Abstract
Genomic analysis of lung adenocarcinomas has revealed that the MGA gene, which encodes a heterodimeric partner of the MYC-interacting protein MAX, is significantly mutated or deleted in lung adenocarcinomas. Most of the mutations are loss-of-function for MGA, suggesting that MGA may act as a tumor suppressor. Here, we characterize both the molecular and cellular role of MGA in lung adenocarcinomas and illustrate its functional relevance in the MYC pathway. Although MGA and MYC interact with the same binding partner, MAX, and recognize the same E-box DNA motif, we show that the molecular function of MGA appears to be antagonistic to that of MYC. Using mass spectrometry-based affinity proteomics, we demonstrate that MGA interacts with a noncanonical PCGF6-PRC1 complex containing MAX and E2F6 that is involved in gene repression, while MYC is not part of this MGA complex, in agreement with previous studies describing the interactomes of E2F6 and PCGF6. Chromatin immunoprecipitation-sequencing (ChIP-seq) and RNA-sequencing (RNA-seq) assays show that MGA binds to and represses genes that are bound and activated by MYC. In addition, we show that, as opposed to the MYC oncoprotein, MGA acts as a negative regulator for cancer cell proliferation. Our study defines a novel MYC/MAX/MGA pathway, in which MYC and MGA play opposite roles in protein interaction, transcriptional regulation and cellular proliferation.
INTRODUCTION
Activation of the MYC pathway is a key driver for diverse cancer types. The MYC gene is frequently amplified in cancer cells, which results in its increased expression (1). Indeed, CCND1, EGFR and MYC are the top three amplified oncogenes based on pan-cancer analysis of 10 different cancer types (2). Many other genomic events, such as chromosomal translocations and super-enhancer amplifications, also activate MYC expression (3-6). The overexpressed MYC protein binds to and activates genes that are promoting cellular proliferation (7,8). In addition, MYC is reported to activate genes that are involved in apoptosis, ribosomal biogenesis, metabolism pathways, genome instability and immune escape, which are involved in tumorigenesis and metastasis (7-9).
The activity of the MYC protein is tightly regulated by a protein interaction network that is centered on the MAX protein (10). MYC interacts with MAX to form a heterodimeric complex, which is essential for MYC to recognize and bind to the E-box DNA sequence (CACGTG) that is enriched in gene promoter regions (11,12). The dimerization of MYC and MAX is mediated through the interaction between their basic region/helix loop helix/leucine zipper (bHLHZ) domains (12). MYC activates gene expression through its association with histone acetyltransferase complexes, the bromodomain protein BRD4 and the transcriptional pause-release complex P-TEFb (13-16). In addition to MYC, MAX also heterodimerizes with another group of proteins containing bHLHZ domains - the MXD family of proteins, including MXD1-4 and MNT. Although the MXD/MAX and MNT/MAX complexes recognize the same E-box DNA sequence as MYC/MAX does, their bindings to DNA result into repression of target genes, which is due to their physical interaction with the repressive complex mSIN3 (17,18).
MGA (MAX gene-associated protein) is a bHLHZ protein that has been reported to interact with MAX and to bind to the E-box DNA sequence (19). MGA, consisting of 3,065 amino acids, is the largest protein in the MAX-interacting protein family (10). In addition to the MAX-interacting bHLHZ domain, MGA contains a T-box domain at the N-terminus, which recognizes the Brachyury binding sequence, suggesting a MAX-independent mechanism for the DNA-binding ability of MGA (19). MGA represses expression of an in vitro MYC-driven luciferase reporter (19), suggesting that MGA may act as a suppressor of MYC target genes. Recent exome-sequencing efforts have revealed that the MGA gene is significantly mutated in lung adenocarcinoma and high risk chronic lymphocytic leukemia (20,21). However, the biological role of MGA in those cancer types, particularly its functional link with the MYC pathway, is largely unknown.
Here, we show that the MGA gene is frequently subject to loss-of-function mutations and copy number deletions in multiple cancer types. We characterize both the molecular and cellular role of MGA in lung adenocarcinoma cells. We integrate results from mass spectrometry-based affinity proteomics, chromatin-immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) approaches to study the gene regulation function of MGA, which reveals that MGA forms a gene repression complex with a noncanonical PCGF6-PRC1 complex and binds to and represses genes that are bound and activated by the MYC oncoprotein. Importantly, we show that, as opposed to MYC, MGA acts as a negative regulator of cancer cell proliferation, supporting its role as a tumor suppressor. Our study helps to elucidate the role of MGA in the MYC oncogenic pathway and supports the definition of a MYC/MAX/MGA pathway that could serve as a potential therapeutic target in the future.
MATERIALS AND METHODS
Genomic analysis of data from The Cancer Genome Atlas (TCGA):
We analyzed sequencing results of TCGA lung adenocarcinoma samples (n=507) and samples from other cancer types on the Pan Cancer c-Bio portal (22,23). We presented analyses of samples with MYC amplification, MGA homozygous deletions or MGA loss-of-function mutations in lung adenocarcinoma, in Figure 1A and S1.
Cell lines:
Cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) project (24) in 2016 and 2017. Cell line identities were verified by SNP genotyping using an Affymetrix SNP array as previously described in the CCLE project (24). Cells were tested negative for mycoplasma using the Lonza MycoAlert Mycoplasma Detection kit at July, 2018. Cells were maintained in RPMI-1640 medium supplemented with 10% heat inactivated FBS and 1% penicillin-streptomycin. Cells lines were passaged every 3 or 4 days and were used for functional assays within 3 months of passages post receipt.
Immunoprecipitation:
We performed immunoprecipitation using antibodies against the endogenous MGA or IgG in HEK293T, A549 and NCI-H23 cells. Around 10 million cells were lysed with NP40 lysis buffer (1% NP40, 150 mM NaCl and 50 mM Tris-HCl pH 8.0) and sonicated by a tip ultrasonic homogenizer. Sonicated cell lysate was first incubated with 2 ug of anti-MGA antibody (Sigma, HPA-042278) or rabbit IgG (Millipore, 12-370) overnight at 4°C and then incubated with mixed Dynabeads A and G (Thermo Fisher) for 2 hours at 4°C. Next, the beads were washed three times with NP40 lysis buffer. Enriched protein was eluted and denatured by NP40 lysis buffer supplemented with LDS sample buffer (Thermo Fisher) and DTT (final concentration: 20 mM).
Quantitative Mass Spectrometry assays:
For immunoprecipitation from HEK293T cells followed by mass spectrometry, we applied the above immunoprecipitation protocol to ~ 100 million cells and used 20 ug of MGA or IgG antibody. After minimal washing to remove non-specifically bound proteins, samples were digested with trypsin, labeled with iTRAQ isobaric mass tag reagents (25), mixed together and analyzed by liquid-chromatography mass spectrometry (affinity proteomics). Briefly, the MGA or IgG pulldown samples were washed twice with 200 μl of 50 mM Tris-HCl (pH 7.5), transferred into fresh 1.5 ml Eppendorf tubes, and washed twice with buffer containing 2 M urea/50 mM Tris (pH 7.5). Samples were incubated in 0.4 μg trypsin in 80 μl of 2 M urea/50 mM Tris supplemented with 1 mM DTT for 1 hour at room temperature while shaking at 1000 g. After short digest, 80 μl of each supernatant was transferred into new tubes. Beads were washed twice with 60 μl of 2M urea/50 mM Tris buffer, and these washes were combined with the supernatant. The combined digestion eluates and washes were spun down at 5000 g for 2 min, transferred into the fresh tubes, and subsequently reduced with 4 mM DTT for 30 min at room temperature. Following reduction, samples were alkylated with 10 mM iodoacetamide for 45 min in the dark at room temperature. Samples were digested overnight with an additional 0.5 ug of trypsin at room temperature, while shaking. Following overnight digestion, samples were acidified (pH <3) with neat formic acid (FA), to a final concentration of 1% FA. Samples were spun down and desalted on C18 StageTips. Briefly, StageTips were conditioned once with 100 μl of MeOH, once with 100 μl of 50% MeCN/0.1% FA, and twice with 0.1% FA. Sample was loaded, washed twice with 0.1% FA, and eluted with 50 ul of 50% MeCN/0.1 % FA. Eluted samples were dried to completion and stored at −80 °C.
Desalted peptides were labeled with iTRAQ reagents as follows. Samples were resuspended in 30 μl of dissolution buffer (0.5 TEAB, pH 8.5) combined with 70 μl of ethanol. Once re-suspended, samples were mixed with iTRAQ reagent, and incubated at room temperature for 1 hour with shaking. Reaction was quenched with 10 ul of 1M Tris-HCl (pH 7.5). Differentially labeled samples were combined, dried down to evaporate ethanol, acidified with neat formic acid to a final concentration of 1%, and desalted on C18 StageTips as described above.
Labeled and desalted peptides were re-suspended in 9 μl of 3% MeCN/0.1% FA and analyzed by online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS) using a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled on-line to a Proxeon Easy-nLC 1000 (Thermo Fisher Scientific). Briefly, 4 μl of each sample was loaded onto a microcapillary column (360 μm outer diameter × 75 μm inner diameter) containing an integrated electrospray emitter tip (10 μm), packed to approximately 22 cm with ReproSil-Pur C18-AQ 1.9 μm beads (Dr. Maisch GmbH) and heated to 50C. Samples were analyzed using a 260 min LC-MS method with the following gradient profile: (min:%B) 0:2; 1:6; 235:30; 244:60; 245:90; 250:90; 251:50; 260:50 (the last two steps at 500 nL/min flow rate). The Q Exactive was operated in the data-dependent mode acquiring HCD MS/MS scans (r = 17,500) after each MS1 scan (r= 70,000) on the top 12 most abundant ions using an MS1 target of 3 × 106 and an MS2 target of 5 × 104. The maximum ion time utilized for MS/MS scans was 120 ms; the HCD-normalized collision energy was set to 28; the dynamic exclusion time was set to 20 s, and the peptide match and isotope exclusion functions were enabled. Charge exclusion was enabled for charge states that were unassigned, 1 and >7.
Immunoblotting:
Cells were lysed by NP40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris-HCl pH 8.0) supplemented with protease and phosphatase inhibitor (Thermo Scientific, 1861281) and denatured by LDS sample buffer (Thermo Fisher) supplemented with 20 mM DTT. Electrophoresis was performed in 3-8% Tris-Acetate gel (for immunoblotting MGA) or 4-12% NuPage Bis-Tris gel in Tris-Acetate SDS or MOPS SDS running buffer respectively. Proteins were transferred to a nitrocellulose membrane (0.2 μm) in Novex Tris-Glycine Transfer Buffer (Thermo Fisher) supplemented with 10% methanol overnight at 4°C at constant voltage of 35V (for transferring MGA) or 2 hours at 70V. Images were taken on a LI-COR instrument. Antibodies that were used for immunoblotting include: MGA (Sigma, HPA-042278), MYC (Cell Signaling, 13987), MAX (Santa Cruz, sc-197), E2F6 (Santa Cruz, sc-53273), HDAC2 (Cell signaling, 5113), HDAC4 (Santa Cruz, sc-46672), CBX3 (Sigma, WH0011335M1) and RNF2 (MBL, D139-3).
MGA expression construct:
For human MGA (NM_001164273.1) cloning, MGA cDNA sequence was subdivided into three fragments using internal restriction sites (1-3168 bp; 3147-6632 bp; 6611-9198 bp). Fragments were amplified by PCR from a cDNA pool using Phusion High-Fidelity DNA Polymerase (Biolabs) following standard protocols. Necessary restriction sites were added to the primer sequences (Table S2). MGA fragments were subcloned in the pGEM-T Easy Vector (Promega, A1360) and then cloned into the pLVX-IRES-ZsGreen1 Vector (Clontech Laboratories) for further analyses.
ChIP-seq analysis:
Chromatin immunoprecipitation-sequencing (ChIP-seq) assays were performed as previously described (26). Antibodies that were used include: MGA (Sigma, HPA-042278), c-MYC (Cell signaling, 13987). The Cell Signaling antibody 13987 (Clone ID: D3N8F) was annotated as an c-MYC specific antibody during our assays but is now noted to also cross react with the N-MYC protein (URL: https://www.cellsignal.com/products/primary-antibodies/c-myc-n-myc-d3n8f-rabbit-mab/13987). Based on the RNA-sequencing results of the A549 cell line from the Cancer Cell Line Encyclopedia (CCLE) project (24) (URL: https://portals.broadinstitute.org/ccle/), c-MYC is the MYC family gene that is predominantly expressed (read counts: 18349) in the lung adenocarcinoma cell line while N-MYC is barely detectable by RNA-seq (read counts: 7, less than 0.1% of c-MYC counts). In addition, siRNA against MYC diminishes the protein signal detected by the antibody (Figure 4A). Thus the ChIP-seq and western-blot signal detected by the cell signaling antibody 13987 is specific to c-MYC in our experiments.
The sequencing libraries were prepared using the NEB ChIP-seq library prep kit (NEB, E6200L) and sequenced on the Illumina MiSeq instrument (50-bp single read sequencing). Sequencing reads were aligned by BWA (27) and the binding sites of MGA and MYC were identified by MACS2 “callpeak” function (28). DNA binding motifs enriched in the MGA binding sites were identified by the Homer de novo motif analysis (29). ChIP-seq tracks (bigWig files) were presented in the Integrative Genomics Viewer (30). The Cistrome Analysis Pipeline (31) was used to generate ChIP-seq heatmaps and to perform ChIP-seq correlation analysis. The averaged ChIP-seq tags at gene promoter regions were calculated by Homer “annotatePeaks” function (29).
RNA sequencing analysis:
RNA was extracted using the Qiagen RNeasy kit with on-column DNase I treatment. 500 ng of RNA for each sample was processed with the NEBNext PolyA mRNA Magnetic Isolation Module (NEB #E7490) and further processed with the NEBNext Ultra Directional RNA Library Prep Kit (NEB E7420S). RNA libraries were then sequenced on an Illumina MiSeq (75-bp paired-end sequencing). Sequencing reads were aligned using the STAR pipeline (32) and quantified by the RSEM pipeline (33). Differential gene expression analysis was performed using the edgeR pipeline (34). GSEA analysis was performed using the Gene Pattern pipeline (35).
RT-qPCR:
Reverse transcription of extracted RNA was carried out using the iScript Reverse Transcription kit (Bio-Rad), and qPCR reactions was performed using the Power SYBR Green PCR MasterMix (Life Technologies) on a CFX384 real-time thermocycler (Bio-Rad), according to manufacturer’s protocols. Primer sequences are available in Table S2.
Competitive cell proliferation assay:
For MGA overexpression experiments, A549 and NCI-H23 cells were transfected either with pLVX-MGA-Zs-Green or empty vector by Lipofectamine LTX Reagent with PLUS Reagent (Thermo Fisher, 15338100), and were then maintained in regular media for 24 hours before cell sorting (J-Ariall SORP) to select transfected cells by GFP signal. GFP-positive cells were then mixed with parental cells and maintained in regular media. The percentage of GFP-positive cells were counted by the flow cytometry analyzer at day 0, 4 and 6.
Public dataset usage:
The following datasets were downloaded from ENCODE (36) and used in the study (MAX ChIP-seq in A549 cells, GEO accession number: GSM935298; E2F6 ChIP-seq in A549 cells, GEO accession number: GSM1010766).
Accession codes:
The newly generated ChIP-seq and RNA sequencing data for this study have been deposited to the Gene Expression Omnibus (GEO) public dataset under the series GSE112188 and GSE112190, respectively.
RESULT
MGA loss-of-function mutations and deletions in lung adenocarcinomas
MGA has been reported to be a significantly mutated gene in lung adenocarcinomas (20 and 37). In the TCGA Pan-Cancer lung adenocarcinoma dataset (38), 27 of the 49 MGA mutations are truncating (Figure 1A). In addition, the MGA gene is subject to copy number deletions (11/507 cases) but not amplifications (Figure S1A), suggesting that MGA may act as a tumor suppressor. These alterations are complementary to MYC genomic activation, where 42 out of 507 (~8%) lung adenocarcinomas acquire amplifications of the MYC gene (Figure S1A, data extracted from the c-Bio portal, portal URL: https://www.cbioportal.org/). As shown in TCGA Pan-Cancer analysis of MYC pathway members including MGA (10), MGA is subject to truncating mutations and copy number deletions in multiple cancer types including uterine corpus endometrial carcinomas (UCEC, 45/509), colorectal carcinomas (CRC, 31/526), and stomach adenocarcinomas (STAD, 19/434), suggesting that MGA may act as a tumor suppressor in multiple lineages (Figure S1B).
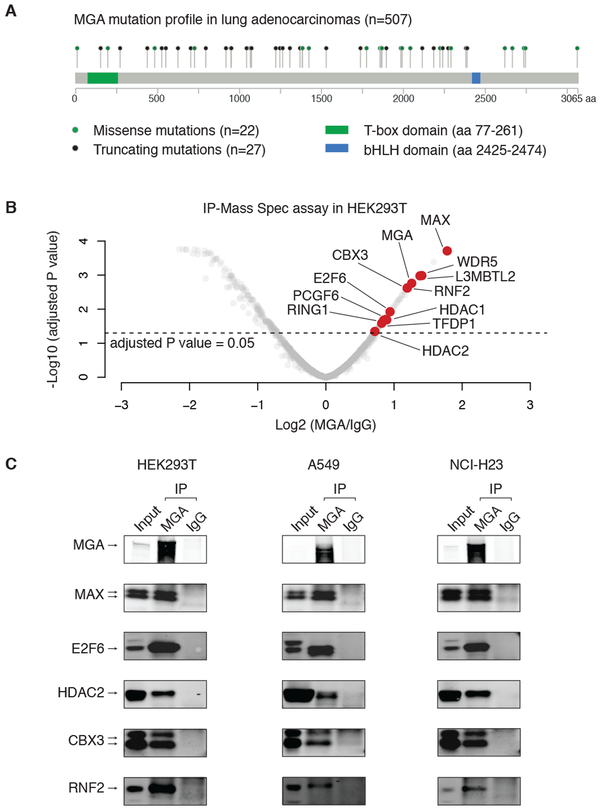
Figure 1: Genomic and proteomic analysis of MGA in lung adenocarcinomas.
A. Mutation profile of MGA in lung adenocarcinomas. Missense (green dots) and truncating mutations (black dots) are highlighted.
B. Immunoprecipitation-mass spectrometry (IP-Mass Spec) results in HEK293T cells (2 biological replicates). We identified proteins that are significantly enriched by the endogenous MGA antibody, relative to IgG control (adjusted P value < 0.05). The proteins that are part of a noncanonical PRC1 (ncPRC1) – the PCGF6-PRC1 complex – are highlighted by red dots.
C. Immunoprecipitation (IP) of MGA and IgG, followed by immunoblotting of MGA-interacting proteins in HEK293T, A549 and NCI-H23 cells.
MGA is part of a noncanonical PRC1 repressive complex
Given the presence of protein-protein interaction domains in MGA, we sought to determine the protein composition of MGA-containing complexes as a clue to MGA function. To do so, we used an anti-MGA antibody bound to beads to immunoprecipitate endogenous MGA from HEK293T cells that are wild-type for the endogenous MGA (validated by Sanger sequencing). As a negative control, the same cell line was treated with non-specific IgG bound to beads. We analyzed the results based on the enrichments of peptide precipitated by anti-MGA antibodies versus IgG controls (2 biological replicates, Figure 1B and Table S1) and identified 84 proteins that are significantly enriched in the MGA immunoprecipitates (adjusted P value < 0.05). MAX, which is known to be the canonical binding partner of MGA (19), and the MGA protein itself are among the most significantly enriched proteins in this immunoprecipitation (Figure 1B).
To understand the MGA-associated protein complex in more depth, we performed David pathway analysis of MGA-interacting proteins (39), which revealed that the MGA complex is enriched in factors involved in transcriptional repression (Figure S2A). Our results were consistent with previous literature in the field. We found that MGA interacts with E2F6 and TFDP1, which form a transcriptional repressing dimer (40), with RING1, RNF2, PCGF6, and L3MBTL2, which are core members of the Polycomb Repressive Complex 1 (PRC1) (41), and with the heterochromatin protein CBX3 (also known as HP1-γ), and the histone deacetylase HDAC1 and HDAC2 (Figure 1B and S2B), all of which are involved in epigenetic and transcriptional repression. The proteins that interact with MGA form a noncanonical PRC1 (ncPRC1) – the PCGF6-PRC1 complex, also known as PRC1-E2F6 (41-44).
To validate the interactome of MGA in the context of lung cancer, we performed immunoprecipitation assays followed by immunoblotting in the lung adenocarcinoma cell lines A549 and NCI-H23, both of which are wild-type for MGA based on RNA sequencing results from the Cancer Cell Line Encyclopedia (CCLE) project (24) (portal URL: https://portals.broadinstitute.org/ccle/). We validated the interaction of endogenous MGA with MAX, E2F6, HDAC2, CBX3 and RNF2 by immunoprecipitating sonicated whole cell extracts with MGA antibodies and IgG controls followed by immunoblotting with protein-specific antibodies (Figure 1C). In contrast, another histone deacetylase factor HDAC4, which was not enriched in the MGA immunoprecipitation-mass spectrometry assay, was not detectable in our immunoprecipitation-western blot validation, consistent with the specificity of our results (Figure S3A). Furthermore, we immunoprecipitated E2F6 and RNF2, separately, using antibodies against the endogenous proteins in NCI-H23 cells and validated their interaction with MGA (Figure S3B). Therefore, our results together with previous studies that identified MGA in the interactomes of E2F6 and PCGF6 (43,45), defined a MGA-ncPRC1 transcriptional repression complex.
MGA co-localizes with MYC and E2F6 in the genome
To understand the gene regulation function of MGA and its involvement in the MYC pathway, we next performed chromatin-immunoprecipitation sequencing (ChIP-seq) assays using antibodies against endogenous MGA and MYC in the lung adenocarcinoma cell line A549. Both MYC and MGA binding sites, defined by the MACS2 ChIP-Seq pipeline using a q-value threshold of < 0.05 (28), are significantly enriched in gene promoter regions (34.5% for MGA, P value < 10−15, Figure 2A), as previously reported for MYC (46). Homer de novo motif analysis (29) of the 9,049 MGA binding sites identified by MACS analysis shows that MGA recognizes the same E-box DNA sequence (centered at CACGTG) as the MYC protein (Figure 2B and S4), which was previously suggested by gel-shifting assays (19). Interestingly, the T-box domain DNA motif was not listed as a significant hit in the analysis (Figure S4), suggesting that the T-box domain is not a major driver for MGA recruitment in these experiments.
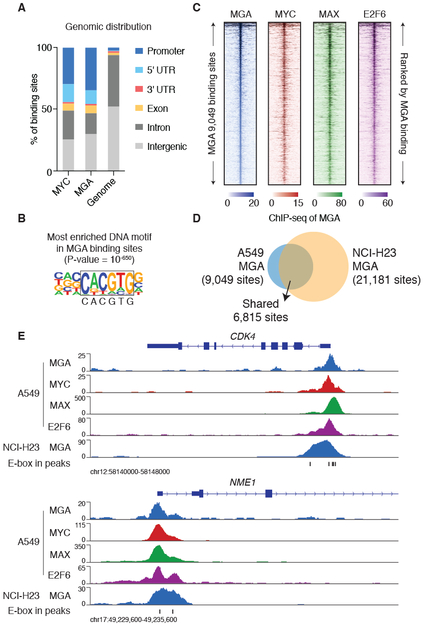
Figure 2: ChIP-seq analyses reveal that MGA co-localizes with MYC and E2F6 in the genome.
A. Genomic distribution of MYC and MGA binding sites in A549 cells.
B. Homer de novo motif analysis identified the top DNA motif enriched in MGA binding sites identified in A549 cells.
C. ChIP-seq signal of MGA, MYC, MAX and E2F6 (A549 cells) at the 9,049 MGA binding sites (top to bottom ranked by MGA ChIP-seq signal ±2.5kb centered at each MGA binding site).
D. Comparison of MGA ChIP-seq binding sites identified in A549 and NCI-H23 cells.
E. ChIP-seq signal of MGA in A549 and NCI-H23 cells, MYC, MAX and E2F6 in A549 cells at the CDK4 and NME1 loci. The E-box DNA motif (CACGTG) positions are indicated.
When we analyzed the ChIP-seq results for MAX and E2F6 from A549 cells derived from the ENCODE project (36), we found that MGA binding sites are enriched for binding of MYC, MAX, and E2F6 (Figure 2C). Statistical analysis shows that the binding intensity of MGA positively correlated with that of MYC (Pearson correlation: r = 0.66), MAX (Pearson correlation: r = 0.49) and E2F6 (Pearson correlation: r = 0.69) across MGA binding sites (Figure S5A). The majority (~75%) of MGA binding sites are also identified by ChIP-seq of MGA in a separate lung adenocarcinoma cell line NCI-H23 that harbors MYC gene amplification (24) (Figure 2D-E). The MGA, MYC, MAX and E2F6 proteins co-localize to the promoter regions of canonical MYC target genes such as CDK4 and NME1 (46,47) (Figure 2E). ChIP-qPCR assays in NCI-H23 cells using primers targeting promoter regions of the AURKA and CDK4 genes validate the colocalization of MYC and MGA (Figure S5B). Taken together, our results showed that MGA forms part of a noncanonical PRC1 complex with MAX and E2F6, and that this MGA-MAX-E2F6 complex binds to the same loci in the genome as the MYC oncoprotein, suggesting that MGA and MYC regulate a similar set of genes but as part of different complexes.
MGA represses MYC target genes
To interrogate the impact of MGA on gene expression, we cloned MGA cDNA into an overexpression vector linked to ZsGreen (an improved GFP variant) via an Internal Ribosome Entry Site (IRES) sequence, which allows us to use GFP signal to monitor MGA protein expression level (more details are described in the Methods section). Two days post transfection, we collected A549 cells carrying the empty-ZsGreen (Control) or MGA-ZsGreen overexpression vector based on positive GFP signal, and immediately performed RNA sequencing (RNA-seq) and MGA ChIP-seq assays. We compared the impact of MGA on gene expression to MGA binding. Based on RNA-seq analysis, we categorized genes into three groups (adjusted P value cutoff = 0.05): 625 MGA-repressed genes, 658 MGA-activated genes, and 17,229 genes that are expressed but unaffected by MGA. We then assessed the MGA binding intensity at gene promoters within ±2kb from transcription start sites (TSS), which showed an overall increase of MGA binding after MGA overexpression (Figure 3A). Genes that are repressed by MGA gained an increased level of MGA binding to their promoters after MGA overexpression, compared to genes that are unaffected by MGA, supporting the notion that MGA binds to and represses these genes (Figure 3A, examples shown in Figure 3B and S6). Interestingly, we also observed a higher level of MGA binding gained at promoters of MGA-activated genes, compared to unaffected genes, suggesting that MGA may play divergent transcriptional regulatory roles for different sets of genes (Figure 3A).
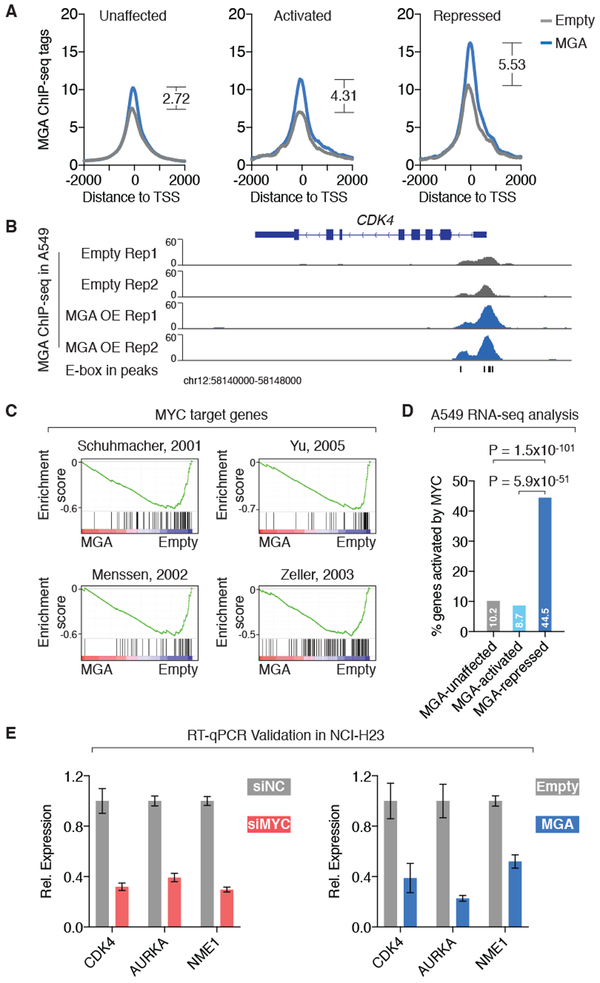
Figure 3: ChIP-seq/RNA sequencing analyses show that MGA binds to and represses MYC target genes.
A. Averaged ChIP-seq signal of MGA in A549 cells with and without MGA overexpression at the transcription start sites (TSS) of genes that are activated, repressed and unaffected by MGA (based on RNA sequencing results in A549 cells with and without MGA overexpression).
B. ChIP-seq signal of MGA in A549 cells with and without MGA overexpression at the CDK4 locus.
C. Gene set enrichment analysis of RNA sequencing results in A549 cells with and without MGA overexpression showed that MGA-repressed genes are enriched in MYC target genes identified by previous studies.
D. Percentage of MGA-regulated genes that are activated by MYC (based on RNA sequencing results from A549 cells with and without siRNA-mediated MYC silencing). P values are derived from fisher exact tests.
E. RT-qPCR assays in NCI-H23 cells validated that either silencing MYC or overexpressing MGA decreased the expression level of the MYC target genes CDK4, AURKA and NME1. Expression level is normalized to controls (siNC or Empty). Error bars: s.d.
Gene set enrichment analysis (GSEA) (35) revealed that MYC target genes, which were identified from previous studies (47-50), are downregulated on the expression level in A549 cells overexpressing MGA, compared to the empty control, suggesting that MGA acts as a repressor for MYC target genes (Figure 3C). In order to validate our finding, we performed RNA sequencing in A549 cells with and without siRNA-mediated MYC silencing. GSEA analysis showed that MGA-repressed and MYC-activated genes are involved in the same pathways such as “G2/M checkpoint”, “MYC targets” and “Glycolysis” (Figure S7A). Indeed, 44.5% of MGA-repressed genes are activated by MYC (Figure 3D). In contrast, 8.7% of MGA-activated or 10.2% MGA-unaffected genes can be activated by MYC (Figure 3D). Most of the MGA-repressed/MYC-activated genes are bound by MGA and E2F6 together, suggesting the role of the PRC1-E2F6 complex in repressing those genes (Figure S7B). Taken together, our results showed that MGA acts as a repressor for the MYC pathway by binding to and repressing genes that are bound and activated by the MYC oncoprotein. We also validated our gene expression results by RT-qPCR assays in a separate lung adenocarcinoma cell line NCI-H23. Indeed, either MYC silencing or MGA overexpression decreased the expression level of canonical MYC target genes including CDK4, AUKRA and NME1 in NCI-H23 cells (Figure 3E).
MGA is a negative regulator of cancer cell proliferation
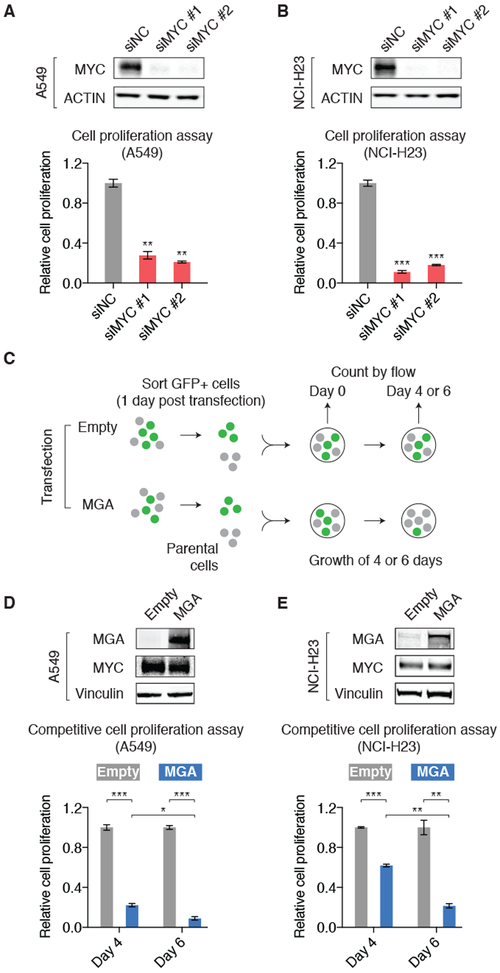
Given the antagonistic functions of MYC and MGA in regulating their target genes, we next aimed to assess their respective roles in cancer cell proliferation. Silencing of MYC by siRNA results in a significant reduction in proliferation of A549 and NCI-H23 cells (Figure 4A-B), consistent with previous reports in other cell lines that MYC acts as a positive regulator for cell proliferation and can be disrupted using siRNA (51). To assess the role of MGA in cell proliferation, we utilized the ZsGreen system and performed competitive cell proliferation assays for A549 and NCI-H23 cells with and without ectopic MGA overexpression. Briefly, cells transfected with the empty-ZsGreen or MGA-ZsGreen vector were collected and counted through GFP-based sorting. The GFP-positive cells were then mixed with the same number of parental A549 or NCI-H23 cells (GFP-negative). Subsequently, the proportion of GFP-positive cells was measured by a flow cytometer over time (including the starting time when the cell populations were mixed) (Figure 4C). We found that, compared to the empty vector control, overexpression of MGA significantly reduced the proliferation rates of A549 and NCI-H23 cells, as measured four days and six days after seeding the cells (Figure 4D-E), suggesting that MGA acts as a repressor for cancer cell proliferation. We also overexpressed the wild-type MGA in the lung adenocarcinoma cell line LXF289 that harbors an MGA truncating mutation (MGA p.Q1067*, based on CCLE database (24) and validated by sanger sequencing) and observed a significant reduction in its cell proliferation as well as expression of MYC-target genes (Figure S8). MGA-overexpressing NCI-H23 cells did not undergo increased apoptosis, as measured by annexin V/propidium iodide staining. A higher proportion of MGA-overexpressing cells was observed in the G1 phase of the cell cycle, as measured by propidium iodide staining in the fixed cells (Figure S9), indicative of cell cycle arrest or delay induced by MGA overexpression. Immunoblots show that overexpression of MGA has little effect on expression level of the MYC protein (Figure 4D-E, top panels), showing that the observed phenotype change is not dependent on modifying MYC expression levels. Our results show that MGA represses cell proliferation in contrast to MYC that is well known to promote cell proliferation.
Figure 4: MGA acts as a repressor for cancer cell proliferation.
A. siRNA-mediated silencing of MYC (verified by immunoblots) decreased the proliferation of A549 cells. Cell number was counted five days post transfection and normalized to the negative control siRNA. P values are derived from t tests. Error bars: s.d., **P<0.01.
B. Same experiment as A, but in NCI-H23 cells, Error bars: s.d., ***P<0.001.
C. Schematic chart explaining the design of competitive cell proliferation assays. A549 and NCI-H23 cells were first transfected with empty-ZsGreen control or MGA-ZsGreen vector. The transfected cells were collected based on GFP signal and mixed with parental cells at 1:1 ratio. The percentage of GFP-positive cells was then counted over time.
D. MGA overexpression (verified by immunoblots) decreased the proliferation of A549 cells. The percentage of GFP-positive cells was counted four and six days post seeding the cells and normalized to the empty-ZsGreen control. P values are derived from t tests. Error bars: s.d., *P<0.05; ***P<0.001.
E. Same experiment as D, but in NCI-H23 cells, Error bars: s.d., **P<0.01; ***P<0.001.
DISCUSSION
In human development and pathogenesis, MYC functions by binding to and regulating its target genes that are involved in varied biological processes. Activation of the MYC pathway is a key driver for diverse cancer types. MYC can be activated by diverse genomic alterations, which commonly result into upregulation of the target genes and promotes tumorigenesis. For example, germ-line genetic variants (52,53), gene amplification (1), chromosomal translocations (3), super-enhancer duplications (4,5), and activation of the upstream Wnt signaling pathway (54) can result into transcriptional activation of the MYC gene. In addition, activation of the RAS pathway also stabilizes MYC on the protein level (55). Furthermore, missense mutations at the amino acid Thr58 block the binding of the E3 ubiquitin ligase FBXW7 to MYC, which enhances the protein stability of MYC and increases its protein abundance in cancer cells bearing this mutation (56).
Here, we provide more analysis of another mechanism that regulates the MYC pathway without altering the transcription or protein level of MYC. Previous in vitro gel-shifting and luciferase reporter assays showed that MGA, another MAX-interacting protein, recognizes the E-box motif and represses reporter expression driven by MYC (19). We integrate ChIP-seq and RNA sequencing assays and show that MGA directly binds to and represses genes that are activated by the MYC oncoprotein on the genome wide scale. In addition, we show that overexpression of MGA inhibits proliferation of lung adenocarcinoma cell lines, which is complementary to previous finding that MGA represses cellular transformation, as measured in rat embryo fibroblasts (19). In tumor samples, the MGA gene is subject to truncating mutations and copy number deletions in multiple cancer types. Our experimental results, complemented by cancer genomics data, suggest that MGA acts as a tumor suppressor by inhibiting the MYC pathway in lung adenocarcinoma. In addition to cell proliferation and transformation, activation of the MYC oncogene has been associated with metastasis in diverse cancer types including lung cancer. For instance, genetically engineered MYC-driven mouse small cell lung cancers often metastasize to livers (57). Given the antagonistic functions of MYC and MGA in gene regulation, it is possible that MGA also plays a role in metastasis, which needs future investigations.
The antagonistic roles of MYC and MGA in gene regulation could be due to their interacting protein complexes. MYC interacts with different types of transcription activators including TRRAP to increase the histone acetylation at gene promoters and P-TEFB to release the paused RNA polymerase II for the target genes (13,15). Previous studies immunoprecipitating E2F6 and PCGF6 identified MGA as an interacting protein (43,45). Our immunoprecipitation of the endogenous MGA followed by mass spectrometry results provided a complete interactome of MGA and showed that, in addition to E2F6 and PCGF6, MGA physically interacts with most members of the noncanonical PRC1 – the PCGF6-PRC1 complex, including multiple epigenetic and transcriptional repressors such as RING1, RNF2, CBX3, HDAC1 and HDAC2, highlighting the importance of the PCGF6-PRC1 complex in the MGA pathway. The PCGF6-PRC1 complex can repress target genes by H2A mono-ubiquitination, H3K9 dimethylation, H3K27 trimethylation and histone de-acetylation (41,44), which may explain the gene repression role of MGA. It has been recently reported that the association with RNF2 and the subsequent H2A mono-ubiquitination are required for the silencing effect of MGA in pluripotent stem cells (58). The PCGF6-PRC1 complex has been described to play important roles in maintenance of embryonic stem cells and suppression of dendritic cells (58,59). Further analysis is needed to assess whether the formation of the MGA-PCGF6-PRC1 complex is required for the repression effect of MYC target genes. Our data suggest that MGA may serve as a sequence-specific DNA-binding factor for recruiting the PCGF6-PRC1 complex to specific targets of the genome, which is in agreement with recent findings that MGA knockdown impairs the binding of PCGF6 to target genes in HEK293 cells and mouse embryonic stem cells (58,60). It remains unanswered which domain or domains of MGA is or are required for the recruitment of the complex and the repression of its target genes. In addition, given our observation that MGA and MYC share a large set of binding sites and target genes, one interesting question that remains unanswered is whether the MGA complex plays an antagonistic role to the MYC complex in the release of RNA polymerase II from the promoter regions of their target genes (15).
Both MYC and MGA function by interacting with the MAX protein. Our study proposes a working model in which the expression level of MYC-target genes is tightly regulated by a balance between the MYC/MAX and MGA/MAX complexes including their associated epigenetic and transcriptional regulators. MYC acts as an accelerator and MGA acts as a brake for driving the expression of their target genes. Either overexpression of MYC or silencing of MGA breaks this balance, leading to aberrant overexpression of the target genes, activation of oncogenic pathways and tumorigenesis. Our results suggest that MGA binds to and represses MYC-target genes. Additionally, the MGA protein may compete with MYC for the interaction with MAX and limit the abundance of the MYC/MAX complex in cells, which may serve as an additional mechanism in suppressing MYC-target genes. Interestingly, the MAX gene is also subject to homozygous inactivating mutations and deletions in small cell lung cancer and gastrointestinal stromal tumors, which results into a complete loss of the MAX protein in those cancers (61,62). One important remaining question is how loss of MAX alters the balance between MYC and MGA and affects their target genes.
The expansion of alterations in the MYC pathway offers the hope of expanding therapeutic options for lung cancer and beyond. Lung cancer is the leading cause of cancer-related mortality worldwide, while around 40% of cases are lung adenocarcinomas (63). Genomic activation of the receptor tyrosine kinases (RTK) and their downstream signal transduction pathways is a key driver for lung adenocarcinomas. Inhibitors of RTK such as EGFR and ALK have also greatly benefited the treatment of patients bearing these alterations (64,65). However, effective and durable therapy of lung adenocarcinoma will likely require combination treatment. The discovery of MGA mutations, along with the previous finding of MYC amplification in lung adenocarcinomas, now defines a new pathway in complementation to the RTK/RAS/RAF pathway, a MYC/MAX/MGA pathway. Roughly 15% of lung adenocarcinoma cases have been detected to harbor either MYC amplification or MGA loss-of-function alterations. Genomic alterations of the MYC/MAX/MGA pathway contribute to epigenetic and transcriptional dysregulation in the genome, which may provide opportunities for novel therapeutic interventions that are targeting epigenetic modification and transcription.
Supplementary Material
Implications:
This study expands the range of key cancer-associated genes whose dysregulation is functionally equivalent to MYC activation and places MYC within a linear pathway analogous to cell cycle or receptor tyrosine kinase/RAS/RAF pathways in lung adenocarcinomas.
ACKNOWLEDGEMENT
We acknowledge research support from the Lung Cancer Research Foundation fund (X.Z.), the 2015 AACR-John and Elizabeth Leonard Family Foundation Basic Cancer Research Fellowship, (X.Z.), National Cancer Institute grants R35CA197568 (M.M.), the Norman R. Seaman Endowment fund (M.M.), the Spain MINECO grants SAF-2014-54571-R (M.S-C) and BES-2015-072204 (P.L.), the Fundación Científica Asociación Española Contra el Cancer grant GCB14-2170 (M.S-C), National Cancer Institute Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 (to S.A.C.) and NIH/NCI U01 CA214125 (to S.A.C), and the European Association for Cancer Research Travel fellowship (P.L.). M. Meyerson is an American Cancer Society Research Professor.
Footnotes
Conflict of interest statement: A.D. Cherniack and M. Meyerson receive commercial research support from Bayer. M. Meyerson has ownership interest in Origimed and also serves as a consultant/advisory board member for Origimed.
REFERENCES
- 1.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battey J, Moulding C, Taub R, Murphy W, Stewart T, Potter H, et al. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983;34:779–87. [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe J-S, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20:1130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90. [DOI] [PubMed] [Google Scholar]
- 9.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018;6:282–300.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–6. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–7. [DOI] [PubMed] [Google Scholar]
- 13.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem. 2002;277:40156–62. [DOI] [PubMed] [Google Scholar]
- 14.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. [DOI] [PubMed] [Google Scholar]
- 15.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–76. [DOI] [PubMed] [Google Scholar]
- 18.Hurlin PJ, Quéva C, Koskinen PJ, Steingrímsson E, Ayer DE, Copeland NG, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Paoli L, Cerri M, Monti S, Rasi S, Spina V, Bruscaggin A, et al. MGA, a suppressor of MYC, is recurrently inactivated in high risk chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1087–90. [DOI] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed Protein Quantitation in Saccharomyces cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Mol Cell Proteomics. 2004;3:1154–69. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Cowper-Sal lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 2012;22:1437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolayeva O, Robinson MD. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol Clifton NJ. 2014;1150:45–79. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 40.Morkel M, Wenkel J, Bannister AJ, Kouzarides T, Hagemeier C. An E2F-like repressor of transcription. Nature. 1997;390:567–8. [DOI] [PubMed] [Google Scholar]
- 41.Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell. 2011;42:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, et al. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 2005;280:1199–208. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa H, Ishiguro K-I, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–6. [DOI] [PubMed] [Google Scholar]
- 44.Qin J, Whyte WA, Anderssen E, Apostolou E, Chen H-H, Akbarian S, et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeller KI, Zhao X, Lee CWH, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99:6274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuhmacher M, Kohlhuber F, Hölzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cappellen D, Schlange T, Bauer M, Maurer F, Hynes NE. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007;8:70–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a Large Chromatin Loop Linked to a Cancer Risk-Associated Single-Nucleotide Polymorphism in Colorectal Cancer Cells. Mol Cell Biol. 2010;30:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. [DOI] [PubMed] [Google Scholar]
- 55.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell. 2017;31:270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Endoh M, Endo TA, Shinga J, Hayashi K, Farcas A, Ma K-W, et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boukhaled GM, Cordeiro B, Deblois G, Dimitrov V, Bailey SD, Holowka T, et al. The Transcriptional Repressor Polycomb Group Factor 6, PCGF6, Negatively Regulates Dendritic Cell Activation and Promotes Quiescence. Cell Rep. 2016;16:1829–37. [DOI] [PubMed] [Google Scholar]
- 60.Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018;14:e1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romero OA, Torres-Diz M, Pros E, Savola S, Gomez A, Moran S, et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 2014;4:292–303. [DOI] [PubMed] [Google Scholar]
- 62.Schaefer I-M, Wang Y, Liang C-W, Bahri N, Quattrone A, Doyle L, et al. MAX inactivation is an early event in GIST development that regulates p16 and cell proliferation. Nat Commun. 2017;8:14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. [DOI] [PubMed] [Google Scholar]
- 64.Kwak EL, Bang Y-J, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mok TS, Wu Y-L, Thongprasert S, Yang C-H, Chu D-T, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.