A distinct class of mitosis regulators, conserved between plant and animal viruses, play active roles in viral pathogenesis.
Abstract
Many animal viral proteins, e.g., Vpr of HIV-1, disrupt host mitosis by directly interrupting the mitotic entry switch Wee1-Cdc25-Cdk1. However, it is unknown whether plant viruses may use this mechanism in their pathogenesis. Here, we report that the 17K protein, encoded by barley yellow dwarf viruses and related poleroviruses, delays G2/M transition and disrupts mitosis in both host (barley) and nonhost (fission yeast, Arabidopsis thaliana, and tobacco) cells through interrupting the function of Wee1-Cdc25-CDKA/Cdc2 via direct protein-protein interactions and alteration of CDKA/Cdc2 phosphorylation. When ectopically expressed, 17K disrupts the mitosis of cultured human cells, and HIV-1 Vpr inhibits plant cell growth. Furthermore, 17K and Vpr share similar secondary structural feature and common amino acid residues required for interacting with plant CDKA. Thus, our work reveals a distinct class of mitosis regulators that are conserved between plant and animal viruses and play active roles in viral pathogenesis.
INTRODUCTION
Normal mitotic cycling, which involves progressive transitions of G1-S-G2-M phases, is essential for proper growth and development of cellular organisms (1–3). However, many animal viruses disrupt the mitotic cycle during their infections to aid viral proliferation in host cells (4). Among the mechanisms used by animal viral proteins to perturb host mitotic cycle, disruption of G2/M transition via directly targeting and interrupting the mitotic entry switch, which is composed of Wee1 kinase, Cdc25 phosphatase, and cyclin-dependent kinase (Cdk1) (5, 6), has attracted substantial research attention in recent years. For example, induction of G2/M arrest by HIV-1 Vpr is associated with its physical interactions with Wee1 and Cdc25C (7, 8). The p17 protein of avian reovirus (ARV) directly binds to Cdk1 and inhibits its kinase activity, thereby disturbing the progression from G2 to M phases (9). Last, several herpesviruses have been shown to perturb host cell G2/M transition by enhancing Cdk1 phosphorylation at the Y15 site (4).
Plant viruses are also known to interfere with host mitotic cycle during their infections, although the molecular mechanisms involved have been studied in only a few cases. For instance, the p12 protein of chrysanthemum virus B induced host cell proliferation by up-regulating the transcription of the genes promoting mitotic cycle (10). The C4 protein of Tomato leaf curl Yunnan virus caused abnormal cell division by increasing the stability of host cyclin D (11). However, to date, there is still no report of disruption of host mitosis by plant viruses through directly targeting and interrupting plant mitotic entry switch, which consists of Wee1-Cdc25-CDKA, with CDKA being the functional homolog of human Cdk1 (12).
Barley yellow dwarf viruses (BYDVs) are a group of related but distinct plant pathogens. They infect more than 150 Poaceae species and cause serious yellow dwarf diseases and economic losses to global cereal crop production (wheat, barley, maize, and oat) (13–15). BYDVs, vectored by several cereal aphid species, have a single-stranded, positive-sense RNA genome and accumulate exclusively in the phloem tissues of host plants (13–15). Dwarfing of host plants is a predominant symptom of BYDV infections, indicating that host cell division and related processes may be targeted and negatively affected by one or more viral proteins. We thus set out to identify potential BYDV protein(s) capable of disrupting host mitosis and to characterize the molecular mechanism behind the disruption. To reach these objectives, we individually expressed all open reading frames (ORFs) carried by the genome of BYDV-GAV, a typical BYDV strain causing a severe wheat yellow dwarf disease (16, 17), in fission yeast (Schizosaccharomyces pombe) cells using an inducible nmt1 (no-message-in-thiamine) transcriptional promoter (18). S. pombe was used in this investigation because (i) the genetic and molecular mechanisms controlling mitosis, including the function of the mitotic entry control Wee1-Cdc25-Cdc2, are well understood in this model species (19) and (ii) it has been successfully used to analyze the cell cycle disruption induced by important animal viral proteins, e.g., HIV-1 Vpr (18, 20).
Our functional test in S. pombe revealed that the 17K protein specified by ORF4, which is conserved in a wide range of cereal-infecting BYDVs and related poleroviruses and functions in virus systemic spread and viral suppression of host RNA silencing (21–23), induced a strong inhibition of cell growth via delaying G2/M transition. More detailed investigations in both host (barley) and nonhost (S. pombe, Arabidopsis thaliana, and Nicotiana benthamiana) cells revealed that 17K directly targeted and interrupted the function of Wee1-Cdc25-CDKA/Cdc2 in plant and fission yeast via protein-protein interactions and alteration of CDKA/Cdc2 phosphorylation. These processes resembled those used by HIV-1 Vpr and ARV p17 in their disruption of animal cell mitosis, although the amino acid sequence identities among 17K, Vpr, and p17 were below 13.7%. Neither the delay of G2/M transition by BYDV 17K nor the similarity in directly targeting and interrupting the mitotic entry switch by animal and plant viral proteins has been studied in the past. Our analyses of the two aspects have shed new light on the repertoire of functions executed by plant virus proteins and on a previously unknown class of mitosis regulators conserved between animal and plant viruses that play active roles in viral pathogenesis.
RESULTS
The 17K protein of BYDV-GAV disrupts fission yeast mitosis
The genome of BYDV-GAV and related BYDV members encodes seven proteins (Fig. 1A). They are involved in viral replication (P1 and P2), virion assembly [P3, the coat protein (CP)], spread in host plants (P3a and P4), transmission by aphid vector (P5), and suppression of RNA silencing (P4 and P6) (13–15, 23, 24). For this work, all BYDV-GAV proteins were expressed in their wild-type (WT) forms or as C-terminal fusion proteins of green fluorescent protein (GFP). All seven GFP fusion proteins were successfully expressed in fission yeast cells (fig. S1A). GFP fusion variants of the P1, P2, P3, P3a, P5, and P6 proteins were found to be distributed throughout the cell, whereas the P4 fusion protein was predominantly located in the nucleus, where it formed aggregates of various sizes associated with the nuclear envelope (NE) (fig. S1B). The same localization pattern was also observed for a P4-GFP fusion ectopically expressed in cultured insect cells and in Arabidopsis roots cells by our previous studies (25, 26). As shown in Fig. 1B, the expression of P4, but not that of the remaining six BYDV-GAV proteins, caused strong inhibition of S. pombe growth. This inhibition also occurred in the cells expressing the GFP fusion of P4 (fig. S1C). P4 is highly conserved in a wide range of cereal-infecting BYDVs and related poleroviruses, with a molecular weight around 17 kDa (hence designated as 17K hereafter) (22, 26). This protein is becoming an important focus of the research on BYDV pathogenesis because of its involvement in systemic virus movement and suppression of RNA silencing in host tissues (21–23).
Fig. 1. Inhibition of fission yeast cell growth and mitosis by BYDV-GAV 17K.
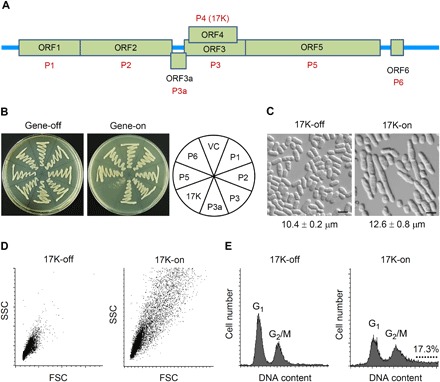
(A) The seven ORFs of BYDV-GAV and their corresponding proteins. (B) Effect of inducible BYDV-GAV protein production on fission yeast colony formation. Gene-off, no production of BYDV-GAV protein; gene-on, inducible production of BYDV-GAV protein. VC, vector control. Cells were streaked onto agar plates, incubated at 30°C, and recorded at 5 days after inoculation. (C) Cell elongation induced by 17K expression in fission yeast. Cell image was captured at 24 hours after 17K induction. Average cell length values of the two cultures differed significantly (P < 0.0001, Student’s t test). Scale bars, 10 μm. (D) Distribution of fission yeast cell lengths in low-nitrogen EMM with or without 17K production as analyzed by forward scatter analysis of 10,000 cells per culture. Cells were collected at 40 hours after 17K induction. FSC, forward scatter; SSC, side scatter. (E) Effect of 17K expression on nuclear DNA content of fission yeast cells as determined by flow cytometry at 40 hours after 17K induction. The dotted line indicates polyploid nuclei in the cells expressing 17K. The datasets shown above were each repeated three times with comparable results obtained. Photo credits: Judit Antal and Zsigmond Benko (Children’s Memorial Institute for Education and Research, Northwestern University Feinberg School of Medicine, Chicago, IL 60614, USA).
The inhibitory effect of 17K on the colony formation of fission yeast (Fig. 1B and fig. S1C) could be the result of cellular growth inhibition or cell death. To differentiate these two possibilities, we measured the growth kinetics of 17K-producing yeast cells. Fission yeast cells were grown under 17K-suppressing and 17K-inducing conditions, respectively, in the liquid Edinburgh minimal medium (EMM). Cellular growth was measured by cell density from 0 to 44 hours after 17K induction. While the 17K-suppressing cells continued to grow into stationary phase, the 17K-producing cells showed substantial growth delay (fig. S1D). Microscopic observation of the 17K-on versus 17K-off cells showed that the induction of 17K expression significantly increased cell lengths (12.6 ± 0.8 μm versus 10.4 ± 0.2 μm) (Fig. 1C). The 17K-mediated cell elongation was verified through a forward scatter analysis in which a total of 10,000 cells were measured (Fig. 1D). Further analysis of cell size distribution indicated that 17K-induced cell elongation increased over time (fig. S1E). Flow cytometry analysis of fission yeast nuclear DNA contents showed that, in the absence of 17K expression, 68.3% of the cells were in the G1 phase and 31.7% of them were in the G2 phase (Fig. 1E, left). In contrast, with 17K expression, there was a clear shift of the cells from G1 (40.6%) to G2/M (42.1%). In addition, a substantial cell population (17.3%) had nuclear DNA content values larger than 2 N (Fig. 1E, right), indicating that 17K affected mitotic G2/M transition and possibly halted the onset of mitosis.
To test this possibility, we analyzed the septation index of 17K-producing cells, which measures the percentage of cells passing mitosis as shown by septum formation between the dividing daughter cells (18). As shown in fig. S1F, the septation index in the 17K-off culture gradually decreased from about 15 to nearly 0% when the cells reached stationary phase after 26 hours of growth; but in the 17K-on culture, the septation index remained to be 20 to 25% at 26 hours, which was correlated with an overall slow cellular growth, suggesting that little or no mitosis occurred during the test period. Collectively, these findings reveal that the 17K protein inhibits fission yeast cell growth by disrupting mitosis, which is caused by the delay of G2/M transition as evidenced by increased cell elongation and accumulation of high nuclear DNA contents in the 17K-expressing cells.
17K suppresses barley growth and disrupts mitosis in root tip cells
To study the effect of 17K on plant mitosis, we mainly used the root samples because plant root tips are convenient materials for monitoring mitosis (2). Barley seedlings, inoculated with the aphids (Schizaphis graminum) carrying BYDV-GAV, were hydroponically cultured to investigate the effect of virus infection on root growth and mitotic cell cycle. The 17K and CP transcripts of BYDV-GAV were detected in both the differentiation and elongation zones (DZ and EZ) of barley primary root tips as early as 2 days post inoculation (DPI), but the virus was not detected in the mitotic zone (MZ) (Fig. 2A). BYDV-GAV infection decreased plant height and became more severe over time (Fig. 2B and fig. S2A). At 7 DPI, it was clear that the infection also reduced the maximum root lengths and total root lengths, and these phenotypes became more severe as the infection progressed (Fig. 2B and fig. S2, B and C).
Fig. 2. Suppression of barley mitosis by 17K.
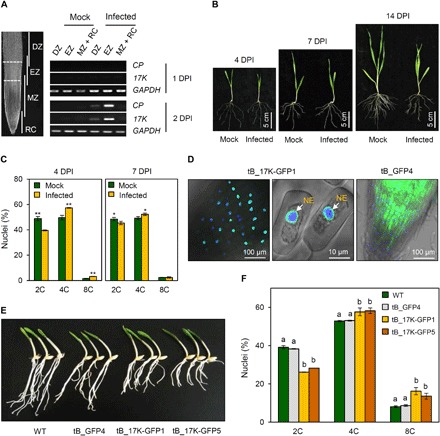
(A) Organization of DZ, EZ, MZ, and root cap (RC) in barley root tips. Dash lines indicate the cuts for preparing DZ, EZ, and MZ + RC samples. Amplification of barley GAPDH gene served as an internal control. (B) Growth of BYDV-GAV–infected barley seedlings and mock controls examined at 4, 7, and 14 DPI, respectively. (C) Analysis of nuclear DNA contents by flow cytometry using root tip cells from BYDV-GAV–infected barley seedlings or mock controls at 4 or 7 DPI. The means (± SE) were calculated from four separated experiments. *P < 0.05 and **P < 0.01 (Student’s t test). (D) Localization of 17K-GFP in transgenic barley line tB_17K-GFP1. (E) The root growth of barley seedlings was inhibited in 17K transgenic lines (tB_17K-GFP1 and tB_17K-GFP5) comprised to WT and GFP transgenic line (tB_GFP4). (F) Flow cytometry analysis of root tip cells from WT and three transgenic barley lines. The means (± SE) were each calculated from three independent assays, with significantly different values labeled by nonidentical letters [P < 0.05, analysis of variance (ANOVA) and least significant difference test (LSD) for multiple comparisons]. Photo credits: Huaibing Jin and Yanjing Zhang (State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China).
Next, we examined subcellular distribution of 17K in the barley roots naturally infected by BYDV-GAV using whole-mount immunofluorescence staining (27) with 17K-specific antibody. Judging from the specific immunofluorescence signals obtained, 17K was mainly located in the nucleus but was also detected in the cytoplasm and along the plasma membrane (fig. S2D). This is consistent with the previous finding of 17K in the nucleus and cytoplasm in BYDV-PAV–infected oat cells by immunogold labeling electron microscopy (22). To examine whether BYDV-GAV infection may affect mitosis, we analyzed barley root tip cells using flow cytometry at two early viral infection time points. At 4 DPI, the proportion of 4C nuclei was significantly higher in BYDV-GAV–infected root tips (57.30%) than mock controls (49.62%), and the percentage of polyploid nuclei (8C) was considerably elevated in BYDV-GAV–infected root tips (3.19%) relative to that in mock controls (1.60%) (Fig. 2C). In contrast, the number of 2C nuclei was substantially higher in the mock controls (48.77%) than in virus-infected plants (39.51%) (Fig. 2C). Similar differences were also observed at 7 DPI (Fig. 2C).
We further tested whether ectopically expressing 17K in barley disrupts mitosis. Two independent transgenic lines expressing 17K-GFP fusion protein (tB_17K-GFP1 and tB_17K-GFP5) and one line expressing free GFP (tB_GFP4) were generated and analyzed. Immunoblot analysis with anti-GFP antibody confirmed the expression of 17K-GFP in tB_17K-GFP1 and tB_17K-GFP5 and free GFP in tB_GFP4, with considerable degradation of 17K-GFP observed in the transgenic barley plants (fig. S2E). The 17K-GFP protein was located mainly in the nucleus (especially the NE) in tB_17K-GFP1 and tB_17K-GFP5, while free GFP was distributed throughout the cell in tB_GFP4 (Fig. 2D).
The root growth of tB_17K-GFP1 and tB_17K-GFP5 seedlings was apparently inhibited compared to WT controls, but such inhibition was not observed in tB_GFP4 seedlings (Fig. 2E). Compared to WT control, the maximum root lengths and total root lengths were significantly reduced in tB_17K-GFP1 and tB_17K-GFP5, but not in tB_GFP4 (fig. S2, F and G). The percentage of 2C nuclei was considerably decreased in tB_17K-GFP1 and tB_17K-GFP5 compared to WT control and tB_GFP4 (Fig. 2F). But the proportion of 4C nuclei was increased in tB_17K-GFP1 (57.62%) and tB_17K-GFP5 (58.19%) relative to WT control (52.82%) and tB_GFP4 (53.07%) (Fig. 2F). The proportion of 8C polyploid nuclei was also elevated in tB_17K-GFP1 (16.19%) and tB_17K-GFP5 (13.59%) compared to WT (8.08%) and tB_GFP4 (8.66%) lines (Fig. 2F). Clearly, BYDV-GAV infection, as well as transgenic expression of 17K alone, decreases barley growth, which is accompanied by accumulation of polyploid cells in the root tips.
17K inhibits nonhost plant growth
A. thaliana and N. benthamiana (tobacco) were chosen as two nonhost species to investigate whether inhibition of growth and disruption of mitosis by 17K is a general phenomenon in plants. For Arabidopsis, we compared WT plants with homozygous and stable transgenic lines including tA_17K-GFP, tA_17K-His, and tA_GFP, which expressed 17K-GFP, histidine-tagged 17K, or free GFP. Transgene expression was under the control of an estradiol (ES)–inducible promoter, and the 17K-GFP fusion was confirmed to be correctly expressed and primarily located in cell nuclei by our previous study (26).
We found that induction of 17K expression severely inhibited the shoot and root growth of Arabidopsis seedlings (Fig. 3, A and B, and fig. S2, H and I). A flow cytometry assay of the root tips showed that tA_17K-GFP had significantly fewer cells with 2C and 4C nuclei than tA_GFP, WT control, and the tA_17K-GFP without 17K induction (Fig. 3C). In contrast, the numbers of polyploid nuclei (8C, 16C, and 32C) were substantially increased in tA_17K-GFP relative to tA_GFP, WT control, and the tA_17K-GFP without 17K expression (Fig. 3C). We further examined the effect of 17K-GFP expression on Arabidopsis mitosis using the G2/M transition marker line pCYCB1;1::GUS (28). It has been reported that down-regulation of CYCB1;1 expression decreases mitosis and promotes endoreduplication (2, 28). Our analysis showed that, relative to the parental marker line pCYCB1;1::GUS, CYCB1;1::GUS expression was severely decreased in the F1 seedlings into which the 17K-GFP transgene was introduced by artificial crossing with the transgenic line tA_17K-GFP (Fig. 3D), thus indicating the delay of G2/M transition by 17K expression in Arabidopsis.
Fig. 3. Inhibition of nonhost plant growth and mitosis by 17K.
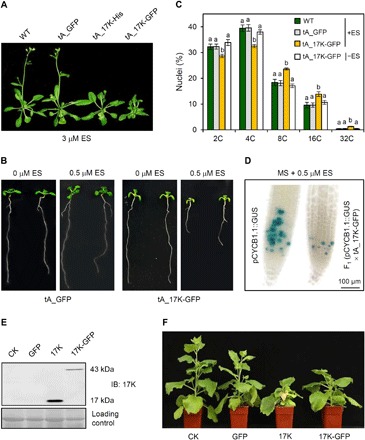
(A) Effects of 17K expression on Arabidopsis growth analyzed using transgenic lines (tA_17K-His and tA_17K-GFP) with inducible expression of 17K by ES and a control line expressing free GFP. (B) Reduction of primary root growth by inducible expression of 17K in transgenic Arabidopsis. (C) Measurement of nuclear DNA contents. DNA contents in the root tip cells of tA_GFP, tA_17K-GFP, or WT control cultured with 0.5 μM ES (+ES) or tA_17K-GFP without ES (−ES) were measured by flow cytometry. The means (± SE) were each calculated from three separate experiments, with significantly different values labeled by nonidentical letters (P < 0.05, ANOVA and LSD for multiple comparisons). (D) Compared to the parental marker line pCYCB1;1::GUS (28), CYCB1,1::GUS signals (represented by the blue precipitates) were substantially decreased in root tips of the F1 seedlings derived from crossing pCYCB1.1::GUS with tA_17K-GFP, with 17K expression induced by ES. (E) Validation of PEBV vector–mediated expression of 17K and 17K-GFP in N. benthamiana using immunoblotting (IB). (F) Inhibition of N. benthamiana growth by 17K or 17K-GFP. CK, mock control (infiltrated with water). The datasets presented were each typical of three independent experiments. Photo credits: Huaibing Jin and Zongliang Xia (State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China).
In the experiment with N. benthamiana, we expressed 17K, 17K-GFP fusion, and GFP using the pea early browning virus (PEBV)–based vector (29). The expression of 17K and 17K-GFP in N. benthamiana was confirmed by immunoblotting with the antibody against 17K (Fig. 3E). Plants expressing 17K or 17K-GFP were dwarfed compared to uninfected controls and those expressing free GFP (Fig. 3F and fig. S2J). Together, the above results demonstrate that ectopic expression of 17K alone can inhibit nonhost plant growth, which is associated with delayed G2/M transition and disrupted mitosis.
17K interacts with Cdc25 and CDKA/Cdc2 but not Wee1
The delay of G2/M transition by 17K expression in fission yeast and plant cells led us to investigate whether 17K may interrupt the function of the highly conserved Wee1-Cdc25-Cdk1/CDKA/Cdc2 mitotic entry control system, which plays a pivotal role in the onset of mitosis in eukaryotic mitotic cycle (5, 6, 12, 30). In principle, entry to mitosis requires the activation of Cdk1/CDKA/Cdc2 at late G2 phase. Before activation, Cdk1/CDKA/Cdc2 is inhibited by Wee1 kinase–catalyzed phosphorylation on its threonine 14 (T14) and tyrosine 15 (Y15) residues, respectively. To trigger mitotic entry, the inhibitory T14/Y15 phosphorylation in Cdk1/CDKA/Cdc2 is removed by the dual-specific phosphatase Cdc25; the activated Cdk1/CDKA/Cdc2, in turn, inhibits Wee1 but further promotes Cdc25 activity, thus generating a bistable switch that permits the transition of cells from G2 to M phases. In addition, another bistable switch composed of B55:PP2A has recently been identified to regulate G2/M transition in mammalian cells (30). However, the conservation and function of this switch in plant cells are still unclear. We therefore focused on investigating the potential effect of 17K on Wee1-Cdc25-CDKA/Cdc2.
As the first step, we tested whether 17K may physically interact with plant Wee1, Cdc25, and CDKA proteins. Plant Wee1 is capable of binding CDKA and inhibiting its activity in vivo, and overexpression of Arabidopsis Wee1 results in plant growth inhibition via G2 arrest (31). Plant Cdc25 has threonine and tyrosine phosphatase activities and can dephosphorylate the CDK complex purified from Arabidopsis (32). Plants are known to express multiple types of CDKs, with the CDKA members having the PSTAIRE cyclin-binding motif being most similar to Cdk1/Cdc2 (fig. S3) (2, 12). Arabidopsis CDKA;1 has multiple functions in both S and M phases and is essential throughout development (33, 34). Unlike Arabidopsis, barley has two closely related PSTAIRE-containing CDK members, HvCDKA1 and HvCDKA2, which are 85.4% identical (fig. S3).
The interaction between 17K and Arabidopsis or barley Cdc25 was first detected using yeast two-hybrid (Y2H) assay (Fig. 4A and fig. S4A) and then verified with luciferase complementation (LC) test in tobacco (Fig. 4A and fig. S4, B and C). The interactions of 17K with Arabidopsis and barley CDKAs were revealed using Y2H and LC and subsequently confirmed by coimmunoprecipitation (Co-IP) experiments carried out in the 17K-GFP–expressing transgenic lines and the tobacco cells expressing 17K-MYC and CDKA;1-FLAG fusions (Fig. 4A and fig. S4, D to K). We also verified the interaction between 17K and HvCDKA in BYDV-GAV–infected barley root cells by Co-IP assay (Fig. 4A and fig. S4L). However, no interaction was found between 17K and Wee1 using the above assays.
Fig. 4. Interruption of mitotic entry switch by 17K in plant cells.
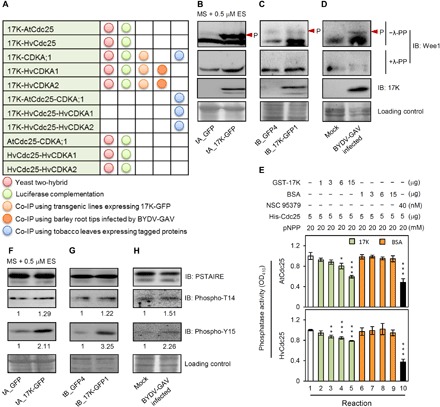
(A) The interactions detected between 17K and components of the mitotic entry switch. (B to D) Effects of 17K expression on Wee1 protein accumulation. Protein samples, extracted from transgenic Arabidopsis seedlings (B), transgenic barley root tips (C), and BYDV-GAV–infected barley root tips (D), were treated with or without λ-PP, followed by immunoblotting with anti-Wee1 antibody. Arrowhead, phosphorylated Wee1. (E) Inhibition of in vitro phosphatase activities of AtCdc25 and HvCdc25 by 17K. Ten reactions were carried out with the reagents listed on top. To facilitate comparison, the OD410 (optical density at 410 nm) absorbance in reaction 1 was set as 1. Each mean (± SE) was calculated from three independent assays. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t test). BSA, bovine serum albumin; pNPP, para-nitrophenyl phosphate. (F to H) Up-regulation of the CDKA with T14 and Y15 phosphorylation by 17K. Protein samples, prepared as detailed in (B) to (D), were subjected to immunoblotting with the antibodies specific for PSTAIRE, phosphor-T14, or phosphor-Y15. Numerical values represent relative intensities of the corresponding bands calculated by ImageJ. The datasets shown were each repeated three times with comparable results obtained.
We further detected interactions among 17K, Cdc25, and CDKA in tobacco cells transiently expressing the three proteins with different tags. The 17K-MYC and Arabidopsis CDKA–hemagglutinin (HA) fusions were present in the precipitates resulted from the pulldown of Arabidopsis Cdc25-YFP (Fig. 4A and fig. S5A). Similar results were obtained among 17K and barley CDKAs and Cdc25 (Fig. 4A and fig. S5, B and C). Although the interaction between Cdc2/Cdk1 and Cdc25 is well established in fission yeast and human (5, 6), this has not been reported in higher plants. We therefore checked the interaction between plant CDKA and Cdc25. Using Y2H and LC assays, positive binding between CDKA and Cdc25 was found for both Arabidopsis and barley proteins (Fig. 4A and fig. S5, D to H).
To explore functional relevance of the observed protein-protein interactions in BYDV-GAV infection, we used a whole-mount immunostaining protocol (27) to determine whether 17K and HvCDKA occur in the same tissues and cells in infected barley root tips. Immunostaining with the 17K antibody revealed that the 17K signals were restricted to the vasculature of the DZ (fig. S5I, arrowhead), which was anticipated because the phloem cells, where BYDVs multiply (13, 14), are located in the vascular bundle. We also detected 17K accumulation in many cells of the EZ and its junction with the MZ (fig. S5I, arrow). However, no 17K was detected in the basal portion of MZ or in the root cap, consistent with the broadly held notion that plant viruses do not typically invade the meristematic tissues of host plants (35). Staining with the anti-PSTAIRE antibody showed that HvCDKA proteins were present in the MZ and in the junction between MZ and EZ in the root tips of BYDV-GAV–infected plants (fig. S5J, arrow). A similar regional distribution pattern of HvCDKA proteins was found in the root tips of mock control barley plants (fig. S5K, arrow). No specific immunostaining signals were observed in two different negative controls (fig. S5, L and M), confirming that the staining results obtained using 17K or PSTAIRE antibodies were specific. This result, together with the fact that the neighboring zones in plant root tips overlap with each other (2, 36), suggest that the cells at the junction of the mitotic and EZs accumulated BYDV-GAV 17K and expressed HvCDKA. In these cells, physical interactions occur among 17K, HvCDKA, and Cdc25, thus making it possible for 17K to disrupt G2/M transition and inhibit mitosis in BYDV-GAV–infected barley roots.
17K affects Wee1 and Cdc25 in different ways
By quantitative reverse transcription polymerase chain reaction (qRT-PCR), we found that Wee1 transcript level was significantly elevated in the Arabidopsis and barley transgenic lines expressing 17K-GFP (fig. S6, A and B). This elevation was also detected in BYDV-GAV–infected barley root tip cells (fig. S6C). We then checked Wee1 protein level by immunoblotting with the antibody specific for plant Wee1. To distinguish between phosphorylated and un-/dephosphorylated forms of Wee1, we prepared two protein samples for each of the analyzed plant materials. The first sample, used for the immunoblotting without λ–protein phosphatase (λ-PP) treatment, was prepared with the addition of protease inhibitor cocktail and PhosSTOP inhibitor in the extraction buffer (Supplementary Materials and Methods). The second sample, used for the immunoblotting with λ-PP treatment, was prepared similarly except that protease inhibitor cocktail and PhosStop inhibitor were omitted to avoid the interference of these inhibitors on λ-PP function. In the immunoblotting without λ-PP treatment (−λ-PP), the phosphorylated and the un-/dephosphorylated forms of Wee1 were up-regulated in the transgenic Arabidopsis and barley lines expressing 17K-GFP and in the barley roots infected by BYDV-GAV compared to their respective controls (Fig. 4, B to D). In the immunoblotting with λ-PP treatment (+λ-PP), the level of un-/dephosphorylated forms of Wee1 was only slightly higher in the presence of 17K than that without 17K expression (Fig. 4, B to D). Notably, the level of un-/dephosphorylated form of Arabidopsis Wee1 varied considerably in the absence and presence of λ-PP treatment, which may be caused by the different protein extraction protocols used and/or λ-PP treatment. In contrast to Wee1, the transcript level of Cdc25 was not significantly affected by 17K expression in either the Arabidopsis and barley transgenic lines expressing 17K-GFP or in the root cells of BYDV-GAV–infected barley (fig. S6, D to F).
Considering that 17K and Cdc25 interacted with each other, we tested whether the catalytic activity of Cdc25 might be inhibited by 17K in two series of in vitro phosphatase assays conducted with bacterially expressed Arabidopsis and barley Cdc25 proteins, respectively. As shown in Fig. 4E, the phosphatase activity of Arabidopsis and barley Cdc25 was substantially inhibited by 17K but not by the control protein bovine serum albumin. As anticipated, Cdc25 activity was drastically reduced in the presence of NSC 95379 (Fig. 4E), a frequently used inhibitor of Cdc25 (32). We also tested whether the presence of 17K might disturb the interaction between CDKA and Cdc25. In the LC assay, the luciferase (LUC) signal generated by CDKA and Cdc25 interaction was significantly decreased in the presence of 17K (fig. S6, G to I). Together, these data indicate that 17K interferes with the function of Wee1 and Cdc25 in different manners and that the presence of 17K negatively affects the interaction between plant (Arabidopsis and barley) CDKA and Cdc25 proteins.
17K expression causes abnormal accumulation of phosphorylated CDKA
We next analyzed the effect of 17K expression on CDKA phosphorylation in plants using anti–phospho-T14 or anti–phospho-Y15 antibodies. As depicted in Fig. 4 (F and G), the T14 and Y15 phosphorylation levels of CDKA increased in the Arabidopsis and barley transgenic lines ectopically expressing 17K-GFP. This abnormal enhancement was also found in the root cells of BYDV-GAV–infected barley plants (Fig. 4H). The amount of CDKA with Y15 phosphorylation was generally higher than that with T14 phosphorylation in the 17K-expressing cells (Fig. 4, F to H). Thus, consistent with the interruption of Wee1-Cdc25-CDKA by 17K, cells expressing 17K abnormally accumulate higher amounts of CDKA with T14 and Y15 phosphorylation.
Silencing HvCDKA aggravates host dwarfing and elevates virus accumulation
To assess the functional importance of 17K effects on Wee1-Cdc25-CDKA in BYDV-GAV–infected host plant, we decreased the expression of HvCDKA using barley stripe mosaic virus (BSMV)–mediated gene silencing (37). The barley plants with or without HvCDKA silencing were further inoculated with the aphids carrying BYDV-GAV or virus-free aphids, resulting in four groups of plants differing in HvCDKA silencing and BYDV-GAV infection. They were then analyzed at 3 weeks after virus inoculation.
As anticipated, HvCDKA transcript and protein levels were generally lower in the plants subjected to gene silencing treatment (+S) than those without such treatment (−S) (fig. S7, A and B). The group of plants with silenced HvCDKA expression and BYDV-GAV infection (i.e., +S/+BYDV-GAV) developed the most severe dwarfing phenotype compared to the other three groups (fig. S7, C and D). Immunoblot analysis confirmed that the protein levels of 17K and CP were higher in the +S/+BYDV-GAV plants than those in the −S/+BYDV-GAV individuals (fig. S7B). These data indicated that silencing HvCDKA expression led to increased BYDV-GAV accumulation, which was further supported by comparing BYDV-GAV CP and 17K transcript levels between these two groups of plants (fig. S7, E and F). Thus, beyond exacerbating the dwarfing phenotype, reducing the expression of HvCDKA by gene silencing also increases BYDV-GAV viral loads in barley.
17K interrupts fission yeast mitotic entry switch Wee1-Cdc25-Cdc2
Because 17K expression in fission yeast cells delayed G2/M transition (Fig. 1), it was necessary to investigate whether the 17K may interrupt fission yeast Wee1-Cdc25-Cdc2. We detected the interaction of 17K with fission yeast Cdc2 and Cdc25 using Y2H (fig. S8, A and B), but again, no interaction was found between 17K and fission yeast Wee1. We also found that the cells expressing 17K accumulated higher levels of phosphorylated and unphosphorylated Wee1 and phosphorylated Cdc2 than those without 17K expression (fig. S8, C and D). Quantitative PCR assay showed that Wee1 transcript level was clearly increased in the fission yeast cells with 17K expression, but Cdc25 transcript level was not significantly affected by 17K expression (fig. S8, E and F).
Furthermore, we carried out genetic tests in several fission yeast cell cycle mutants to explore the functional involvement of Wee1-Cdc25-Cdc2 in 17K-induced delay of G2/M transition. The induction of abnormal cell elongation, and hence delay of G2/M transition, by 17K in fission yeast occurred in the mutant strain cdc2-3w but not in cdc2-1w (fig. S8G). Because cdc2-1w is partially resistant to Cdc2 phosphorylation and cdc2-3w is tolerant to Cdc2 dephosphorylation (38), this result indicates that efficient phosphorylation of Cdc2 is required for 17K-induced delay of G2/M transition in fission yeast. In the test with the temperature-sensitive wee1-50 mutant strains (38), the induction of cell elongation by 17K expression was gradually weakened as temperature rose from 25.5° (permissive to Wee1 function) to 30° (semipermissive) or 36.5°C (nonpermissive) (fig. S8H), suggesting that induction of abnormal cell elongation (and delay of G2/M transition) by 17K expression in fission yeast requires the function of Wee1.
Similarities among BYDV-GAV 17K, HIV-1 Vpr, and ARV p17 in the disruption of mitosis
The delay of G2/M transition and disruption of mitosis by BYDV-GAV 17K via directly targeting mitotic entry switch resembles that reported for HIV-1 Vpr and ARV p17 (see Introduction). This prompted us to investigate whether there might exist structural and mechanistic similarities among 17K, Vpr, and p17 in mitosis disruption. Amino acid sequence identities were low (less than 13.7%) among the three proteins, which propelled us to investigate whether they might share similar secondary structural features. Previously, we predicted that the N-terminal region of 17K contained mainly four α helices (25, 26). Here, we verified this prediction using secondary structure modeling (Fig. 5A, left). Furthermore, circular dichroism (CD) analysis indicated that recombinant 17K was predominantly helical (Fig. 5B and fig. S9A). The secondary structure of Vpr, as revealed by previous CD and nuclear magnetic resonance studies (39), was also largely helical with three α helices in the N-terminal part (Fig. 5A, middle). The secondary structure of ARV p17 is still unknown, but our computational modeling indicated that the main secondary structural feature of p17 included four α helices in its N-terminal portion (Fig. 5A, right). Vpr and p17 could interact with the CDKAs of tobacco, Arabidopsis, and barley in Y2H assay, and their N terminal region was sufficient for the interaction (fig. S9B). When ectopically expressed in tobacco using the PEBV vector, Vpr, and p17, as well as Vpr-GFP and p17-GFP fusion proteins, inhibited plant growth (Fig. 5C and fig. S9, C and D), which were accompanied by increased polyploid cells (Fig. 5D). This result led us to test whether 17K might negatively affect animal cell mitosis by ectopically expressing 17K in the human embryonic kidney (HEK)–293Ta cells (fig. S9E). 17K expression inhibited the growth of HEK-293Ta cells, which was associated with enlarged cell size (Fig. 5, E and F). Measurement of nuclear DNA contents at two time points (12 and 24 hours after transfection) showed that 17K expression significantly elevated the proportion of the cells at G2/M compared with the vector control (Fig. 5G).
Fig. 5. Similarities among the secondary structures of BYDV-GAV 17K, HIV-1 Vpr, and ARV p17 and in the inhibition of tobacco growth.
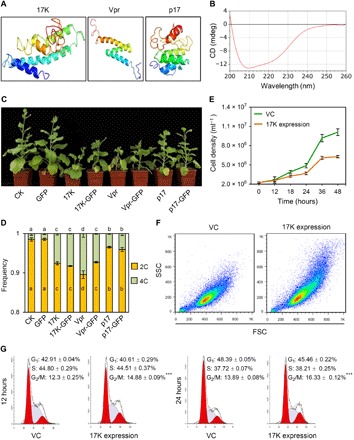
(A) Secondary structure of 17K, Vpr, or p17 modeled with QUART software (47). (B) CD spectrum of recombinant 17K. Two negative peaks around 208 and 222 nm indicated that 17K was mainly composed of α helices. (C) Inhibition of tobacco growth by 17K, 17K-GFP, Vpr, Vpr-GFP, p17, or p17-GFP expressed using PEBV vector. (D) Nuclear DNA contents in young leaves of the tobacco plants displayed in (C). The means (± SE) were each calculated from three independent flow cytometry assays, with significantly different means labeled by nonidentical letters (P < 0.05, ANOVA and LSD for multiple comparisons). (E to G) Effects of 17K expression on HEK-293Ta cell proliferation (E), cell size distribution (analyzed by flow cytometry with 30,000 cells per culture) (F), and cell cycle progression at 12 and 24 hours after transfection (G). The means (± SE) in (G) were each calculated from four separated experiments. ***P < 0.001 (Student’s t test). The datasets presented were representative of at least three separate experiments. Photo credits: Huaibing Jin (State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China).
On the basis of the data presented above, we examined whether there might be amino acid residues that were important for the disruption of mitosis by both 17K and Vpr. Amino acid sequence alignment revealed 11 conserved residues between the two proteins (Fig. 6A). The two leucine residues L22 and L67 and the arginine amino acid R88 in Vpr have previously been found involved in the delay of G2/M transition by this protein (40–42). Therefore, we mutated the three residues to alanines in both Vpr and 17K and tested the resulting mutants, i.e., 17K-L48A/L96A/R117A and Vpr-L22A/L67A/R88A, in tobacco as described above. Compared to WT Vpr and 17K, the inhibition of tobacco growth by 17K-L48A/L96A/R117A and Vpr-L22A/L67A/R88A was substantially reduced, which was paralleled by significant decreases in the proportion of polyploid cells (Fig. 6, B to D). In line with this finding, 17K-L48A/L96A/R117A and Vpr-L22A/L67A/R88A failed to interact with the CDKAs of tobacco, Arabidopsis, and barley, although their WT counterparts did so effectively (Fig. 6E). These results, together with the data reported in Fig. 5, suggest the existence of structural and mechanistic similarities in the disruption of mitosis by 17K, Vpr, and p17.
Fig. 6. Analysis of the three residues conserved between 17K and Vpr and their requirement for efficient disruption of mitosis.
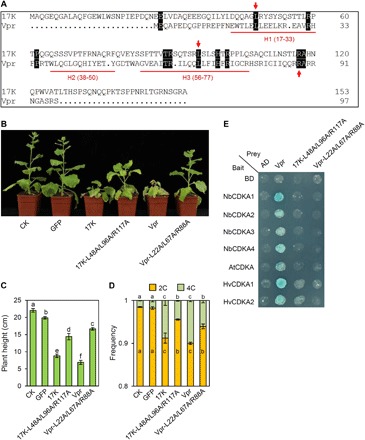
(A) Amino acid sequence alignment of BYDV-GAV 17K and HIV-1 Vpr. H1 (17-33), H2 (38-50), and H3 (56-77), respectively indicate the three α helices of Vpr. Red arrows mark the three residues that have been reported to function in Vpr-induced G2/M arrest (40–42). (B and C) Decreased inhibition of tobacco growth by the 17K mutant 17K-L48A/L96A/R117A and the Vpr mutant Vpr-L22A/L67A/R88A. 17K, Vpr, and their corresponding mutants were expressed in N. benthamiana using the PEBV vector. The graph was taken at 4 weeks after agroinfiltration. (D) Nuclear DNA contents in the young leaves of the tobacco plants shown in (B). The means (± SE) were each calculated from three separated flow cytometry assays, with significantly different means labeled by dissimilar letters (P < 0.05, ANOVA and LSD for multiple comparisons). (E) The 17K mutant 17K-L48A/L96A/R117A and the Vpr mutant Vpr-L22A/L67A/R88A failed to interact with the CDKAs of N. benthamiana, Arabidopsis, and barley in Y2H assays. As a control, WT Vpr showed positive interactions with the same set of CDKAs. The datasets presented above were each typical of three independent assays. Photo credits: Huaibing Jin (State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China).
DISCUSSION
In this study, we demonstrated the delay in G2/M transition and disruption of mitosis caused by BYDV-GAV 17K, and revealed the underlying mechanism of these effects using a variety of molecular, biochemical, and genetic approaches. Parallel analysis of BYDV-GAV–infected barley host plants not only supported the findings in nonhost species (e.g., fission yeast and Arabidopsis) and transgenic lines but also revealed their significance in the induction of host dwarfism by BYDVs in nature. Furthermore, we found a high similarity in the mechanism underlying the disruption of mitosis by BYDV-GAV 17K, HIV-1 Vpr, and ARV p17 and discovered a previously unidentified class of plant and animal viral proteins that disrupt mitosis by directly targeting and interrupting the conserved mitotic entry switch Wee1-Cdc25-Cdk1.
A new function identified for BYDV 17K
Most plant viruses encode one or more proteins that assist their spread in host tissues (43). More recently, a number of plant virus proteins have been shown to have RNA silencing suppression activity to counteract host defense (44). Several viral proteins, including the 17K of BYDVs, can perform both functions (43, 44). Here, we demonstrate that expression of 17K induces a delay in G2/M transition and disruption of mitosis in both host and nonhost organisms through interrupting the conserved mitotic entry switch Wee1-Cdc25-Cdk1, thus identifying a new function for the 17K protein conserved in a wide range of cereal-infecting BYDVs and related poleroviruses. Apart from BYDV-GAV 17K, none of the others have been reported to disrupt mitosis in both host and nonhost species by perturbing the mitotic entry switch. Hence, the 17K protein of BYDVs is unique from previously studied plant viral proteins. This expands our understanding of the spectrum of functions performed by plant virus proteins.
Because 17K can act as a viral suppressor of RNA silencing (VSR) in host cells (23), it will be interesting and important to investigate whether the mitosis disruption function of 17K may be linked with its VSR activity in future research. In a preliminary analysis, we found that the 17K-L48A/L96A/R117A mutant, whose mitosis disruption function was clearly compromised (Fig. 6, B to E), did not differ substantially from WT 17K in VSR activity (fig. S9, F to H). Thus, it is unlikely that the mitosis disruption function of 17K is closely linked with its VSR activity. Nevertheless, more work is still needed to examine whether the debilitation of 17K’s VSR activity may negatively affect its disruption of host mitosis.
In the host plants naturally infected by BYDV, 17K was found present in the nucleus and cytoplasm and along the plasma membrane in this work (fig. S2D) and by a previous study (22). The finding of 17K in the cytoplasm and along the membrane is in line with its role in assisting BYDV movement in host plants (21, 22). The presence of 17K in the nucleus facilitates its disruption of mitosis and suppression of RNA silencing in BYDV-infected host tissues. Therefore, the existence of 17K in multiple subcellular locations is consistent with its multifunctionalities in BYDV infections.
17K disrupts mitosis by delaying G2/M transition through directly targeting and interrupting the mitotic entry switch Wee1-Cdc25-Cdk1/CDKA/Cdc2
We found that abnormal accumulation of polyploid nuclei occurred in the fission yeast (Fig. 1E), barley (Fig. 2F), Arabidopsis (Fig. 3C), tobacco (Fig. 5D), and human cells (Fig. 5G) ectopically expressing 17K (or 17K-GFP) and in the root cells naturally infected by BYDV-GAV (Fig. 2C), suggesting that delay of G2/M transition is a common step in the disruption of mitosis by 17K in both host and nonhost organisms. This proposition is additionally supported by abnormally increased cell size in the fission yeast and human cells expressing 17K (Figs. 1, C and D, and 5F) and by the decrease of pCYCB1;1::GUS expression in the 17K-expressing Arabidopsis roots (Fig. 3D).
We further reveal that the impediment of G2/M transition in barley and Arabidopsis cells by 17K is triggered by directly interrupting Wee1-Cdc25-CDKA via multiple means, including protein-protein interactions and alteration of CDKA phosphorylation. First, 17K may up-regulate Wee1’s function. This is supported by an elevated transcript level of Wee1 and enhanced accumulation of both phosphorylated and unphosphorylated forms of Wee1 upon 17K expression (Fig. 4, B to D, and fig. S6, A to C). Second, it is highly likely that 17K lowers Cdc25’s phosphatase activity in vivo because of its capabilities to physically bind to plant Cdc25 (Fig. 4A and fig. S4, A to C) and to inhibit Cdc25’s catalytic activity in vitro (Fig. 4E). Third, 17K may interfere with CDKA’s activities via its direct binding to CDKA (Fig. 4A and fig. S4, D to L). This is supported by the result that 17K negatively affected the interaction between CDKA and Cdc25 in plant cells (fig. S6, G to I). Together, these processes may bring multiple abnormalities to Wee1-Cdc25-CDKA, leading to abnormal accumulation of phosphorylated CDKA, inadequate activation of CDKA, and thus the delay of G2/M transition. A direct evidence supporting the above scenario is the finding of higher levels of CDKA with T14 and Y15 phosphorylation in the 17K-expressing Arabidopsis and barley cells (Fig. 4, F to H). The observation that the CDKA with Y15 phosphorylation accumulated to a larger extent than that with T14 phosphorylation in 17K-expressing cells points to the possibility that the two types of phosphorylated CDKA may not have identical roles in the interruption of Wee1-Cdc25-CDKA and delay of G2/M transition by 17K.
Many of the negative effects exerted by 17K on the mitotic entry control system found in plant cells were also observed in fission yeast, suggesting that the mechanism underlying the interruption of mitotic entry switch by 17K is conserved in plant and fission yeast cells. Furthermore, analysis of the mutant fission yeast strains cdc2-1w, cdc2-3w, and wee1-50 clearly demonstrated the critical importance of the step of Cdc2 phosphorylation by Wee1 in the delay of G2/M transition by 17K (fig. S8, G and H). This type of experiment is difficult to perform in plants because of the lack of well-defined mitotic mutants. However, the findings made with cdc2-1w, cdc2-3w, and wee1-50 strains lend support to the functional significance of the interruptions of Wee1, Cdc25, and CDKA by 17K in its delay of G2/M transition in plant cells. Consequently, the availability of fission yeast mitotic mutants will assist further dissection of the questions that remained to be answered behind the interruption of Wee1-Cdc25-CDKA/Cdc2. For example, how does 17K up-regulate Wee1 expression and the accumulation of both phosphorylated and unphosphorylated Wee1? What are the full consequences of 17K binding to Cdc25 and CDKA/Cdc2 on their activities? The answer of these questions may also enrich the understanding of the regulation of mitotic cycle under stress conditions, which is still understudied in plant cells (12).
17K is an important and early trigger of host dwarfism
From the data gathered in this work and the points discussed above, we propose that the 17K protein of BYDVs is very likely an important and early trigger of host dwarfism. The evidence supporting this proposition includes (i) 17K was the only one of the seven BYDV-GAV proteins with mitosis suppression activity in fission yeast (Fig. 1); (ii) 17K, whether transgenically expressed alone or naturally expressed in BYDV-GAV–infected cells, inhibited barley root growth through disrupting G2/M transition and mitosis via interrupting the mitotic entry control system (Figs. 2 and 4 and figs. S4 to S6); and (iii) the perturbation of G2/M transition and mitosis by 17K occurred early during BYDV-GAV infection because the virus was found present in host root tips at 2 DPI (Fig. 2A). BYDV-GAV 17K may also affect mitotic cycle in other parts of infected barley (e.g., shoot apex), and complex physiological changes may subsequently occur in the host as BYDV-GAV infection progresses. Although these disruptions may also contribute to the full development of host symptoms, they represent later events because the root has long been known to be the first organ plant viruses spread to after entering host cells (35, 43). Thus, early viral spread to the root and subsequent interruption of mitotic entry control system in the root tip by 17K are critical events in the induction of host dwarfing symptom by BYDVs.
Consistent with the triggering of host dwarfism by 17K via interrupting the mitotic entry control system Wee1-Cdc25-CDKA, we found that silencing HvCDKA expression aggravated the dwarfing symptom of BYDV-GAV–infected plants and increased viral accumulation (fig. S7). It is possible that silencing HvCDKA expression leads to further decrease of HvCDKA activity on top of the reduction conferred by viral 17K, thus exacerbating the disruption of mitosis and the inhibition of host cell and organ growth by BYDV-GAV. Disruption of host cell mitosis is beneficial to the multiplication of animal viruses because of reducing the interference of cell division on viral replication and spread (4). This may also explain increased viral accumulation in the HvCDKA-silenced and BYDV-GAV–infected barley. On the basis of this reasoning and the points discussed above, we speculate that disruption of mitosis by 17K may contribute to BYDV proliferation in naturally infected host plants through decreasing the perturbance of cell division and related processes on viral replication and spread.
Lastly, it is worthwhile to point out that the important role of 17K in triggering host (barley) dwarfism may need a full BYDV infection or additional BYDV-encoded proteins because transgenic expression of 17K-GFP alone in barley did not cause marked inhibition of plant growth (Fig. 2E). Considerable degradation of 17K-GFP was observed in transgenic barley lines (fig. S2E). But the transgenically expressed 17K-GFP was found to be relatively more stable in Arabidopsis by our previous study (26) and in this work (Fig. 4B); the 17K and 17K-GFP ectopically expressed in N. benthamiana were also fairly stable (Fig. 3E). Moreover, the higher stability of 17K-GFP (17K) in Arabidopsis and N. benthamiana appeared to result in stronger plant growth inhibition (Figs. 3, A, B, and F ; 5C; and 6B). Therefore, the disruption of mitosis and the resultant growth inhibition by 17K in host and nonhost plants may be regulated in a complex manner, which requires more studies to be fully understood.
Convergence in the interruption of mitotic entry control system by plant and animal viral proteins
Although there are increasing interests in studying the effects of plant virus infection on host cell division and growth (10, 11), to our knowledge, BYDV-GAV 17K represents the first plant viral protein that is found to disrupt mitosis in both host and nonhost organisms by delaying G2/M transition via directly targeting and interrupting the conserved mitotic entry switch Wee1-Cdc25-Cdk1. The mechanism that we revealed for 17K resembled those reported for HIV-I Vpr and ARV p17 in their disruption of animal cell mitosis, suggesting convergence in the interruption of mitotic entry switch by the animal and plant viruses whose pathogenesis involves debilitation of host mitosis.
The structural basis underpinning the interruption of mitotic entry switch by 17K, Vpr, and p17 may involve α helices in the N-terminal region and critical amino acid residues of the three viral proteins. This reasoning is supported by (i) a shared secondary structural feature among 17K, Vpr, and p17, i.e., possession of three or four α helices in the N-terminal region (Fig. 5, A and B); (ii) sufficiency of the N-terminal region for interacting with plant CDKA proteins (fig. S9B); and (iii) decreased potency of 17K and Vpr to inhibit plant growth, to cause polyploid cells, and to interact with plant CDKA proteins after mutating the three residues conserved between the two proteins (Fig. 6, B to E). Our results, plus previous knowledge on the mitotic disruption properties of Vpr and p17 (4, 9, 18), suggest that 17K, Vpr, and p17 may represent a previously unidentified class of mitotic regulators that are conserved between plant and animal viruses and play active roles in viral pathogenesis.
In summary, our study reveals that a plant viral protein (BYDV 17K) disrupts mitosis in both host and nonhost cells through directly targeting and interrupting the fundamental mitotic entry switch Wee1-Cdc25-Cdk1/CDKA/Cdc2 and that BYDV 17K contributes to host dwarfing symptom via its disruption of mitosis. The new insights obtained on 17K, together with previous findings on its involvement in viral systemic spread and suppression of host RNA silencing (21–23), highlight 17K as a key for systematically dissecting the molecular pathogenesis of BYDV infection in future research. Our work also exposes commonality in the molecular mechanism used by plant and animal viral proteins to interrupt host cell mitosis, thus providing a valuable direction for comparative analysis of the pathogenesis between plant and animal viruses. The capacity to interfere with the function of mitotic entry control system in fission yeast, plant, and human cells may prompt the use of 17K for further resolving the molecular processes governing eukaryotic cell cycle control under normal or stressed conditions.
MATERIALS AND METHODS
Fission yeast strains, transgenic plants, human cell line, gene constructs, and antibodies
All fission yeast strains, transgenic Arabidopsis and barley lines, human cell culture, and tobacco plants used in this study were listed in table S1. The gene constructs and PCR primers were provided in table S1. A total of 12 antibodies were used in this study. Eleven of them were purchased commercially or provided by other researchers, i.e., mouse monoclonal antibody to GFP (Sigma-Aldrich, 11814460001), mouse monoclonal antibody to FLAG tag (Sigma-Aldrich, F3165), mouse monoclonal antibody to MYC tag (Sigma-Aldrich, C3956), rat monoclonal antibody to HA tag (Roche, 11867423001), mouse monoclonal antibody to the PSTAIRE motif (Sigma-Aldrich, P7962), rabbit polyclonal antibody to phospho–Cdc2-Thr14 (Abcam, ab58509), rabbit polyclonal antibody to phospho–Cdc2-Tyr15 (Cell Signaling Technology, 9111), rabbit polyclonal antibody to plant Wee1 (Abcam, ab137377), rabbit polyclonal antibody to fission yeast Wee1 (Abcam, ab233540), rabbit polyclonal antibody to plant actin protein (EASYBIO, BE0027), and mouse monoclonal antibody to BYDV-GAV CP (provided by X. Zhou, Chinese Academy of Agricultural Sciences). The mouse polyclonal antibody to BYDV-GAV 17K was developed in this study using His-tagged 17K purified as described before (26). The preimmune antiserum used as a negative antibody control in the whole-mount immunostaining assay (fig. S5L) was taken from the mouse before immunization with His-tagged 17K. Details on the generation and analysis of recombinant fission yeast strains and transgenic plants were provided in the Supplementary Materials.
Flow cytometry assay
For fission yeast, the cells carrying pYZ1N-17K were inoculated into two fresh aliquots of low-nitrogen EMM (2.5 μM ammonium chloride) without or with thiamine at a density of 5 × 105 cells/ml. After 48 hours of growth, the cells were collected, fixed with 7 ml of 95% ethanol, treated with ribonuclease A (RNase A) (5 mg/ml) in 50 mM sodium citrate (pH 7.0), and stained with propidium iodine. The nuclear DNA fluorescence in individual cells was measured using a FACScan equipped with the LYSYS II software (BD Biosciences, San Jose, CA, USA).
For barley and Arabidopsis samples, the root tips were collected from the desired genotypes and chopped on ice with a razor blade in 400-μl nuclei isolation buffer [20 mM Mops, 30 mM Na citrate, 45 mM MgCl2, and 1% Triton X-100 (pH 7.0)]. The suspension was filtered through a 40-μm nylon mesh. After 15 min of digestion with RNase A (1 μg in 10 μl) at 25°C, the samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/ml) to facilitate the measurement of nuclear DNA fluorescence using a FACSAria III Fusion Flow Cytometer (BD Biosciences, San Jose, CA, USA). In the case of tobacco, the immature leaves were collected from the plants inoculated with different PEBV constructs and processed for flow cytometry assay as described above.
The HEK-293Ta cells, collected at 12 and 24 hours after 17K expression (see below), respectively, were treated in 1 ml of 70% ethanol at 4°C overnight. The fixed cells were washed three times with 1× phosphate-buffered saline (PBS) buffer (pH 7.4) and digested using RNase (100 μg/ml) in 1× PBS buffer (pH 7.4) for 30 min, followed by staining with propidium iodide. The cells were subjected to flow cytometry assay in a FACSAria II flow cytometer (BD Biosciences, San Jose, CA, USA), with the resulting data analyzed using the ModFit software (Verity Software House, Topsham, ME, USA).
Y2H assay
For Y2H, complementary DNA sequences of the appropriate genes obtained by RT-PCR were cloned into the pB42AD and pLexA vectors, respectively (table S1). Analysis of protein-protein interactions was performed according to the Yeast Protocols Handbook (www.takarabio.com/). Briefly, the bait and prey constructs were cotransformed into the yeast strain EGY48 [MATa, his3, trp1, ura3, and LexAop(x6)-LEU1 Plus p8op-lacZ] using the lithium acetate method. The positive colonies were screened on the SD-Ura/-His/-Trp medium plates at 30°C for 3 days, which were further verified on the SD/Gal/Raf/-Ura/-His/-Trp/-Leu medium containing X–β-Gal.
LC assay
For LC assay (45), the coding sequences of the genes under study were amplified by RT-PCR and cloned into the pCAMBIA1300-nLUC or pCAMBIA1300-cLUC vectors (table S1), with the aim to express N-terminal or C-terminal luciferase fusion proteins. To detect protein-protein interactions, the constructs were transformed into the Agrobacterium strain GV3101 individually, with the desired Agrobacterium strains combined and introduced onto N. benthamiana leaves through infiltration. At 60 hours after infiltration, the leaves were detached and treated with the luciferase substrate in darkness. After 10-min reaction, the signals were detected in a luminescence imaging system (CHEMIPROHT 1300B/LND, 16 bits; Roper Scientific) with an exposure time of 10 min.
Co-IP assay
Co-IP assay was performed using the root tips of barley plants infected by BYDV-GAV, transgenic Arabidopsis and barley plants with 17K expression, N. benthamiana leaves expressing 17K-MYC and Arabidopsis CDKA;1-FLAG, or N. benthamiana leaves expressing different combinations of 17K-MYC, CDKA-HA, and Cdc25-YFP. Total proteins were extracted from the desired tissues using the lysis buffer [10 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride] containing 1× protease inhibitor cocktail (Roche Diagnostics Corp). The mixture was centrifuged for 15 min (18,200g) at 4°C. The supernatant was filtered through Miracloth (Merck Millipore, 475855) and incubated with the 17K antibody cross-linked with Protein A/G (Bimake, B23201), GFP-Trap_A beads (Chromotek, gta-20), or anti-FLAG magnetic beads (Bimake, B26101) at 4°C overnight. The beads were recovered and washed four times with the washing buffer [10 mM tris-HCl (pH 7.5), 150 mM NaCl, and 0.5 mM EDTA]. The proteins were eluted by boiling in 2× SDS sample buffer for 5 min, which were then separated by 12% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting with appropriate antibodies.
Ectopic expression using PEBV vector
The PEBV-based vector was used to express 17K, 17K-GFP, 17K-L48A/L96A/R117A, Vpr, Vpr-GFP, Vpr-L22A/L67A/R88A, p17, or p17-GFP in N. benthamiana plants. As a control, free GFP was also expressed using PEBV. The coding sequences of Vpr, Vpr-GFP, p17, p17-GFP, Vpr-L22A/L67A/R88A, and 17K-L48A/L96A/R117A were synthesized (Beijing Shengyuan Kemeng Gene Biotechnology Co. Ltd.), followed by cloning into PEBV vector using the primers listed in table S1. For efficient expression in N. benthamiana plants, the codons of WT and mutant Vpr and p17 coding sequences were optimized in the website of Integrated DNA Technologies (https://sgstage.idtdna.com/pages). Cloning of 17K, 17K-GFP, or GFP into PEBV vector was accomplished likewise by PCR with specific primers (table S1). The recombinant PEBVs expressing 17K, 17K-GFP, Vpr, Vpr-GFP, p17, p17-GFP, or GFP were each introduced into the leaves of N. benthamiana plants via agroinfiltration (29). The same number of plants was infiltrated with sterile water as controls. The treated plants were examined for morphology, plant height, and accumulation of 17K, 17K-GFP, Vpr-GFP, p17-GFP, or GFP at 4 weeks after infiltration. Detection of 17K, 17K-GFP, Vpr-GFP, p17-GFP, or GFP was executed by immunoblotting with relevant antibodies.
Cdc25 phosphatase activity assay
The coding sequences of Arabidopsis and barley Cdc25 were each amplified by RT-PCR with specific primers (table S1), followed by cloning into the bacterial expression vector pET32a. The resulting constructs were expressed in the Escherichia coli BL21 (DE3) cells, with the histidine-tagged Cdc25 purified using nickel affinity chromatography (Invitrogen, K95001). BYDV-GAV 17K was expressed using the bacterial expression vector pGET-4T-1, with the GST-17K recombinant protein purified using a MagneGST protein purification system (Promega, V8603). The purified proteins were quantified using the Bradford assay and checked by SDS-PAGE. Cdc25 phosphatase activity assay was conducted as detailed previously (32). Briefly, AtCdc25 or HvCdc25 (5 μg) together with different amounts of 17K (1, 3, 6, and 15 μg) were incubated with 20 mM para-nitrophenyl phosphate in 1 ml of reaction buffer [25 mM tris-HCl, 250 mM NaCl, and 5 mM 2-mercaptoethanol (pH 7.6)] for 30 min at 30°C, with each reaction terminated by the addition of 10 μl of 3 N NaOH. The resulting absorbance was measured by a spectrophotometer at 410 nm. Bovine serum albumin was used as a control. The specific Cdc25 inhibitor, NSC 95379 (32), was also included in the assay as a positive control of the inhibition of Cdc25 activity.
Whole-mount immunostaining
Whole-mount immunostaining of barley root tips was conducted essentially as described previously (27). Briefly, the root tips, collected from BYDV-GAV–infected barley plants at 7 DPI or the mock-inoculated controls, were fixed in 1× microtubule stabilizing buffer (MTSB) containing 2% formaldehyde and 0.1% Triton X-100 for 30 min at 37°C, followed by overnight at 4°C. Then, the samples were treated by a series of alcohol solutions with the concentration gradually decreased from 100 to 20%. Cell walls were digested with a mixture of 0.2% Driselase and 0.15% macerozyme in 2 mM MES buffer (pH 5.0) for 1 hour at 37°C, and plasma membranes were permeabilized with 3% IGEPAL CA-630 and 10% dimethyl sulfoxide in 1× MTSB buffer for 30 min at 37°C. Root tips were placed in a Fast Red TR/Naphthol AS-MX solution overnight at 4°C to quench endogenous alkaline phosphatase activities. Afterward, the samples were blocked in 1× MTSB containing 2% bovine serum albumin overnight at 4°C. Subsequently, the root tips were treated with the primary antibody (anti-17K or anti-PSTAIRE) at 1:500 dilution in the blocking buffer at 4°C overnight and then the secondary antibody diluted in the blocking buffer (1:1000) at 4°C overnight. For examining subcellular distribution of 17K in BYDV-GAV–infected barley cells, the secondary antibody used was goat anti-mouse immunoglobulin G (IgG) (H+L) conjugated with daylight 488 (Invitrogen, 35502), with the resultant fluorescence signals examined under a Zeiss LSM 710 confocal microscope. The immune-stained root tips were treated with DAPI (Invitrogen, D1306) to visualize the nucleus as described before (26). For detecting 17K and HvCDKA proteins in the different regions of BYDV-GAV–infected barley root tips, the secondary antibody used was goat anti-mouse IgG conjugated with alkaline phosphatase (Sigma-Aldrich, A3562), with the signals developed using the bromochloroindolyl phosphate–nitro blue tetrazolium substrate (Promega, S3771) by incubation at 4°C. The color development was monitored under a light microscope and stopped by washing the samples with distilled water. After mounting in the Glycerol Jelly Mounting Medium, the specimens were examined and photographed using the EVOS XL microscope (Life Technologies, NY, USA). The background color in the captured images was light yellow due to the reaction of endogenous alkaline phosphatases with the Fast Red TR/Naphthol AS-MX substrate in the quenching step. The positive signals derived from specific antigen-antibody reaction were dark purple.
BSMV-mediated silencing of HvCDKA
BSMV-mediated silencing of HvCDKA was carried out as described by Yuan et al. (37). Details of the experiment were described in the Supplementary Materials.
Ectopic expression of 17K protein in HEK-293Ta cells
The codons of 17K protein coding sequence were optimized for efficient expression in human cells as described above. The resultant sequenced was synthesized commercially (Beijing Shengyuan Kemeng Gene Biotechnology Co. Ltd.) and amplified by PCR using the primers with built-in Xba I or Bam HI sites (table S1), followed by cloning into the vector pCDH-CMV-MCS-EF1-copGFP (System Biosciences), which was designed to concomitantly express target protein and free GFP in the transfected cells (46). The recombinant construct and the empty vector control were introduced into HEK-293Ta cells, respectively. The transfected cells were sampled at desired time points for flow cytometry analysis with the aid of GFP fluorescence (46).
Protein modeling and CD assay
The amino acid sequences of 17K and p17 were subjected to protein modeling using the QUARK online software (https://zhanglab.ccmb.med.umich.edu/QUARK/), which is a computer algorithm for ab initio protein structure prediction (47). For CD analysis, a histidine-tagged 17K protein was expressed in the bacterial strain BL21(DE3) pLysS and purified using nickel affinity chromatography (Invitrogen, K95001). The resultant protein was analyzed with the Chirascan-plus CD spectrometer (Applied Photophysics Ltd., Leatherhead, UK).
Statistical analyses
Numerical data were presented as means ± SE. Statistical analysis of the data was conducted using either Student’s t test (for pairwise comparisons) or a combination of one-way analysis of variance (ANOVA) and least significant difference test (LSD, for multiple comparisons) in the Statistical Package for the Social Sciences (SPSS) program (SPSS Inc., Chicago, IL, USA).
Accession numbers
The nucleic acid sequences used in this study can be found in GenBank, Arabidopsis Genome Initiative, or Solanaceae Genomics Network under the following accession numbers: NC_004666 (BYDV-GAV), NP_001342995 (S. pombe Cdc2), NP_587933 (S. pombe Wee1), NP_001342797 (S. pombe Cdc25), AT3G48750 (Arabidopsis CDKA;1), AT1G02970 (Arabidopsis Wee1), AT5G03455 (Arabidopsis Cdc25), AK370270 (barley CDKA1), AK373637 (barley CDKA2), AK372043 (barley Wee1), AK356881 (barley Cdc25), NP_001777 (human Cdk1), Niben101Scf00745g02009 (N. benthamiana CDKA1), Niben101Scf02217g05009 (N. benthamiana CDKA2), Niben101Scf06290g03007 (N. benthamiana CDKA3), Niben101Scf06698g01004 (N. benthamiana CDKA4), CAD26723.1 (HIV-1 Vpr), and AAK18187.1 (ARV p17).
Supplementary Material
Acknowledgments
We thank all members of the Zhao laboratory and the Wang laboratory for discussions and support of this project. We thank X. Zhou for providing the antibody to BYDV-GAV CP and X. Wang for constructive help on this study. Funding: This work was supported by the Ministry of Science and Technology of China (2017YFD0101000 and 2017YFD0100600 to D.W.) and an intramural start-up fund from Chicago Children’s Memorial Institute of Education and Research, Northwestern University Feinberg School of Medicine and the University of Maryland Medical Center (to R.Y.Z.). Author contributions: H.J., Z.D., Y.Z., R.Y.Z., and D.W. conceived and designed the experiments. H.J., Z.D., Y.Z., J.A., Z.X., Y.W., Y.G., X.Z., X.H., Y.C., Q.S., K.Z., R.E.E., Z.B., C.F., G.L., D.R., J.L., S.B., R.Y.Z., and D.W. performed the experiments and analyzed the data. H.J., R.Y.Z., and D.W. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaba3418/DC1
REFERENCES AND NOTES
- 1.Dominguez-Brauer C., Thu K. L., Mason J. M., Blaser H., Bray M. R., Mak T. W., Targeting mitosis in cancer: Emerging strategies. Mol. Cell 60, 524–536 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Polyn S., Willems A., De Veylder L., Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 23, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Novák B., Heldt F. S., Tyson J. J., Genome stability during cell proliferation: A systems analysis of the molecular mechanisms controlling progression through the eukaryotic cell cycle. Curr. Opin. Syst. Biol. 9, 22–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Y., Sanyal S., Bruzzone R., Breaking bad: How viruses subvert the cell cycle. Front. Cell. Infect. Microbiol. 8, 396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hégarat N., Rata S., Hochegger H., Bistability of mitotic entry and exit switches during open mitosis in mammalian cells. Bioessays 38, 627–643 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Domingo-Sananes M. R., Kapuy O., Hunt T., Novák B., Switches and latches: A biochemical tug-of-war between the kinases and phosphatases that control mitosis. Phil. Trans. R. Soc. Lond. B Biol. Sci. 366, 3584–3594 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamata M., Watanabe N., Nagaoka Y., Chen I. S. Y., Human immunodeficiency virus type 1 Vpr binds to the N lobe of the Wee1 kinase domain and enhances kinase activity for Cdc2. J. Virol. 82, 5672–5682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goh W. C., Manel N., Emerman M., The human immunodeficiency virus Vpr protein binds Cdc25C: Implications for G2 arrest. Virology 318, 337–349 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Chiu H. C., Huang W. R., Liao T. L., Chi P. I., Nielsen B. L., Liu J. H., Liu H.-J., Mechanistic insights into avian reovirus p17-modulated suppression of cell-cycle CDK/cyclin complexes and enhancement of p53 and cyclin H interaction. J. Biol. Chem. 293, 12542–12562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukhovitskaya N. I., Solovieva A. D., Boddeti S. K., Thaduri S., Solovyev A. G., Savenkov E. I., An RNA virus-encoded zinc-finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell 25, 960–973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei Y., Yang X., Huang C., Zhang X., Zhou X., Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη-mediated phosphorylation in Nicotiana benthamiana. PLOS Pathog. 14, e1006789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Veylder L., Beeckman T., Inzé D., The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8, 655–665 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Miller W. A., Rasochová L., Barley yellow dwarf viruses. Annu. Rev. Phytopathol. 35, 167–190 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Gray S., Gildow F. E., Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41, 539–566 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Ali M., Anwar S., Shuja M. N., Tripathi R. K., Singh J., The genus Luteovirus from infection to disease. Euro. J. Plant Patho. 151, 841–860 (2018). [Google Scholar]
- 16.Jin Z., Wang X., Chang S., Zhou G., The complete nucleotide sequence and its organization of the genome of barley yellow dwarf virus-GAV. Sci. China C Life Sci. 47, 175–182 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Liu Y., Chen L., Zhao D., Wang X., Zhang Z., Wheat resistome in response to barley yellow dwarf virus infection. Funct. Integr. Genomics 13, 155–165 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y., Cao J., O'Gorman M. R., Yu M., Yogev R., Effect of human immunodeficiency virus type 1 protein R (vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J. Virol. 70, 5821–5826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood E., Nurse P., Sizing up to divide: Mitotic cell-size control in fission yeast. Annu. Rev. Cell Dev. Biol. 31, 11–29 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Li G., Poulsen M., Fenyvuesvolgyi C., Yashiroda Y., Yoshida M., Simard J. M., Gallod R. C., Zhao R. Y., Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 114, E376–E385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chay C. A., Gunasinge U. B., Dinesh-Kumar S. P., Miller W. A., Gray S. M., Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein read-through domain and 17-kDa protein, respectively. Virology 219, 57–65 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Nass P. H., Domier L. L., Jakstys B. P., D’Arcy C. J., In situ localization of barley yellow dwarf virus-PAV 17-kDa protein and nucleic acids in oats. Phytopathology 88, 1031–1039 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Fusaro A. F., Barton D. A., Nakasugi K., Jackson C., Kalischuk M. L., Kawchuk L. M., Vaslin M. F. S., Correa R. L., Waterhouse P. M., The luteovirus P4 movement protein is a suppressor of systemic RNA silencing. Viruses 9, E294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smirnova E., Firth A. E., Miller W. A., Scheidecker D., Brault V., Reinbold C., Rakotondrafara A. M., Chung B. Y.-W., Ziegler-Graff V., Discovery of a small non-AUG-initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. PLOS Pathog. 11, e1004868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K., Xia Z., Zhang Y., Wen Y., Wang D., Brandenburg K., Harris F., Phoenix D. A., Interaction between the movement protein of barley yellow dwarf virus and the cell nuclear envelope: Role of a putative amphiphilic α-helix at the N-terminus of the movement protein. Biopolymers 79, 86–96 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Xia Z., Wang Y., Du Z., Li J., Zhao R. Y., Wang D., A potential nuclear envelope-targeting domain and an arginine-rich RNA binding element identified in the putative movement protein of the GAV strain of barley yellow dwarf virus. Funct. Plant Biol. 35, 45–50 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Pasternak T., Tietz O., Rapp K., Begheldo M., Nitschke R., Ruperti B., Palme K., Protocol: An improved and universal procedure for whole-mount immunolocalization in plants. Plant Methods 11, 50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P., Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Constantin G. D., Krath B. N., MacFarlane S. A., Nicolaisen M., Johansen I. E., Lund O. S., Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J. 40, 622–631 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Hutter L. H., Rata S., Hochegger H., Novák B., Interlinked bistable mechanisms generate robust mitotic transitions. Cell Cycle 16, 1885–1892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Schutter K., Joubès J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman D. T., Kushnir S., Inzé D., De Veylder L., Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19, 211–225 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landrieu I., da Costa M., De Veylder L., Dewitte F., Vandepoele K., Hassan S., Wieruszeski J.-M., Faure J.-D., Van Montagu M., Inzé D., Lippenset G., A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 13380–13385 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dissmeyer N., Weimer A. K., Pusch S., De Schutter K., Kamei C. L. A., Nowack M. K., Novák B., Duan G.-L., Zhu Y.-G., De Veylder L., Schnittger A., Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowack M. K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A. K., De Winter F., Yang F., Schnittger A., Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22, 1030–1040 (2012). [DOI] [PubMed] [Google Scholar]
- 35.R. Hull, Plant Virology (San Diego Academic Press, ed. 5, 2013). [Google Scholar]
- 36.Schiefelbein J. W., Benfey P. N., The development of plant roots: New approaches to underground problems. Plant Cell 3, 1147–1154 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan C., Li C., Yan L., Jackson A. O., Liu Z., Han C., Yu J., Li D., A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLOS ONE 6, e26468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enoch T., Nurse P., Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication cell. Cell 60, 665–673 (1990). [DOI] [PubMed] [Google Scholar]
- 39.Morellet N., Bouaziz S., Petitjean P., Roques B. P., NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 327, 215–227 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Barnitz R. A., Chaigne-Delalande B., Bolton D. L., Lenardo M. J., Exposed hydrophobic residues in human immunodeficiency virus type 1 Vpr helix-1 are important for cell cycle arrest and cell death. PLOS ONE 6, e24924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolton D. L., Lenardo M. J., Vpr cytopathicity independent of G2/M cell cycle arrest in human immunodeficiency virus type 1-infected CD4+ T cells. J. Virol. 81, 8878–8890 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., Lu Y., Ratner L., Arginine residues in the C-terminus of HIV-1 Vpr are important for nuclear localization and cell cycle arrest. Virology 242, 414–424 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Heinlein M., Plant virus replication and movement. Virology 479-480, 657–671 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Prasad A., Sharma N., Muthamilarasan M., Rana S., Prasad M., Recent advances in small RNA mediated plant virus interactions. Crit. Rev. Biotechnol. 39, 587–601 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.-M., Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H., Zhu S., Mo Y.-Y., Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 19, 439–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu D., Zhang Y., Toward optimal fragment generations for ab initio protein structure assembly. Proteins 81, 229–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/20/eaba3418/DC1


