Complementary clues from enzyme and genome evolution reveal how a mint turned into catmint.
Abstract
Catnip or catmint (Nepeta spp.) is a flowering plant in the mint family (Lamiaceae) famed for its ability to attract cats. This phenomenon is caused by the compound nepetalactone, a volatile iridoid that also repels insects. Iridoids are present in many Lamiaceae species but were lost in the ancestor of the Nepetoideae, the subfamily containing Nepeta. Using comparative genomics, ancestral sequence reconstructions, and phylogenetic analyses, we probed the re-emergence of iridoid biosynthesis in Nepeta. The results of these investigations revealed mechanisms for the loss and subsequent re-evolution of iridoid biosynthesis in the Nepeta lineage. We present evidence for a chronology of events that led to the formation of nepetalactone biosynthesis and its metabolic gene cluster. This study provides insights into the interplay between enzyme and genome evolution in the origins, loss, and re-emergence of plant chemical diversity.
INTRODUCTION
Plants from the genus Nepeta L. are commonly known as catmint or catnip due to their ability to modify the behavior of cats. Catnip affects approximately two-thirds of domestic cats and many wild felid species including lions, tigers, and ocelots (1) and induces playful actions such as rolling over, cheek rubbing, and pawing (2). The responsible agents are nepetalactones, volatile metabolites thought to mimic cat pheromones (Fig 1A). Most likely, the adaptive function of nepetalactones in Nepeta is to protect against herbivorous insects (3), not to stimulate cats; notably, nepetalactones can repel insects with efficiencies comparable to the synthetic repellent N,N-diethyl-meta-toluamide (DEET) (4). Furthermore, Nepeta species produce different nepetalactone stereoisomers, with their ratio affecting the effectiveness of insect repellence (4).
Fig. 1. Iridoid and nepetalactone biosynthesis.
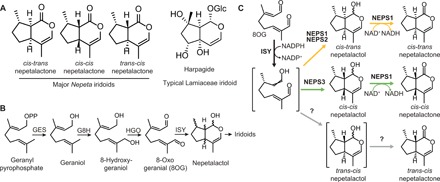
(A) Volatile nepetalactone stereoisomers found in Nepeta. Harpagide is a typical nonvolatile iridoid glucoside found in other iridoid-producing Lamiaceae. (B) Early iridoid biosynthetic pathway in plants: geraniol synthase (GES), geraniol 8-hydroxylase (G8H), 8-hydroxygeraniol oxidoreductase (HGO), and iridoid synthase (ISY). (C) Knowledge of nepetalactone biosynthesis in N. mussinii as reported in Lichman et al. (9). The crossed double bond depiction in the ISY product refers to unknown or undefined stereochemistry.
Nepetalactones are iridoids, a class of atypical monoterpenes that act as defensive compounds in some flowering plants. Iridoid biosynthesis begins with hydrolysis of geranyl pyrophosphate, catalyzed by geraniol synthase (GES), followed by oxidation, catalyzed by geraniol 8-hydroxylase (G8H) and 8-hydroxygeraniol oxidoreductase (HGO), yielding the precursor 8-oxogeranial (8OG) (Fig. 1B) (5). The first committed step into the iridoid pathway is the reductive cyclization of 8OG into nepetalactol, catalyzed by iridoid synthase (ISY) (6).
We initiated elucidation of the biosynthetic basis of the three major nepetalactone stereoisomers found in Nepeta species and determined that Nepeta ISYs are not responsible for determining nepetalactone stereochemistry (7). Instead, ISYs catalyze the reduction of 8OG but do not control the subsequent cyclization. These enzymes yield a reactive intermediate that, without partner enzymes present, spontaneously cyclizes to form a mixture of products including cis-trans-nepetalactol (8, 9).
In Nepeta mussinii, several enzymes act in combination with ISY to control the formation of nepetalactone stereoisomers (Fig. 1C) (9). Three nepetalactol-related short-chain reductase/dehydrogenases (NEPS1, NEPS2, and NEPS3) were identified. NEPS1 is a dehydrogenase, catalyzing the formation of cis-trans-nepetalactone or cis-cis-nepetalactone from the corresponding nepetalactol isomer. In contrast, NEPS2 and NEPS3 appear to act as cyclases: NEPS2 was found to promote the formation of cis-trans-nepetalactol, while NEPS3 promotes the formation of cis-cis-nepetalactol, an isomer only present in trace quantities in the uncatalyzed cyclization. The enzymes responsible for the formation of the trans-cis-nepetalactone isomers have not been identified, yet other NEPS homologs are hypothesized to play this role.
Iridoids are widespread in the asterid subgroup of flowering plants including the mint family (Lamiaceae) to which Nepeta belongs (Fig. 2A) (10). Ancestral state reconstructions indicate that iridoid biosynthesis was present in the most recent common ancestor of Lamiaceae (Fig. 2) (11). Most iridoid producers in Lamiaceae produce iridoid glycosides that act as a defense against chewing insects [e.g., harpagide; Fig. 1A] (10, 12). However, the ability to produce iridoids appears to have been lost in the ancestor of the Nepetoideae lineage (a large subclade of mints), and mono- and sesquiterpene volatiles usurped them as key defensive compounds (10). However, iridoid biosynthesis re-emerged in one genus of Nepetoideae—Nepeta, primarily in the form of the volatile nepetalactones (Fig. 2) (10). Nepeta therefore has emerged as an important model to investigate the loss and subsequent evolutionary re-emergence of a major class of defensive compounds.
Fig. 2. Lamiaceae and ISY.
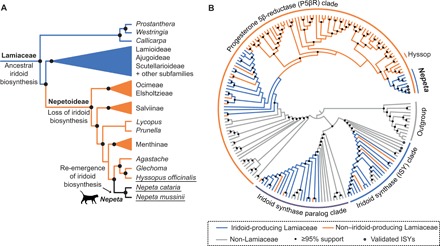
(A) Phylogenetic tree of Lamiaceae species as reported in (10); black circles, bootstrap support ≥99%. (B) Cladogram of the PRISE gene family. See fig. S15 for annotated phylogram.
To uncover how Nepeta forms nepetalactones and how it re-evolved the ability to produce iridoids, we sequenced the genomes of two iridoid-producing species [Nepeta cataria L. and Nepeta mussinii Spreng. ex. Henckel (syn. Nepeta racemosa Lam.)] along with the closely related, non–iridoid-producing hyssop (Hyssopus officinalis L.). This comparative genomic approach, combined with phylogenetic analysis and enzymology of reconstructed ancestral biosynthetic enzymes, provides experimental evidence for a sequence of molecular and genomic events that led to the re-emergence of iridoids and the evolution of novel chemistry.
RESULTS
The early iridoid pathway in Nepeta and hyssop
The metabolite profiles of N. cataria, N. mussinii, and H. officinalis were analyzed to determine iridoid content (figs. S1 to S7). The major volatile components of Nepeta leaves and flowers are nepetalactones, which are absent from hyssop tissues. We also tentatively identified soluble iridoid glycosides (e.g., 1,5,9-epi-deoxyloganic acid) in Nepeta tissues, but there was no evidence of iridoid glycosides in hyssop tissues.
Next, genome assemblies of the three species (N. cataria, N. mussinii, and H. officinalis) were generated (tables S1 to S4). The presence of highly conserved gene copies in N. cataria indicated the presence of multiple duplicated genes, consistent with a tetraploid genome, in contrast with hyssop and N. mussinii which are diploids (table S3). Gene candidates (GES, G8H, and HGO; Fig. 1C) encoding biosynthetic enzymes that yield the iridoid precursor 8OG were identified by similarity to the previously characterized sequences from the nonmint Catharanthus roseus (L.) G. Don of Apocynaceae, producer of the iridoid-derived anticancer compound vinblastine (figs. S8 to S10 and table S5) (5), and through tissue-specific coexpression patterns (figs. S11 to S13). The encoded enzyme candidates were recombinantly produced and analyzed for activity in vitro (fig. S14). Candidate enzymes from both Nepeta species demonstrated expected activities. We were also able to detect activity for the G8H candidate from hyssop, while GES and HGO candidates could not be heterologously produced with sufficient quantity or purity for biochemical assay. Thus, their activities remain uncharacterized. Notably, GES, G8H, and HGO genes in hyssop were lowly expressed in all tissues, which may account for the absence of iridoids in hyssop (fig. S13).
Geraniol is detected in the roots of hyssop (fig. S3). However, in hyssop, the GES candidate gene is not expressed in roots and its transcripts are only present in flowers, albeit at very low levels (fig. S13). The origin of this geraniol is therefore unclear. It may arise from a pathway that does not require GES, such as the Nudix pathway found in rose flowers (13).
ISY, which acts after HGO (Fig. 1C), catalyzes the first committed step in all known plant iridoid pathways (6, 7). ISY belongs to the PRISE (progesterone 5β-reductase/ISY) enzyme family (14). Hyssop does not contain a copy of ISY; no Nepetoideae transcriptomes or genomes contain genes that phylogenetically group with previously characterized ISYs (Fig. 2B and fig. S15). In contrast, all iridoid-producing Lamiaceae species outside of Nepetoideae have a putative ISY gene in this clade. This suggests that the loss of iridoid biosynthesis in the entire Nepetoideae is due, in part, to the loss of the ISY gene. Although ISY genes were identified in Nepeta, they are not orthologous with other previously characterized ISYs but instead are present in a separate clade of putative progesterone 5β-reductase orthologs (P5βRs; Fig. 2B). This finding strongly suggests that Nepeta regained iridoid biosynthesis through the parallel evolution of a novel ISY enzyme.
Identification of nepetalactone gene clusters in Nepeta
Having identified the major components of the iridoid pathway in Nepeta and hyssop, we used our three genome assemblies to analyze the genomic organization of these genes. Within the Nepeta genomes, iridoid biosynthetic enzymes are organized in syntenic gene clusters containing ISY, NEPS homologs, and major latex protein–like genes (MLPL) (Fig. 3A). Clusters of functionally related nonhomologous genes are a feature of a subset of plant metabolic pathways, although the exact role of these genomic clusters is currently unclear (15). Two gene clusters were identified in N. cataria and one in N. mussinii. As N. cataria is a tetraploid and N. mussinii a diploid, this distribution indicates that the cluster formation predated the emergence of either species. The cluster in N. mussinii also features GES, which is absent from both N. cataria clusters.
Fig. 3. Metabolic gene clusters in Nepeta.
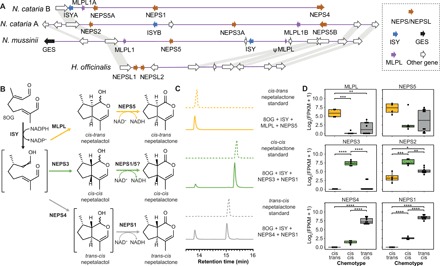
(A) Genomic organization and syntenic relationships of nepetalactone biosynthesis gene cluster in Nepeta. Gray polygons show syntenic relationships. (B) Biosynthetic pathway of nepetalactone stereoisomer formation based on combination of in vitro assay data and in planta gene expression. The crossed double bond depiction in the ISY product refers to unknown or undefined stereochemistry. (C) In vitro multienzyme cascade assays demonstrating selective formation of nepetalactone stereoisomers (see also figs. S16 to S20). (D) Differential expression of nepetalactone biosynthetic genes in N. mussinii individuals with distinct nepetalactone stereo-chemotypes. Significant differences determined by analysis of variation with Tukey’s post hoc test (n = 4 to 10; ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05) (see also fig. S21).
Although the core ISY, NEPS, and MLPL gene content is conserved, the gene order, orientation, and NEPS content differ between the three clusters. However, it is unclear whether this reduced local synteny is unique to this region or reflects a genome-wide trend. In the syntenic location in the hyssop genome, a NEPS-like gene is present but no PRISE/ISY or GES gene was identified. Gene expression analyses in N. cataria and N. mussinii revealed that clustered and nonclustered iridoid biosynthetic genes, including the previously unreported MLPL and NEPS homologs (NEPS4 and NEPS5), are coordinately expressed across different tissues (figs. S11 and S12).
Elucidation of nepetalactone biosynthesis pathway
The enzymes encoded by the ISY, NEPS, and MLP-like (MLPL) genes were biochemically characterized. Using different combinations of NEPSs and MLPL in the presence of ISY and 8OG led to the formation of the three distinct nepetalactone stereoisomers, indicating that NEPS and MLPL work in combination with ISY to control the stereochemical mode of 8OG cyclization (Table 1; Fig. 3, B and C; and figs. S16 to S20).
Table 1. NEPS and MLPL in vitro activities.
Summary of activities observed in in vitro assays with ISY and 8OG (Fig. 3 and figs. S16 to S20). Cyclisation refers to formation of nepetalactol isomers from the reactive ISY product; dehydrogenation refers to formation of nepetalactone isomers from corresponding nepetalactols. Ticks (✓), activity observed; question mark (?), possible activity, not verified; tilde (~), trace activity; double tick (✓✓), in vitro activity supported by in planta gene expression profiling data (only tested for N. mussinii).
| Enzyme | Cyclization | Dehydrogenation | |||||
| cis-trans | cis-cis | trans-cis | cis-trans | cis-cis | trans-cis | ||
| Nm | NEPS1 | ? | ✓ | ✓ | ✓✓ | ||
| Nc | NEPS1 | ? | ✓ | ✓ | ✓ | ||
| Nm | NEPS2 | ||||||
| Nc | NEPS2 | ||||||
| Nm | NEPS3 | ✓✓ | ~ | ||||
| Nc | NEPS3A | ✓ | ✓ | ||||
| Nm | NEPS3B | ✓ | ~ | ||||
| Nm | NEPS4 | ✓✓ | ✓ | ? | ~ | ||
| Nc | NEPS4 | ✓ | ✓ | ? | ~ | ||
| Nm | NEPS5 | ? | ✓✓ | ✓ | ✓ | ||
| Nc | NEPS5 | ? | ✓ | ✓ | ✓ | ||
| Nm | MLPL | ✓✓ | |||||
| Nc | MLPLA | ✓ | |||||
| Nc | MLPLB | ✓ | |||||
The previously observed cis-cis cyclase activity of NmNEPS3 was also obtained here, together with the characterization of two new NEPS3 enzymes from N. cataria (9). As expected, both NcNEPS3A and NcNEPS3B formed cis-cis-nepetalactol (Table 1 and fig. S18). While NmNEPS3 and NcNEPS3B do form trace quantities of cis-cis-nepetalactone, NcNEPS3A produced it in greater amounts (fig. S18). This indicates that it has comparatively higher dehydrogenase activity, forming cis-cis-nepetalactone from cis-cis-nepetalactol.
The previously unreported NEPS4 appears to be the missing trans-cis-cyclase, and NEPS1 acts as the partner dehydrogenase. This is shown for enzymes from N. cataria and N. mussinii (fig. S19). Incubation of only NmNEPS4 or NcNEPS4 with ISY, but not the other NEPS, causes the formation of trans-cis-iridodials and cis-trans-nepetalactone. When NEPS1, a dehydrogenase, is added, trans-cis-nepetalactone formation occurs. These results indicate that NEPS4 can cyclize the reactive ISY product into trans-cis-nepetalactol. This intermediate is released from the NEPS4 active site. However, it is unstable and, in the absence of a dehydrogenase, it will decay to trans-cis-iridodial (16). We have previously shown that trans-cis-iridodial is not a substrate for NEPS1 (9). Therefore, when NEPS1 is present, it must be taking up the reactive trans-cis-nepetalactol before its decay and oxidizing it into the stable trans-cis-nepetalactone. In this extraordinary sequence of reactions, there are two unstable intermediates moving between active sites. NEPS4 also has residual dehydrogenase activity, although this activity appears to be selective for cis-trans-nepetalactol over trans-cis-nepetalactol. This suggests that NEPS4 has separate binding modes for its cyclization and dehydrogenation activities.
We previously demonstrated that NEPS1 could catalyze the dehydrogenation of cis-trans-nepetalactol and cis-cis-nepetalactol (9). Here, we validate these results with NmNEPS1 and NcNEPS1 (Table 1 and figs. S18 and S20). We also report the additional NEPS1 dehydrogenase activity on trans-cis-nepetalactol when tested with NEPS4 (Table 1 and fig. S19). The newly reported NEPS5s, from both N. cataria and N. mussinii, are dehydrogenases with similar behavior to NEPS1 (Table 1 and figs. S18 and S20). They appear to catalyze formation of cis-trans-, cis-cis- and trans-cis-nepetalactones, although the latter two only in conjunction with NEPS3 or NEPS4 cyclases, respectively.
Our previous observation of NmNEPS2 promoting the cis-trans-nepetalactol formation at the expense of side products was not confirmed here with NmNEPS2 and NcNEPS2. Instead, we did not detect any specific notable activity for NEPS2, either alone or in conjunction with other enzymes.
However, we have identified additional enzymes that appear to promote formation of cis-trans-nepetalactol when tested in conjunction with ISY. This appears to be not a NEPS but the MLPL proteins found in the gene clusters (fig. S17A). Incubation of NmMLPL with ISY caused a significant increase in cis-trans-nepetalactol content compared to an ISY alone or a BSA control (fig. S17C). NcMLPLA and NcMLPLB appear to have similar behaviors. Furthermore, addition of MLPL to ISY and NEPS5 reactions reduces the formation of minor cis-cis- and trans-cis-nepetalactone isomers (fig. S20B). MLPLs are part of the PR-10 family of proteins; three enzymes from this family were reported to be involved in benzylisoquinoline alkaloid biosynthesis (17). They typically catalyze energetically undemanding reactions that have appreciable “spontaneous” rates in buffer or solvent. We expect that the addition of MLPL will enhance the efficiency of recombinant systems which use ISY to produce cis-trans-nepetalactol (18).
The NEPS and MLPL enzymes that catalyze the formation of nepetalactone isomers in Nepeta are found to exhibit both multiple and overlapping activities in vitro (Table 1). To determine the contribution of these enzymes to nepetalactone isomers in planta, we compared gene expression of NEPSs and MLPL genes across accessions of N. mussinii with distinct nepetalactone stereo-chemotypes (Fig. 3D, fig. S21, and table S6). The expression levels of the NEPS and MLPL genes largely correlated with in planta chemistry and were concordant with in vitro activities, suggesting that NEPS and MLPL are responsible for all three nepetalactone stereoisomers observed in N. cataria and N. mussinii.
Cyclase in planta expression patterns were correlated with in vitro activities: MLPL, NEPS3, and NEPS4 expression were highest in plants primarily producing cis-trans-, cis-cis-, and trans-cis-nepetalactone, respectively (Fig. 3D). Despite multiple activities in vitro, expression of NEPS1 appeared to be associated primarily with trans-cis-nepetalactone production. NEPS5 expression was high in cis-trans-nepetalactone producers, suggesting that it is the key cis-trans dehydrogenase. However, there was also high NEPS5 expression in a portion of cis-cis- and trans-cis-producers, suggesting that it might not be dedicated to cis-trans-nepetalactone production. NEPS2 was expressed in all plants but was highest in cis-cis-nepetalactone producers. This may indicate that it has an as-yet unknown role in this pathway. Curiously, no dehydrogenase was associated with cis-cis-nepetalactone production. It is possible that NEPS1 and NEPS5, both expressed at low levels in cis-cis-nepetalactone producers, share responsibility for the cis-cis dehydrogenation step. However, the role of NEPS2 and the missing dedicated cis-cis dehydrogenase remain two unresolved aspects of the nepetalactone biosynthetic pathway.
Evolution of ISY in Nepeta
With phylogenetic, genomic, and enzymatic information in-hand, we developed a model of iridoid evolution in Nepeta that integrates enzyme and genome evolution. One model for the emergence of metabolic novelty involves an ancestral enzyme with a minor side activity; following gene duplication, this side activity is optimized in one paralog, yielding a novel enzyme with a dedicated biosynthetic role (19, 20). ISY is a member of the PRISE family, and N. cataria and N. mussinii contain three different types of PRISE homologs: ISYs, P5βRs, and intermediate sequences that we named secondary ISYs (SISYs) (Fig. 4, A and B, and fig. S22). NmSISY is full length and has low expression levels (fig. S12), while NcSISYA and NcSISYB are silent, fragmented pseudogenes. ISYs are located in the aforementioned gene cluster (hereafter, NEPS locus), whereas P5βRs and SISYs are found as tandem duplicates in a different genomic region (P5βR locus) (Fig. 4A and table S7).
Fig. 4. Evolution of ISY in Nepeta.
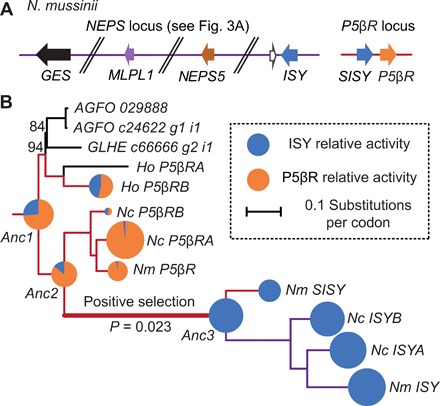
(A) Genomic location of three PRISE homologs in N. mussinii. Similar genome structures are present in N. cataria (table S7 and Fig. 3A). (B) Phylogram of PRISEs from Nepetinae (subtribe containing Nepeta). The highly diverged pseudogenes NcSISYA and NcSISYB were excluded from the phylogenetic inference to improve model support. Pie charts show in vitro enzyme activities of extant PRISEs from hyssop and Nepeta and reconstructed ancestral PRISEs. For enzyme activities, the area of each chart sector is proportional to relative activity; total chart area is proportional to the sum of relative activities. HoP5βRA failed to express in sufficient quantity to assay. All branches have >99% support unless noted, see fig. S22 for alignment of sequences and fig. S23 for annotated phylogram. Ho, H. officinalis; Nm, N. mussinii; Nc, N. cataria; AGFO, Agastache foeniculum; GLHE, G. hederacea.
To examine how the novel ISY gene emerged from a P5βR clade in Nepeta, we used ancestral sequence reconstruction (ASR) to infer the sequences of PRISE genes present at key events in the evolution of the Nepeta lineage. ASR has been previously used within the context of plant natural product biosynthesis to investigate the evolution of methyltransferases from promiscuous ancestors (21) and chalcone isomerase from a nonenzymatic ancestor (22). We targeted three key nodes for investigation: Nepetinae (Anc1), Nepeta (Anc2), and the SISY-ISY duplication (Anc3). We used a single reconstruction at each node. These were well supported, with 95, 87, and 80% of codons having a posterior probably above 0.95 for Anc1, Anc2, and Anc3, respectively. Anc2 has six codons with ambiguous amino acid assignments, where the posterior probably of the most likely codon is below 0.6 and the next most likely codon encodes a different amino acid. Anc3 has just one of these ambiguous positions, and Anc1 has none.
We measured the catalytic activity of extant and predicted ancestral PRISEs for reduction of progesterone (a putative substrate of P5βRs) (23) and 8OG, the substrate of ISY (Fig. 4B, fig. S24, and table S8). Predicted ancestral PRISEs in Nepetinae (Anc1) and Nepeta (Anc2) were capable of catalyzing both reactions. Anc3, in contrast, demonstrated no detectable P5βR activity and a significant increase in ISY activity relative to Anc2.
Furthermore, we analyzed the PRISE tree to detect any branches in which genes may have been under positive selection. Selection can be detected by comparing ratios of nonsynonymous (dN) to synonymous mutations (dS) at each codon. Of the five branches investigated (fig. S23 and table S9), positive selection was only detected along the branch from Anc2 to Anc3. Along this branch, 5 of 376 total codons were found to be under selection (dN/dS > 1). Residues identified with particularly significant indications of selection include a lysine to phenylalanine transition (NcISYA position 150, P = 0.999), an aspartic acid to lysine transition (NcISYA position 177, P = 0.944) and a case where a serine residue had switched codons, via a threonine or cytosine (NcISYA position 86, P = 0.988). Recent analysis of the serine codon switching phenomenon indicates that it is driven by selection (24).
Overall, these ancestral reconstructions and positive selection analyses support the model of a gene duplication of a PRISE ancestor with minor ISY side activity, followed by selective pressure to form a dedicated iridoid biosynthetic enzyme. While this hypothesis fits a standard model of enzyme evolution through duplication of promiscuous ancestors, we acknowledge that further work is required to unravel the molecular basis for this transition. More reconstructions at each node would provide greater confidence in the quantitative differences between ancestral activities. Furthermore, mutagenesis experiments would be useful to validate predictions made by both the ASR and positive selection analyses and would, therefore, provide more insight into the evolution of ISY in Nepeta.
Assembly of a nepetalactone gene cluster
In addition to the re-evolution of ISY from a PRISE ancestor, the crucial innovation required for nepetalactone biosynthesis was the NEPS enzymes, which act in partnership with ISY to control the profile of stereoisomers formed from the cyclization reaction. These enzymes, despite having diverse cyclase and dehydrogenase activities (Table 1 and Fig. 3), form a single clade that is unique to the Nepeta lineage (fig. S25). We compared the evolution and diversification of NEPS with the emergence of ISY activity by generating chronograms of PRISEs and NEPSs (Fig. 5A and figs. S26 to S28).
Fig. 5. Enzyme and genome evolution in Nepeta iridoid biosynthesis.
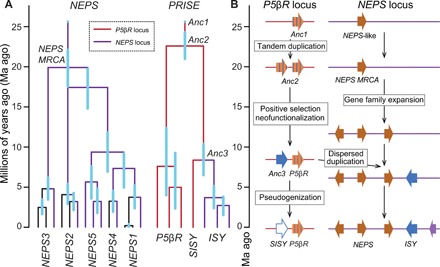
(A) Selected Nepeta clades from chronograms of NEPS and PRISE genes. Blue bars are 95% confidence intervals. Colored branches represent genomic location. Complete chronograms can be found in figs. S27 and S28. (B) Proposed chronology of events in Nepeta nepetalactone biosynthesis evolution.
To account for changes in mutation rates across branches, such as those caused by changing selective pressure, we used a relaxed clock model within a penalized likelihood (PL) framework, which enables each branch to have a separate rate. We also constrained both chronograms at three nodes, including Nepetinae (i.e., the common ancestor of hyssop and Nepeta), to gain more accurate divergence time estimates at our nodes of interest.
These chronograms reveal that the most recent common ancestor of the NEPS genes (22 to 18 Ma ago) was present at the time of, or just after, the Anc2 duplication event (24 to 21 Ma ago). Furthermore, key diversification events of the NEPS genes were concurrent with the selection and evolution of ISY activity (23 to 9 Ma ago). This timing supports a model where ISY and NEPS catalytic activities coevolved.
It is possible to infer the genomic location of the ancestral PRISE genes and thereby address whether gene clustering preceded or followed evolution of nepetalactone biosynthesis. Given the location of extant SISYs and P5βRs at the P5βR locus (Fig. 4A and table S7), we can infer that PRISE ancestors Anc1, Anc2, and Anc3 were also located in this locus (Fig. 5A). Approximately 23 Ma ago, Anc2 at the P5βR locus underwent tandem duplication and neofunctionalization; one descendant maintained P5βR activity, leading to extant P5βRs, while the other acquired ISY functionality through positive selection leading to Anc3 (Fig. 5B). Concurrent with this Anc2 duplication event, within the NEPS locus, the NEPS common ancestor underwent a series of gene duplications. The ISY-like Anc3 then duplicated into the NEPS locus via a dispersed duplication (e.g., transposition or translocation, 9 Ma ago). The original copy of Anc3 at the P5βR locus became redundant, leading to reduction of expression and eventual pseudogenization, as observed in the form of SISY in current N. mussinii and N. cataria genomes. Thus, evolution of ISY activity occurred before the formation of the gene cluster (Fig. 5B).
The speciation of both N. mussinii and N. cataria occurred after gene cluster formation (i.e., the movement of ISY into the NEPS locus), as supported by the presence of the gene clusters and evidence of tandem SISY/P5βR in the N. mussinii genome and both N. cataria subgenomes, indicating a common ancestor with these features. A comparison of species and gene divergence times also supports a chronology where speciation (~5.5 Ma ago) (fig. S26) succeeded the ISY clustering (~9 Ma ago) (Fig. 5A).
The location of GES also supports a chronology where evolution of enzyme activity precedes gene cluster formation. In N. mussinii, the functional GES is clustered with ISY and NEPS in the NEPS locus, while a pseudogenized GES is present at a different locus (table S10). The equivalent syntenic loci in N. cataria contain functional GESs, suggesting that in the N. mussinii lineage (i.e., after speciation of N. cataria and N. mussinii), a functional GES moved into the NEPS/ISY gene cluster.
Through a simultaneous examination of enzyme and genome evolution, it is possible to provide insight into the formation and divergence of plant metabolic gene clusters. In particular, this study of Nepeta iridoid clusters highlights how biosynthetic genes were recruited into a gene cluster after they gained function. This chronology of clustering following functionalization has previously been proposed in the evolution of other plant metabolic gene clusters (25, 26). In the Nepeta genomes, the presence of the pseudogenized SISYs and GES provides clear experimental evidence supporting this mechanism of cluster formation.
DISCUSSION
The evolution of nepetalactone biosynthesis in Nepeta is not simply a re-emergence of iridoid biosynthesis in this lineage; it also represents an evolutionary innovation. Nepetalactone is an example of repeated evolution with a twist; while nepetalactones maintain the iridoid structure, there are key differences compared to other iridoids found in the mint family. Most notably, NEPS1 and NEPS5 oxidize the lactol of the ISY product to form lactone, while iridoids in the rest of the mint family are glucosylated at this position (Fig. 1A). Thus, nepetalactone iridoids in Nepeta function as volatiles rather than the two-component defense role adopted by glycosylated iridoids (12). In addition, while the rest of the Lamiaceae produce only a single iridoid scaffold stereoisomer, NEPS3 and NEPS4 allow Nepeta to produce different ratios of nepetalactone stereoisomers, which results in changes to the insect repellent properties (4). Many non-iridoid volatile monoterpenes (such as 3-pinanone produced in hyssop) demonstrate general insect repellent properties (27). Nepetalactone is produced de novo by aphids as a sex pheromone (28); thus, its production by Nepeta may mediate as-yet unknown interactions with aphids (29). Such a scenario could account for the strong selective pressure required for the evolution of nepetalactone biosynthesis, which represents an important future topic of research.
Nepeta also produces glycosylated iridoids including 1,5,9-epideoxyloganic acid and nepetaside (figs. S5 and S6). While it is unclear when these iridoids emerged in Nepeta relative to the nepetalactones, ISY activity is required for their formation. Biosynthesis of these Nepeta-specific iridoid glycosides may also require the activity of NEPS: Nepetaside has a lactone moiety, which points to dehydrogenase activity, and 1,5,9-epideoxyloganic acid has cis-cis stereochemistry, indicating the involvement of a cyclase. This suggests that Nepeta iridoid glycoside biosynthesis requires both ISY and NEPS and therefore may have arisen during, or soon after, the emergence of nepetalactones. Further elucidation of glycoside biosynthesis in Nepeta is required to test this hypothesis.
The biosynthesis of the iridoid cat attractant nepetalactone in Nepeta provides a rich system for investigation of the dynamics of metabolic and chemical defense evolution. The loss of iridoid biosynthesis in the early evolution of the mint subclade Nepetoideae appears to be due to the interplay of regulatory changes (reductions in expression of upstream genes) and gene loss (deletion of ISY). Its re-emergence in Nepeta appears to have been driven by the coevolution of ISY and NEPS, with chronograms that date the divergences of PRISEs and NEPSs genes consistent with the timing required for this model. The presence of pseudogenes, in combination with phylogenetic analyses, ASR, and selection analyses together provide a molecular snapshot indicating that the evolution of iridoid biosynthesis in Nepeta preceded the formation of the iridoid gene cluster. This proposed chronology suggests that gene clusters are not the cradle of plant metabolic novelty but may instead emerge subsequent to enzyme evolution while the nascent metabolism is under strong positive selection. This study highlights the complementarity of enzyme and genome evolution in elucidating the emergence of novel chemistry in plants.
MATERIALS AND METHODS
Tissue metabolite analysis by GC-MS
For metabolite analysis of tissues by gas chromatography–mass spectrometry (GC-MS), two approaches were taken. Internal pools of volatiles were analyzed after their extraction from a tissue by dichloromethane as described previously (10). Compound identification was achieved using electron impact (EI) spectra, picking best-matched compounds in the National Institute of Standards and Technology Standard Reference Database EI library. Identification of nepetalactone isomers in Nepeta tissues was performed as previously described (7). The proportions of nepetalactone isomers present was determined by identifying nepetalactone peaks using verified standards and then dividing isomer peak area by total nepetalactone peak area.
Extraction and analysis of iridoid glycosides by liquid chromatography–MS
Frozen tissues were homogenized using a TissueLyser II (Qiagen, UK), and 50 mg of the homogenate was suspended in 1 ml of methanol. Samples were extracted by vortex for 2 min followed by ultrasonication in a bath at room temperature for 5 min. For each tissue, three extracts were collected from the same biological tissue. The extracts were centrifuged and diluted with aqueous formic acid (0.1%, v/v and 1:5, v/v). Last, the supernatant was filtered through a hydrophilic polytetrafluoroethylene (PTFE) 0.22-μm membrane (Merck, Feltham, UK) for nontargeted tandem mass analysis using high-resolution MS.
Shimadzu IT-ToF-MS (ion-trap time-of-flight mass spectrometer) was used to analyze the extracts. Identification and chromatographic separation of the compounds on this instrument were performed on a Shimadzu Nexera X2 UPLC system (Shimadzu Corp., Kyoto, Japan). Each sample (4-μl injection) was eluted through a Kinetex XB-C18 2.6-μm column (100 mm by 2.1 mm, 100 Å; Phenomenex, UK) at a column temperature of 40°C and a flow rate of 0.5 ml min−1. The mobile phase consisted of (A) aqueous formic acid (0.1%, v/v) and (B) acetonitrile. A linear gradient used was as follows: 0 to 10 min, 26% B; 10 to 11 min, 26 to 95% B; 11 to 12 min, 95 to 2% B; 12 to 15 min, 2% B. Optimized MS conditions were as follows: negative ion mode; negative electrospray voltage, −3.5 kV; CDL (curve desolvation line) temperature, 250°C; block heater temperature, 300°C; nebulizing gas (N2) flow, 1.5 liter min−1; drying gas (N2) pressure, 100 kPa. Full-scan MS (mass spectrum) readings were acquired in the mass/charge ratio (m/z) range of 100 to 1200 for MS and m/z 100 to 1000 for MS2. The MS2 data were collected in an automatic data-dependent manner using collision-induced dissociation of the most abundant singly charged species in a scan, with an exclusion time of 0.8 s. Argon was used as the collision gas, and the collision energy was set at 50%. Before data acquisition, the instrument was calibrated with sodium trifluoroacetic acid clusters (CF3CO2Na) against the entire mass range (m/z 100 to 1200 Da) specified for the instrument. Data acquisition and processing were performed using LCMSsolution (version 3.80, Shimadzu Corp., Kyoto, Japan).
The occurrence of iridoid glucosides was tentatively confirmed by qualitative analysis using a combined set of criteria. This included retention time (chromatographic separation), accurate mass spectra, tandem mass [loss of hexose sugar: Δ162 and other characteristic iridoid fragments (30)], and photodiode array. All major chromatography peaks with a signal-to-noise ratio of 3:1 or higher that could be recognized as iridoid glucoside/s were taken into consideration. The results were compared with previous reports for isolation and spectral identification of iridoid glucosides from the studied species (31).
Genome sequencing, assembly, and annotation
DNA was extracted from young leaves of N. cataria, N. mussinii, and H. officinalis using the Qiagen DNeasy Plant Mini Kit or with a modified cetyl trimethylammonium bromide (CTAB) method (32). For each species, multiple Illumina-compatible paired-end libraries and mate-pair libraries were constructed and sequenced on the Illumina platform as previously described (tables S1 and S2) (33). Paired-end reads were cleaned by removing adapters and low-quality sequences using Cutadapt (34), while mate-pair libraries were processed using NextClip (35). ALLPATHS-LG (v52488) (36) was used to generate an assembly of each genome. For H. officinalis, chromosome-scale assemblies were generated using Proximo Hi-C scaffolding as described previously (37), and gaps were filled using PBJelly (v15.8.24) (38) with reads greater than 1 kb. The gap-filled pseudomolecules were error-corrected using the Illumina whole-genome shotgun sequencing reads with Pilon (v1.22) (39). Genome assembly quality was assessed using Benchmarking Universal Single-Copy Orthologs v3.0.2 (40); final metrics of the three assemblies including quality assessment results demonstrate high-quality assemblies for these three species (table S3).
Genomes were annotated for repetitive sequences and protein-coding genes as described previously (41). Briefly, custom species-specific repeat libraries were constructed using RepeatModeler (v1.0.8; http://repeatmasker.org/), and putative protein-coding genes were removed using ProtExcluder (v1.1) (42). RepeatMasker (v4.0.6) was used with the custom species-specific repeat libraries in addition to Viridiplantae RepBase (43) repeats to mask the cognate genome. To provide transcript evidence for annotation, N. cataria, N. mussinii, and H. officinalis were grown in growth chambers until plants reached maturity. Diverse tissues (closed flower buds, open flowers, immature leaf, mature leaf, petiole, root, and stem) were collected from each species. Total RNA was isolated, treated with TURBO DNA-free DNase (Thermo Fisher Scientific, Waltham, MA), libraries were constructed using the Illumina TruSeq Stranded mRNA Library Preparation Kit (Illumina, CA) and sequenced on the Illumina platform generating paired-end 150-nt reads. Reads were cleaned using Cutadapt (v1.15) (34), aligned to the cognate genome using TopHat2 (v2.1.1) (44), and genome-guided transcript assemblies were generated using Trinity (v2.6.6) (45). Species-specific genome-guided leaf RNA-sequencing (RNA-seq) assemblies were used to train Augustus (v3.1) (46). Gene structures were improved using additional genome-guided transcript assemblies using PASA2 (v2.0.2; https://github.com/PASApipeline) (47) generating a set of working models. Expression abundances [fragments per kb exon model per million mapped reads (FPKM)] were determined from the TopHat2 alignments using Cufflinks2 (v2.2.1; table S4) (48). Functional annotation of the gene models was determined using BLAST search results against the predicted Arabidopsis thaliana proteome (TAIR10; Arabidopsis.org), Swiss-Prot entries (release 2015_08), and Pfam search results were generated using HMMER v3.1b2 (49) with a cutoff of 1 × 10−5. From the working gene models, a high confidence gene set was identified on the basis of expression in any RNA-seq library greater than 0 FPKM or that had a match to a Pfam domain. Partial gene models and models with matches to transposable element-related PFAM domains were excluded from the high confidence model set.
Identification of syntenic loci
All-versus-all BLASTP of the predicted proteins was performed with BLAST+ (v2.7.1) with an E-value cutoff of 1 × 10−5 and a maximum of five alignments reported. MCScanX (git commit 7b61f32) (50) was run with the default parameters to generate putative collinear blocks that were refined with manual curation using the BLASTP results.
N. mussinii diversity panel
Different accessions (genotypes) of N. mussinii were obtained from Herbal Haven (Saffron Walden, UK) and maintained in glasshouses. To determine the chemotype of each accession, plants were cut back, and after 2 weeks, young leaves were isolated. Three samples, each from different branches, were taken from each plant. Leaf tissue (approximately 100 mg) was frozen in liquid nitrogen, homogenized with a ball mill with a tungsten bead (27 Hz, 30 s, two repetitions) and then split for parallel RNA isolation and metabolite profiling.
Nepetalactone stereoisomer content was analyzed by GC-MS as described previously (7). In all samples, there was a single isomer accounting for >60% of nepetalactone content, and this major isomer was used to define the sample chemotype.
Total RNA was extracted using a standard CTAB protocol (51), and RNA-seq libraries constructed and sequenced as described above with the exception that single-end 50-nt reads were generated (table S6). Reads were cleaned using Cutadapt (v1.14) (34) and aligned using TopHat2 (v2.1.1) (44) to the N. mussinii reference genome appended with the previously reported NmNEPS3 sequence (9); expression abundances were calculated using Cufflinks (v2.2.1) (48). To provide comparable expression abundances from the gene expression atlas of the reference genotype used for assembly and annotation, 50 nt from read 1 of the N. mussinii reference gene expression atlas paired-end reads (see above) was clipped using Cutadapt (34) and processed in parallel with the N. mussinii diversity panel libraries.
Gene expression analysis
Unless otherwise stated, the expression matrix in FPKM was log-transformed using the formula Log2(FPKM + 1), and analyses were made using the stats library in the R platform (52). For examination of N. mussinii diversity panel, analysis of variance models was used to fit log-transformed expression levels [log2(FPKM + 1)] for every gene across the three different chemotype categories. Tukey’s test followed by Benjamini-Hochberg multiple test correction was then used to identify P values for each comparison. Correlations between cis-trans-nepetalactone, MLPL, and NEPS5 were further examined using Spearman’s rank correlation.
Self-organizing maps (SOM) were performed on the Z-scores of the log-transformed expression matrix using the som function in the kohonen library (53). The Z-score was calculated by subtracting the mean expression of each gene and dividing by their SD, resulting in each gene having a mean expression of 0 and an SD of 1. To avoid boundary effects, the SOM was set to have a 20 × 20 toroidal grid, with a hexagonal topology. To determine internodal similarity, the codebook vectors of the nodes were clustered via hierarchical clustering, and enrichment of biosynthetic genes in the selected cluster was calculated using a hypergeometric test. Tissue-specific expression patterns were analyzed and presented using hierarchically clustered heatmaps, using the pheatmap library (54). For both internodal clustering in SOMs and heatmaps, hierarchical clustering followed a complete linkage method of Euclidean distances.
Identification and cloning of genes
Genes of interest were identified by blast searching gene models with previously characterized iridoid biosynthesis enzymes from Nepeta (NEPSs and PRISEs) and Catharanthus (GES, G8H, and HGOA) (5, 7, 9, 10). The genomic locations and tissue expression patterns of the genes were used to assess whether the genes were genomically clustered and/or coexpressing with other iridoid biosynthetic genes.
Physical complementary DNA libraries were formed from young leaf RNA using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) with oligo d(T)20 primers according to kit instructions. Genes were cloned into pOPINF Escherichia coli expression vectors as previously described (9), with the exception of G8Hs which were cloned into a pESC-Leu2d vector for expression in yeast (55). Genes with closely related homologs were first cloned using unique UTRs (untranslated regions), and thereafter, the coding region was amplified for addition to the expression vector. PRISE and GES genes were obtained as N-terminal truncates to improve soluble expression. Predicted ancestral sequences, along with selected genes that could not be cloned or recombinantly expressed, were obtained via synthetic genes (Twist BioScience). Synthetic genes were codon-optimized for E. coli using COOL (56), optimizing for codon context and GC content. Synthetic genes were subcloned from the shuttle vector (pTwist Amp High Copy) into pOPINF, except NcNEPS4 which was provided in a pET28(+) expression vector and used directly. Hyssop GES and HGOA did not express in pOPINF and were cloned into pOPINJ (glutathione S-transferase tag) to improve protein solubility but did not express with this plasmid either.
All cloned and subcloned genes were sequenced by Sanger Sequencing (Eurofins Genomics). A few nucleotide differences were observed between genomic and cloned sequences.
Enzyme expression and purification in E. coli
For expression of NEPSs and MLPLs, E. coli expression strain cells (SoluBL21) containing the plasmids of interest were grown overnight [LB medium, 10 ml, with carbenicillin (100 μg/ml)]. 2xYT medium [100 ml, with carbenicillin (100 μg/ml)] was inoculated with overnight culture (5%, v/v) and grown at 37°C until OD600 (optical density at 600 nm) = 0.5. The culture was then grown at 18°C until OD600 = 0.6 to 0.8, and protein production was induced with addition of isopropyl-β-d-thiogalactopyranoside (500 μM). The cells were incubated at 18°C for 16 hours before harvesting by centrifugation (4000g, 10 min). If the pellets were not used immediately, they were washed in phosphate-buffered saline before storage at −20°C.
Cell pellets were resuspended in BugBuster MasterMix [10 ml, with cOmplete EDTA-free protease inhibitors (Roche)], incubated at 4°C for 20 min, and then centrifuged (35,000g, 20 min). Ni-nitrilotriacetic acid agarose (1 ml; Qiagen) was washed in binding buffer [50 mM tris-HCl (pH 8), 50 mM glycine, 5% (v/v) glycerol, 0.5 M NaCl, 20 mM imidazole, and 1 mM dithiothreitol (DTT)] and added to the supernatant lysate, and the mixture was incubated at 4°C for 1 hour, gently rocking. The mixture was centrifuged (1000g, 1 min), and the supernatant was discarded. The Ni-NTA pellet was washed three times with 10 ml of binding buffer, and then, elution buffer [50 mM tris-HCl (pH 8), 50 mM glycine, 5% (v/v) glycerol, 0.5 M NaCl, 500 mM imidazole, and 1 mM DTT] was added (2.5 ml). The mixture was centrifuged (1000g, 1 min), and the supernatant was collected and filtered. The buffer was exchanged with sample buffer [20 mM Hepes (pH 7.5) and 150 mM NaCl] using a PD-10 column (GE Healthcare). The proteins were aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C. SDS–polyacrylamide gel electrophoresis and spectrophotometric analysis (absorbance at 280 nm) were used to check purity and approximate quantity of protein.
NmNEPS4 and NcNEPS4 were grown as in 1 liter of cultures and purified by ÄKTA as previously described (6). HGOA and GES were purified as described above but with 1 liter of cultures and lysis by sonication (2 min total, 2 s on, 3 s off; amplitude, 40% pulses; Sonics Vibra-Cell). PRISEs were obtained as described above but in 1 liter of culture, with lysis by cell disruption (two passes, 25 kPi, Constant Systems) and four buffer exchanges using centrifugal filtration [Amicon Ultra 10-kDa molecular weight cutoff (Merck)]. Of the PRISE proteins examined, only HoP5βRA failed to express in adequate quantities for activity assays. GES and HGOA from hyssop failed to express adequately for enzyme assays, despite attempts to optimize conditions.
GC-MS general method
For GC-MS analysis of enzyme assays, samples were injected in split mode (2 μl; split ratio, 5:1) at an inlet temperature of 220°C on a Hewlett Packard 6890 GC-MS equipped with a 5973 mass selective detector and an Agilent 7683B series injector and autosampler. Separation was performed on a Zebron ZB5-HT-INFERNO column (5% phenyl methyl siloxane; length, 35 m; diameter, 250 μm) with guard column. Helium was used as mobile phase at a constant flow rate of 1.2 ml/min and an average velocity of 37 cm/s. Two temperature runs were used for detection: Method 1 (GES, G8H, and NEPS assays): After 5 min at 80°C, the column temperature was increased to 110°C at a rate of 2.5 K/min then to 280°C at 120 K/min and kept at 280°C for another 4 min. Method 2 (HGOA assays): After an initial temperature at 60°C, the column temperature was increased to 100°C at a rate of 20 K/min then to 160°C at 2 K/min, then another increase to 280°C at 100 K/min, and maintained for 4 min. A solvent delay of 5 min was allowed before collecting MS spectra at a fragmentation energy of 70 eV. Chemically characterized standards were used to identify compounds by retention time and EI spectra.
End-point enzyme assays
End-point assays using 8OG as a substrate were performed as described previously (6). For assays with ISY, NEPS, and MLPL (100 μl): buffer [0.1 M 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.5)], ISY (250 nM), NEPS/MLPL/BSA (typically 2 μM each unless noted), NAD+ (5 mM, added in the presence of NEPSs), NADPH (1 mM), and 8OG (0.5 mM, added last) were mixed and incubated for 3 hours at 30°C. The reactions were extracted into 100 μl of ethyl acetate [with 1 or 10 μM (+)-camphor internal standard], and the organic fraction was analyzed by GC-MS.
For quantification, GC-MS chromatograms were analyzed using OpenChrom Lablicate Edition (version 1.4.0.202001031827). Peaks were detected using Peak Detector First Derivative tool and integrated using Peak Max Integrator. Peaks of interest were identified by comparing retention times and EI spectra to chemical characterized standards. Peak areas were normalized across runs using the (+)-camphor internal standard and presented as areas relative to 1 μM camphor.
Activity of purified GES was determined by incubating GES (4.43 μM), DTT (1 mM), MgCl2 (20 mM), MnCl2 (500 μM), glycerol (10%, v/v), geranyl pyrophosphate (0.1 mM), and MOPS buffer (10 mM, 100 μl total) at 30°C for 1 hour. Activity of purified HGOA was determined by incubating enzyme (7 μM), NAD(P)+ (2 mM), 8-hydroxygeraniol (0.5 mM), NaCl (25 mM), and Hepes buffer (12.5 mM, 100 μl total) at 30°C. Reactions were quenched by mixing 100 μl of ethyl acetate, centrifuging to separate and collecting the top organic layer for analysis by GC-MS.
Activities of G8H enzymes were determined using yeast feeding assays. Saccharomyces cerevisiae (pep4KO) cells containing the appropriate vectors were used to inoculate 2 × 2 ml of SC-Leu media with 2% (v/v) glycerol. Cultures were incubated for 48 hours at 30°C, pooled, and then pelleted at 3500g for 5 min. The cultures were washed twice with 10 ml of water, pelleting each time at 3500g for 5 min. SC-Leu (2 ml) with 2% (v/v) galactose was added to each culture, which was then split into four tubes. To three of these aliquots, 0.5 mM of geraniol was added, and to one, an equal volume of analytical grade ethanol was added. The aliquots were incubated for 24 hours at 30°C. Reactions were quenched by adding 200 μl of 3:1 ethyl acetate:acetone solution. The mixture was vortexed and centrifuged, and the top organic layer was collected and filtered for GC-MS analysis.
PRISE activities
Progesterone 5β-reductase activities were measured as initial rates using GC-MS. Reactions (400 μl total, n = 3 per enzyme) consisted of enzymes (500 nM), NADPH (1 mM), progesterone (400 μM), dimethyl sulfoxide (4%, v/v), and MOPS buffer [20 mM (pH 7.5)]. Components were mixed and incubated at 30°C. Samples (100 μl) were taken from reactions at 2 and 4 hours, extracted into ethyl acetate (100 μl), and analyzed by GC-MS (see method below). Total ion chromatograms were analyzed Agilent MassHunter Qualitative analysis. Product peaks were identified by retention time comparison to an authentic standard (5β-dihydroprogesterone) and integrated. Peak areas were converted to concentrations using a product standard curve, and initial linear rates were calculated for each replicate. Analysis of variation was used to compare activities across enzymes (log10 transformation, “aov” function in R), and results were grouped using Tukey’s post hoc test (α = 0.05, “HSD.test” function in R).
ISY activities were measured on a FLUOStar Omega multiplate reader (BMG Labtech) using plate kinetics mode and absorbance at 340 nm (25°C, positioning delay 0.2 s, 22 flashes per well). Reactions (200 μl total) consisted of enzymes (5 to 300 nM), NADPH (100 μM), 8OG (0 to 100 μM), and MOPS buffer [20 mM (pH 7.5)]. Components were preincubated for 2 min before the addition of 8OG and then mixed by pipetting. NADPH consumption was measured, and the initial linear rate was calculated (1 to 20 min, “lm” function in R). For activity measurements (100 μM 8OG, n = 3 to 6 per enzyme), analysis of variation was used to compare activities across enzymes (log10 transformation, “aov” function in R), and results were grouped using Tukey’s post hoc test (α = 0.05, “HSD.test” function in R). For kinetic analyses (0, 1.5625, 3.125, 6.25, 12.5, 25, 50, or 100 μM 8OG, n = 21 to 32 per enzyme), the Michaelis-Menten or substrate inhibition models were fit to rates (“nls” function in R).
For pie chart depiction of PRISE activities, mean values were calculated for each enzyme and then normalized by dividing means by the maximum observed rates (ISY activity = NmISY and P5βR activity = NcP5βRA). The area of each pie chart section is proportional to the normalized activity, and thus, the total area of each chart is proportional to the sum of the two normalized activities.
Early iridoid gene phylogenetics
For early iridoid enzymes, genes of interest were identified using previously characterized iridoid biosynthesis enzymes from Catharanthus (GES, CRO_T119458; G8H, CRO_T133061; HGOA, CRO_T107879) (7). Lamiaceae orthogroups containing the Catharanthus genes of interest were identified (G8H, OG0000115; GES, OG0011057; HGOA, OG0003405) (10). Genome gene models for Hyssopus and Nepeta were searched by blast for related sequences. Near-identical and fragments of sequences were removed from the analysis. Open reading frames were extracted, aligned by translation using MAFFT (Auto settings, v 7.388) (57), and phylogenies were inferred from codons using iQTree v1.6.9 (58) with ModelFinder (59) together with UFBoot2 (×1000) (60) and SH-aLRT supports (×1000). Outgroups were selected on the basis of known species relationships. For G8H, the phylogeny of the large orthogroup was initially inferred using FastTree (61) before the clade most closely related to the specified Catharanthus G8H sequence was identified. The expression patterns of Nepeta genes of interest were used to identify which genes are most likely involved in iridoid biosynthesis. Phylograms were depicted using Geneious Prime 2019 or iTOL 4.4.2 (62).
PRISE phylogenetics
Sequences corresponding to PRISE homologs were obtained through orthogroup analysis of Lamiaceae and reference transcriptomes (OG0002182) (10). Homologs were identified from the Nepeta and Hyssopus genomes by blast searching gene model coding sequences. The NcSISYB pseudogene did not appear in the gene models. To identify NcSISYB, NmSISY was used as a query to blast the N. cataria genome sequence. The reversed sequence in the region of g3325 had notable sequence similarity with the 5′ region of NmSISY and NcSISYA. There also appeared to be some sequence similarity with the 3′ of NcSISYA downstream of a string of NNNs adjacent to g3324. This region appears to be a SISY pseudogene, NcSISYB.
Extra PRISE sequences were identified by blast searches of National Center for Biotechnology Information (NCBI) nr databases, Mentha transcriptomes (http://langelabtools.wsu.edu/mgr/), sequence read archive (SRA) Lamiaceae transcriptomes, and “skim” genomes. For SRA-derived transcriptomes, raw reads were downloaded from NCBI SRA for Isodon rubescans (SRR714241), Ocimum americanum (SRR2029848 and SRR2029849), Ocinum teniflorum (SRR1177846), Phlomis purpurea (SRR1920173), Salvia miltiorrhiza (SRR1745640), and Salvia sclarea (SRR983807). Adapters and low-quality bases were removed using Cutadapt (v1.8.1) (34) requiring a minimum base quality of 20 and a minimum size of 20 nt. De novo transcript assemblies were generated using Trinity (v20140717) (45) using the default parameters, and only transcripts with length equal and greater than 150 base pairs (bp) were retained for subsequent analyses. For generation of skim genomes for Agastache foeniculum, Melissa officinalis, Perilla frutescens, Lycopus americanus, Glechoma hederacea, and Collinsonia canadensis, approximately 30 million 2 × 150 bp reads were generated for each mint species using the HiSeq 4000 platform. Adapters and low-quality bases were removed using Cutadapt (v1.8.1) requiring a minimum base quality of 20 and a minimum size of 20 nt. De novo genome assemblies were generated using Abyss (v1.9.0) with a kmer size of 65 (63).
Open reading frames for PRISE enzymes were extracted from the transcripts, and stop codons were removed. Near-identical sequences were removed from the analysis. Sequences ROMY_c75305_g1_i2 and PRMI_c25272_g1_i1 were obtained by searching raw isoform transcriptomes (10). A sequence labeled ORVU_c87504_g1_i1 was removed from the analysis as it was revealed to be a contamination from ROMY_c75305_g1_i2. NcSISYA and NcSISYB were not included in the analysis, because their inclusion caused instability in the tree topology. For PRISE genes from Nepeta, the sequenced verified cloned gene sequences were used in phylogenetic analysis. For hyssop PRISEs, the original genomic sequences and not the cloned gene sequences were used in the phylogenetic analyses, as this analysis was performed before cloning of these genes.
Translated sequences were aligned using MAAFT (G-INS-i, v7.017) (57). The codon-based phylogeny was inferred using iQTree v1.6.1 (58) with ModelFinder (59). The output tree was then used to realign the codon sequences using PRANK (64) before the phylogeny was inferred again using IQ-TREE v1.6.1 with ModelFinder together with ultrafast bootstraps (UFBoot2, ×1000) (60) and SH-aLRT supports (×1000). The cladogram was depicted using the iTOL 4.4.2 (62) and Adobe Illustrator.
Ancestral sequence reconstruction
For prediction of ancestral sequences and analysis of positive selection, a PRISE subtree was isolated with PEVO_c56018_g1_i3 as an outgroup. Short sequences (<1159 nucleotides) were removed, and the phylogeny was again inferred using iQTree v1.6.9 with ModelFinder together with UFBoot2 (60) (×1000) and SH-aLRT supports (×1000). This tree was used for ASR with FastML using default codon settings (65). For Anc1 and Anc2, the most probable sequences were selected for protein assays. For Anc3, the most probable sequence was used with the exception of codon 283. The amino acid at codon 283 is equivocal: p(TTG, Leu) = 0.38 and p(TTC, Phe) = 0.34. The Anc3 used had a TTC (Phe) in this position, as it was based on a preliminary iteration of the gene phylogeny. Phe is present in this equivalent position in Anc1, Anc2, and Nepeta P5βRs, whereas Leu is present in all Nepeta ISYs. Therefore, the presence of Phe-283 in the tested Anc3 is very unlikely to be responsible for its ISY-like nature. For expression and protein production in E. coli, sequences were truncated (starting at S/NVAL) and codon-optimized.
Positive selection analysis
The focused PRISE gene phylogeny used for ASR was also used for positive selection analysis using PAML 4.9i (66). The CodeML software implemented in PAML can test for positive selection in codon alignments along gene lineages and in specific sites. This is achieved by modeling ratios of synonymous mutation rates to nonsynonymous mutation rates (dN/dS = ω). First, branch lengths were optimized using the M0 model (single ω across alignment and phylogeny) with codon model 2 (F3X4). These branch lengths were used as initial values to fit a variety of different codon models to the data using the M0 model. A total of 11 codon models were tested (parameters: CodonFreq = 0, 1, 2, 3, 6, or 7 and estFreq = 0 or 1) and compared using Bayesian information criteria (BIC) (n = 376, number of informative codon patterns). The fMutSel codon model with observed codon frequencies (CodonFreq = 7, estFreq = 0) was the best fit according to BIC and was used for positive selection tests (67).
The branch-site model A test 2 was used to detect positive selection along specified branches in the phylogeny (parameters: model = 2, NSites = 2). For H0, ω2 for the selected branch is fixed at 1 (fix_omega = 1, omega = 1), whereas for H1, ω2 is estimated (fix_omega = 0). A likelihood ratio test was used to determine whether ω2 > 1 is significant (positive selection). Five separate branches were tested for positive selection, and the P values were corrected using Benjamini-Hochberg adjustment.
NEPS sequences and phylogenetics
Sequences corresponding to NEPS and NEPS-like homologs were obtained through orthogroup analysis of Lamiaceae and reference transcriptomes (OG0004576) (10). NEPS and NEPS-like genes within Nepeta and hyssop were identified by blasting gene models (blastn) with NcNEPS1 and NmNEPSL1 (g21780). As described above for the PRISEs: published transcriptomes, SRA-derived transcriptomes, and skim genomes were queried to identify more NEPS homologs identified. Open reading frames were identified, and short sequences were removed (<580 bp). Sequences were aligned (translation align, MAAFT) (57), and a maximum-likelihood tree was inferred (iQTree) (58). The clade containing NEPS and NEPS-like sequences was subsampled, keeping a sequence from Catharanthus as an outgroup (CRO_T131109). The codon sequences were realigned with PRANK (64), and the tree was inferred with iQTree (58).
To check thoroughly for NEPS genes outside Nepeta, NcNEPS1 was used as a query to blast (blastn) other databases. Reasonable blast hits (>50% coverage, >60% pairwise identity) were obtained, aligned with MAAFT to NEPS and NEPS-like genes from the Lamiaceae, and a tree was inferred using FastTree (61). The placement of the sequences within the phylogeny was used to determine whether these sequences were within the NEPS clade or were merely NEPS-like sequences from the wider SDR110C family. Numerous databases were searched: Callicarpa americana (gene models, unpublished from Mint Evolutionary Genomics Consortium); Salvia hispanica (contigs, unpublished from Mint Evolutionary Genomics Consortium); hyssop (genome, presented here); Mentha longifolia (genome scaffolds); S. miltiorrhiza (genome scaffolds); skim genomes as described above for A. foeniculum, M. officinalis, P. frutescens, L. americanus, G. hederacea, and C. canadensis (unpublished, from Mint Evolutionary Genomics Consortium); Tectona grandis (chromosome assembly) and Dracocephalum tanguticum (reads identified by blast searching NCBI SRA). No databases other than those from Nepeta contained sequences which placed phylogenetically within the NEPS clade, indicating that NEPS evolved within the lineage of Nepeta or, more strictly, along the stem lineage from the Nepetinae common ancestor to the last common ancestor of N. mussinii and N. cataria.
Molecular dating analyses
We identified the largest subclades in each of our PRISE and NEPS ML trees (figs. S22 and S28) that could be reliably rooted with at least one outgroup sequence from Lamiales. Sequences comprising these PRISE and NEPS subclades, respectively, were parsed from a corresponding multiple sequence alignment (methods above) and manually edited as necessary to remove gaps in the subsampled alignment. A new ML tree was inferred from each alignment with RAxML version 8.2.10 (68) and the GTRGAMMA model. The resulting PRISE and NEPS trees were rooted with sequences from Petrea volubilis L. (PEVO_c39703_g1_i1) and Lancea tibetica Hook.f. & Thomson (LATI_c23199_g1_i1) + Aureolaria pectinata (Nutt.) Pennell (AUPE_c43669_g1_i1), respectively, and used for divergence time estimation. All dating analyses used a relaxed clock approach, as implemented in treePL (69) with PL (70). Optimization parameters for each analysis were identified using the “prime” option in treePL, followed by a “thorough” analysis using those parameters. A smoothing parameter for each analysis was determined by cross-validation. We used the following age constraints for each estimation procedure: a minimum and maximum age of 25.31 and 25.63 Ma for the crown node of Nepetinae; a minimum age of 47.8 Ma for the crown node of Nepetoideae; a minimum and maximum age of 59.99 and 70.94 Ma, respectively, for the crown node of Lamiaceae; and a maximum age of 107 Ma for the root node, representing the Lamiales crown.
The age constraint for Nepetinae corresponded to the lower and upper bounds of the 95% confidence interval of estimated ages for the Nepetinae crown derived from a Lamiaceae divergence time analysis (methods below). The age constraint for Nepetoideae corresponded to the fossil pollen of Ocimum (71), whereas the remaining constraints corresponded to the lower and upper bounds of the 96% highest posterior density interval of estimated ages for the Lamiaceae crown (72) and the oldest estimated age reported for the Lamiales crown (73), respectively. We conducted additional dating analyses to provide 95% confidence intervals [e.g., see (74)] for ages estimated at internal nodes in the PRISE and NEPS trees. Model parameters and branch lengths were re-estimated with RAxML for both ML topologies using a bootstrap approach with 1000 replicates, producing a set of 1000 bootstrap phylograms with identical topologies for each dataset. We also performed a similar procedure using a species tree and corresponding data matrix for Lamiaceae (10) to provide a secondary age calibration for the crown node of Nepetinae in the PRISE and NEPS divergence time analyses. However, given the robust support for the species tree topology and large matrix size, we used only 100 bootstrap replicates for the analysis. All bootstrap trees were dated using the approach described above (i.e., excluding use of the Nepetinae constraint in the species tree analyses), and age statistics at all internal nodes were summarized across each tree set with TreeAnnotator version 1.10.4 (http://beast.community/treeannotator).
Supplementary Material
Acknowledgments
We thank N. Hernandez-Lozada for comments on the manuscript. We also thank Z. Yang for advice on selection analysis. We are grateful to P. Brett and L. Hill for metabolomics support. Funding: Funds for this study were provided by a grant to C.R.B., N.D., S.E.O’C., D.S., and P.S.S. from the National Science Foundation Plant Genome Research Program (IOS-1444499). C.R.B. is also supported from Hatch Funds (MICL02431). B.R.L. is supported by a UK Research and Innovation Future Leaders Fellowship (MR/S01862X/1). Author contributions: B.R.L., C.R.B., and S.E.O’C. conceived and designed the project, with contributions from other authors. B.R.L., M.O.K., L.K.H., and C.R.-L. performed tissue metabolite analysis. B.V. and J.C.W. grew plants and isolated and sequenced DNA and RNA. J.P.H. and D.Z. assembled and annotated genomes. L.P. characterized early iridoid enzymes. B.R.L. and M.O.K. characterized NEPS and PRISE enzymes. B.R.L. performed basic phylogenetic tree inference, selection analysis, and ASR. G.T.G., M.S., and T.J.K. performed molecular clock analysis with input from D.E.S. and P.S.S. All authors analyzed data. B.R.L., C.R.B., and S.E.O’C. wrote the manuscript, with contributions from all other authors. Competing interests: B.R.L. and S.E.O’C. have filed patent applications covering the use of some genes reported here (WO2019224536). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Raw sequence reads are available in the National Center for Biotechnology Information under BioProject IDs PRJNA359989, PRJNA529676, PRJNA529674, and PRJNA557218. The three genome assemblies, annotated genes/transcripts/peptides/functional annotation, expression matrices, gff files, gene sequences, primer sequences, gene alignments, phylogenetic trees, FastML output, dating analysis data and LC-MS files are available at the Dryad Digital Repository (doi:10.5061/dryad.88tj450). GenBank accession numbers related to this work are MT108261 to MT108293.
SUPPLMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaba0721/DC1
REFERENCES AND NOTES
- 1.Tucker A. O., Tucker S. S., Catnip and the catnip response I. Econ. Bot. 42, 214–231 (1988). [Google Scholar]
- 2.Bol S., Caspers J., Buckingham L., Anderson-Shelton G. D., Ridgway C., Buffington C. A., Schulz S., Bunnik E. M., Responsiveness of cats (Felidae) to silver vine (Actinidia polygama), Tatarian honeysuckle (Lonicera tatarica), valerian (Valeriana officinalis) and catnip (Nepeta cataria). BMC Vet. Res. 13, 70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisner T., Catnip: Its raison d’etre. Science 146, 1318–1320 (1964). [DOI] [PubMed] [Google Scholar]
- 4.Birkett M. A., Hassanali A., Hoglund S., Pettersson J., Pickett J. A., Repellent activity of catmint, Nepeta cataria, and iridoid nepetalactone isomers against Afro-tropical mosquitoes, ixodid ticks and red poultry mites. Phytochemistry 72, 109–114 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Miettinen K., Dong L., Navrot N., Schneider T., Burlat V., Pollier J., Woittiez L., van der Krol S., Lugan R., Ilc T., Verpoorte R., Oksman-Caldentey K.-M., Martinoia E., Bouwmeester H., Goossens A., Memelink J., Werck-Reichhart D., The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5, 3606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geu-Flores F., Sherden N. H., Courdavault V., Burlat V., Glenn W. S., Wu C., Nims E., Cui Y., O’Connor S. E., An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492, 138–142 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Sherden N. H., Lichman B., Caputi L., Zhao D., Kamileen M. O., Buell C. R., O’Connor S. E., Identification of iridoid synthases from Nepeta species: Iridoid cyclization does not determine nepetalactone stereochemistry. Phytochemistry 145, 48–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kries H., Kellner F., Kamileen M. O., O’Connor S. E., Inverted stereocontrol of iridoid synthase in snapdragon. J. Biol. Chem. 292, 14659–14667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lichman B. R., Kamileen M. O., Titchiner G. R., Saalbach G., Stevenson C. E. M., Lawson D. M., O’Connor S. E., Uncoupled activation and cyclisation in catmint reductive terpenoid biosynthesis. Nat. Chem. Biol. 15, 71–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mint Evolutionary Genomics Consortium , Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant 11, 1084–1096 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Stull G. W., Schori M., Soltis D. E., Soltis P. S., Character evolution and missing (morphological) data across Asteridae. Am. J. Bot. 105, 470–479 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Dobler S., Petschenka G., Pankoke H., Coping with toxic plant compounds - The insect’s perspective on iridoid glycosides and cardenolides. Phytochemistry 72, 1593–1604 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Magnard J. L., Roccia A., Caissard J. C., Vergne P., Sun P., Hecquet R., Dubois A., Hibrand-Saint Oyant L., Jullien F., Nicolè F., Raymond O., Huguet S., Baltenweck R., Meyer S., Claudel P., Jeauffre J., Rohmer M., Foucher F., Hugueney P., Bendahmane M., Baudino S., Biosynthesis of monoterpene scent compounds in roses. Science 349, 81–83 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Petersen J., Lanig H., Munkert J., Bauer P., Müller-Uri F., Kreis W., Progesterone 5β-reductases/iridoid synthases (PRISE): Gatekeeper role of highly conserved phenylalanines in substrate preference and trapping is supported by molecular dynamics simulations. J. Biomol. Struct. Dyn. 34, 1667–1680 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Nützmann H. W., Huang A., Osbourn A., Plant metabolic clusters – From genetics to genomics. New Phytol. 211, 771–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liblikas I., Santangelo E. M., Sandell J., Baeckström P., Svensson M., Jacobsson U., Unelius C. R., Simplified isolation procedure and interconversion of the diastereomers of nepetalactone and nepetalactol. J. Nat. Prod. 68, 886–890 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Gabur I., Chawla H. S., Snowdon R. J., Parkin I. A. P., Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 132, 733–750 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Brown S., Clastre M., Courdavault V., O’Connor S. E., De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 3205–3210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moghe G. D., Last R. L., Something old, something new: Conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 169, 1512–1523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christ B., Xu C., Xu M., Li F.-S., Wada N., Mitchell A. J., Han X.-L., Wen M.-L., Fujita M., Weng J.-K., Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 10, 3206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R., O’Donnell A. J., Barboline J. J., Barkman T. J., Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. U.S.A. 113, 10613–10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaltenbach M., Burke J. R., Dindo M., Pabis A., Munsberg F. S., Rabin A., Kamerlin S. C. L., Noel J. P., Tawfik D. S., Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol. 14, 548–555 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Bauer P., Munkert J., Brydziun M., Burda E., Müller-Uri F., Gröger H., Muller Y. A., Kreis W., Highly conserved progesterone 5β-reductase genes (P5βR) from 5β-cardenolide-free and 5β-cardenolide-producing angiosperms. Phytochemistry 71, 1495–1505 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rogozin I. B., Belinky F., Pavlenko V., Shabalina S. A., Kristensen D. M., Koonin E. V., Evolutionary switches between two serine codon sets are driven by selection. Proc. Natl. Acad. Sci. U.S.A. 113, 13109–13113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuba Y., Nguyen T. T., Wiegert K., Falara V., Gonzales-Vigil E., Leong B., Schäfer P., Kudrna D., Wing R. A., Bolger A. M., Usadel B., Tissier A., Fernie A. R., Barry C. S., Pichersky E., Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25, 2022–2036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto K., Fujita M., Shenton M. R., Akashi S., Sugawara C., Sakai A., Horie K., Hasegawa M., Kawaide H., Mitsuhashi W., Nojiri H., Yamane H., Kurata N., Okada K., Toyomasu T., Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 87, 293–304 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Pohlit A. M., Lopes N. P., Gama R. A., Tadei W. P., Andrade-Neto V. F., Patent literature on mosquito repellent inventions which contain plant essential oils—A review. Planta Med. 77, 598–617 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Dawson G., Griffiths D. C., Janes N. F., Mudd A., Pickett J. A., Wadhams L. J., Woodcock C. M., Griffiths D. C., Janes N. F., Mudd A., Pickett J. A., Wadhams L. J., Woodcock C. M., Identification of an aphid sex pheromone. Nature 325, 614–616 (1987). [Google Scholar]
- 29.Koczor S., Szentkirályi F., Birkett M. A., Pickett J. A., Voigt E., Tóth M., Attraction of Chrysoperla carnea complex and Chrysopa spp. lacewings (Neuroptera: Chrysopidae) to aphid sex pheromone components and a synthetic blend of floral compounds in Hungary. Pest Manag. Sci. 66, 1374–1379 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Li C., Zhang X. L., Xue X. Y., Zhang F. F., Xu Q., Liang X. M., Structural characterization of iridoid glucosides by ultra-performance liquid chromatography/electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. 22, 1941–1954 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Formisano C., Rigano D., Senatore F., Chemical constituents and biological activities of Nepeta species. Chem. Biodivers. 8, 1783–1818 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W., Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population-dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao D., Hamilton J. P., Pham G. M., Crisovan E., Wiegert-Rininger K., Vaillancourt B., Della Penna D., Buell C. R., De novo genome assembly of Camptotheca acuminata, a natural source of the anti-cancer compound camptothecin. Gigascience 6, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 35.Leggett R. M., Clavijo B. J., Clissold L., Clark M. D., Caccamo M., NextClip: An analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics 30, 566–568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gnerre S., Maccallum I., Przybylski D., Ribeiro F. J., Burton J. N., Walker B. J., Sharpe T., Hall G., Shea T. P., Sykes S., Berlin A. M., Aird D., Costello M., Daza R., Williams L., Nicol R., Gnirke A., Nusbaum C., Lander E. S., Jaffe D. B., High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 108, 1513–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bickhart D. M., Rosen B. D., Koren S., Sayre B. L., Hastie A. R., Chan S., Lee J., Lam E. T., Liachko I., Sullivan S. T., Burton J. N., Huson H. J., Nystrom J. C., Kelley C. M., Hutchison J. L., Zhou Y., Sun J., Crisà A., Ponce de León F. A., Schwartz J. C., Hammond J. A., Waldbieser G. C., Schroeder S. G., Liu G. E., Dunham M. J., Shendure J., Sonstegard T. S., Phillippy A. M., Van Tassell C. P., Smith T. P., Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 49, 643–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.English A. C., Richards S., Han Y., Wang M., Vee V., Qu J., Qin X., Muzny D. M., Reid J. G., Worley K. C., Gibbs R. A., Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLOS ONE 7, e47768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C. A., Zeng Q., Wortman J., Young S. K., Earl A. M., Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Zhao D., Hamilton J. P., Bhat W. W., Johnson S. R., Godden G. T., Kinser T. J., Boachon B., Dudareva N., Soltis D. E., Soltis P. S., Hamberger B., Buell C. R., A chromosomal-scale genome assembly of Tectona grandis reveals the importance of tandem gene duplication and enables discovery of genes in natural product biosynthetic pathways. Gigascience 8, giz005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell M. S., Law M., Holt C., Stein J. C., Moghe G. D., Hufnagel D. E., Lei J., Achawanantakun R., Jiao D., Lawrence C. J., Ware D., Shiu S. H., Childs K. L., Sun Y., Jiang N., Yandell M., MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanke M., Morgenstern B., AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haas B. J., Wortman J. R., Ronning C. M., Hannick L. I., Smith R. K. Jr., Maiti R., Chan A. P., Yu C., Farzad M., Wu D., White O., Town C. D., Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 3, 7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. R. Eddy, HMMER user’s guide (2010); ftp://selab.janelia.org/pub/software/hmmer3/.
- 50.Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., Lee T. H., Jin H., Marler B., Guo H., Kissinger J. C., Paterson A. H., MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meisel L., Fonseca B., González S., Baeza-Yates R., Cambiazo V., Campos R., Gonźalez M., Orellana A., Retamales J., Silva H., A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol. Res. 38, 83–88 (2005). [DOI] [PubMed] [Google Scholar]
- 52.R Core Team, R: A language and environment for statistical computing. (2018); www.r-project.org/.
- 53.Wehrens R., Buydens L. M. C., Self- and super-organizing maps in R: The kohonen Package. J. Stat. Softw. 21, (2007). [Google Scholar]
- 54.R. Kolde, pheatmap: Pretty Heatmaps (2019).
- 55.Dang T. T. T., Franke J., Tatsis E., O’Connor S. E., Dual catalytic activity of a cytochrome p450 controls bifurcation at a metabolic branch point of alkaloid biosynthesis in Rauwolfia serpentina. Angew. Chem. Int. Ed. 56, 9440–9444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chin J. X., Chung B. K., Lee D., Codon Optimization OnLine ( COOL ): a web-based multi-objective optimization platform for synthetic gene design. Bioinformatics 30, 2210–2212 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability article fast track. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoang D. T., Chernomor O., Von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price M. N., Dehal P. S., Arkin A. P., FastTree 2—Approximately maximum-likelihood trees for large alignments. PLOS ONE 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M., Birol I., ABySS: A parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loytynoja A., Goldman N., Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320, 1632–1635 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Ashkenazy H., Penn O., Doron-Faigenboim A., Cohen O., Cannarozzi G., Zomer O., Pupko T., FastML: A web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Yang Z., Nielsen R., Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25, 568–579 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith S. A., O’Meara B. C., TreePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Sanderson M. J., Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Kar R. K., On the Indian origin of Ocimum (Lamiaceae): A palynological approach. Palaeobotanist 43, 43–50 (1996). [Google Scholar]
- 72.Yao G., Drew B. T., Yi T. S., Yan H. F., Yuan Y. M., Ge X. J., Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol. Phylogenet. Evol. 98, 184–200 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Janssens S. B., Knox E. B., Huysmans S., Smets E. F., Merckx V. S. F. T., Rapid radiation of Impatiens (Balsaminaceae) during Pliocene and Pleistocene: Result of a global climate change. Mol. Phylogenet. Evol. 52, 806–824 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Sanderson M. J., r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Li B., Cantino P. D., Olmstead R. G., Bramley G. L. C., Xiang C.-L., Ma Z.-H., Tan Y.-H., Zhang D.-X., A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep. 6, 34343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li B. O., Olmstead R. G., Two new subfamilies in Lamiaceae. Phytotaxa 313, 222–226 (2017). [Google Scholar]
- 77.Vining K. J., Johnson S. R., Ahkami A., Lange I., Parrish A. N., Trapp S. C., Croteau R. B., Straub S. C. K., Pandelova I., Lange B. M., Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Mol. Plant 10, 323–339 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., Xu J., Li Y., Song C., Wang B., Sun W., Shen G., Zhang X., Qian J., Ji A., Xu Z., Luo X., He L., Li C., Sun C., Yan H., Cui G., Li X., Li X., Wei J., Liu J., Wang Y., Hayward A., Nelson D., Ning Z., Peters R. J., Qi X., Chen S., Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant 9, 949–952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaba0721/DC1


