R. Lu, X. Zhao, J. Li, P. Niu, B. Yang, H. Wu, W. Wang, H. Song, B. Huang, N. Zhu, Y. Bi, X. Ma, F. Zhan, L. Wang, T. Hu, H. Zhou, Z. Hu, W. Zhou, L. Zhao, J. Chen, Y. Meng, J. Wang, Y. Lin, J. Yuan, Z. Xie, J. Ma, W.J. Liu, D. Wang, W. Xu, E.C. Holmes, G.F. Gao, G. Wu, W. Chen, W. Shi, W. Tan, Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet 395 (2020) 565-574. doi:10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed]
- 17.Park M., Thwaites R.S., Openshaw P.J.M. COVID-19: Lessons from SARS and MERS. Eur. J. Immunol. 2020;50:308–311. doi: 10.1002/eji.202070035. [DOI] [Google Scholar]
- 18.Chen C., Zhang X.R., Ju Z.Y., He W.F. Advances in the research of cytokine storm mechanism induced by Corona Virus Disease 2019 and the corresponding immunotherapies. Zhonghua Shao Shang Za Zhi. 2020;36:E005. doi: 10.3760/cma.j.cn501120-20200224-00088. [DOI] [PubMed] [Google Scholar]
- 19.Applegate W.B., Ouslander J.G. COVID-19 Presents High Risk to Older Persons. J. Am. Geriatr. Soc. 2020 doi: 10.1111/jgs.16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y.H., Qin Y.Y., Lu Y.Q., Sun F., Yang S., Harypursat V., Tang S.Q., Huang Y.Q., He X.Q., Zeng Y.M., Li Y., Xu X.L., Zhao T., Chen Y.K. Effectiveness of glucocorticoid therapy in patients with severe novel coronavirus pneumonia: protocol of a randomized controlled trial. Chin. Med. J. (Engl) 2020 doi: 10.1097/cm9.0000000000000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apel A., Ofran Y., Wolach O., Shimony S., Ram R., Levi I., Zektser M., Koren-Michowitz M. Safety and efficacy of blinatumomab: a real world data. Ann. Hematol. 2020 doi: 10.1007/s00277-019-03854-0. [DOI] [PubMed] [Google Scholar]
- 22.Nebelsiek T., Beiras-Fernandez A., Kilger E., Mohnle P., Weis F. Routine use of corticosteroids to prevent inflammation response in cardiac surgery. Recent. Pat. Cardiovasc. Drug Discov. 2012;7:170–174. doi: 10.2174/157489012803832829. [DOI] [PubMed] [Google Scholar]
- 23.Valdes I., Lazo L., Hermida L., Guillen G., Gil L. Can Complementary Prime-Boost Immunization Strategies Be an Alternative and Promising Vaccine Approach Against Dengue Virus? Front. Immunol. 2019;10:1956. doi: 10.3389/fimmu.2019.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews J.D., Chesson J.M., McCaw J.M., McVernon J. Understanding influenza transmission, immunity and pandemic threats. Influenza Other Respir. 2009;3:143–149. doi: 10.1111/j.1750-2659.2009.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]