Abstract
p62 is a multifunctional protein involved in multiple cellular processes including proliferation, drug sensitivity and autophagy-associated cancer cell growth. However, the role of p62 in colon cancer remains controversial. Here we investigated the expression of p62 protein in colon cancer and its clinical significance.
Patients with colon adenocarcinoma who underwent resection at the Third Affiliated Teaching Hospital of Xinjiang Medical University (Affiliated Cancer Hospital) were retrospectively analyzed. The expression of p62 protein in tumor tissues and adjacent normal tissues was detected by immunohistochemistry and western-blotting. Real-time quantitative polymerase chain reaction was used to detect the expression level of p62 messenger ribonucleic acid in specimens. Progression-free survival (PFS) and overall survival (OS) were assessed using Kaplan-Meier method and the log-rank test.
A total of 85 colon cancer patients were enrolled, including 55 (64.71%) patients with high p62 expression, and 30 (35.29%) patients with low p62 expression. The transcription and expression level of p62 in colon cancer tissues were higher than those in adjacent normal tissues (P < .01). High expression of p62 was an independent risk factor for the poor prognosis (PFS and OS) of colon cancer.
p62 may be a potential indicator of determining the progression and prognosis evaluation of colon cancer.
Keywords: colon cancer, immunohistochemistry, indicators, p62 protein
1. Introduction
Colon cancer represents one of the most common malignancies worldwide and a common cause of morbidity and mortality.[1] Colon cancer brings about metastases in 60% of the cases, with involvement of the liver (40%), lung (15%), or both (10%).[2] Increased treatment advances in the past 20 years,[3] and development of chemotherapeutic regimens combined with surgical resection has made it possible to treat the metastatic colon cancer, half of patients develop local or distant recurrence. The development and progression of colon cancer are complex and results of combined actions of multiple factors and pathways. Numerous studies have tried to elucidate the mechanisms, which, however, are still poorly understood. Therefore, it is precisely important to identify colon cancer at an early stage. Identification of molecular markers remains crucial for designing novel and efficient treatment strategy.
Over the last 2 decades numerous molecular biomarkers for colon cancer have been extensively studied. There are several screening strategies for detection of colon cancer which can basically be classified as noninvasive and invasive imaging techniques. But recent studies have also highlighted that they usually lack of proper sensitivity and specificity to screen colon cancer.[4] Therefore, it is required to find out more reliable and novel biomarkers for the early detection of colon cancer. Globally, enormous research efforts have been given at present for identifying molecular markers based on deoxyribonucleic acid (DNA), ribonucleic (RNA), or protein to develop novel, noninvasive biomarker detection methods for colon cancer in blood and stool. Therefore, in future identification of a new biomarker will help to the development of a valid, sensitive and reliable clinical assay that can be served as an alternative diagnostic tool to conduct treatment selection to the deadly disease.
p62/sequestosome1 is a multifunctional ubiquitinated binding protein[5] and involved in signaling pathways of many cell life activities, including autophagy,[6] and its abnormal expression and regulation are closely associated with development and progression of malignant tumors, such as lung cancer, prostatic cancer, and endometrial cancer.[7] Many studies found correlations between p62 protein expression and cancer, but no direct links have been reported. It has been reported that an abundance of p62 protein is associated with breast tumors and liver cirrhosis.[8] p62 overexpression in breast tumors and regulation by prostate-derived Ets factor in breast cancer cells. Aberrant expression of fetal RNA-binding protein p62 in liver cancer and liver cirrhosis,[9] and p62 has been identified as an important Nuclear factor-κB (NF-κB) mediator in tumorigenesis. A study by Mathew et al showed autophagy suppresses tumorigenesis through elimination of p62.[10] p62 and the sequestosome, a novel mechanism for protein metabolism and may play a significant role in an ubiquitination-mediated regulatory mechanism during cell proliferation and differentiation. Those findings indicate that p62 may be involved in various tumors and may serve as an indicator factor. While there is little research to prove the relationship between p62 and colon cancer.
To investigate the expression levels of p62 protein and p62 mRNA in colon cancer patients and to compare these expressive levels with clinicopathological characteristics. Expression of p62 protein were detected in colon cancer tissues and adjacent normal tissues by immunohistochemistry, Real-time quantitative polymerase chain reaction (RT-qPCR), and Western bolt in this study. In addition, an investigation was performed with respect to the role of p62 in the progression and prognosis of colon cancer.
2. Methods
2.1. Patients
Patients with colon adenocarcinoma undergoing resection at the Third Affiliated Teaching Hospital of Xinjiang Medical University (Affiliated Cancer Hospital) from October 2013 to October 2016 were enrolled in this retrospective cohort study. The inclusion criteria were:
-
(1)
Aged over 18 years;
-
(2)
Histopathologically confirmed primary colon adenocarcinoma;
-
(3)
Treatment-naïve;
-
(4)
Patients with complete medical records and follow-up results.
The exclusion criteria were:
-
(1)
Patients with previous history of other malignant tumors in 5 years;
-
(2)
Patients who have received radiotherapy or chemotherapy before operation.
This study was approved by the Ethics Committee of The Third Affiliated Teaching Hospital of Xinjiang Medical University (Affiliated Cancer Hospital) on October 11, 2017. Ethics approval number is W-2017010. Informed consent was given and signed by all patients.
2.2. Pathological examination
Specimens were collected in 20 minutes in vitro to ensure fresh specimens. Tumor tissues were obtained at the cancer foci, and normal colon tissue was obtained as the control at the area about 15 cm away from the distal or proximal side of the tumor focus. After obtaining specimens, each specimen was divided into 2 parts. One was fixed with formaldehyde, and the paraffin section was prepared for immunohistochemical detection of the expression level of p62 protein. Another 1 was immediately stored in liquid nitrogen for RT-qPCR to detect the expression level of p62 protein mRNA and Western bolt to detect the expression level of p62 protein.
All obtained tumor specimens and tissue specimens around the tumor were histopathologically examined. The pathological diagnosis of colon cancer was based on the World Health Organization (2000) Standard,[11] and the staging was in accordance with the American Joint Committee on Cancer cancer staging manual (2016 Version 8).[12]
2.3. Immunohistochemical staining
The paraffin-embedded tissues were continuously sliced at a thickness of 4 μm. After the baking was completed, it was routinely dewaxed and hydrated, and sections were placed into the citrate antigen retrieval solution, and underwent microwave heating to boiling to perform antigen retrieval. The prepared 3% hydrogen peroxide was added to the sectioned tissue to block endogenous peroxidase, and incubated for 15 minute at room temperature. After washing for 3 times with 0.1 M phosphate-buffered saline, the primary antibody (diluted by 1:100, Sigma Company) was incubated overnight in the refrigerator at 4°C. Horseradish peroxidase-labeled secondary antibody IgG polymer (Southern Biotech Company) was added dropwise and incubated at 37°C for 30 minute. The above steps were washed with phosphate buffer for 5 minute × 3 times. Diaminobenzidine was used for developing, and hematoxylin was used for counterstaining; conventional dehydration, mounting, and microscopic photographing were performed. The positive control was a positive control picture taken with the reagent, and the negative control was a section in which phosphate-buffered saline was added to replace the primary antibody. Other procedures of the negative control were completely consistent with those in the detected tissues.
For p62 staining, markedly yellow or brownish yellow granules in the cytoplasm were considered as positive staining. The staining results were evaluated by standard comprehensive positive cell rate and staining intensity. p62 staining intensity was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The proportion of p62-positive cells was evaluated as 0% to 100%. The final results were scored using quick (Q) score, which was calculated as the percentage of positive cells (P) multiplied by the intensity (I) (Q = P × I; maximum = 300).[13] The median Q score of was used as the cut off value to classify each cancer as low and high p62 expression.
2.4. mRNA detection
RT-qPCR was used to detect the mRNA level of p62. The primers were designed by Sangon Biotech (Shanghai) Co., Ltd. as follows: p62 upstream primer, 5′-CCAGAGAGTTCCAGCACAGA-3′; downstream primer, 5′-CCGACTCCATCTGTTCCTCA-3′; β-actin upstream primer, 5′-AGCGAGCATCCCCCAAAGTT-3′; downstream primer, 5′-GGGCACGAAGGCTCATCATT-3′. After taking tissues from liquid nitrogen, trizol was added and ground to homogenate, and total RNA was extracted. cDNA synthesis was performed using Synergy Brands, Inc. Green I fluorescent dye RT-qPCR detection method. Three parallel-controlled reaction wells were set for each sample. Actin was used as the reference housekeeping gene for RT- PCR analysis. After the reaction, the computer automatically analyzed the fluorescent signal and converted it to the initial copy number and ct value. The 2−ΔΔCt value was the relative expression level of mRNA.[14]
2.5. Detection of protein expression
Western blot: The tissues were removed from the liquid nitrogen and cut up; the lysate (containing PMSF and cocktail) was added and repeatedly ground into a homogenate. After the lysate acted on tissues for 30 minute, the homogenate was aspirated and centrifuged to collect supernatants. After measuring the protein concentration by BCA method, 5 × loading buffer was added, denatured at 100°C for 10 minute, and cooled to room temperature. For equal amounts of specimens, sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 12% of mass fraction was used to separate proteins. After electrophoresis, the protein was transferred to a pure nitrocellulose blotting membrane and blocked with 5% skim milk; it was blocked with primary antibody for 1 hour, and blocked again with horseradish peroxidase-labeled secondary antibody blocking solution for 1 hour. Electrogenerated chemiluminescence reagent was added; developing and photograph were performed. The results were recorded by analyzing the gray values in the gel imaging analysis system.
2.6. Follow-up
All patients were followed up by telephone after surgery until December 2018. The overall survival (OS) was defined as the time from operative treatment to death or the last follow-up. Progression-free survival (PFS) was defined as the length of time from treatment to disease progression, death, or last follow-up. Serum carcino-embryonic antigen, chest and abdomen pelvic computed tomography, colonoscopy in outpatients and inpatients were detected.
2.7. Statistical analysis
Statistical analysis were performed by using SPSS software 19.0 (IBM Corp., Armonk, NY). Categorical variables were expressed as frequencies and percentages, and comparisons of ratios were made using the Chi-square test or Fisher exact test. Continuous variables were tested for normality. Normally distributed continuous variables were expressed as means ± standard deviation and were compared between 2 groups using paired-sample t tests. Nonnormally distributed continuous variables were presented as median (range) and compared between groups using the Wilcoxon test. Cox proportional hazard model was used for multivariate analysis. PFS and OS were assessed using Kaplan-Meier method and the log-rank test. P < .05 was considered statistically significant.
3. Results
3.1. Baseline characteristics
A total of 85 colon cancer patients were enrolled in this study. Among them, 55 (64.71%) patients showed high p62 expression, and 30 (35.29%) patients showed low p62 expression. No significant differences were found in age (P = .118) and gender (P = .763) between the 2 groups. (Table 1)
Table 1.
Clinicopathological features of patients with different p62 protein expression level in tumorous tissue.
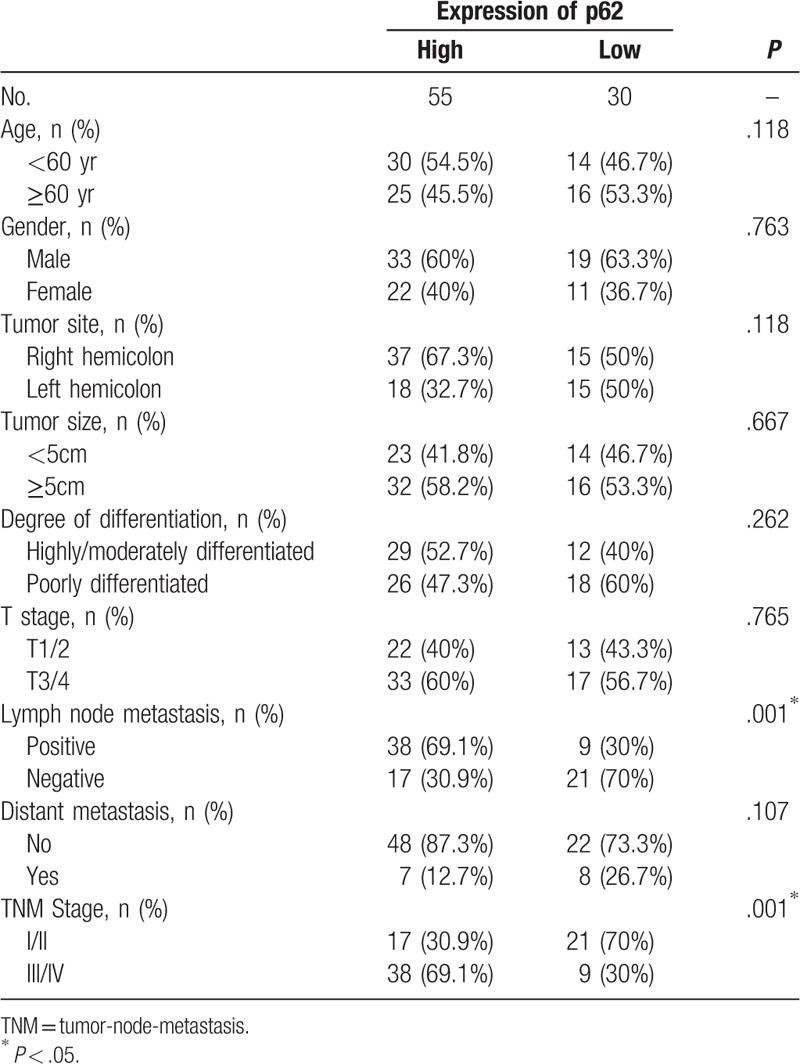
The differences in lymph node metastasis between high p62 expression group and low p62 expression group were significant (P = .001). In addition, patients with high p62 expression were more likely to have advanced diseases (tumor-node-metastasis stage III/IV) compared to those with low p62 expression (P = .001). However, there was no significant relationship between p62 expression and the other clinical pathological features of the patients, such as tumor site and size, and degree of differentiation (all P > .05) (Table 1).
3.2. Correlation between p62 expression and clinical characteristics
The p62 expression in colon cancer specimens and adjacent non-tumor tissue were showed in Figure 1. Particles sharing brown color indicated to p62 protein expression occurred in some parts of gland parietes. It is clearly that staining of normal tissues was weak, and a small amount of stained p62 was distributed in the cytoplasm, while colon cancer tissues showed strongly positive staining of p62; in addition to the cytoplasm, it was also distributed in some cell nucleuses (×400 magnification). Among the 85 adjacent normal tissues, 9 (10.59%) showed high expression and 76 (89.41%) showed low expression. The expression of p62 in colon cancer was significantly higher than that in adjacent normal tissues (Fig. 2), and the difference was statistically significant (1.058 ± 0.2720 vs 0.3536 ± 0.1014, P < .01).
Figure 1.
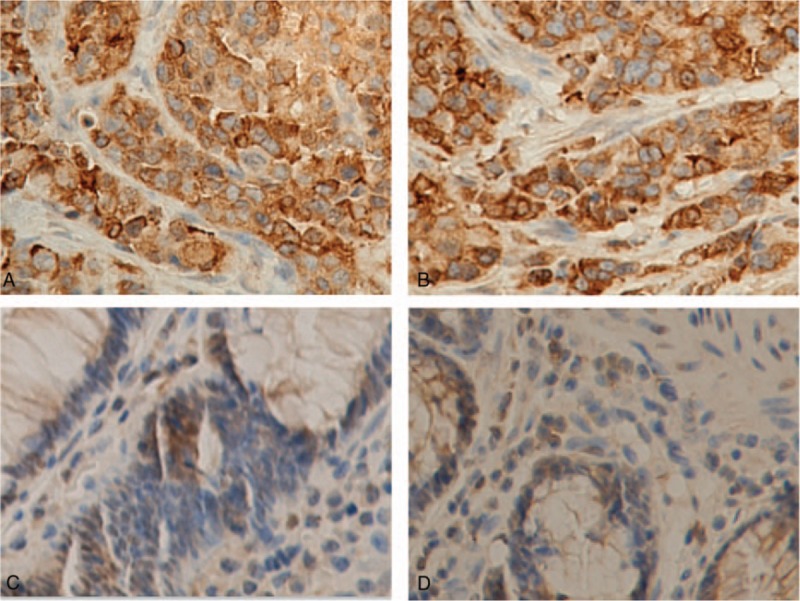
Examples of p62 immunostaining in colon adenocarcinomas and adjacent normal tissues. Tumor tissue (A) and adjacent normal tissue (C) for a 56-yr-old male patients with moderately differentiated adenocarcinoma of the ascending colon (pT3N1M0, IIIB). Tumor tissue (B) and adjacent normal tissue (D) for a 68-yr old female patients with poorly differentiated adenocarcinoma of the descending colon (pT4aN2aM0, IIIC). (A) and (B) showed their tumors with strong immunostaining and (C) and (D) demonstrated their adjacent normal tissues with negative p62 staining (×400 magnification).
Figure 2.
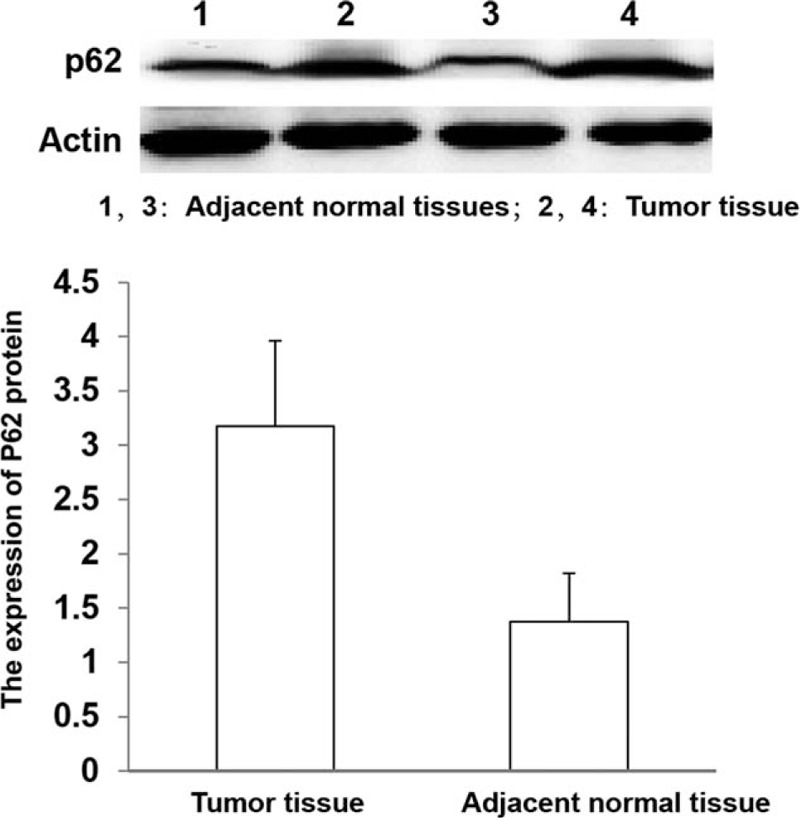
Western blot analysis of p62 protein expression in colon cancer and adjacent normal tissues (P < .01).
The relative expression level of p62 mRNA in colon cancer tissues was significantly higher than that in distant normal tissues (3.1752 ± 0.7865 vs 1.3753 ± 0.4431, P < .01).
3.3. p62 expression and survival analysis
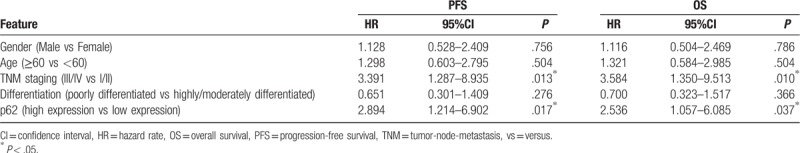
Kaplan-Meier analysis found that the PFS in patients with high expression of p62 was 27.61 ± 2.50 months, which was shorter than that of those with lower expression (44.22 ± 3.44 months, P < .01) (Fig. 3). The OS of patients with low expression of p62 was 52.37 ± 3.03 months, which was significantly prolonged than that of patients with higher expression (37.70 ± 2.13 months, P < .05). Multivariate analysis showed that tumor-node-metastasis staging III/IV (PFS: HR = 3.391, 95% CI: 1.287–8.935, P = .013; OS:HR = 3.584, 95% CI: 1.350–9.513, P = .010) and high expression of p62 (PFS: HR = 2.894, 95% CI: 1.214–6.902, P = .017; OS: HR = 2.536, 95% CI: 1.057–6.085, P = .037) were independent prognostic factors affecting PFS and OS of patients with colon cancers (Table 2).
Figure 3.
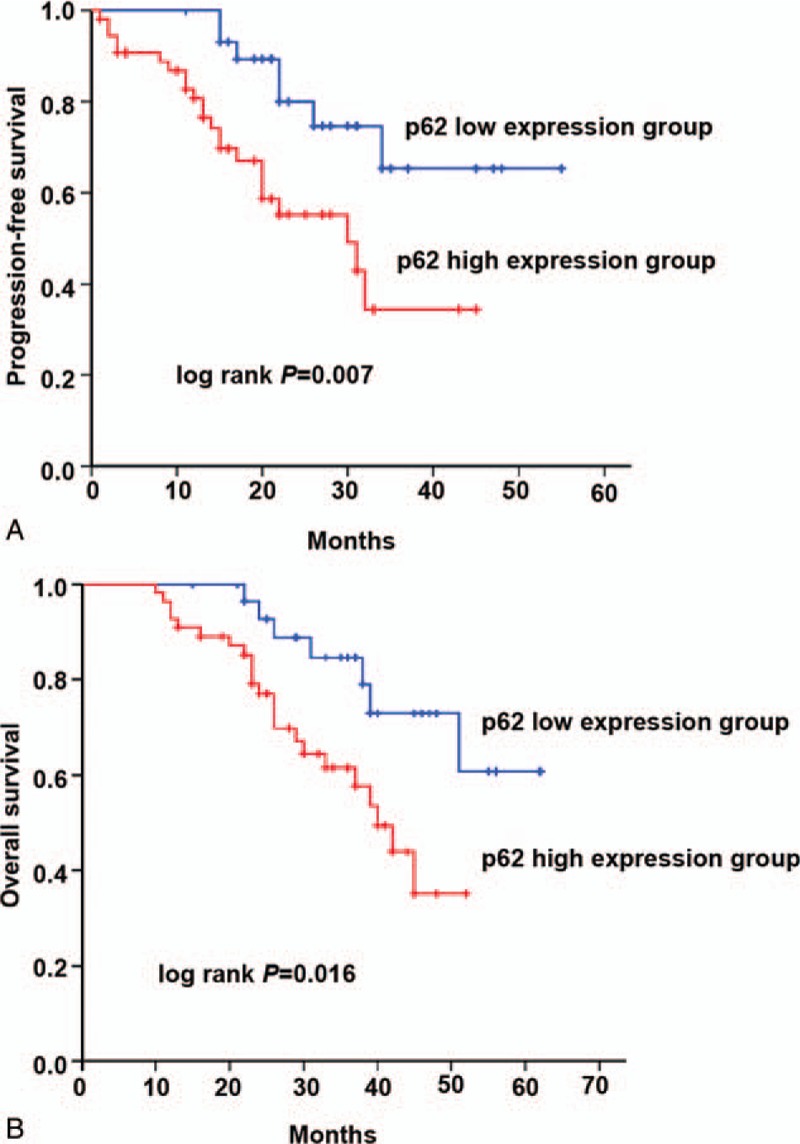
Relation between the expression of p62 in patients with colon cancer and PFS (A) and OS (B). PFS = progression-free survival, OS = overall survival.
Table 2.
Analysis of influencing factors of PFS and OS in patients with colon cancer by using COX multivariate regression analysis.
4. Discussion
In the present study, we demonstrated that normal colon tissues express lower levels of p62 when compared with the patient-matched primary tumor samples. High expression levels of p62 are associated with short progression-free survival of patients with colon cancer. The study provides evidence for the p62 protein as a potential indicator of determining the progression and prognosis evaluation of colon cancer.
p62 has a wide range of functions, acting as a substrate and regulating proteins in mammalian selective autophagy,[5] and it links degrading substrates to autophagy structures,[15] thereby mediating the degradation of substrates such as protein polymers, damaged mitochondria, and invading microorganisms by the autophagy pathway. Furthermore, p62 regulates cell death by binding to and activating caspase 8.[16] p62 can also regulate life activities of cells by activating the transcription factor NF-κB and then activating TRAF.[4] p62 acts as a substrate during cell autophagy and is degraded by the autophagy pathway. Abnormal accumulation of p62 is suggestive of autophagy dysfunction in cancer cells.[17,18] Autophagy can remove damaged or degraded organelles, some structurally abnormal proteins and macromolecular substances, and prevent the invasion of harmful substances. In this way, it can maintain the stability of the genome, induce cell senescence, and inhibit tumor development and progression.[19]
The findings of this study demonstrate that the expression levels of p62 mRNA and protein in colon cancer tissues were significantly higher than that in adjacent normal tissues. Its suggests that abnormal autophagy activity may play a promoting role in the development and metastasis of colon cancer, as supported by previous studies in colon cancer[20,21] and other types of cancers.[22,23] In addition, abnormal aggregation of p62 in cells activate 2 active oxygen scavenging systems, Nuclear factor erythroid 2– related factor 2 and NF-κB, which enhance the viability of cancer cells under stress conditions.[24,25] The accumulation of p62 in cells can also activate oncogene signals, such as mammalian target of rapamycin,[26] contributing to carcinogenesis.
The expression levels of p62 were unrelated to clinical indicators such as sex, age, ethnicity, tumor site, tumor size, histological type, degree of differentiation, and tumor invasion depth, but were associated with lymph node metastasis. The positive rate of lymph node was 69.1% (38/55) in patients with high expression of p62, and 30.0% (9/30) in those with low expression. Studies of the expression of p62 in human liver cancer and its significance found that high expression of p62 in liver cancer tissues was positively correlated with clinical staging and survival.[27,28] The rate of high expression of p62 was 92.3% in patients of stage III and IV, and 58.3% in those of stage I and II. The rates of recurrence-free survival and OS in patients with high expression of p62 were worse than those in patients with low expression of p62, suggesting the high expression of p62 was related to patient prognosis. High expression of p62 was also observed in patients with oral squamous cell carcinoma, which was associated with higher malignancy and worse prognosis.[29] In the tissue microarray and immunohistochemical detections of 12,427 prostate cancer tissues,[30] the expression of p62 was markedly related to Gleason grading of tumor histology, clinical staging and lymph node metastasis. Immunohistochemical detections on human ovarian cancer and normal ovarian tissues showed the positive rate of p62 in ovarian cancer tissues and normal ovarian tissues was 71.67% and 11.67%, respectively.[31] The difference had statistical significance. In addition, the the positive expression of p62 was associated with chemotherapy sensitivity of ovarian cancer patients, suggesting it may be a molecular marker for the prediction of chemotherapy sensitivity. In the present study, the PFS and OS in patients with high expression of p62 were significantly shorter than those in patients with low expression of p62. Cox multivariate regression analysis showed that the expression of p62 and clinical staging were independent prognostic factors for colon cancer; Patients with high expression of p62 had poor prognosis. Nakayama et al[32] showed that p62 was an important prognostic factor in colon cancer since p62 promotes cancer cell proliferation. Niklaus et al[33] indicated that high p62 expression was associated with autophagy activation and unfavorable colon cancer prognosis.
The above findings suggested that p62 plays a role in regulating development and progression of colon cancer and may be a potential molecular marker for diagnosis and prognosis evaluation of colon cancer. However, its specific molecular mechanism is still not clear. Further studies are needed to determine the detailed mechanisms of p62 in orchestrating tumor development, especially the role of p62 in facilitating tumor pre-metastatic niche formation and metastasis. In addition, more samples need to be collected as the validation set to conclude the result.
In conclusion, the results in this study suggest that p62 mRNA and protein are abnormally highly expressed in tumor tissues of patients with colon cancer. High expression of p62 is an independent risk factor for the prognosis of colon cancer. Detection of p62 level may be a potential indicator of disease progression and prognosis evaluation.
Author contributions
Conceptualization: Xiyan Wang.
Funding acquisition: Bing Zhao.
Investigation: Cheng Lei, Bing Zhao, Lin Liu, Xiangyue Zeng, Zhen Yu.
Methodology: Cheng Lei, Lin Liu, Xiangyue Zeng, Zhen Yu.
Writing – original draft: Cheng Lei, Bing Zhao.
Writing – review and editing: Xiyan Wang.
Footnotes
Abbreviations: NF-κB = nuclear factor-κB, OS = overall survival, PFS = progression-free survival, RT-qPCR = real-time quantitative polymerase chain reaction.
How to cite this article: Lei C, Zhao B, Liu L, Zeng X, Yu Z, Wang X. Expression and clinical significance of p62 protein in colon cancer. Medicine. 2020;99:3(e18791).
The study was supported by the Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2017D01C412).
The authors have no conflicts of interest to disclose.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Eadens MJ, Grothey A. Curable metastatic colorectal cancer. Curr Oncol Rep 2011;13:168–76. [DOI] [PubMed] [Google Scholar]
- [3].De Rosa M, Pace U, Rega D, et al. Genetics, diagnosis and management of colorectal cancer. Oncol Rep 2015;34:1087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sanz L, Diaz-Meco MT, Nakano H, et al. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J 2000;19:1576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131–45. [DOI] [PubMed] [Google Scholar]
- [6].Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res 2012;66:457–62. [DOI] [PubMed] [Google Scholar]
- [7].Iwadate R, Inoue J, Tsuda H, et al. High expression of p62 protein is associated with poor prognosis and aggressive phenotypes in endometrial cancer. Am J Pathol 2015;185:2523–33. [DOI] [PubMed] [Google Scholar]
- [8].Thompson HG, Harris JW, Wold BJ, et al. p62 overexpression in breast tumors and regulation by prostate-derived Ets factor in breast cancer cells. Oncogene 2003;22:2322–33. [DOI] [PubMed] [Google Scholar]
- [9].Lu M, Nakamura RM, Dent ED, et al. Aberrant expression of fetal RNA-binding protein p62 in liver cancer and liver cirrhosis. Am J Pathol 2001;159:945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137:1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Selby JV, Friedman GD, Quesenberry CP, Jr, et al. A case-control study of screening sigmoidoscopy and mortality from colorectal cancer. N Engl J Med 1992;326:653–7. [DOI] [PubMed] [Google Scholar]
- [12].Amin MB, Edge SB, Greene FL, et al. AJCC Cancer Staging Manual. 8th ed.New York: Springer; 2017. [Google Scholar]
- [13].Charafe-Jauffret E, Tarpin C, Bardou VJ, et al. Immunophenotypic analysis of inflammatory breast cancers: identification of an ’inflammatory signature’. J Pathol 2004;202:265–73. [DOI] [PubMed] [Google Scholar]
- [14].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [15].Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007;131:1149–63. [DOI] [PubMed] [Google Scholar]
- [16].Jin Z, Li Y, Pitti R, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009;137:721–35. [DOI] [PubMed] [Google Scholar]
- [17].Pugsley HR. Assessing autophagic flux by measuring LC3, p62, and LAMP1 co-localization using multispectral imaging flow cytometry. J Vis Exp 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu WJ, Ye L, Huang WF, et al. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol Biol Lett 2016;21:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen HY, White E. Role of autophagy in cancer prevention. Cancer Prev Res 2011;4:973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burada F, Nicoli ER, Ciurea ME, et al. Autophagy in colorectal cancer: an important switch from physiology to pathology. World J Gastrointest Oncol 2015;7:271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu Y, Yao J, Xie J, et al. The role of autophagy in colitis-associated colorectal cancer. Signal Transduct Target Ther 2018;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yun CW, Lee SH. The roles of autophagy in cancer. Int J Mol Sci 2018;19: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marinkovic M, Sprung M, Buljubasic M, et al. Autophagy modulation in cancer: current knowledge on action and therapy. Oxid Med Cell Longev 2018;2018:8023821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Komatsu M. Potential role of p62 in tumor development. Autophagy 2011;7:1088–90. [DOI] [PubMed] [Google Scholar]
- [25].Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. Front Oncol 2012;2:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lau A, Wang XJ, Zhao F, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 2010;30:3275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xiang X, Qin HG, You XM, et al. Expression of P62 in hepatocellular carcinoma involving hepatitis B virus infection and aflatoxin B1 exposure. Cancer Med 2017;6:2357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mizuno Y, Shimada S, Akiyama Y, et al. DEPDC5 deficiency contributes to resistance to leucine starvation via p62 accumulation in hepatocellular carcinoma. Sci Rep 2018;8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu JL, Chen FF, Lung J, et al. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer 2014;111:944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Burdelski C, Reiswich V, Hube-Magg C, et al. Cytoplasmic accumulation of sequestosome 1 (p62) is a predictor of biochemical recurrence, rapid tumor cell proliferation, and genomic instability in prostate cancer. Clin Cancer Res 2015;21:3471–9. [DOI] [PubMed] [Google Scholar]
- [31].Li S, Wei Y. Association of HMGB1, BRCA1 and P62 expression in ovarian cancer and chemotherapy sensitivity. Oncol Lett 2018;15:9572–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakayama S, Karasawa H, Suzuki T, et al. p62/sequestosome 1 in human colorectal carcinoma as a potent prognostic predictor associated with cell proliferation. Cancer Med 2017;6:1264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Niklaus M, Adams O, Berezowska S, et al. Expression analysis of LC3B and p62 indicates intact activated autophagy is associated with an unfavorable prognosis in colon cancer. Oncotarget 2017;8:54604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]


