Summary
Clostridioides difficile is a Gram-positive, spore-forming, toxin-producing anaerobe pathogen, and can induce nosocomial antibiotic-associated intestinal disease. While production of toxin A (TcdA) and toxin B (TcdB) contribute to the main pathogenesis of C. difficile, adhesion and colonization of C. difficile in the host gut are prerequisites for disease onset. Previous cell wall proteins (CWPs) were identified that were implicated in C. difficile adhesion and colonization. In this study, we predicted and characterized Cwp22 (CDR20291_2601) from C. difficile R20291 to be involved in bacterial adhesion based on the Vaxign reverse vaccinology tool. The ClosTron-generated cwp22 mutant showed decreased TcdA and TcdB production during early growth, and increased cell permeability and autolysis. Importantly, the cwp22 mutation impaired cellular adherence in vitro and decreased cytotoxicity and fitnessovertheparentstraininamouseinfectionmodel. Further more,lactate dehydrogenase cytotoxicity assay, live-deadcells taining and transmission electron microscopy confirmed the decreased cell viability of the cwp22 mutant. Thus, Cwp22 is involved in cell wall integrity and cellviability,which could affect most phenotypes of R20291.Our data suggest that Cwp22 is an attractive target for C. difficile infection therapeutics andprophylactics.
Introduction
Clostridioides difficile (formerly Clostridium difficile) (Lawson et al., 2016; Oren and Garrity, 2018) is a Gram-positive, spore-forming, toxin-producing, anaerobic bacterium that has established itself as a leading cause of nosocomial antibiotic-associated diarrhoea in developed countries (Sebaihia et al., 2006). Symptoms of C. difficile infection (CDI) range from mild diarrhoea, intestinal inflammation to pseudomembranous colitis (Lessa et al., 2012). Recently, morbidity and mortality rates of CDI have been increasing steadily, causing over 500,000 infections per year in the United States alone with an estimated cost of $1–3 billion (Dubberke and Olsen, 2012; Lessa et al., 2015).
C. difficile has a number of virulence factors. Among them are two large potent exotoxins, toxin A (TcdA) and toxin B (TcdB) that are recognized as the major virulence factors of C. difficile (Lyras et al., 2009; Kuehne et al., 2010). These toxins can disrupt the actin cytoskeleton of intestinal epithelial cells through glucosylation of the Rho family of GTPases to induce mucosal inflammation and the symptoms associated with CDI (Peniche et al., 2013). Other important virulence traits that impact host infection and colonization include: (i) cell wall proteins (CWPs) (Biazzo et al., 2013); (ii) tissue degradative proteases (Seddon et al., 1990; Seddon and Borriello, 1992; Borriello, 1995); (iii) flagella and fimbriae (Edwards et al., 2000; Stevenson et al., 2015; Lorenz et al., 2019).
To date, 29 CWP-coding genes have been identified in C. difficile. The family of CWPs play an important role in composing the outer layer of the bacterial cell, and are likely to be involved in C. difficile colonization and pathogenesis (Biazzo et al., 2013; Bradshaw et al., 2018). In the family of CWPs, 12 of 29 CWP-coding genes are clustered in the same region, named after the S-layer protein (SlpA) coding gene slpA (Cwp1), whereas the remaining 17 coding genes are distributed throughout the genome (Bradshaw et al., 2018). Among them, SlpA is an abundant protein that was first studied. The S-layer protein contains two mature SLPs derived from a common precursor. The high-molecular-weight SLP (about 47 kDa) acts as a binding subunit, and the low-molecular-weight SLP (about 36 kDa) modulates colonization and pathogenesis (Takeoka et al., 1991; Calabi et al., 2001). The role of the C. difficile S-layer in colonization, immunity, virulence and viability has been well-studied (Calabi et al., 2002; Pechine et al., 2005; Ausiello et al., 2006; Pechine et al., 2007; Kirk et al., 2017). Several other CWPs such as Cwp84, Cwp2, Cwp66, and CwpV have been investigated, and play important roles in host cell adhesion and immune system evasion during CDI (Waligora et al., 2001; Emerson et al., 2009; Kirby et al., 2009; Bradshaw et al., 2017; Wydau-Dematteis et al., 2018). Kirby et al. (Kirby et al., 2009) identified a surface-associated protease, Cwp84, which cleaves SlpA precursor into mature form. Bradshaw et al. (Bradshaw et al., 2017) determined the structure of Cwp2 and demonstrated that cwp2 mutation resulted in increased TcdA release and impaired cellular adherence in vitro. Cwp66, which works as an adhesin, and CwpV, which confers phase-variable phage infection resistance, were also characterized (Waligora et al., 2001; Emerson et al., 2009; Sekulovic et al., 2015). Recently, a new surface protein Cwp19, which is a novel lytic transglycosylase involved in stationary-phase autolysis and can affect toxin release in C. difficile, was also identified (Wydau-Dematteis et al., 2018). Biazzo et al. (Biazzo et al., 2013) analysed 14 of the other 17 CWP genes scattered throughout the C. difficile genome, and identified the conserved DNA sequences and protein expression of Cwp13, CwpV, Cwp16, Cwp18, Cwp19, Cwp20, Cwp22, Cwp24 and Cwp25, suggesting that these CWPs may possess important functions in C. difficile. However, the roles of some CWPs, such as Cwp22, Cwp24, and Cwp25, in vivo are still not very clear.
The biochemical function of Cwp22 (CD630_27130) as an L, D-transpeptidase has been demonstrated in C. difficile 630 (Peltier et al., 2011; Sutterlin et al., 2018). Here, we identified the homologous protein (CDR20291_2601) from the epidemic strain C. difficile R20291 (referred hereafter as R20291) using a reverse vaccinology method and studied the role of this protein in vivo by phenotypically characterizing a strain with an insertional inactivation of the cwp22 gene (CDR20291_2601, referred hereafter as 2601 gene). The cwp22 mutation results in decreased toxin production in the bacteria’s early growth, delayed sporulation and decreased motility. Moreover, the cwp22 mutation impaired cellular adherence in vitro, and decreased cytotoxicity and fitness compared with the parent strain in a mouse infection model. Furthermore, cell viability assay through lactate dehydrogenase (LDH) cytotoxicity detection, live-dead cell staining and transmission electron microscopy (TEM) analysis revealed that the cell autolysis and the cell wall permeability of the mutant were increased. Taken together, Cwp22 is an important CWP involved in cell wall integrity and permeability, affecting most phenotypes of R20291, which could also be a new potential target for C. difficile vaccine development.
Results
Bioinformatic identification and analysis of putative cell wall protein Cwp22
Vaxign is a web-based reverse vaccinology tool that uses comparative genomic sequence analysis to predict vaccine candidates based on different criteria such as cellular localization and adhesion probability (He et al., 2010). Using R20291 as the seed strain, Vaxign analysis predicted 31 C. difficile proteins to be cell membrane bound, likely to be adhesins, and conserved in other 12 genomes. Among these proteins is YP_003219080.1 (Cwp22), a putative CWP of 653 amino acids with a predicted molecular weight of 71.97 kDa and a pI of 8.89. Cwp22 is encoded by the CDR20291_ 2601 (2601) gene in strain R20291 (NCBI Entrez Gene ID of 8,468,749).
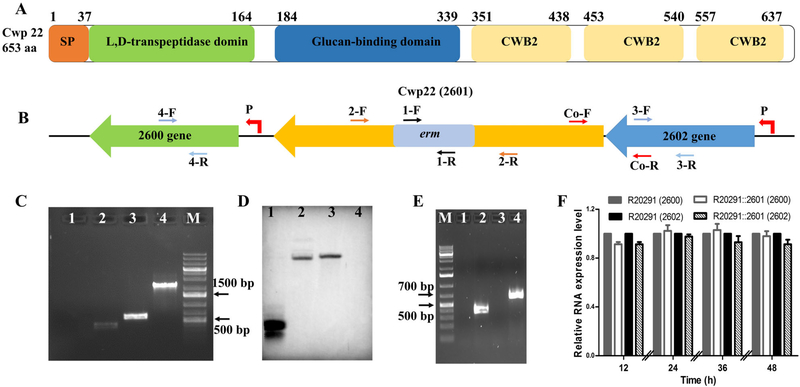
Based on conserved domain analysis, Cwp22 has a putative 37-amino acid signal sequence and three domains (Fig. 1A). The putative N-terminal catalytic domain belongs to the Erfk (YkuD) superfamily (COG1376) with L,D-transpeptidase activity, the C-terminal domain contains three tandem repeats of the cell wall binding motif CWB2 (pfam04122), and the Glucan-binding domain belongs to the COG5263 superfamily, which plays role of carbohydrate transport and metabolism. The similar domains of Cwp22 (CD630_27130) in C. difficile 630 were identified and characterized in vitro in previous work (Peltier et al., 2011; Sutterlin et al., 2018).
Fig. 1.
Construction and identification of cwp22 mutant.
A. Schematic representation of Cwp22 structure. Cwp22 contains a 37 amino acid signal peptide, and three main domains that are N-terminal catalytic domain, C-terminal three repeats cell wall binding motif CWB2 (pfam04122) and 8 type 1 cell wall binding (CWB1- glucan binding domain) repeats.
B. Up and down stream genes of 2601 (cwp22). P indicates promoter.
C. Identification of cwp22 mutation. M: 1 kb DNA ladder; 1: ermB gene verification with R20291 genome using primer 1-F/R; 2: ermB gene verify-cation with R20291::2601 genome using primer 1-F/R; 3: correct insertion verification with R20291 genome using primer 2-F/R; 4: correct insertion verification with R20291::2601 genome using primer 2-F/R.
D. Verification of single insertion mutation by Southern blot analysis. 1: ermB gene used as a positive control; 2: R20291::2601 genome digested with EcoRI and XbaI; 3: R20291::2601 genome digested with EcoRI and Hind III; 4: R20291 genome digested with EcoRI and Hind III used as a negative control.
E. Verification of 2601 truncation and co-transcription of 2601 and 2602 genes. 1: Test of genomic contamination in total R20291 RNA using 16 s primers; 2: Test of 2601 and 2602 co-transcription with R20291 cDNA as template using primer Co-F/R; 3: Test of 2601 gene transcription with R20291::2601 cDNA as template using primer 2-F/R; 4: Test of 2601 gene transcription with R20291 cDNA as template using primer 2-F/R.
F. Test of polar effect of 2601 gene inactivation on up and down stream genes. Primers 2-F/R, 3-F/R and 4-F/R were used to detect the transcription of 2601, 2602 and 2600 genes respectively. Experiments were independently repeated thrice. Bars stand for mean ± SEM. One-way ANOVA was used for statistical significance.
Construction of cwp22 mutant and complementation strain
To analyse the role of Cwp22, the ClosTron system was used to inactivate the 2601 gene. Insertion of the Group II intron into the 2601 gene was verified by intron and 2601gene-specific primers 1-F/R and 2-F/R (Fig. 1B and C). The single chromosomal insertion of the intron was further confirmed by Southern blot analysis (Fig. 1D). Meanwhile, reverse transcription PCR (RT-PCR) and real-time quantity PCR (RT-qPCR) were conducted using primers 2-F/R, 3-F/R, 4-F/R and Co-F/R to confirm truncation of cwp22 and no polar effect of cwp22 mutation on up-/downstream genes (Fig. 1E and F).
Effects of cwp22 mutation on growth profile and toxin expression
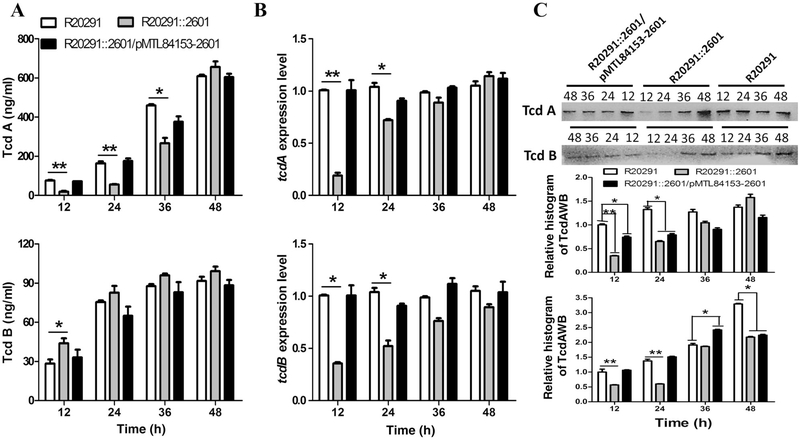
The effect of the cwp22 mutation on R20291 growth was first analysed in BHIS medium. Supporting Information Figure S1A showed that R20291::2601 reached higher cell density (OD600) at the stationary phase. However, when the mutant entered into the late stationary phase, the turbidity of the mutant cultures (16–36 h) decreased faster than R20291 indicating that the mutant autolyzed faster than the parent (Supporting Information Fig. S1B and C).
To assay the effect of the cwp22 mutation on toxin production, the toxin concentration of culture supernatants collected at 12, 24, 36 and 48 h of postinoculation was determined by ELISA. Our preliminary data showed that there is no significant difference in toxin expression of R20291 VS R20291/pMTL84153 and R20291::2601 VS R20291::2601/pMTL84153 (data not shown). Therefore, we just compared R20291, R20291::2601 and R20291::2601/ pMTL84153–2601. Figure 2A showed that the TcdA concentration of R20291::2601 supernatants was significantly lower than that of R20291 before 36 h (at 12 h: 3.1-fold less, 24 h: 2.9-fold less, 36 h: 0.7-fold less), and reached a similar level at 48 h compared with R20291. Interestingly, the TcdB concentration of R20291::2601 was 1.5-fold of that of R20291 at 12 h with significant difference, and reached a similar level after 12 h.
Fig. 2.
Effect of cwp22 mutation on toxin expression.
A. Determination of toxin concentration in C. difficile supernatants.
B. Determination of toxin expression on transcription level.
C. Determination of intracellular toxin concentration by western blot and histogram analysis. Experiments were independently repeated thrice.
Bars stand for mean ± SEM (*P < 0.05, **P < 0.01). One-way ANOVA was used for statistical significance.
To analyse the expression of the tcdA and tcdB genes, RT-qPCR was performed. As shown in Fig. 2B, tcdA (tcdB) transcription in R20291 was about 5.2 (2.8)-fold (*P < 0.05) at 12 h, 1.5 (1.9)-fold (*P < 0.05) at 24 h, 1.1 (1.3)-fold at 36 h and 0.87 (1.1)-fold at 48 h of those in R20291::2601. The transcription level of the tcdA gene was consistent with the ELISA results of the supernatants. Surprisingly, the transcription of the tcdB gene was not consistent with the ELISA results. To further check the production of TcdA and TcdB, the intracellular TcdA and TcdB were measured by Western blot analysis (Fig. 2C). We tried to use anti-Escherichia coli RNA polymerase β antibody (no available antibody for Gram-positive bacteria) to detect the RNA polymerase β subunit of C. difficile, which could be used as a control protein for Western blot analysis, but the antibody did not work in C. difficile. Therefore, we detected the total LDH concentration (supernatants and intracellular fluids) of C. difficile by ELISA, and used it as the calibration protein for Western blot histogram analysis. Figure 2C showed that the intracellular TcdA/TcdB production of the mutant was lower than the parent, which was consistent with the transcriptional analysis results. These results indicated that the cell wall permeability and composition of the cwp22 mutant might be altered, which could affect toxin release.
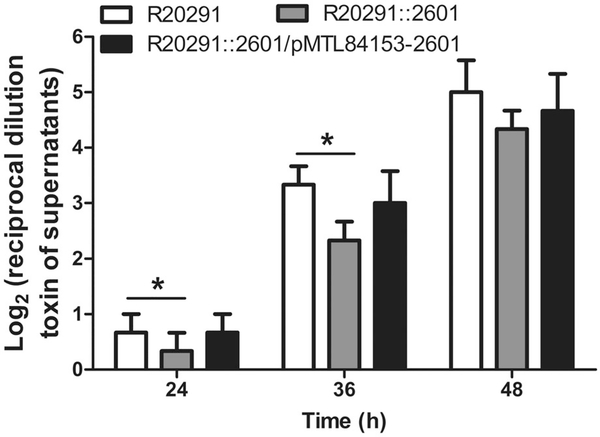
Total toxin production was further evaluated by measuring cytotoxic titre of C. difficile culture supernatants. To assay toxin titre, CT26 cells were exposed to 24, 36 and 48 h of postincubated C. difficile supernatants respectively. Figure 3 showed that the relative cytotoxic titre of R20291 supernatants was 2.0-fold that of R20291::2601 supernatants at 24 h (*P < 0.05), 1.4 fold at 36 h (*P < 0.05) and reached a similar cytotoxicity level at 48 h. Results indicated that the cytotoxicity of the cwp22 mutant decreased compared with the wild type before 36 h, and reached the same level at 48 h.
Fig. 3.
Effect of cwp22 mutation on cytotoxicity of C. difficile supernatants on CT26 cells. The CT26 cells were exposed to twofold serial dilutions of C. difficile culture supernatants. The cytotoxic titre was defined as the reciprocal of the highest dilution that caused 50% CT26 cells rounding. Purified Tcd B and BHIS media were used as positive and negative controls respectively. Experiments were independently repeated thrice. Bars stand for mean ± SEM (*P < 0.05).One-way ANOVA was used for statistical significance.
Effects of cwp 22 mutation on cell wall integrity
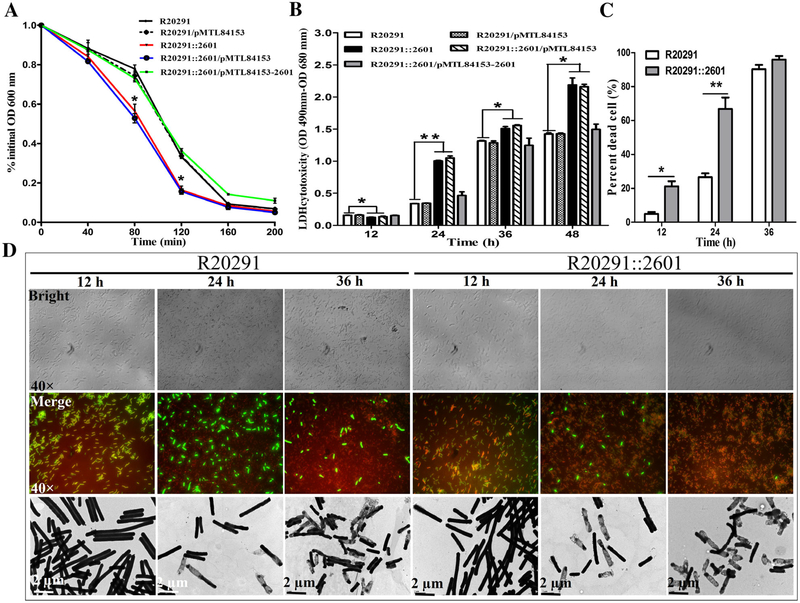
In order to determine if the cwp22 mutation affects C. difficile autolysis, a Triton X-100 autolysis assay was conducted. As shown in Fig. 4A, R20291::2601 lysed significantly faster than R20291 at 80 and 120 min of incubation (*P < 0.05), suggesting that mutation of cwp22 decreased bacteria resistance to Triton X-100, and increased autolysis of R20291.
Fig. 4.
Effect of cwp22 mutation on cell wall integrity.
A. Triton X-100 autolysis assay.
B. LDH cytotoxicity assay.
C. Percent of dead cells.
D. Detection of cell viability. Top panel: images from the bright field; middle panel: images from the merged green/red staining with CFDA and PI respectively; bottom panel: images of TEM (80 kv). Experiments were independently repeated thrice. Bars stand for mean ± SEM (*P < 0.05, **P < 0.01). Student’s unpaired t-test was used for two groups comparison. One-way ANOVA was used for comparison of more than two groups..
To check whether the cell wall of R20291::2601 was altered, we detected the LDH cytotoxicity of C. difficile strains. Figure 4B showed the permeability of R20291::2601 was higher than R20291 with a significant difference after 12 h of incubation (*P < 0.05), suggesting that the cell wall permeability of the cwp22 mutant might be changed.
To further analyse the effect of the cwp22 mutation on the cell, we analysed C. difficile cell viability through live-dead cell staining and TEM. Four areas of cells (>400 cells) on slide were counted with microscope soft-ware (dead bacteria were dyed as red colour with propidium iodide (PI), and live bacteria were dyed as green colour with CDFA), and the percent of ghost cells accounted in total cells was calculated (Fig. 4C). As shown in Fig. 4C and D, significantly more empty cells of R20291::2601 were found before 36 h incubation (*P < 0.05), indicating more cell wall permeability and cell autolysis of the mutant than R20291.
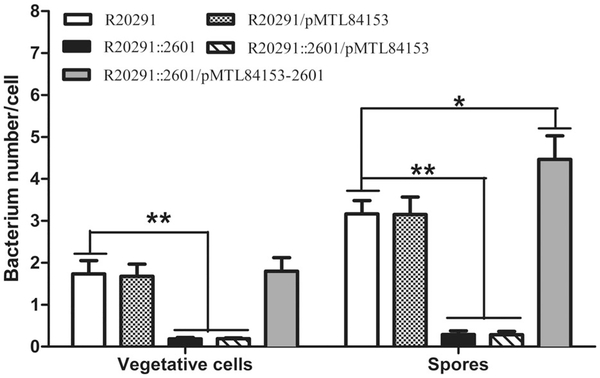
cwp22 mutation reduces bacterial adhesion in vitro
The ability of vegetative cells and spores to adhere to HCT-8 cells in vitro was assayed. Figure 5 showed that for vegetative cells, the mean adhesion of R20291 was 1.73 ± 0.30 bacteria/cell, while R20291::2601 was 0.19 ± 0.01, which decreased by 89% (**P < 0.01), the complementation strain was 1.8 ± 0.31; for spores, the mean adhesion of R20291 was 3.17 ± 0.30, while R20291::2601 was 0.28 ± 0.02, which decreased by 91% (**P < 0.01), the complemented strain was 4.47 ± 0.94 (*P < 0.05). These data suggested that the adherence of the cwp22 mutant was significantly lower than the parent in both vegetative cells and spores.
Fig. 5.
Effect of cwp22 mutation on adhesion of C. difficile vegetative cells and spores.
Adhesion ability of vegetative cells and spores of C. difficile was determined on HCT-8 cells. Experiments were independently repeated thrice. Bars stand for mean ± SEM (*fiPcance.< 0.05, **P < 0.01). One-way ANOVA was used for statistical signiance.
Effects of cwp22 mutation on biofilm formation, motility and spore resistance to heat/ethanol
To further characterize the effect of cwp22 mutation on C. difficile physiology, the ability of C. difficile strains to form biofilms, motility and spore resistance to heat/ethanol was analysed. Supporting Information Figure S2 showed the biofilm of R20291::2601 decreased 45% (**P < 0.01) at 24 h and 15% (*P < 0.05) at 72 h compared to R20291. To assay the motility, swarming and swimming abilities of C.difficiles trains were determined at 48 hand 24 hincubation respectively. Supporting Information Figure S3 showed the diameter of the swarming zone of R20291::2601 (15.15 ± 1.2 mm), decreased by 23% (*p < 0.05) compared with R20291 (19.70 ± 1.6 mm). Meanwhile, the diameter of the swimming zone of the mutant (13.94 ± 2.1 mm,) decreased by 38% (*p < 0.05) compared with R20291 (22.33 ± 2.9 mm). The sucrose gradient-purified C. difficile spores (Racine and Vary, 1980) were used to detect the resistance to heat (65C) and ethanol (100%). Supporting Information Figure S4 showed that there was no significant difference insporeresistancebetweenR20291::2601andR20291.
Effects of cwp22 mutation on sporulation and germination
Sporulation and germination of C. difficile strains were analysed. R20291::2601 (5.23% ± 2.21% at 24 h, *P < 0.05; 36.67% ± 4.37% at 48 h, *P < 0.05) showed significantly delayed sporulation compared with R20291 (21.33% 2.41% at 24 h; 87.23% 3.12%) at 24 h (Supporting Information Fig. S5A), while at 72 h, the sporulation ratio of the mutant was almost the same as the parent. As shown in Supporting Information Fig.S5B and 5C, there is no significant difference in germination ratio between R20291::2601 and R20291.
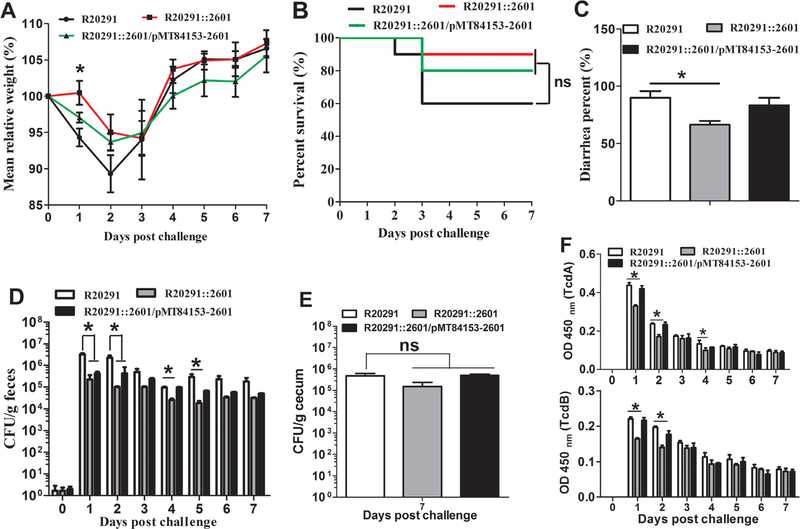
Evaluation of cwp22 mutation on C. difficile virulence and adhesion in mouse model of C. difficile infection
To evaluate whether cwp22 mutation affects bacterial virulence and adhesion in vivo, a mouse model of CDI was performed. Thirty mice (n = 10 per group) were challenged with R20291, R20291::2601 or R20291::2601/pMT84153–2601 spores (1 × 106 spores/mouse) via gavage after antibiotic treatment. The R20291::2601 infection group lost less weight compared with the R20291 infection group, and there was a significant difference at day postchallenge 1 (Fig. 6A). Figure 6B showed that 40% of mice succumbed to severe disease within 3 days in the R20291 infection group compared 10% mortality in the group infected with R20291::2601 (no significant difference with log-rank analysis). Meanwhile, 90% of mice developed diarrhoea in the R20291 infection group versus 70% in the mutant infection group (Fig. 6C). As shown in Fig. 6, the CFU of the R20291::2601 infection group decreased both in faecal samples (Fig. 6D) and cecum (Fig. 6E) compared with the R20291 infection group, and there was a significant difference in faecal samples at days postchallenge 1, 2,4 and 5, while no significant difference in adherence to the cecum was detected. To analyse the persistence ratio of the complementation plasmid in R20291::2601, the number of R20291::2601/pMT84153–2601 in faecal samples were counted (Supporting Information Table S2). The results showed that the 2601 gene was helpful to maintain pMTL84153–2601 in R20291::2601 and there was no significant difference in plasmid persistence at days postchallenge 1 compared with the original charged spores (100%). While the complementation plasmid pMTL84153-2601 in R20291::2601 was not very stable after days postchallenge 2 without antibiotic selection, the complementation still can be reached to a certain extent.
Fig. 6.
Evaluation of cwp22 mutation on C. difficile virulence in mice.
A. Mean relative weight changes.
B. Survival curves.
C. Diarrhoea percentages.
D. C. difficile in faeces.
E. C. difficile in cecum at postchallenge day 7.
F. Toxin titre of faecal samples. Bars stand for mean ± SEM (*P < 0.05). One-way ANOVA was used for statistical significance. Animal survivals were analysed by Kaplan–Meier survival analysis with a log-rank test of significance.
To analyse toxin level in the gut, the titre of TcdA and TcdB in faeces was measured (Fig. 6F). A comparison between the cwp22 mutant and the wild-type strain revealed significant toxin decrease in mutant faeces at days postchallenge 1 (TcdA: 27% less, *P < 0.05; TcdB: 30% less, *P < 0.05), 2 (TcdA: 29% less, *P < 0.05; TcdB: 31% less, *P < 0.05) and 4 (TcdA: 18% less, *P < 0.05). The above results indicated that the cwp22 mutation impaired the colonization and pathogenesis ability of R20291.
Discussion
In this study, we reported the identification and characterization of a putative surface protein Cwp22 from C. difficile R20291. Our data showed that Cwp22 was involved in several cellular processes of C. difficile such as toxin production, sporulation, bacteria motility and cell viability. Notably, the cwp22 inactivation increased cell permeability and autolysis, impaired cellular adherence in vitro, decreased cytotoxicity with significant differences and decreased virulence over the wild strain in the mouse infection model. Our results indicated that Cwp22 could be a new potential target for CDI therapeutics and prophylactics.
The major virulence factors of C. difficile are two exotoxins, TcdA and TcdB (Voth and Ballard, 2005). The toxin encoding genes tcdA and tcdB are located in a 19.6 kb pathogenicity locus (PaLoc), which also contains three additional genes, tcdC, tcdR and tcdE (Braun et al., 1996; Mani and Dupuy, 2001). TcdC is an antagonist of TcdR that negatively regulates TcdR-containing RNA polymerase holoenzyme (Dupuy et al., 2008). While previous studies showed that TcdC might have a moderate role in regulating toxin expression, it is not a major determinant of the hypervirulence of C. difficile (Murray et al., 2009; Bakker et al., 2012; Martin-Verstraete et al., 2016). tcdR has been shown to encode an RNA polymerase sigma factor that positively regulates both toxin genes and its own gene (Moncrief et al., 1997; Mani et al., 2002). In this study, our data suggested that both TcdA and TcdB expression were decreased before 24 h of postinoculation; these results prompted us to examine the tcdR transcription. The transcription analysis of tcdR showed that the transcription of tcdR in R20291::2601 decreased 2.3-folds (*P < 0.05) at 12 h, 1.2-folds (*P < 0.05) at 24 h and 0.5-fold at 36 h (*P < 0.05) compared with the wild strain, and reached a similar expression level after 48 h of postinoculation (Supporting Information Fig. S6A). TcdR acts a positive regulator of toxin expression, and is regulated by many other network regulators, such as CcpA, CodY and σD (El Meouche et al., 2013; Martin-Verstraete et al., 2016). El Meouche et al. (El Meouche et al., 2013) demonstrated that SigD could positively regulate toxin expression via direct control of tcdR. In our study, the sigD expression of the cwp22 mutant decreased 6.2-folds (**P < 0.01) at 12 h and 0.8-fold (*P < 0.05) at 24 h compared with the wild-type strain, and reached a similar expression level after 36 h of postinoculation (Supporting Information Fig. S6B). In El Meouche’s study, they also confirmed that SigD is implicated in the positive regulation of C. difficile motility as reported previously (Aubry et al., 2012). We also demonstrated that both the swarming (decreased by 23%, *P < 0.05) and swimming (decreased by 38%, *P < 0.05) abilities of the cwp22 mutant decreased compared with the wild strain (Supporting Information Fig. S3). As shown in Supporting Information Fig. S7, the motility-related gene fliC of mutant was expressed twofold less (*P < 0.05) than the wild-type strain at 24 h through RT-qPCR analysis. Toxin release is the key factor in pathogenesis, while the mechanism of toxin transportation is still unclear. TcdE, the holin-like protein coded by tcdE, has been identified as being involved in toxin release (Govind and Dupuy, 2012; Olling et al., 2012; Govind et al., 2015). However, the results of the TcdE studies from different groups were controversial. On the one hand, the function of TcdE in toxin release in C. difficile JIR8094 and R20291 cultured in tryptoneyeast (TY) broth was confirmed (Govind and Dupuy, 2012; Govind et al., 2015). On the other hand, Olling et al. (Olling et al., 2012) identified that the release of toxin from C. dificile 630Δerm is not affected by the inactivation of the tcdE gene. Recently, it was identified that Cwp19 acts as a novel lytic tansglycosylase, which is involved in toxin release through stationary-phase autolysis in C. difficile 630Δerm (Wydau-Dematteis et al., 2018). In that study, Wydau-Dematteis et al. (Wydau-Dematteis et al., 2018) proved that TcdE and bacteriolysis were coexisting mechanisms for toxin release, with their relative contributions in vitro depending on growth conditions (works in BHIS not in TY). In this study, we also detected the increase of autolysis and cell permeability of the cwp22 mutant compared with the parent strain. Taken together, we propose that the decreased expression of the sigD gene contributed to the low expression of toxin and motility of R20291::2601 though the cell permeability was increased. As the regulation of toxin production is a complex response of C. difficile to particular nutrient availability, the transcriptome analysis of R20291::2601 would provide us new knowledge on the regulation map of the cwp22 mutation on toxin production as well as some other pleiotropic phenotype changes, such as sporulation and germination.
Cwp22, a putative CWP, is composed of a β-sandwich and a conserved active site consisting of a (Y/L)XXHG(S/T) motif followed by SXGC(I/V)R(M/L) (Bradshaw et al., 2018). It contains an ErfK (YkuD) domain followed by 8 type 1 cell wall binding (CWB1- Clucon binding domain) repeats and 3 type 2 cell wall binding domains (CWB2), which mediate the adherence of Cwp22 to the cell wall through interaction with the anionic polymer PSII (Willing et al., 2015; Bradshaw et al., 2018). While L,D-transpeptidase enzyme activity of Cwp22 in R20291 was predicated through bioinformatics analysis with NCBI’s Conserved Domain Database (CDD), the Cwp22 protein from R20291 was successfully expressed and purified (data not shown), and the enzyme activity analysis is under way in our group. The ErfK domain is found in L,D-transpeptidase (Cwp22), which are involved in peptidoglycan crosslinking like D,D-tanspeptidase. Previous studies showed that the peptidoglycan of C. difficile contains an unusually high (73%) content of 3 ! 3 cross-links generated by L,D-transpeptidases compared with all other firmicutes, and the L,D-transpeptidases could confer resistance to β-lactam antibiotics (Biarrotte-Sorin et al., 2006; Peltier et al., 2011). Interestingly, we did not detect a significant MIC difference for β-lactam antibiotics between the cwp22 mutant and R20291 (data not shown), which indicates that some other enzyme could also play the same role of L,D-transpeptidase in R20291. Recently, Peltier et al. (Peltier et al., 2011) and Sutterlin et al. (Sutterlin et al., 2018) identified and purified three L,D-transpeptidases, CD630_29630 (LdtCd1), CD630_27130 (LdtCd2) and CD630_30070 (LdtCd3), in C. difficile 630. Among them, LdtCd2 and LdtCd3 can catalyse the formation of 3 ! 3 cross-links (L,D-transpeptidase activity), while LdtCd1 displays only L,D-carboxypeptidase activity (Sutterlin et al., 2018). They demonstrated that the inactivation of the ldtCd1, ldtCd2 and ldtCd1 plus ldtCd2 could result in a 22%, 15% and 28% decrease in the proportion of muropeptide dimers containing 3 ! 3 cross-links respectively. Meanwhile, they also found that the proportion of 4 ! 3 cross-links was reduced by 4%, 4% and 3% compared with the wild type strain with unknown reasons in the ldtCd1, ldtCd2 and ldtCd1 plus ldtCd2 mutation strains respectively. These observations showed that each of the ldtCd1, ldtCd2 or ldtCd1 could result in significant decrease in the abundance of 3 ! 3 cross-links and in overall peptidoglycan reticulation, which could reconstruct the bacterial cell wall constitution (Peltier et al., 2011; Sutterlin et al., 2018). Based on the protein sequences of L,D-transpeptidases found in C. difficile 630, we also searched three homologous proteins that were predicated as L,D-transpeptidases in R20291: are CDR20291_2797 (99.36% identity to LdtCd1), CDR20291_2601 (Cwp22, 99.22% identity to LdtCd2) and CDR20291_2843 (98.59% identity to LdtCd3). Though Cwp22, as LdtCd2, has been identified in previous studies that could change cell wall composition, the pleiotropic roles of Cwp22 in C. difficile were not demonstrated until now. In this present study, our results showed that the cwp22 mutation could result in increased cell autolysis, decreased cell viability and adherence to HCT-8 cells in vitro and the mouse gut in vivo and decreased pathogenesis in mice, which confirmed that the CWP Cwp22 (LdtCd2) mutation could indirectly reconstruct the cell wall. We also tried to extract the CWP with low PH method (Calabi et al., 2001) to check the constitution change of the R20291::2601 cell wall compared with R20291, but we did not get a clear significant difference result by running a normal SDS-PAGE gel (data not shown). The cell wall proteomics analysis combined with LC–MS analysis could be used to highlight the subtle changes of the R20291::2601 cell wall in future studies.
Over the past decade, CDI has become a serious problem in the developed world, and results in an estimated 29,000 deaths and an estimated cost of $1–3 billion in the United States alone (Dubberke and Olsen, 2012; Lessa et al., 2015; Zhu et al., 2018). However, many aspects of CDI remain unclear, in particular, the mechanisms of C. difficile colonization in the gut. Although CWPs involved in bacteria colonization have been identified, more CWPs need to be studied for their important roles in CDI and potential application in developing new therapeutics and prophylactics in C. difficile. Like other bacteria, C. difficile also possess multiple adhesins, and several CWPs have been previously characterized, including SlpA, Cwp2, Cwp6, CwpV, Cwp66, Cwp84 and Cwp19 (Waligora et al., 2001; Wright et al., 2005; Kirby et al., 2009; Dang et al., 2010; Reynolds et al., 2011; Bradshaw et al., 2017; Ferreira et al., 2017; Wydau-Dematteis et al., 2018). Although the exact role of each CWP in pathogenesis remains to be further elucidated, antibodies to many CWPs have been found in serum samples from CDI patients, and investigational vaccines targeting Cwp84 (Pechine et al., 2011; Sandolo et al., 2011) have been developed, indicating that certain CWPs are surface exposed in vivo and could be developed into vaccines (Pechine et al., 2005; Wright et al., 2008; Biazzo et al., 2013). CWPs and adhesins are favourable protective antigens for vaccine development against infection with Gram-positive bacterial pathogens (He et al., 2014; Ong et al., 2017). A recent study of manually annotated protective vaccine antigens from over 10 Gram-positive bacteria found 56.8% of these protective antigens are adhesins or adhesin-like proteins. In addition, 19.8% of the protective antigens in Gram-positive bacteria were found to be located in the cell wall, and 87.5% of these protective cell wall antigens are also adhesins (Ong et al., 2017). Cwp22, like the other CWPs Cwp2, Cwp6, CwpV, Cwp22, Cwp19 and Cwp84, is also an abundant and conserved protein in C. difficile through gel free analysis of the extracts (Ferreira et al., 2017), suggesting that it is required for some cellular processes. Meanwhile, Cwp22 (CDR20291_2601 gene) ranks top of candidates including cwp84 and slpA, which was predicated by Vaxign tool for exploring novel potential surface proteins with potential adhesion activity. Therefore, as a CWP, Cwp22 is a very promising protective vaccine antigen. An immune response against Cwp22 would block its pleiotropic functions and lead to possible effective protection against CDI.
In conclusion, we characterized the surface protein Cwp22 in R20291, and detected the pleiotropic functions of Cwp22. Cwp22 is an attractive target for vaccine development, and the evaluation of the capacity of Cwp22 to induce a protective immune response is under way in our group.
Experimental procedures
Comparative genomic analysis of C. difficile genomes
Using the Vaxign reverse vaccinology tool (He et al., 2010), we systematically analysed all proteins in the genome of C. difficile R20291 in terms of cellular localization, adhesin probability, transmembrane helices, sequence conversation with the genomes of other 12 C. difficile strains, sequence similarity to human and mouse proteins and protein length. These other 12 strains are strains 630, BI1, ATCC 43255, CD196, CIP 107932, QCD-23 m63, QCD-32 g58, QCD37×79, QCD-63q42, QCD-66c26, QCD-76w55 and QCD97b34. Protein-conserved domain analysis was performed using the NCBI’s CDD (Marchler-Bauer et al., 2017).
Bacterial strains, plasmids and culture conditions
Table 1 lists the strains and plasmids used in this study. C. difficile strains were cultured in BHIS (brain heart infusion broth supplemented with 0.5% yeast extract and 0.1% cysteine) at 37C in an anaerobic chamber (90% N2, 5% H2, 5% CO2). For production of spores, C. difficile strains were cultured in Clospore medium as described (Perez et al., 2011). E. coli DH5α and E. coli HB101 were grown aerobically at 37C in LB media (1% tryptone, 0.5% yeast extract, 1% NaCl). Antibiotic selection was used when needed: for E. coli (per millilitre), 15 μg of chloramphenicol; for C. difficile (per millilitre), 15 μg thiamphenicol, 50 μg kanamycin, 250 μg D-cycloserine, 8 μg cefoxitin and 20 μg lincomycin.
Table 1.
Bacterial strains and plasmids utilized in this study.
| Strain or plasmid | Genotype or phenotype | Reference |
|---|---|---|
| E. coli DH5α | Cloning host | NEB |
| E. coli HB101 | Conjugation donor | (Williams et al., 1990) |
| C. difficile R20291 | Clinical isolate; ribotype 027 | (Stabler et al., 2009) |
| R20291::2601 | R20291 2601::EMr | This work |
| R20291::2601/pMTL84153 | R20291::2601 containing empty plasmid pMTL84153 | This work |
| R20291::2601/pMTL84153-2601 | R20291 2601 gene complemented with pMTL84153-2601 | This work |
| R20291/pMTL84153 | R20291 containing empty plasmid pMTL84153 | This work |
| Plasmids | ||
| pMTL007C-E2 | ClosTron plasmid (ColE1, pCD6, catP) | (Heap et al., 2010) |
| pMTL007C-E2-2601/1483a | pMTL007C-E2 derivative retargeted to 2601(Cwp22) gene | This work |
| pMTL84151 | E. coli-C. difficile shuttle plasmid (pCD6, catP, ColE1 + tra, MCS) | (Heap et al., 2009) |
| pMTL82153 | E. coli-C. difficile shuttle plasmid (pCD6, catP, ColE1 + tra, MCS) | (Heap et al., 2009) |
| pMTL84153 | E. coli-C. difficile shuttle plasmid (pCD6, catP, ColE1 + tra, Pfdx + MCS) | This work |
| pMTL84153-2601 | Complement vector, pMTL84153 containing 2601 gene | This work |
DNA manipulations and chemicals
DNA manipulations were carried out according to standard techniques (Chong, 2001). Recombinant plasmids were conjugated into C. difficile according to the method described earlier (Heap et al., 2010). The DNA markers, T4 DNA ligase, restriction enzymes, PCR product purification kit, DNA gel extraction kit, First-strand cDNA synthesis kit and SYBR Green RT-qPCR kit were purchased from Thermo Fisher Scientific (Waltham, MA). Plasmid DNA, chromosomal DNA and total RNA were isolated using QIAGEN column (Qiagen, UK). PCRs were performed with the high-fidelity DNA polymerase NEB Q5 (New England, UK). Primers (Supporting Information Table S1) were synthesized by IDT (Coralville, IA). All chemicals were purchased from Sigma (St. louis, MO) unless those stated otherwise.
Construction of cwp22 mutant and complementation strains
The ClosTron system was used for inactivation of the 2601 gene (cwp22) as described previously (Heap et al., 2010). The 353 bp retarget intron was designed on the website (http://clostron.com/clostron2.php?), and was synthesized and cloned into the plasmid pMTL007C-E2, producing pMTL007C-E2–2601, with services of ATUM company (https://www.atum.bio/eCommerce/login), and subsequently was conjugated into R20291. Successful transconjugants were selected with selective plate BHISTKC (15 μg ml−1 thiamphenicol, 50 μg ml−1 kanamycin, 8 μg ml−1 cefoxitin). Subsequent correct intergrants (R20291::2601) were selected on BHIS-Lm (20 μg ml−1 lincomycin) plates.
The 2601 gene, a 1962 bp fragment, was amplified with primers Re-F/R. The PCR products flanked with SacI-BamHI restriction enzyme sites were digested, and then cloned into pMTL84153 plasmid, which was constructed from pMTL84151 and pMTL82153, yielding the complemented plasmid pMTL84153–2601, and subsequently was conjugated into R20291::2601 and verified by PCR, yielding the complemented strain R20291::2601/ pMTL84153–2601. The empty plasmid pMTL84153 was also conjugated into R20291 and R20291::2601 as negative controls respectively.
Confirmation of the cwp22 mutation by PCR and southern hybridization
PCR was performed to confirm the insertion of the targetron in the right position with primers 1-F/R and 2-F/R. Meanwhile, RT-PCR and RT-qPCR were conducted using primers 2-F/R, 3-F/R and 4-F/R to confirm truncation of cwp22 mRNA and no polar effect of cwp22 mutation on up-/downstream of genes respectively. All RT-qPCRs were repeated in triplicate, independently. Data analysis was conducted by using the comparative CT (2−ΔΔCT) method with16s rRNA as control. Primers Co-F/R were used to verify the co-transcription of 2602 and 2601 genes. Southern blotting was used to identify the single copy insertion of the targetron in the genome (Waligora et al., 2001). For Southern hybridization, the genome (5 μg) of R20291 and R20291::2601 were digested with either EcoRI-HindIII or EcoRI-XbaI. The specific probe was synthesized using the intron/ermB sequence as a template with the DIG High Prime DNA Labelling and Detection Starter kit I (Sigma, St. louis, MO), generated using the oligonucleotide primer Perm.
Growth profile, toxin expression and cytotoxicity assay
C. difficile strains were cultured to an optical density of OD600 of 0.8 in BHIS, and then diluted to an OD600 of 0.2. One millilitre of culture dilution was inoculated into 100 ml BHIS, followed by measuring OD600 for 48 h.
For determination of toxin concentration in C. difficile cultures, 10 ml of C. difficile cultures were collected at 12, 24, 36 and 48 h postinoculation. The OD600 of cultures were adjusted to the same value with fresh BHIS. Then the cultures were centrifuged at 4C, 12000 × g for 5 min, filtered with 0.22 μm filter and used for ELISA. For the intracellular toxin analysis, the centrifuged C. difficile pellets were washed three times with PBS, and resuspended in 2 ml of PBS with 200 μl of 0.2 mm glass beads. Afterward, the bacteria were vortexed at 4C for 20 min, following centrifuged at 4C, 12,000 × g for 5 min. Then, the supernatants from the bacteria lysis were normalized to the same protein concentration with BCA (bicinchoninic acid) protein assay, and used for intracellular toxin detection by Western blot analysis and ELISA. Anti-TcdA (PCG4.1, Novus Biologicals, Centennial, CO) and anti-TcdB (AI, Gene Tex, USA) were used as coating antibodies for ELISA, and HRP-Chicken anti-TcdA and HRP-Chicken anti-TcdB (Gallus Immunotech, Shirley, MA) were used as detection antibodies in both ELISA and Western blot analysis.
For toxin transcription analysis, cultures of C. difficile strains were collected at 12, 24, 36 and 48 h of postinoculated respectively. Then, the total RNA was extracted with TRIzol reagent. The transcription of tcdA and tcdB was determined through RT-qPCR with primers tcdA-F/R and tcdB-F/R respectively. All RT-qPCRs were repeated in triplicate, independently. Data analysis was conducted by using the comparative CT (2−ΔΔCT) method with16s rRNA as a control.
To determine cytotoxicity of C. difficile cultures, cytotoxic titers of culture supernatants were determined according to the revised protocol (Winston et al., 2016). Briefly, 1 ml of a 24, 36 and 48 h BHIS cultured strains were collected and adjusted to the same OD600. Then, the cultures were centrifuged at 4C, 12,000 × g for 10 min, and filtered with 0.22 μm filters. Afterwards, the supernatants were serially diluted by twofold with PBS, and 50 μl of supernatants were added into 50 μl of CT-26 cells with 95% confluence (105/well) in a 96-well plate, followed incubating overnight at 37C/5% CO2. Cell morphology alterations were monitored and imaged under a microscope after overnight incubation. The cytotoxic titre was defined as the reciprocal of the highest dilution that caused 50% CT26 cell rounding (Winston et al., 2016). CT26 cells treated with purified Tcd B and BHIS media were used as positive and negative controls respectively.
Cell autolysis, LDH cytotoxicity and cell viability analysis
To determine Triton X-100 induced-autolysis, C. difficile strains were cultured to an OD600 of 0.8 to log phase, and then 5 ml of each culture was collected and washed with 50 mM potassium phosphate buffer (pH 7.0). The pellets were resuspended in a final volume of 2.5 ml of 50 mM potassium phosphate buffer containing 0.01% of Triton X-100. Afterwards, the bacteria were incubated anaerobically at 37C, and the OD600 was detected at 0, 0.5, 1, 2, 4 and 6 h. The lysis percent was shown as % initial OD600.
For the LDH cytotoxicity analysis, the supernatants from different strains were collected, and filtered with 0.22 μm filters as described above, then the LDH concentration of the supernatants was detected with the Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher, Waltham, MA) according to the instructions of the manufacturer.
For cell viability analysis, the live-dead cell staining was performed (Fuller et al., 2000; Stiefel et al., 2015). Briefly, 12, 24, 36 and 48 h postincubated C. difficile strains were collected and cell number was normalized to 108 CFU ml−1 respectively. Then 1 ml of each strain cultures was centrifuged at 4C, 5,000 × g for 10 min, and washed with PBS for three times. Afterwards, the bacteria were resuspended in 100 μl of 0.1 mM sodium phosphonate buffer. The chemical 5(6)-CFDA (5-(and-6)-carboxyfluorescein diacetate) was used to dye live C. difficile, and the PI was used to dye dead bacteria. The final concentration of 50 mM 5(6)-CFDA and 200 ng ml−1 of PI was used to co-dye C. difficile strains, following addition of the dye mixture, and C. difficile cells were incubated at 4C overnight for monitoring under a fluorescence microscope. The CFDA and PI were excited at 495 and 538 nm respectively. To further detect the cell viability change of C. difficile strains, TEM analysis was performed. Specimens were prepared according to the previous method used in C. difficile (Baban et al., 2013; Calderon-Romero et al., 2018), and detected by JEM-1400 (Jeol Ltd., Tokyo, Japan) TEM. Briefly, 1 ml of bacterial cultures were collected and fixed with 2.5% gluteraldehyde. After fixation, samples were post-fixed with 1% osmium tetroxide (EMS) and dehydrated with a graded series of ethanol (30, 50 and 70), followed resuspended in 70% ethanol for TEM detection directly.
Adhesion of C. difficile vegetative cells and spores to HCT-8 cells
The adhesion ability of vegetative cells and spores was evaluated with HCT-8 cells (ATCC CCL-244) (Janvilisri et al., 2010). Briefly, cells were grown to 95% confluence (2 × 105/well) in a 24-well plate, followed by infection with 6 × 106 of vegetative cells (log phase) or spores at a multiplicity of infection of 30:1, and cultured in the anaerobic chamber at 37∘ C for 1 h. After incubation, the infected cells were washed and suspended in RPMI media, and plated on BHIS plates with 0.1% TA to enumerate the adhered vegetative cells or spores. The ability of C. difficile strains to adhere to HCT-8 cells was calculated as follows: CFU adhered vegetative cells or spores/total cell numbers.
Evaluation of virulence of R20291 and cwp22 mutant in the mouse model of C. difficile infection
C57BL/6 female mice (6 weeks old) were purchased from Charles River Laboratories, Cambridge, MA. All studies were approved by the Institutional Animal Care and Use Committee of University of South Florida. The experimental design and antibiotic administration were performed as previously described (Sun et al., 2011). Briefly, 30 mice were divided into three groups in six cages. Group 1 was challenged with R20291 spores, group 2 with R20291::2601 spores and group 3 with R20291::2601/pMTL84153–2601 spores respectively. Mice were given an orally administered antibiotic cocktail (kanamycin 0.4 mg ml−1, gentamicin 0.035 mg ml−1, colistin 0.042 mg ml−1, metronidazole 0.215 mg ml−1 and vancomycin 0.045 mg ml−1) in drinking water for 4 days. After 4 days of antibiotic treatment, all mice were given autoclaved water for 2 days, followed by one dose of clindamycin (10 mg kg−1, intraperitoneal route) 24 h before spores challenge (Day 0). Afterwards, mice were orally challenged with 106 of spores by gavage, and monitored daily for a week for changes in weight, diarrhoea, mortality and other symptoms of the disease.
Enumeration of C. difficile in faeces and cecum tissues, and determination of toxin levels in faeces
Faecal pellets were collected from postinfection day 0 to day 7, and stored in −70∘C. To enumerate C. difficile numbers, faeces were diluted into PBS at a final concentration of 0.1 g ml−1. Then, 100 μl of the faecal solution was added to 900 μl of absolute ethanol, and kept at room temperature for 1 h to inactivate vegetative cells. After that, faecal samples were serially diluted and plated on BHIS-CCT (250 μg ml−1 D-cycloserine, 8 μg ml−1 cefoxitin, 0.1% TA). The plates were incubated at 37∘C in the anaerobic chamber for 24–48 h, and then the colonies were counted and expressed as CFU/g faeces. To determine toxin tilter in faecal samples, 0.1 g ml−1 of faecal samples were diluted two times with PBS, and then the concentration of TcdA and TcdB was measured by ELISA. To determine C. difficile adherence to the cecum, the intact cecum of the mice was collected on day 7, and then weighted and homogenized in PBS at a final concentration of 10 mg ml−1 (Baban et al., 2013). Then, cecum samples were serially diluted and plated on BHIS-CCT plates, and the colonies were counted and expressed as CFU/g cecum.
Statistical analysis
The reported experiments were carried out in independent biological triplicates except animal experiments, and each sample was additionally taken in technical triplicates. Animal survivals were analysed by Kaplan–Meier survival analysis. Student’s unpaired t-test was used for two groups comparison. One-way analysis of variance (ANOVA) was used for more than two groups comparison. Results are expressed as mean ± standard error of the mean. Differences were considered statistically significant if P < 0.05 (*).
Supplementary Material
Fig. S1 Effect of cwp22 mutation on C. difficile growth.
(A) C. dificile strains were cultured in BHIS and monitored by measuring OD600 for 48 h. (B) Growth ratio plotted into time from 16 to 36 h. (C) Slope of growth ratio from 16 to 36 h. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S2 Effect of cwp22 mutation on C. difficile biofilm formation.
Biofilm of C. difficile strains were detected at 14 and 72 h respectively. Briefly, C. difficile strains were cultured in 48-well plate with RCM (Reinforced Clostridial Medium) at 37 °C. After 24 or 72 h culture, the biofilm at the bottom of well was fixed with 2.5% GA for 30 min, followed by dying with 0.25% (w/v) crystal violet for 10 min. Then the wells were washed with PBS, followed by addition of acetone to dissolve the crystal violet. The absorbance was recorded at OD570. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05, **P < 0.01). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S3 Effect of cwp22 mutation on C. difficile motility.
C. difficile strains were cultured to an OD600 of 0.8. Then 2 μl of each cultures were penetrated into soft BHIS agar (0.175%) plate for swimming analysis, alternatively 2 μl of each cultures were dropped onto 0.3% BHIS agar plate for swarming analysis. The swimming assay plates were cultured for 24 h and the swarming assay plates were culture for 48 h respectively. (A) Swarming assay. 1: R20291::2601 (15.15 ± 1.2 mm, *P < 0.05); 2: R20291::2601/pMTL84153 (13.94 ± 0.7 mm, *P < 0.05); 3: R20291::2601/pMTL84153–2601 (18.18 ± 2.2 mm); 4: R20291 (19.70 ± 1.6 mm). (B) Swimming assay. 1: R20291::2601 (13.94 ± 2.1 mm, *P < 0.05); 2: R20291::2601/ pMTL84153 (12.73 ± 1.8 mm, *P < 0.05); 3: R20291::2601/pMTL84153–2601 (19.09 ± 0.3 mm); 4: R20291 (22.33 ± 2.9 mm). Experiments were independently repeated thrice.(*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S4 Effect of cwp22 mutation on C. difficile spores resistance to heat and ethanol.
C. difficile spores were collected from 2 weeks Clospore media cultured bacteria and purified with a sucrose gradient layer (50%, 45%, 35%, 25%, 10%). To determine spore resistance, 1 × 106 spores were treated with ethanol (100% v/v) and heat (65°C) for 0–6 h at 37°C respectively. Spores treated at 0, 0.5, 1, 2, 4, and 6 h were plated on BHIS plates with 0.1% TA. Percentage of spore viability following treatment was calculated as follows: post treated CFU / untreated CFU. (A) Spores resistance to heat (65°C). (B) Spores resistance to 100% ethanol. Experiments were independently repeated thrice. Bars stand for mean ± SEM. One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S5 Effect of cwp22 mutation on C. difficile sporulation and germination.
(A) Sporulation assay. C. difficile strains were cultured in Clospore media for 5 days. Afterwards, the CFU/ml of cultures from 24, 48 and 72 h incubation were counted on BHIS plates with 0.1% TA to detect sporulation ratios. The sporulation ratio was calculated as CFU (65 °C heated) / CFU (no heated). (B) Germination assay. The heated purified spores were diluted to an OD600 0f 1.0 in the germination buffer [10 mM Tris (pH 7.5), 150 mM NaCl, 100 mM glycine, 10 mM TA] to detect germination ratio. The value of OD600 was monitored immediately (0 min, t0), and was detected once every 2 min (tx) for 20 min at 37 °C. The germination ratio was calculated as OD600 (tx) / OD600 (T0). The boiled spores (red line, 100 °C, 20 min) were used as a negative control. (C) DPA release assay. The heated purified spores were diluted into germination buffer supplemented with 800 μM of TbCl3 (Terbium(III) chloride). The DPA released from spores was monitored with Multi-Mode Reader (excitation, 270 nm; emission, 545 nm) The boiled spores (100 °C, 20 min) were used as positive control (red line), and the spores in germination buffer without TA were used as a negative control (green line). Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S6 Effect of cwp22 mutation on C. difficile transcription of sigD and tcdR.
(A) tcdR gene transcription analysis. (B) sigD gene transcription analysis. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S7 Effect of cwp22 mutation on C. difficile transcription of fliC.
Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Table S1. Primers utilized in this study.
Table S2. Complementation plasmid maintenance assay.
Acknowledgements
This work was supported in part by National Institutes of Health grants (K01-DK092352, R21-AI113470, R03-DK112004 and R01-AI132711 to X. Sun, and R01-AI081062 to Y. He).
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
References
- Aubry A, Hussack G, Chen WX, KuoLee R, Twine SM, Fulton KM, et al. (2012) Modulation of toxin production by the llagellar regulon in Clostridium difficile. Infect Immun 80: 3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausiello CM, Cerquetti M, Fedele G, Spensieri F, Palazzo R, Nasso M, et al. (2006) Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microb Infect 8: 2640–2646. [DOI] [PubMed] [Google Scholar]
- Baban ST, Kuehne SA, Barketi-Klai A, Cartman ST, Kelly ML, Hardie KR, et al. (2013) The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 8: e73026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker D, Smits WK, Kuijper EJ, and Corver J (2012) TcdC does not significantly repress toxin expression in Clostridium difficile 630 Delta Erm. PLoS One 7: e43247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, and Mayer C (2006) Crystal structure of a novel beta lactam insensitive peptidoglycan transpeptidase. J Mol Biol 359: 533–538. [DOI] [PubMed] [Google Scholar]
- Biazzo M, Cioncada R, Fiaschi L, Tedde V, Spigaglia P, Mastrantonio P, et al. (2013) Diversity of cwp loci in clinical isolates of Clostridium difficile. J Med Microbiol 62: 1444–1452. [DOI] [PubMed] [Google Scholar]
- Borriello SP (1995) Virulence factors of Clostridium difficile. Microb Ecol Health 8: 183–183. [Google Scholar]
- Bradshaw WJ, Roberts AK, Shone CC, and Acharya KR (2018) The structure of the S-layer of Clostridium difficile. J Cell Commun Signal 12: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw WJ, Kirby JM, Roberts AK, Shone CC, and Acharya KR (2017) Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro. Febs J 284: 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Hundsberger T, Leukel P, Sauerborn M, and vonEichelStreiber C (1996) Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181: 29–38. [DOI] [PubMed] [Google Scholar]
- Calabi E, Calabi F, Phillips AD, and Fairweather NF (2002) Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun 70: 5770–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, et al. (2001) Molecular characterization of the surface layer proteins from Clostridium difficile. Mol Microbiol 40: 1187–1199. [DOI] [PubMed] [Google Scholar]
- Calderon-Romero P, Castro-Cordova P, Reyes-Ramirez R, Milano-Cespedes M, Guerrero-Araya E, Pizarro-Guajardo M, et al. (2018) Clostridium difficile exosporium cysteine-rich proteins are essential for the morphogenesis of the exosporium layer, spore resistance, and affect C. difficile pathogenesis. PLoS Pathog 14: e1007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L (2001) Molecular cloning - a laboratory manual, 3rd edition. Science 292: 446–446. [Google Scholar]
- Dang THT, de la Riva L, Fagan RP, Storck EM, Heal WP, Janoir C, et al. (2010) Chemical probes of surface layer biogenesis in Clostridium difficile. ACS Chem Biol 5: 279–285. [DOI] [PubMed] [Google Scholar]
- Dubberke ER, and Olsen MA (2012) Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55: S88–S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy B, Govind R, Antunes A, and Matamouros S (2008) Clostridium difficile toxin synthesis is negatively regulated by TcdC. J Med Microbiol 57: 685–689. [DOI] [PubMed] [Google Scholar]
- Edwards RA, Schifferli DM, and Maloy SR (2000) A role for Salmonella fimbriae in intraperitoneal infections. Proc Natl Acad Sci U S A 97: 1258–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Meouche I, Peltier J, Monot M, Soutourina O, PestelCaron M, Dupuy B, and Pons JL (2013) Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 8: e83748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JE, Reynolds CB, Fagan RP, Shaw HA, Goulding D, and Fairweather NF (2009) A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol Microbiol 74: 541–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira TG, Moura H, Barr JR, Domingues RMCP, and Ferreira ED (2017) Ribotypes associated with Clostridium difficile outbreaks in Brazil display distinct surface protein profiles. Anaerobe 45: 120–128. [DOI] [PubMed] [Google Scholar]
- Fuller ME, Streger SH, Rothmel RK, Mailloux BJ, Hall JA, Onstott TC, et al. (2000) Development of a vital fluorescent staining method for monitoring bacterial transport in subsurface environments. Appl Environ Microbiol 66: 4486–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind R, and Dupuy B (2012) Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 8: e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind R, Fitzwater L, and Nichols R (2015) Observations on the role of TcdE isoforms in Clostridium difficile toxin secretion. J Bacteriol 197: 2600–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xiang Z, and Mobley HL (2010) Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol 2010: 297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Racz R, Sayers S, Lin Y, Todd T, Hur J, et al. (2014) Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res 42: D1124–D1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Pennington OJ, Cartman ST, and Minton NP (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Meth 78: 79–85. [DOI] [PubMed] [Google Scholar]
- Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, and Minton NP (2010) The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Meth 80: 49–55. [DOI] [PubMed] [Google Scholar]
- Janvilisri T, Scaria J, and Chang YF (2010) Transcriptional profiling of Clostridium difficile and Caco-2 cells during infection. J Infect Dis 202: 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, and Shone CC (2009) Cwp84, a surface associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J Biol Chem 284: 34666–34673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JA, Gebhart D, Buckley AM, Lok S, Scholl D, Douce GR, et al. (2017) New class of precision antimicrobials redefines role of Clostridium difficile S-layer in virulence and viability. Sci Transl Med 9: eaah6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, and Minton NP (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467: 711–U797. [DOI] [PubMed] [Google Scholar]
- Lawson PA, Citron DM, Tyrrell KL, and Finegold SM (2016) Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40: 95–99. [DOI] [PubMed] [Google Scholar]
- Lessa FC, Gould CV, and McDonald LC (2012) Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 55: S65–S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa FC, Winston LG, McDonald LC, and Difficil EIPC (2015) Burden of Clostridium difficile infection in the United States. New Engl J Med 372: 2369–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Preusse M, Bruchmann S, Pawar V, Grahl N, Pils MC, et al. (2019) Importance of flagella in acute and chronic Pseudomonas aeruginosa infections. Environ Microbiol 21: 883–897. [DOI] [PubMed] [Google Scholar]
- Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, et al. (2009) Toxin B is essential for virulence of Clostridium difficile. Nature 458: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani N, and Dupuy B (2001) Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci USA 98: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, et al. (2002) Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol 184: 5971–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, et al. (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45: D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Verstraete I, Peltier J, and Dupuy B (2016) The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 8: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief JS, Barroso LA, and Wilkins TD (1997) Positive regulation of Clostridium difficile toxins. Infect Immun 65: 1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray R, Boyd D, Levett PN, Mulvey MR, and Alfa MJ (2009) Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of Toxin B or Toxin A. BMC Infect Dis 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, et al. (2012) Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 52: 92–100. [DOI] [PubMed] [Google Scholar]
- Ong E, Wong MU, and He Y (2017) Identification of new features from known bacterial protective vaccine antigens enhances rational vaccine design. Front Immunol 8: 1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, and Garrity GM (2018) Notification of changes in taxonomic opinion previously published outside the IJSEM. Int J Sys Evol Microbiol 68: 2137–2138. [DOI] [PubMed] [Google Scholar]
- Pechine S, Deneve C, Le Monnier A, Hoys S, Janoir C, and Collignon A (2011) Immunization of hamsters against Clostridium difficile infection using the Cwp84 protease as an antigen. Fems Immunol Med Mic 63: 73–81. [DOI] [PubMed] [Google Scholar]
- Pechine S, Gleizes A, Janoir C, Gorges-Kergot R, Barc MC, Delmee M, and Collignon A (2005) Immunological properties of surface proteins of Clostridium difficile. J Med Microbiol 54: 193–196. [DOI] [PubMed] [Google Scholar]
- Pechine S, Janoir C, Boureau H, Gleizes A, Tsapis N, Hoys S, et al. (2007) Diminished intestinal colonization by Clostridium difficile and immune response in mice after mucosal immunization with surface proteins of Clostridium difficile. Vaccine 25: 3946–3954. [DOI] [PubMed] [Google Scholar]
- Peltier J, Courtin P, El Meouche I, Lemee L, ChapotChartier MP, and Pons JL (2011) Clostridium difficile has an original peptidoglycan structure with a high level of N-acetylglucosamine deacetylation and mainly 3–3 cross-links. J Biol Chem 286: 29053–29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peniche AG, Savidge TC, and Dann SM (2013) Recent insights into Clostridium difficile pathogenesis. Curr Opin Infect Dis 26: 447–453. [DOI] [PubMed] [Google Scholar]
- Perez J, Springthorpe VS, and Sattar SA (2011) Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int 94: 618–626. [PubMed] [Google Scholar]
- Racine FM, and Vary JC (1980) Isolation and properties of membranes from Bacillus megaterium spores. J Bacteriol 143: 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CB, Emerson JE, de la Riva L, Fagan RP, and Fairweather NF (2011) The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation promoting function. PLoS Pathog 7: e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandolo C, Pechine S, Le Monnier A, Hoys S, Janoir C, Coviello T, et al. (2011) Encapsulation of Cwp84 into pectin beads for oral vaccination against Clostridium difficile. Eur J Pharm Biopharm 79: 566–573. [DOI] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. (2006) The multidrug resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38: 779–786. [DOI] [PubMed] [Google Scholar]
- Seddon SV, and Borriello SP (1992) Proteolytic activity of Clostridium difficile. J Med Microbiol 36: 307–311. [DOI] [PubMed] [Google Scholar]
- Seddon SV, Hemingway I, and Borriello SP (1990) Hydrolytic enzyme production by Clostridium difficile and its relationship to toxin production and virulence in the hamster model. J Med Microbiol 31: 169–174. [DOI] [PubMed] [Google Scholar]
- Sekulovic O, Bedoya MO, Fivian-Hughes AS, Fairweather NF, and Fortier LC (2015) The Clostridium difficile cell wall protein CwpV confers phase variable phage resistance. Mol Microbiol 98: 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson E, Minton NP, and Kuehne SA (2015) The role of flagella in Clostridium difficile pathogenicity. Trends Microbiol 23: 275–282. [DOI] [PubMed] [Google Scholar]
- Stiefel P, Schmidt-Emrich S, Maniura-Weber K, and Ren Q (2015) Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol 15: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang H, Zhang Y, Chen K, Davis B, and Feng H (2011) Mouse relapse model of Clostridium difficile infection. Infect Immun 79: 2856–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin L, Edoo Z, Hugonnet JE, Mainardi JL, and Arthur M (2018) Peptidoglycan cross-linking activity of L,D-transpeptidases from Clostridium difficile and inactivation of these enzymes by beta lactams. Antimicrob Agents Chemother 62: e01607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeoka A, Takumi K, Koga T, and Kawata T (1991) Purification and characterization of S layer proteins from Clostridium difficile GAI 0714. J Gen Microbiol 137: 261–267. [DOI] [PubMed] [Google Scholar]
- Voth DE, and Ballard JD (2005) Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18: 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waligora AJ, Hennequin C, Mullany P, Bourlioux P, Collignon A, and Karjalainen T (2001) Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun 69: 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Young DI, and Young M (1990) Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J Gen Microbiol 136: 819–826. [DOI] [PubMed] [Google Scholar]
- Willing SE, Candela T, Shaw HA, Seager Z, Mesnage S, Fagan RP, and Fairweather NF (2015) Clostridium difficile surface proteins are anchored to the cell wall using CWB2 motifs that recognise the anionic polymer PSII. Mol Microbiol 96: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JA, Thanissery R, Montgomery SA, and Theriot CM (2016) Cefoperazone treated mouse model of clinically relevant Clostridium difficile strain R20291. Jove-J Vis Exp 118: e54850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Drudy D, Kyne L, Brown K, and Fairweather NF (2008) Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J Med Microbiol 57: 750–756. [DOI] [PubMed] [Google Scholar]
- Wright A, Wait R, Begum S, Crossett B, Nagy J, Brown K, and Fairweather N (2005) Proteomic analysis of cell surface proteins from Clostridium difficile. Proteomics 5: 2443–2452. [DOI] [PubMed] [Google Scholar]
- Wydau-Dematteis S, El Meouche I, Courtin P, Hamiot A, LaiKuen R, Saubamea B, et al. (2018) Cwp19 is a novel lytic transglycosylase involved in stationary phase autolysis resulting in toxin release in Clostridium difficile. MBio 9: e00648–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu DL, Sorg JA, and Sun XM (2018) Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Effect of cwp22 mutation on C. difficile growth.
(A) C. dificile strains were cultured in BHIS and monitored by measuring OD600 for 48 h. (B) Growth ratio plotted into time from 16 to 36 h. (C) Slope of growth ratio from 16 to 36 h. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S2 Effect of cwp22 mutation on C. difficile biofilm formation.
Biofilm of C. difficile strains were detected at 14 and 72 h respectively. Briefly, C. difficile strains were cultured in 48-well plate with RCM (Reinforced Clostridial Medium) at 37 °C. After 24 or 72 h culture, the biofilm at the bottom of well was fixed with 2.5% GA for 30 min, followed by dying with 0.25% (w/v) crystal violet for 10 min. Then the wells were washed with PBS, followed by addition of acetone to dissolve the crystal violet. The absorbance was recorded at OD570. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05, **P < 0.01). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S3 Effect of cwp22 mutation on C. difficile motility.
C. difficile strains were cultured to an OD600 of 0.8. Then 2 μl of each cultures were penetrated into soft BHIS agar (0.175%) plate for swimming analysis, alternatively 2 μl of each cultures were dropped onto 0.3% BHIS agar plate for swarming analysis. The swimming assay plates were cultured for 24 h and the swarming assay plates were culture for 48 h respectively. (A) Swarming assay. 1: R20291::2601 (15.15 ± 1.2 mm, *P < 0.05); 2: R20291::2601/pMTL84153 (13.94 ± 0.7 mm, *P < 0.05); 3: R20291::2601/pMTL84153–2601 (18.18 ± 2.2 mm); 4: R20291 (19.70 ± 1.6 mm). (B) Swimming assay. 1: R20291::2601 (13.94 ± 2.1 mm, *P < 0.05); 2: R20291::2601/ pMTL84153 (12.73 ± 1.8 mm, *P < 0.05); 3: R20291::2601/pMTL84153–2601 (19.09 ± 0.3 mm); 4: R20291 (22.33 ± 2.9 mm). Experiments were independently repeated thrice.(*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S4 Effect of cwp22 mutation on C. difficile spores resistance to heat and ethanol.
C. difficile spores were collected from 2 weeks Clospore media cultured bacteria and purified with a sucrose gradient layer (50%, 45%, 35%, 25%, 10%). To determine spore resistance, 1 × 106 spores were treated with ethanol (100% v/v) and heat (65°C) for 0–6 h at 37°C respectively. Spores treated at 0, 0.5, 1, 2, 4, and 6 h were plated on BHIS plates with 0.1% TA. Percentage of spore viability following treatment was calculated as follows: post treated CFU / untreated CFU. (A) Spores resistance to heat (65°C). (B) Spores resistance to 100% ethanol. Experiments were independently repeated thrice. Bars stand for mean ± SEM. One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S5 Effect of cwp22 mutation on C. difficile sporulation and germination.
(A) Sporulation assay. C. difficile strains were cultured in Clospore media for 5 days. Afterwards, the CFU/ml of cultures from 24, 48 and 72 h incubation were counted on BHIS plates with 0.1% TA to detect sporulation ratios. The sporulation ratio was calculated as CFU (65 °C heated) / CFU (no heated). (B) Germination assay. The heated purified spores were diluted to an OD600 0f 1.0 in the germination buffer [10 mM Tris (pH 7.5), 150 mM NaCl, 100 mM glycine, 10 mM TA] to detect germination ratio. The value of OD600 was monitored immediately (0 min, t0), and was detected once every 2 min (tx) for 20 min at 37 °C. The germination ratio was calculated as OD600 (tx) / OD600 (T0). The boiled spores (red line, 100 °C, 20 min) were used as a negative control. (C) DPA release assay. The heated purified spores were diluted into germination buffer supplemented with 800 μM of TbCl3 (Terbium(III) chloride). The DPA released from spores was monitored with Multi-Mode Reader (excitation, 270 nm; emission, 545 nm) The boiled spores (100 °C, 20 min) were used as positive control (red line), and the spores in germination buffer without TA were used as a negative control (green line). Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S6 Effect of cwp22 mutation on C. difficile transcription of sigD and tcdR.
(A) tcdR gene transcription analysis. (B) sigD gene transcription analysis. Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Fig. S7 Effect of cwp22 mutation on C. difficile transcription of fliC.
Experiments were independently repeated thrice. Bars stand for mean ± SEM, (*P < 0.05). One-way analysis of variance (ANOVA) was used for statistical significance.
Table S1. Primers utilized in this study.
Table S2. Complementation plasmid maintenance assay.