A novel B cell subset that produces proangiogenic cytokines is increased in pathologies associated with angiogenesis.
Abstract
B cells contribute to immune responses through the production of immunoglobulins, antigen presentation, and cytokine production. Several B cell subsets with distinct functions and polarized cytokine profiles have been reported. In this study, we used transcriptomics analysis of immortalized B cell clones to identify an IgG4+ B cell subset with a unique function. These B cells are characterized by simultaneous expression of proangiogenic cytokines including VEGF, CYR61, ADM, FGF2, PDGFA, and MDK. Consequently, supernatants from these clones efficiently promote endothelial cell tube formation. We identified CD49b and CD73 as surface markers identifying proangiogenic B cells. Circulating CD49b+CD73+ B cells showed significantly increased frequency in patients with melanoma and eosinophilic esophagitis (EoE), two diseases associated with angiogenesis. In addition, tissue-infiltrating IgG4+CD49b+CD73+ B cells expressing proangiogenic cytokines were detected in patients with EoE and melanoma. Our results demonstrate a previously unidentified proangiogenic B cell subset characterized by expression of CD49b, CD73, and proangiogenic cytokines.
INTRODUCTION
The function of B cells has long been thought to be limited to the generation of immunoglobulin-producing plasma cells. However, B cells can exert a more diverse range of immune effector and regulatory functions. Distinct functional B cell subsets have been identified on the basis of their cytokine production profiles. Immunosuppressive B regulatory (reg) cells (1) and other potential B cell subsets, such as B effector 1 (Be1) and Be2 cells, as well as interleukin-17 (IL-17)–producing B cells, have been reported (2, 3). B cells can secrete a wide range of cytokines and accumulate in chronic inflammatory areas and around tumor cells. Their interaction with tissue cells and tumor cells and their contribution to tissue remodeling remain largely open questions.
Angiogenesis is an essential physiological process that occurs during embryogenesis, normal tissue development, and repair after injury. Through a controlled series of events, angiogenesis allows new vessels to grow from preexisting vessels to meet the physiological needs of tissues (4). Angiogenesis also plays a role in tumor growth (5) and is involved in tissue remodeling in chronic inflammatory conditions such as asthma and eosinophilic esophagitis (EoE) (6).
A wide range of secreted molecules promotes this process through direct interaction with vascular endothelial cells. These include vascular endothelial growth factors (VEGFs), fibroblast growth factors (FGFs), platelet-derived growth factors (PDGFs), hepatocyte growth factors, axon guidance factors, and angiopoietins (7). Other factors, including cysteine-rich angiogenic inducer 61 (CYR61) (8), andromedullin (ADM) (9), and midkine (MDK) (10), have also been reported to promote angiogenesis. Free adenosine also promotes angiogenesis both through its direct mitogenic effects on endothelial cells and through induction of proangiogenic factors such as VEGF, IL-8, and FGF from vascular and immune cells (11). Extracellular adenosine 5′-triphosphate (ATP) levels are significantly elevated in inflamed and hypoxic tissues. This extracellular ATP can be rapidly hydrolyzed by ectonucleotidases. CD39 is an ectonucleoside triphosphate diphosphohydrolase and is the rate-limiting enzyme in the conversion of ATP and adenosine 5′-diphosphate into adenosine 5′-monophosphate (AMP) (12). AMP is then converted to adenosine by ecto-5′-nucleotidase (NT5E), also known as CD73. CD39 is expressed on most peripheral (>90%) B cells and monocytes and can be expressed on a subset of CD4+ T cells, cytotoxic T cells, and natural killer (NK) cells (12) while CD73 is expressed by 75% of the B cells and a subset of CD8+ T cells, CD4+ T cells, and NK cells (12).
CD49b, also known as integrin subunit alpha 2 (ITGA2), is expressed as a part of the α2β1 integrin heterodimer on platelets, NK cells, T cells, and fibroblasts. α2β1 functions as a collagen receptor. The β1 subunit (CD29) is expressed on most of the hematopoietic and nonhematopoietic cells. CD49b and LAG-3 coexpression has been reported as a signature of type 1 regulatory (Tr1) T cells (13).
Immunoglobulin G4 (IgG4) can be regarded as an anti-inflammatory immunoglobulin isotype because of its low affinity for binding to Fcγ receptors, its inability to fix complement, and its functional monovalency, which results from its rearrangement of immunoglobulin heavy chains by a mechanism called Fab arm exchange (14, 15). No murine antibody isotype exists that shares these characteristics with human IgG4; therefore, mouse models are not suitable to study this immunological mechanisms associated with IgG4. IgG4 appears to play a role in the induction and maintenance of immune tolerance to allergens by blocking allergen-specific IgE, and allergen-specific IgG4 antibodies are significantly increased during the course of allergen-specific immunotherapy and in high-dose allergen–exposed individuals, such as beekeepers and cat owners (16, 17).
Besides its potential role in immune tolerance to allergens, IgG4 has also been associated with pathological conditions. EoE, melanoma, and IgG4-related diseases are associated with structural tissue remodeling including angiogenesis. Enhanced IgG4 responses have been reported in patients with EoE where high amounts of IgG4 antibodies against food allergens associated with EoE were found. These patients did not show elevated specific IgE levels (18). Moreover, tissue IgG4 levels were later found to correlate with other disease parameters including esophageal eosinophil counts, as well as IL-4, IL-10, and IL-13 expression (19). Increased angiogenesis has been reported in the esophageal mucosa of patients with pediatric EoE (20). IgG4-expressing tissue-infiltrating B cells were found near tumors in patients with melanoma, and these tumor-associated B cells were polarized to produce IgG4. Both tumor-specific and nonspecific IgG4 antibodies blocked IgG1-mediated tumoricidal functions. Moreover, serum IgG4 was inversely correlated with patient survival and disease-free survival (21, 22).
In this study, we identify B cells that express IgG4 and produce proangiogenic cytokines and have the capacity to promote angiogenesis. We further show that these cells are characterized by the expression of CD49b and CD73. Cells with this phenotype are elevated in circulation of patients with EoE and melanoma and are present in affected tissues. Thus, our findings reveal a previously unidentified proangiogenic B cell subset that is associated with tissue remodeling in chronic inflammatory conditions and tumor angiogenesis.
RESULTS
A subset of B cells promotes angiogenesis
Using a previously described method of B cell immortalization (23), we generated a set of 27 memory B cell clones, of which 10 were IgG4+ and 17 were IgG1+ from eight individuals. The generation and characteristics of the clones are shown in fig. S1 and table S1, respectively. To determine the cytokine expression profile of each individual clone, B cell clones were stimulated for 4 hours with anti–B cell receptor (BCR), after which RNA expression was analyzed using next-generation sequencing. B cell clones were clustered on the basis of their expression of the top 100 most variably expressed genes encoding secreted immunomodulatory proteins (fig. S2). This approach resulted in the identification of a cluster of B cells showing marked up-regulation of cytokines with known angiogenesis-promoting characteristics. All of the B cells that comprised this “proangiogenic” cluster were IgG4+ (Fig. 1A). Differential expression analysis [false discovery rate (FDR) < 0.01, log2 fold change > 0.5] between the proangiogenic cluster and the other nonangiogenic B cell clones showed a significant up-regulation of genes encoding cytokines that are known to promote angiogenesis, tissue remodeling, and wound healing such as VEGFA, PDGFA, CYR61, bone morphogenic protein 2 (BMP2), slit homolog 2 (SLIT2), ADM2, FGF2, nephroblastoma overexpressed (NOV), secreted phosphoprotein 1 (SPP1), PDGFC, hepatoma-derived growth factor-related protein 3 (HDGFRP3), tumor necrosis factor receptor superfamily member 11B (TNFRSF11B), CXCL8 (encoding IL-8), MDK, and others (Fig. 1, A and B). Several genes with pleiotropic or unknown effects on angiogenesis, including IL-36 receptor antagonist (IL36RN), IL36B, IL-1 receptor antagonist (IL1RN), VGF, and transforming growth factor β2 (TGFB2), were up-regulated as well. Expression of IL10, IL16, semaphorin-4A (SEMA4A), C-C motif chemokine 22 (CCL22), and TNF-related apoptosis–inducing ligand (TNFSF10) was down-regulated in proangiogenic clones. While the effects of IL-10, CCL22, and IL-16 on angiogenesis remain unclear, as both pro- and antiangiogenic effects have been reported, TNFSF10 and SEMA4 have been shown to negatively regulate angiogenesis (24, 25). The cytokine expression profile of this proangiogenic cluster was remarkably stable, as indicated by real-time polymerase chain reaction (PCR) analysis performed after 3 weeks of culture with CD40L and IL-21 followed by 4 hours of stimulation with anti-BCR (Fig. 1, B and C). Expression of the proangiogenic cytokines VEGF-A, CYR61, FGF2, and IL-8 could also be induced in primary peripheral B cells (fig. S3). The culture conditions used for the expansion of immortalized B cell clones (CD40L + IL-21) in combination with anti-BCR stimulation most effectively induced the production of these cytokines in primary B cells (fig. S3).
Fig. 1. A subset of B cells promotes angiogenesis.
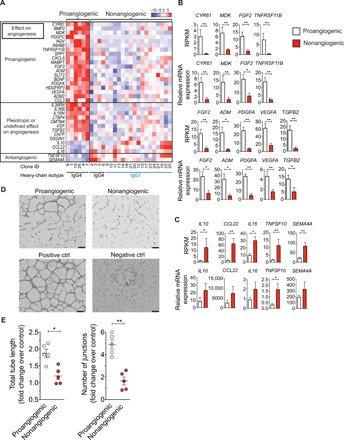
(A) Heat map showing gene-scaled (z score) log2 normalized counts of genes encoding secreted immunomodulatory proteins that are differentially expressed between proangiogenic B and nonangiogenic B cell clones (FDR < 0.01, log2 fold change > 0.5). The top box indicates genes with known proangiogenic effects, the middle box indicates genes with unknown or pleiotropic effects on angiogenesis, and the bottom box indicates genes with known anti-angiogenic effects. (B and C) Reads per kilobase million (RPKM) expression values from normal goat serum data (top) and real-time qPCR gene expression after prolonged (>3 weeks) in vitro expansion (bottom) of proangiogenic (n = 5) and nonangiogenic (n = 5) clones (mean ± SEM). *P < 0.05 and **P < 0.01, Mann-Whitney test. (B) Genes that were up-regulated in proangiogenic clones. (C) Genes that were down-regulated in proangiogenic clones. (D) Representative images of HUVEC tube formation assay to quantify proangiogenic effect of B cell clones (scale bars, 400 μm). Negative control, IMDM +2% FCS; positive control, EGM medium with growth factors. (E) Quantitative analysis of rate of HUVEC tube formation induced by supernatants of pro- and nonangiogenic B cell clones (mean ± SEM). *P < 0.05 and **P < 0.01, Mann-Whitney test.
To assess the functional capacity of proangiogenic B cell clones, we tested their potential to promote tube formation of human umbilical vein endothelial cells (HUVECs) (26). Supernatants collected from anti-BCR–stimulated proangiogenic B cell clones induced an increase of the total tube length (average increase, 1.9-fold over control) and number of junctions (average increase, 4.9-fold over control) in the tube formation assay (Fig. 1, D and E, and fig. S4), demonstrating their functional capacity to promote angiogenesis.
Proangiogenic B cells are characterized by expression of CD49b and CD73
To determine surface markers associated with proangiogenic B cells, we looked at differentially expressed genes encoding cell surface proteins. This resulted in the identification of 20 surface markers that were significantly differentially expressed (FDR < 0.01, log2 fold change > 0.5) between pro- and nonangiogenic B cell clones (Fig. 2A). Of these genes, PVRL2 (encoding CD112), NT5E (encoding CD73), CD276, ITGA2 (encoding CD49b), IL1R1 (encoding CD121a), and CDH2 (encoding CD325) showed the most uniform differential expression profile with high expression on proangiogenic clones and low expression on nonangiogenic clones. Consistently up-regulated surface expression of CD49b and CD73 was observed on proangiogenic B cell clones by flow cytometry (Fig. 2B). CD49b and CD73 were also both expressed on a subset of peripheral B cells, while peripheral B cells did not express CD112, CD325, and CD276, and all B cells were positive for CD53 (Fig. 2C). On the basis of these data, CD49b and CD73 represented potential surface markers for the identification of proangiogenic B cells.
Fig. 2. Proangiogenic B cells are characterized by expression of CD49b and CD73.
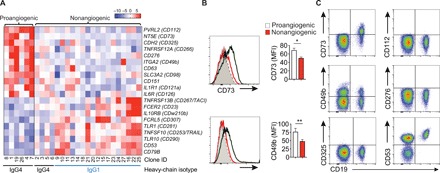
(A) Heat map showing gene-scaled (z score) log2 normalized counts of CD marker–encoding genes that are differentially expressed between proangiogenic B and nonangiogenic B cell clones (FDR < 0.01, log2 fold change > 0.5). (B) Flow cytometry analysis of CD73 and CD49b surface expression on proangiogenic (black line) (n = 5) and nonangiogenic (red line) B cell clones (n = 20) (mean ± SEM). Grey dotted line indicates isotype control. *P < 0.05 and **P < 0.01, Mann-Whitney test. (C) Flow cytometry analysis of surface expression of CD73 and CD49b on freshly isolated peripheral blood B cells.
CD73+CD49b+ B cells form a distinct population among circulating B cells
Staining of CD49b and CD73 on peripheral B cells from healthy individuals revealed a distinct CD73+CD49b+ population (Fig. 3A). Real-time quantitative PCR (qPCR) mRNA expression analysis of proangiogenic cytokines by B cell populations sorted based on surface expression of CD49b and CD73 showed that the expression of TGFB2, MDK, FGF2, CYR61, and VEGFA was up-regulated in CD73+CD49b+ B cells compared to CD73−CD49b− B cells (Fig. 3B). Surface expression of CD39 as well as the VEGF receptor FLT1 was higher on CD73+CD49b+ B cells (Fig. 3C). The frequency of CD49b+ B cells was significantly increased after 3 days of in vitro stimulation of total B cells with CD40L + IL-21, whereas B cell stimulation with CD40L + IL-21 led to a reduction of CD73+ B cells (Fig. 3D).
Fig. 3. CD49b+CD73+ B cells form a distinct population of B cells and express proangiogenic cytokines.
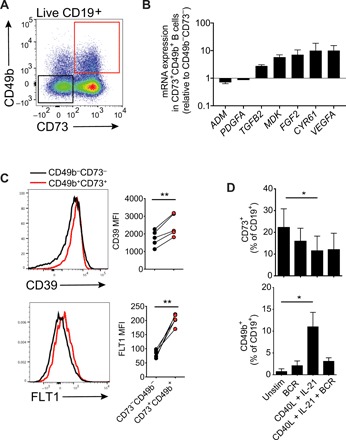
(A) Gating of CD49b+CD73+ B cells in PBMCs of healthy donor. (B) mRNA expression of proangiogenic cytokines in B cell populations sorted based on their expression of CD49b and CD73 (n = 4). (C) Flow cytometric analysis of CD39 and FLT1 expression on CD49b+CD73+ B cells stained directly ex vivo. (D) Effect of 3-day in vitro stimulation of primary B cells on the expression of CD49b and CD73 (n = 4).
Proangiogenic B cells show increased frequencies in circulation and are present in esophageal tissue of patients with EoE
To demonstrate an in vivo relevant function for proangiogenic B cells, we first investigated tissue biopsies and peripheral blood of patients with EoE. We observed a 3.2-fold increase in the median frequency of circulating CD73+CD49b+ B cells in patients with EoE compared to healthy controls (Fig. 4A). Furthermore, there was a moderate positive correlation between the frequency of circulating CD73+CD49b+ B cells and the number of eosinophils in lamina propria and stromal tissue of esophagus in patients with EoE (Fig. 4B). Moreover, transcriptional activity of several proangiogenic cytokine genes was significantly up-regulated in EoE esophageal biopsies compared to control esophageal biopsies. These included VGF, PDGFA, CYR61, FGF2, and MDK, while TGFB2, VEGFA, and ADM showed a trend toward higher expression (Fig. 4C). A subgroup of five patients with EoE showed much higher up-regulation of these cytokines. No correlation was found between the proangiogenic cytokine expression levels and the frequency of circulating CD73+CD49b+ B cells or the number of eosinophils in esophageal tissue of patients with EoE. Purified CD73+CD49b+ B cells isolated from peripheral blood of patients with EoE showed higher transcriptional levels of VEGFA, TGFB2, AMD, and FGF2 than CD73−CD49b− cells (Fig. 4D). Confocal microscopy staining of esophageal biopsies demonstrated that CD20+ B cells and/or CD138+ plasma cells were present in 50% (9 of 18) of EoE esophageal biopsies (Fig. 4E). VEGF-A+ B cells were detected in four of nine biopsies that contained B cells (Fig. 4, F and H). In seven of these nine tissues (78%), IgG4+ B cells or plasma cells were detected. Some of these B cells expressed CYR61, as well as CD49b and CD73 (Fig. 4, G and H).
Fig. 4. Proangiogenic B cell frequency is elevated in patients with EoE and correlates with esophageal eosinophil counts.
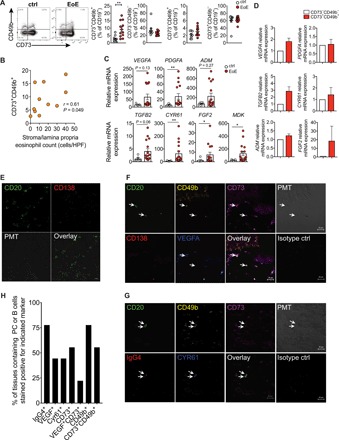
(A) Frequencies of circulating CD73+CD49b+, CD73−CD49b−, CD73−CD49b+, and CD73+CD49b− B cells in patients with EoE (n = 12) compared to healthy controls (n = 10). Dot plots show representative stainings of CD49b and CD73 among CD19+ live cells from healthy controls or patients with EoE. (B) Correlation of circulating CD73+CD49b+ B cell frequencies and stroma/lamina propria eosinophil count. HPF, high-power field. (C) mRNA expression of proangiogenic cytokines in esophageal biopsies of patients with EoE (n = 18). (D) mRNA expression of proangiogenic cytokines in purified peripheral CD73−CD49b− and CD73+CD49b+ B cells from patients with EoE (n = 4). (E) Confocal microscopy staining for CD20 and CD138 on an esophageal biopsy from a patient with EoE. (F) Confocal microscopy staining of CD20, CD49b, CD73, CD138, and VEGF on EoE esophageal biopsy. (G) Confocal microscopy staining of CD20, CD49b, CD73, IgG4, and CYR61 on EoE esophageal biopsy. (H) Frequencies of tissues stained positive for B cells expressing different markers. The percentage of tissues in which CD20+ or CD138+ cells expressing indicated markers were detected is shown (n = 9).
Proangiogenic B cells show increased frequencies in circulation and are present in tumor tissue of patients with melanoma
Because angiogenesis is essential in tumor growth, we assessed the potential expression of proangiogenic B cells in the clearly defined, easily accessible, and relatively common tumor melanoma. First, we assessed the frequency of proangiogenic B cells in peripheral blood of patients with advanced metastatic melanoma. There was a significantly increased frequency of CD73+CD49b+ B cells in the circulation of these patients compared to healthy controls (Fig. 5A). Single-cell suspensions were prepared from surgically excised metastatic melanoma lesions from three patients with melanoma, and the expression of CD73 and CD49 was analyzed using flow cytometry. The frequencies of CD73+CD49b+ B cells were generally lower among tumor-infiltrating B cells compared to circulating B cells (Fig. 5B). This may indicate loss of surface markers due to enzymatic treatment required for cell isolation from tissue samples or down-regulation of CD49b upon migration into the tissue. Alternatively, CD73+CD49b+ cells may be induced by the tumor microenvironment to migrate and accumulate in the blood.
Fig. 5. Proangiogenic B cells in patients with melanoma.
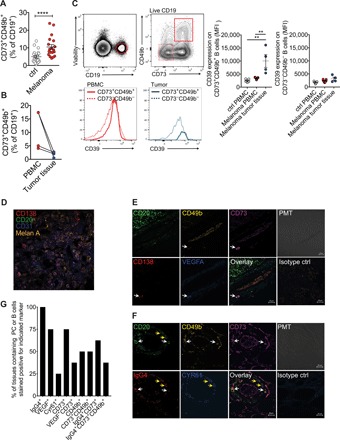
(A) Frequency of circulating CD73+CD49b+ B cells in patients with melanoma (n = 19) compared to healthy controls (n = 20). (B) Frequency of CD73+CD49b+ B cells in matched PBMC and tumor-derived single-cell suspensions. (C) Expression level of CD39 on CD73−CD49b− and CD73+CD49b+ B cells from PBMCs and tumor tissue. (D) Confocal microscopy staining for CD20, CD138, and Melan A on melanoma tumor tissue. (E) Confocal microscopy staining of CD20, CD49b, CD73, IgG4, and VEGFA on melanoma tumor tissue. Arrows indicate CD138+ plasma cells positive for CD49b, CD73, and VEGFA. (F) Confocal microscopy staining of CD20, CD49b, CD73, IgG4, and CYR61 on melanoma tumor tissue. White arrows indicate CD20+ B cells positive for CD49b, CD73, and IgG4. Yellow arrows indicate CD20+ B cells stained positive for IgG4 and CYR61. (G) Frequencies of tissues stained positive for B cells expressing different markers. The percentage of tissues in which CD20+ or CD138+ cells expressing indicated markers were detected is shown (n = 8).
The expression level of CD39 was significantly up-regulated on melanoma tumor-infiltrating CD73+CD49+ B cells compared to peripheral CD73+CD49b+ B cells from healthy controls as well as patients with melanoma (Fig. 5C). Coculture of B cells with the melanoma cell line FM55 alone did not significantly alter the expression of CD49b or CD73, while BCR stimulation down-regulated CD49b expression induced by IL-21 and CD40L in the presence or absence of FM55 (fig. S5).
Tumor-infiltrating B cells were detected in the vicinity of CD31+ blood vessels in eight of the nine analyzed tumor tissues at varying frequencies (Fig. 5D). VEGF-A+ and CD73+ B cells were detected in 75% (six of eight) of the tumor sections (Fig. 5, E and G). However, we could observe VEGF-A+CD73+ double-positive B cells only in three of these six (50%) tissues (Fig. 5, E and G). CD49b+ B cells were detected in 50% of the tissues that contained infiltrating B cells. All samples that contained CD49b+ cells also contained CD49b+CD73+ B cells (Fig. 5, E to G). IgG4+ B cells were detected in all of the tumor tissues that contained B cells (eight of nine) (Fig. 5, F and G). Of the tissues that stained positive for IgG4+ B cells, 62.5% contained IgG4+CD73+ B cells and 37.5% of the tissues that stained positive for IgG4+ B cells contained IgG4+CD73+CD49b+ B cells (Fig. 5G).
DISCUSSION
An increasing number of studies demonstrate immunoregulatory or immune effector functions of B cells that extend beyond antibody production. Here, we report a population of human B cells that is capable of producing a wide range of proangiogenic cytokines and inducing angiogenesis.
A major limitation to detailed analysis of human B cells is their limited capacity to be cultured in vitro. This limitation can be largely overcome through transduction of B cells with BCL6 and BCL-XL, resulting in the formation of immortalized B cells that can be expanded in vitro in the presence of IL-21 and CD40L for extended periods of time (23, 27). This method enables the generation and functional characterization of B cell clones. Transcriptomics analysis of IgG1- and IgG4-switched memory B cell clones revealed a distinct clustering based on the expression of cytokine genes. A cluster of IgG4+ B cell clones expressed a wide range of proangiogenic cytokines including VEGFA, PDGFA, TGFB2, CYR61, BMP2, SLIT2, ADM2, FGF2, NOV, SPP1, PDGFC, NAMPT, HDGFRP3, CCL3, TNFRSF11B, IL8, and MDK. Functional experiments demonstrated that these clones were able to promote in vitro angiogenesis.
Differential expression analysis between pro- and nonangiogenic B cell clones identified CD49b and CD73 coexpression as a potential surface marker combination to characterize circulating B cells with a proangiogenic function. CD49b (also known as α2β1 integrin) is an integrin alpha subunit that is expressed on many cell types including NK T cells, NK cells, fibroblasts, and platelets. CD49b, when coexpressed together with LAG-3, has also been identified as a marker for identification of Tr1 cells (13).
Extracellular adenosine can be generated by the ectonucleotidases CD73 and CD39. CD39 is ubiquitously expressed by B cells, whereas CD73 is expressed on a subset of human B cells, both in the peripheral blood and in the germinal centers (28, 29). We found that CD73+ B cells, independent of the expression of CD49b, expressed higher levels of CD39 than CD73− B cells. Although CD39 was not up-regulated on proangiogenic B cell clones, we observed a significantly higher expression level of CD39 in tumor-infiltrating CD73+CD49b+ B cells in patients with melanoma compared to peripheral CD73+CD49b+ B cells. This suggests that tumor-infiltrating CD73+CD49b+ B cells have an increased capacity to convert ATP to adenosine as it has been reported that CD39hi B cells produce elevated levels of adenosine (30). Under normal physiological conditions, the levels of extracellular ATP are very low because it is localized in the cytosol. However, extracellular ATP levels can increase significantly in certain conditions such as inflammation, hypoxia, and malignancy (12). This ATP can be converted to free adenosine by cells that coexpress CD73 and CD39. Free adenosine can bind to A2a and A2b receptors and thereby promote angiogenesis and fibrosis and enhance suppressive function of Tregs, Tr1 cells, and myeloid-derived suppressor cells (12, 31). Therefore, strategies targeting CD39 and CD73 are currently being investigated as potential therapeutic interventions to restore antitumor immunity (12).
Recent findings have revealed that certain B cells may promote tumor progression. Different mechanisms seem to be involved in this process. B cell–deficient mice have been reported to mount strong cytotoxic antitumor responses against D5 mouse melanoma cells and MCA304 sarcoma cells, while wild-type mice failed to control tumor growth (32). The proposed responsible mechanism was that B cell–derived IL-10 could suppress interferon-γ production by CD8+ T cells and NK cells. Breg cells producing IL-10, IL-35, and TGF-β have also been suggested to promote tumor progression though the suppression of T cell responses (1). A particular subset of plasma cells expressing IgA, IL-10, and PD-L1 has been shown to interfere with T cell–dependent immunogenic chemotherapy (33).
Angiogenesis plays a key role in physiological processes such as tissue development and wound healing. Moreover, it also plays a critical role in tumor metastasis and tissue remodeling in chronic inflammatory conditions such as EoE (6). We found an increased frequency of peripheral CD49b+CD73+ B cells in patients with melanoma and EoE. The frequencies of these cells in single-cell suspensions derived from melanoma tumor tissues were lower than in circulation. It remains unclear whether the detection of these cells is affected by the enzymatic treatment applied during the preparation of tissues or that the expression of particularly CD49b is down-regulated in the tumor microenvironment and under chronic inflammation in EoE.
There is increasing evidence that B cells can have a multitude of roles in regulating the tumor microenvironment and can have both pro- and antitumor effects (34). B cells can have a tumoricidal function primarily through the production of tumor antigen–specific immunoglobulins. In addition, B cells can act as antigen-presenting cells and contribute to survival and proliferation of tumor-infiltrating T cells (34). The fact that the proangiogenic cytokine–producing B cell clones were IgG4-switched reinforces the previously reported connection between IgG4 production and tumor-promoting capacity (21). Tumor antigen–reactive IgG4 antibody production and IgG4 irrespective of its tumor specificity were both found to impair the tumoricidal capacity of IgG1 antibodies specific for the melanoma-associated antigen, CSPG4 (chondroitin sulfate proteoglycan 4). Moreover, IgG4+ B cells were significantly increased in melanoma lesions compared to healthy skin, and serum IgG4 levels were inversely correlated with patient survival (21, 22). In addition to the findings in melanoma, IgG4 antibodies specific to culprit food proteins were found at high titers in adult and pediatric patients with EoE (18, 35). Granular deposits of IgG4 antibodies and IgG4+ plasma cells were detected in the lamina propria of patients with EoE (18). Patients with EoE also demonstrate clear signs of tissue remodeling in the esophagus including increased fibrosis, vascularity, and vascular activation (20, 36).
We have previously reported that IL-10–producing naive B cells are more prone to switch to IgG4-producing plasma cells than B cells that do not produce IL-10 (37). These cells were enriched among CD73−CD25+CD71+ B cells. Here, we report the identification of a B cell population that is characterized by the expression of IgG4, CD73, and CD49b; is capable of producing many proangiogenic cytokines; and exerts a proangiogenic function but does not produce IL-10. The expression of CD73 in these two IgG4-associated B cell populations is also linked to these functions. Namely, the CD73− fraction includes immune regulatory B cells, whereas CD73+ fraction includes proangiogenic B cells. As observed in the present study, multiple functions of memory B cells are reflected in different clusters of IgG4 memory B cells, indicating that not all of the cells that switch to IgG4 develop a proangiogenic phenotype. It would be interesting to further investigate the factors that differentiate these types of IgG4-switched B cells in further studies. IgG4+CD73+CD49b+ B cell clones produced a wide range of proangiogenic factors and facilitated endothelial tube formation. Elevated frequencies of CD73+CD49b+ B cells were detected in patients suffering from melanoma and EoE, two model diseases with a demonstrated link to IgG4 and angiogenesis. Together, our findings demonstrate a previously unidentified proangiogenic B cell subset characterized by the expression of CD49b and CD73.
MATERIALS AND METHODS
Generation and characterization of B cell clones
For the generation of B cell clones, peripheral blood mononuclear cells (PBMCs) from bee venom–exposed healthy or allergic individuals were isolated. Switched IgG+ (gated as CD19+IgM−IgA−) memory B cells were sorted using a FACSAria III (BD Biosciences, Franklin Lakes, NJ, USA) (fig. S1A). Cells were cocultured for 36 hours in IMDM (Iscove’s Modified Dulbecco’s Medium) (Gibco) [supplemented with 8% fetal calf serum (FCS) and penicillin/streptomycin] with γ-irradiated (50 gray) mouse L cell fibroblasts stably expressing CD40L (CD40L-L cells, 105 cells ml−1) cells and recombinant IL-21 (Miltenyi Biotec) and immortalized using a retroviral vector containing BCL6, BCL-XL, and GFP as described previously (23). Single memory B cells were sorted and expanded with CD40L and IL-21. With this approach, we obtained 17 IgG1 clones, 10 IgG4 clones, 3 IgG3 clones, and 2 IgG2 clones. We continued with focusing on IgG4 and IgG1 clones because we had significant numbers from these two subgroups. We were primarily interested in IgG4 clones that specifically clustered based on angiogenesis genes. To demonstrate their clonality, RNA was isolated from individual expanded B cell clones (100,000 cells per clone). RNA was isolated using RNeasy Plus Micro Kit (Qiagen, Hilden, Germany). BCR sequence analysis was performed as described elsewhere (38).
RNA sequencing and data analysis
B cell clones were stimulated with goat anti-human IgG + IgM (H + L) F(ab′) fragment (10 μg/ml; BCR stimulation) (Jackson ImmunoResearch, Cambridgeshire, UK) for 4 hours. Live GFP+ B cells were sorted using a FACSAria III (BD Biosciences, Franklin Lakes, NJ, USA). RNA was isolated using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany).
Total RNA samples (1 μg) were ribosome-depleted and then reverse-transcribed into double-stranded cDNA, with actinomycin added during first-strand synthesis. cDNA samples were fragmented, end-repaired, and polyadenylated before ligation with TruSeq adapters. The adapters contain the index for multiplexing. Fragments containing TruSeq adapters on both ends were selectively enriched by means of PCR. The quality and quantity of the enriched libraries were validated with a Qubit (1.0) Fluorometer and Bioanalyzer 2100 (Agilent). The product was a smear with an average fragment size of approximately 360 base pairs (bp). The libraries were normalized to 10 nM in 10 mM tris-Cl (pH 8.5) with 0.1% Tween 20.
A TruSeq SR Cluster Kit v4-cBot-HS or a TruSeq PE Cluster Kit v4-cBot-HS (Illumina) was used for cluster generation by using 8 pM of pooled normalized libraries on the cBOT. Sequencing was performed on an Illumina HiSeq 2500 single-end 126-bp device by using TruSeq SBS Kit v4-HS (Illumina). Raw data are available at the Gene Expression Omnibus (GEO; accession no. GSE110278).
After quality control of raw reads with FastQC (version 0.10.0; Babraham Institute, Cambridge, UK), whole transcriptome quantification was performed using RSEM (version 1.2.18) (39) using the Ensembl version 75 annotations of the GRCh37.p13 human genome assembly. Differential expression analysis between conditions was performed using edgeR likelihood ratio test method (version 3.14.0) (40) on Trimmed Mean of M values (TMM) normalized counts. The Benjamini-Hochberg procedure (41) was used to adjust p values and genes with an FDR < 0.01 and a log2 fold change > 0.5 were considered differentially expressed. For clustering, TMM normalized counts were log2-transformed and converted to row scaled z scores (41). Then, a distance matrix was calculated using Euclidean distance and hierarchical clustering was performed using the “complete linkage” clustering algorithm. Cluster analysis did not reveal a clustering that was biased on the basis of donor type or antigen specificity (fig. S2 and table S1). The set of CD (cluster of differentiation) markers was obtained from UniProt (www.uniprot.org/docs/cdlist.txt) (42), and the set of genes encoding secreted immunomodulatory proteins (including cytokines and chemokines) was obtained from the Immunology Database and Analysis Portal (ImmPort) (www.immport.org/immport-open/public/reference/genelists) (43). Heat map plots were generated with the ComplexHeatmap R/bioconductor package (44). The GEO accession number for the global gene transcriptional analysis reported here is GSE129616.
Patient selection and sample collection
Peripheral blood samples were obtained from patients with EoE, melanoma, and healthy controls. PBMCs were isolated and cryopreserved. Peripheral blood and tissue samples from patients with melanoma were obtained from the University of Zürich and the Cantonal Hospital St. Gallen Biobanks. EoE patient samples were from the Swiss Eosinophilic Esophagitis Cohort Study. Recruitment of patients, documentation of informed consent, collection of blood and tissue specimens, and experimental measurements were carried out with ethical approval from the Ethikkommission Ostschweiz (EKOS 16/079), the Ethikkommission Zürich (EK no. 647), and the Ethikkommission Nordwest- und Zentralschweiz (EKNZ 2015-388). Single-cell suspensions from resected tumor tissue from patients with melanoma were obtained using a tumor dissociation kit, human (Miltenyi Biotec, Bergisch Gladbach, Germany). Patient characteristics are listed in table S2 (melanoma) and table S3 (EoE).
Tube formation assay
HUVECs (Angioproteomie, Boston, MA, USA) were seeded at a density of 5000 cells/cm2 on a culture flask coated with Speed Coating Solution (PeloBiotech, Planegg, Germany) and grown until confluence in Endothelial Cell Growth Medium, EGM-2 (Lonza, Basel, Switzerland) containing 2% FCS (Sigma-Aldrich) and growth supplements. HUVECs were trypsinized and resuspended in B cell culture supernatants obtained from B cell clones that were stimulated and cultured for 3 days in IMDM in the presence of CD40L + IL-21 (25 ng/ml) and anti-BCR (10 μg/ml; Jackson ImmunoResearch Europe, Newmarket Suffolk, UK). As positive control, HUVECs were cultured on the matrigel in EGM-2 medium, and as negative control, cells were cultured in IMDM (Gibco) supplemented with 2% FCS and penicillin/streptomycin. HUVECs were plated at a density of 2 × 104 per well in triplicates in a 48-well plate coated with 100 μl of Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning, Wiesbaden, Germany) and incubated for 22 hours at 37°C in an atmosphere containing 5% CO2. After incubation, medium was removed and plates were washed with PBS. Formation of capillary-like structures was observed using an LSM 510 META confocal laser-scanning inverted microscope (Zeiss, Oberkochen, Germany). Phase-contrast images of a representative area were taken at ×10 magnification and processed with Photoshop CC 2017 (Adobe Systems). Tube formation was quantified using ImageJ 1.51h (Wayne Rasband, National Institute of Health, USA) and Angiogenesis Analyzer plugin (Gilles Carpentier, Universite Paris Est Creteil Val de Marne, France). The method for quantification of tube-like structures is illustrated in fig. S4.
Flow cytometry
Immunophenotyping was performed on PBMCs from patients and healthy individuals. The antibodies used for staining are listed in table S4. Zombie yellow (BioLegend, San Diego, CA) was used for dead cell exclusion. Samples were measured by multicolor flow cytometry (FACSAria III, BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using FlowJo v10.0.7 (Tree Star, Ashland, OR).
Bead suspension array
Immunoglobulin heavy-chain isotypes of B cell clones were determined by analysis of cell culture supernatants of B cell clones using the Bio-Rad Bio-Plex Pro Human Isotyping Assay (Bio-Rad, Hercules, CA).
Real-time qPCR
EoE tissue biopsies were stored in RNAlater RNA stabilization reagent (Qiagen, Hilden, Germany) and stored at −80°C. RNA was isolated from EoE esophageal biopsies using a Precellys 24 tissue homogenizer according to the manufacturer’s instructions (Bertin Technologies SAS, Montigny-le-Bretonneux, France). RNA from B cells was isolated using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany). mRNA was reverse-transcribed with the Revert Aid RT Kit (Thermo Fisher Scientific, Waltham, MA USA). Real-time qPCR was performed on a 7900HT Fast Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using the Maxima Sybr Green/Rox qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) protocol. The primers used are listed in table S5. Results were normalized to EF1a (eukaryotic translation elongation factor 1 alpha) expression for each sample. Gene expression levels, shown in arbitrary units, were calculated by subtracting EF1a cycle threshold (Ct) from the gene Ct to obtain the ΔCt value. Arbitrary units for each sample = 10,000 × (2−ΔCt).
Confocal microscopy
Paraffin-embedded melanoma sections underwent deparaffinization and rehydration (2 × 10 min in xylol, 2 × 3 min in 100% isopropanol, 2 × 3 min in 96% isopropanol, 3 min in 70% isopropanol, and 2 × 5 min in H2O), followed by antigen retrieval by boiling the sections in sodium citrate buffer [10 mM sodium citrate and 0.05% Tween 20 (Sigma-Aldrich) in PBS (pH 6)] in a pressure cooker for 4 min. The frozen EoE samples were cut with a microtome (Leica CM3050S) with a thickness of 7 μm and stored at −80°C until use. Before staining, sections were defrosted and dried by air and fixated with 4% paraformaldehyde for 10 min and washed three times with PBS for 5 min.
For both frozen and paraffin sections, nonspecific binding was blocked by incubating for 25 min with Perm/blocking buffer (1% BSA, 0.2% Triton X-100, and 10% goat serum in PBS). Then, sections were sequentially stained with the following combinations of antibodies: (I) CYR61 (followed by secondary staining with goat anti-mouse IgG-AF405), CD20 (followed by secondary staining with goat anti-rabbit IgG-AF488), CD73 (followed by secondary staining with goat anti-mouse IgG2b-AF594), IgG4-Dylight650 and CD49b-AF546, (II) CD138 (followed by secondary staining with goat anti-mouse IgG-AF405), CD20 (followed by secondary staining with goat anti-rabbit IgG-AF488, CD73 (followed by secondary staining with goat anti-mouse IgG2b-AF594), VEGF-A-Dylight633 and CD49b-AF564, (III) matching isotype controls. Details regarding these antibodies and dilutions can be found in table S6. All antibodies were incubated for 45 min at room temperature, and in between antibodies, they were washed three times for 5 min with PBS. After staining, sections were coverslipped with ProLong Diamond (Invitrogen, Carlsbad, CA, USA) and dried for 24 to 72 hours before images were taken. Images were acquired and analyzed with a ZEISS LSM780 confocal microscope and the software ZEN 2012.
Images were acquired with a Zeiss LSM 780 confocal microscope (Carl Zeiss, Oberkochen, Germany) in lambda mode with a 40× oil immersion objective with the lasers (405 to 488 nm). Full spectrum data range was used, and the used fluorophores were first added to the database as single staining; furthermore, autofluorescence of the different tissues was measured. After acquiring the images, linear unmixing was performed, with the fluorophores and the autofluorescence. Then, histogram stretching was performed to enhance the quality of the pictures. PMT pictures were made at the same spot as pictures with the laser lines to provide an overview of the tissue. From each tissue, four images of areas containing CD20+ B cells or CD138+ plasma cells were analyzed. In each tissue, the presence of IgG4+, VEGF+, Cyr61+, CD73+, VEGF+CD73+, CD49b+, and CD73+CD49b+ cells was determined. If cells expressing these markers were detected in a tissue, it was scored as positive. The percentage of positive tissues was calculated as the ratio between tissues positively stained for the markers and the total tissues analyzed.
Supplementary Material
Acknowledgments
Funding: This project was supported by the Swiss National Science Foundation (PMPDP3_151326 to E.G., PP00P3_157448 to L.F., 320030–159870 and 310030_179428 to M.A., and 32473B to A.S.), the Promedica Stiftung (1406/M and 1412/M to E.G.), the Swiss Cancer Research Foundation (KFS-4243-08-2017 to E.G.), and the Christine Kühne-Center for Allergy Research and Education (CK-CARE). Author contributions: W.v.d.V., C.A.A., and M.A. designed the study. W.v.d.V., A.G., K.J., D.V., O.F.W., B.R., U.O., M.H., B.S., M.v.S., D.H., A.W., T.K., and R.J.F.G. collected data. H.S. provided the technique and material for B cell immortalization. F.C.-G. and G.T. analyzed transcriptomics data. A.S., D.I., Y.-T.C., C.F., E.G., and L.F. collected patient data and samples. W.v.d.V., C.A.A., and M.A. drafted the manuscript with input from the other authors. All authors approved the final submitted version of this paper. Competing interests: A patent application related to this work was filed by H.S. (US2010/0113745A1 filed on 9 December 2006 by the Academic Medical Center of the University of Amsterdam). H.S. owns stock in AIMM Therapeutics. The authors declare that they have no other competing interests H.S. owns stock in AIMM Therapeutics. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The reagents needed for B cell immortalization are available through a material transfer agreement with AIMM therapeutics, which can be found on www.aimmtherapeutics.com or can be requested via email (info@aimmtherapeutics.com). The GEO accession number for the global gene transcriptional analysis reported in this paper is GSE129616.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaaz3559/DC1
REFERENCES AND NOTES
- 1.van de Veen W., Stanic B., Wirz O. F., Jansen K., Globinska A., Akdis M., Role of regulatory B cells in immune tolerance to allergens and beyond. J. Allergy Clin. Immunol. 138, 654–665 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Harris D. P., Haynes L., Sayles P. C., Duso D. K., Eaton S. M., Lepak N. M., Johnson L. L., Swain S. L., Lund F. E., Reciprocal regulation of polarized cytokine production by effector B and T cells. Nat. Immunol. 1, 475–482 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Bermejo D. A., Jackson S. W., Gorosito-Serran M., Acosta-Rodriguez E. V., Amezcua-Vesely M. C., Sather B. D., Singh A. K., Khim S., Mucci J., Liggitt D., Campetella O., Oukka M., Gruppi A., Rawlings D. J., Trypanosoma cruzi trans-sialidase initiates a program independent of the transcription factors RORγt and Ahr that leads to IL-17 production by activated B cells. Nat. Immunol. 14, 514–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jour G., Ivan D., Aung P. P., Angiogenesis in melanoma: An update with a focus on current targeted therapies. J. Clin. Pathol. 69, 472–483 (2016). [DOI] [PubMed] [Google Scholar]
- 5.De Palma M., Biziato D., Petrova T. V., Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 17, 457–474 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Nhu Q. M., Aceves S. S., Tissue remodeling in chronic eosinophilic esophageal inflammation: Parallels in asthma and therapeutic perspectives. Front. Med. 4, 128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neufeld G., Kessler O., Pro-angiogenic cytokines and their role in tumor angiogenesis. Cancer Metastasis Rev. 25, 373–385 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Babic A. M., Kireeva M. L., Kolesnikova T. V., Lau L. F., CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 95, 6355–6360 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oehler M. K., Hague S., Rees M. C., Bicknell R., Adrenomedullin promotes formation of xenografted endometrial tumors by stimulation of autocrine growth and angiogenesis. Oncogene 21, 2815–2821 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Choudhuri R., Zhang H.-T., Donnini S., Ziche M., Bicknell R., An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 57, 1814–1819 (1997). [PubMed] [Google Scholar]
- 11.Auchampach J. A., Adenosine receptors and angiogenesis. Circ. Res. 101, 1075–1077 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allard B., Longhi M. S., Robson S. C., Stagg J., The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 276, 121–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagliani N., Magnani C. F., Huber S., Gianolini M. E., Pala M., Licona-Limon P., Guo B., Herbert D. R., Bulfone A., Trentini F., Di Serio C., Bacchetta R., Andreani M., Brockmann L., Gregori S., Flavell R. A., Roncarolo M.-G., Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat. Med. 19, 739–746 (2013). [DOI] [PubMed] [Google Scholar]
- 14.van der Neut Kolfschoten M., Schuurman J., Losen M., Bleeker W. K., Martínez-Martínez P., Vermeulen E., den Bleker T. H., Wiegman L., Vink T., Aarden L. A., De Baets M. H., van de Winkel J. G., Aalberse R. C., Parren P. W., Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science 317, 1554–1557 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Davies A. M., Sutton B. J., Human IgG4: A structural perspective. Immunol. Rev. 268, 139–159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Veen W., Akdis M., Role of IgG4 in IgE-mediated allergic responses. J. Allergy Clin. Immunol. 138, 1434–1435 (2016). [DOI] [PubMed] [Google Scholar]
- 17.van de Veen W., Wirz O. F., Globinska A., Akdis M., Novel mechanisms in immune tolerance to allergens during natural allergen exposure and allergen-specific immunotherapy. Curr. Opin. Immunol. 48, 74–81 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Clayton F., Fang J. C., Gleich G. J., Lucendo A. J., Olalla J. M., Vinson L. A., Lowichik A., Chen X., Emerson L., Cox K., O’Gorman M. A., Peterson K. A., Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology 147, 602–609 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg C. E., Mingler M. K., Caldwell J. M., Collins M. H., Fulkerson P. C., Morris D. W., Mukkada V. A., Putnam P. E., Shoda T., Wen T., Rothenberg M. E., Esophageal IgG4 levels correlate with histopathologic and transcriptomic features in eosinophilic esophagitis. Allergy 73, 1892–1901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persad R., Huynh H. Q., Hao L., Ha J. R., Sergi C., Srivastava R., Persad S., Angiogenic remodeling in pediatric EoE is associated with increased levels of VEGF-A, angiogenin, IL-8, and activation of the TNF-α–NFκB pathway. J. Pediatr. Gastroenterol. Nutr. 55, 251–260 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Karagiannis P., Gilbert A. E., Josephs D. H., Ali N., Dodev T., Saul L., Correa I., Roberts L., Beddowes E., Koers A., Hobbs C., Ferreira S., Geh J. L., Healy C., Harries M., Acland K. M., Blower P. J., Mitchell T., Fear D. J., Spicer J. F., Lacy K. E., Nestle F. O., Karagiannis S. N., IgG4 subclass antibodies impair antitumor immunity in melanoma. J. Clin. Invest. 123, 1457–1474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karagiannis P., Villanova F., Josephs D. H., Correa I., Van Hemelrijck M., Hobbs C., Saul L., Egbuniwe I. U., Tosi I., Ilieva K. M., Kent E., Calonje E., Harries M., Fentiman I., Taylor-Papadimitriou J., Burchell J., Spicer J. F., Lacy K. E., Nestle F. O., Karagiannis S. N., Elevated IgG4 in patient circulation is associated with the risk of disease progression in melanoma. Oncoimmunology 4, e1032492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwakkenbos M. J., Diehl S. A., Yasuda E., Bakker A. Q., van Geelen C. M., Lukens M. V., van Bleek G. M., Widjojoatmodjo M. N., Bogers W. M., Mei H., Radbruch A., Scheeren F. A., Spits H., Beaumont T., Generation of stable monoclonal antibody–producing B cell receptor–positive human memory B cells by genetic programming. Nat. Med. 16, 123–128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito D., Kumanogoh A., The role of Sema4A in angiogenesis, immune responses, carcinogenesis, and retinal systems. Cell Adh. Migr. 10, 692–699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na H.-J., Hwang J.-Y., Lee K.-S., Choi Y. K., Choe J., Kim J.-Y., Moon H.-E., Kim K.-W., Koh G. Y., Lee H., Jeoung D., Won M.-H., Ha K.-S., Kwon Y.-G., Kim Y.-M., TRAIL negatively regulates VEGF-induced angiogenesis via caspase-8-mediated enzymatic and non-enzymatic functions. Angiogenesis 17, 179–194 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Ponce M. L., Tube formation: An in vitro matrigel angiogenesis assay. Methods Mol. Biol. 467, 183–188 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Kwakkenbos M. J., van Helden P. M., Beaumont T., Spits H., Stable long-term cultures of self-renewing B cells and their applications. Immunol. Rev. 270, 65–77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saze Z., Schuler P. J., Hong C.-S., Cheng D., Jackson E. K., Whiteside T. L., Adenosine production by human B cells and B cell–mediated suppression of activated T cells. Blood 122, 9–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schena F., Volpi S., Faliti C. E., Penco F., Santi S., Proietti M., Schenk U., Damonte G., Salis A., Bellotti M., Fais F., Tenca C., Gattorno M., Eibel H., Rizzi M., Warnatz K., Idzko M., Ayata C. K., Rakhmanov M., Galli T., Martini A., Canossa M., Grassi F., Traggiai E., Dependence of immunoglobulin class switch recombination in B cells on vesicular release of ATP and CD73 ectonucleotidase activity. Cell Rep. 3, 1824–1831 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Figueiró F., Muller L., Funk S., Jackson E. K., Battastini A. M., Whiteside T. L., Phenotypic and functional characteristics of CD39high human regulatory B cells (Breg). Oncoimmunology 5, e1082703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Z.-w., Dong K., Zhang H.-z., The roles of CD73 in cancer. Biomed. Res. Int. 2014, 460654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue S., Leitner W. W., Golding B., Scott D., Inhibitory effects of B cells on antitumor immunity. Cancer Res. 66, 7741–7747 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Shalapour S., Font-Burgada J., Di Caro G., Zhong Z., Sanchez-Lopez E., Dhar D., Willimsky G., Ammirante M., Strasner A., Hansel D. E., Jamieson C., Kane C. J., Klatte T., Birner P., Kenner L., Karin M., Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S.-s., Liu W., Ly D., Xu H., Qu L., Zhang L., Tumor-infiltrating B cells: Their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 16, 6–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuyler A. J., Wilson J. M., Tripathi A., Commins S. P., Ogbogu P. U., Kruzsewski P. G., Barnes B. H., McGowan E. C., Workman L. J., Lidholm J., Rifas-Shiman S. L., Oken E., Gold D. R., Platts-Mills T. A. E., Erwin E. A., Specific IgG4 antibodies to cow's milk proteins in pediatric patients with eosinophilic esophagitis. J. Allergy Clin. Immunol. 142, 139–148.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aceves S. S., Newbury R. O., Dohil R., Bastian J. F., Broide D. H., Esophageal remodeling in pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol. 119, 206–212 (2007). [DOI] [PubMed] [Google Scholar]
- 37.van de Veen W., Stanic B., Yaman G., Wawrzyniak M., Söllner S., Akdis D. G., Rückert B., Akdis C. A., Akdis M., IgG4 production is confined to human IL-10–producing regulatory B cells that suppress antigen-specific immune responses. J. Allergy Clin. Immunol. 131, 1204–1212 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Jackson K. J., Wang Y., Collins A. M., Human immunoglobulin classes and subclasses show variability in VDJ gene mutation levels. Immunol. Cell Biol. 92, 729–733 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A pratical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 42.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharya S., Dunn P., Thomas C. G., Smith B., Schaefer H., Chen J., Hu Z., Zalocusky K. A., Shankar R. D., Shen-Orr S. S., Thomson E., Wiser J., Butte A. J., ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 180015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaaz3559/DC1


