Abstract
To explore the association between epidermal growth factor (EGF) 61A/G polymorphism and lung cancer.
All eligible case-control studies published up to August, 2019 were identified by searching PubMed, The excerpta medica database, China Academic Journals Full-text Database, China Biology Medicine, China National Knowledge Infrastructure, China Science and Technology Journal Database, and Wanfang databases. Two researchers independently identified the literature, extracted data, and evaluated quality according to inclusion and exclusion criteria. Meta-analysis was performed by Stata 15.0.
A total of 6 studies is included, including 1487 cases and 2044 control subjects. Compared with allele A, allele G was considered to have no association with the risk of lung cancer, odds ratio = 1.07 (95% confidence interval: 0.98–1.15). GG recessive genotype, GG + GA dominant genotype, GG homozygote genotype and GA heterozygote genotype were found out that all of them are not associated with the risk of lung cancer. No association between EGF 61A/G polymorphism and lung cancer was found out by ethnical subgroup analysis. However, in view of the limitations of this study, such as the results of quantitative and sensitivity analysis may be lack of accuracy, so the conclusions of allele model and recessive gene model should be made carefully.
It suggested that there was no association between polymorphism of EGF 61A/G and susceptibility of lung cancer.
Keywords: epidermal growth factor, gene polymorphism, lung cancer, meta-analysis
1. Introduction
According to the data of global statistical report, up to now, the incidence and mortality of lung cancer rank first among all kinds of malignant tumors, seriously threatening the quality of life and life span of human beings. [1] In the past 30 years, the incidence of lung cancer had increased nearly 4 times. According to the World Health Organization forecast, the annual incidence of lung cancer in China will be as high as 1 million by 2025. [1] The main causes of lung cancer are genetic and environmental factors, but the specific pathogenesis has not yet been elucidated. At present, it is believed that smoking is the main cause of lung cancer, [2] but not all patients with lung cancer smoke, which also suggests that genetic variation is also involved in the occurrence and development of lung cancer. Now the general point is that the occurrence and development of lung cancer is the result of the interaction between environment and gene. [3] As an endocrine growth factor, epidermal growth factor (EGF) plays an important role in regulating cell proliferation, differentiation, and vascular production by binding to specific epidermal grow factor receptor.[4,5,6] EGF is located on chromosome 4. There is a correlation between the functional polymorphism of 61 A/G locus and susceptibility of various malignant tumors, such as gastric cancer, liver cancer, colorectal cancer, and so on.[6,7,8] In the study of Peng et al[6,7,8] on EGF 61A/G polymorphism and susceptibility of gastric cancer, the results of a meta-analysis of 6 case-control studies showed that compared with AA genotypes, GG and GG + GA genotypes increased the risk of gastric cancer, especially among Asian people. The correlation between EGF 61 A/G locus and lung cancer susceptibility has also been studied, but the conclusions are not consistent. In this study, case-control studies on EGF 61A/G polymorphism and lung cancer risk were collected for meta-analysis, and the correlation between EGF 61A/G polymorphism and lung cancer risk was comprehensively evaluated.
2. Materials and methods
2.1. Literature retrieval
All studies from establishment to publication at August, 2019 were identified by searching PubMed, The excerpta medica database, China Academic Journals Full-text Database, China Biology Medicine, China National Knowledge Infrastructure, China Science and Technology Journal Database, and Wanfang databases. The research object is limited to human beings and language is not limited. English and Chinese retrieval is based on “epidermal growth factor or EGF,” “polymorphism or SNP or variant,” and “lung carcinoma/cancer,” supplemented by literature review.
2.2. Inclusion and exclusion criteria
2.2.1. Inclusion criteria
Study on the correlation between polymorphism of EGF 61A/G and susceptibility of lung cancer;
Case-control study;
Data in the literature are complete, or statistical indicators odds ratio (OR) and 95% confidence interval (CI) can be provided directly or indirectly.
2.2.2. Exclusion criteria
Literature on case-only studies, case reports, reviews, and comments;
Republished and incomplete literature;
Control group in the literature not satisfied with Hardy-Weinberg (H-W) genetic balance.
2.3. Data extraction
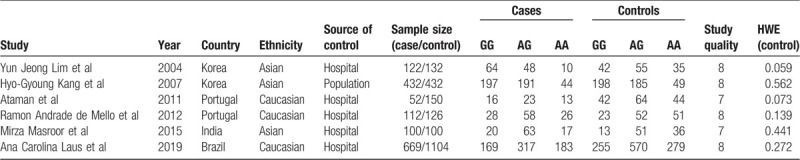
Two researchers independently identified the literature and extracted data. When disagreements arise, they are resolved through discussion or with the assistance of a third researcher. Data extraction included author, publication year, ethnicity, source of control, total number of cancer group and control group, distribution of genotypes, and H-W balance of control group. If P < .05, it is considered that it was not in accordance with H-W balance.
2.4. Literature quality evaluation
Newcastle–Ottawa scale criteria was used. [9] Star-level criteria was used to evaluate 3 parts.
Study objects selection of case group and control group;
Comparability of study objects between case group and control group;
Exposure to risk factors.
Studies with scores greater than or equal to 7 were identified as high-quality studies.
2.5. Statistical methods
Stata 15.0 was used for meta-analysis. OR and its 95% CI were selected as the combined effect quantities. Heterogeneity test was conducted for the included studies, Q test and I statistics were used. If P > .05 or I 2 < 50%, it suggests that there is no heterogeneity among the studies. Fixed-effect model was used for combined analysis. [10] On the contrary, the random-effect model was adopted. Sensitivity analysis excludes individual literature in turn and then re-conducts meta-analysis to estimate the size of the comprehensive effect. Publication bias was quantitatively detected by Egger regression method, if P < .05, it was considered that there was publication bias.
3. Results
3.1. Literature retrieval
A total of 6 relevant literature was found out by searching, all of which could extract data. Therefore, this study included 6 papers, including 1487 cases and 2044 controls. All the 6 papers were in English. Two papers were about Korean population,[11,12] 2 about Portuguese population,[13,14] 1 about Indian population, [15] and 1 about Brazilian population. [16] Except for 1 literature in which the control group came from the community, the rest came from the hospital. Through H-W genetic balance test, the results indicated that in the test of the 6 literature control groups, P were greater than .05, which satisfied the genetic balance. The specific process of literature screening can be found in Figure 1, and the data characteristics of literature can be found in Table 1.
Figure 1.
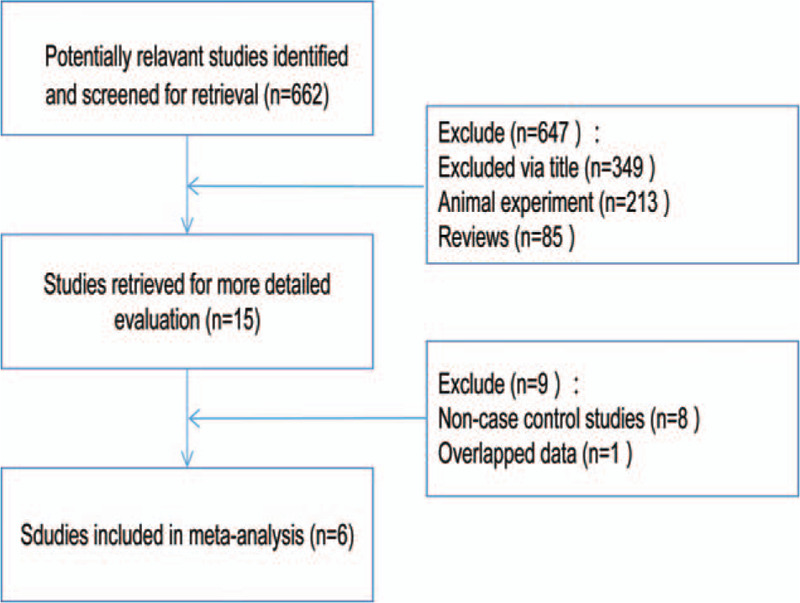
A flow diagram of the study selection process.
Table 1.
Characters of included studies.
3.2. Heterogeneity test
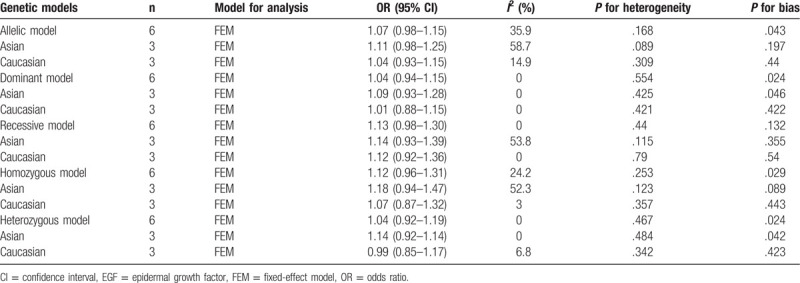
The risk between the polymorphism of EGF 61A/G and the susceptibility of lung cancer can be seen in Table 2. Heterogeneity of EGF 61A/G polymorphism was tested in 5 genetic models, as well as in ethnic subgroups. The results showed that the P value of heterogeneity test of all models was greater than .05, so the fixed effect model was selected for analysis.
Table 2.
Results of meta-analysis for EGF 61A/G polymorphism and lung cancer risk.
3.3. Results of meta-analysis
Table 2 shows the combined OR value of lung cancer and the risk of EGF 61A/G locus polymorphism. In allele comparison (G vs A), OR = 1.07 (95% CI: 0.98–1.15); in dominant genetic model (GG + GA vs AA), OR = 1.04 (95% CI: 0.94–1.15); in recessive genetic model (GG vs GA + AA), OR = 1.13 (95% CI: 0.98–1.30); in homozygote model (GG vs AA), OR = 1.12 (95% CI: 0.96–1.31). In the heterozygote model (GA vs AA), OR = 1.04 (95% CI: 0.92–1.19). The 95% CI of all OR values contains 1, indicating that the difference is not statistically significant. Subgroup analysis of races showed that the results of Asian and Caucasian analysis also failed to produce a statistically significant genetic model. Five gene models and forest graphs of subgroup analysis are shown in Figure 2. This indicates that the polymorphism of EGF 61A/G cannot be considered to be associated with the risk of lung cancer.
Figure 2.
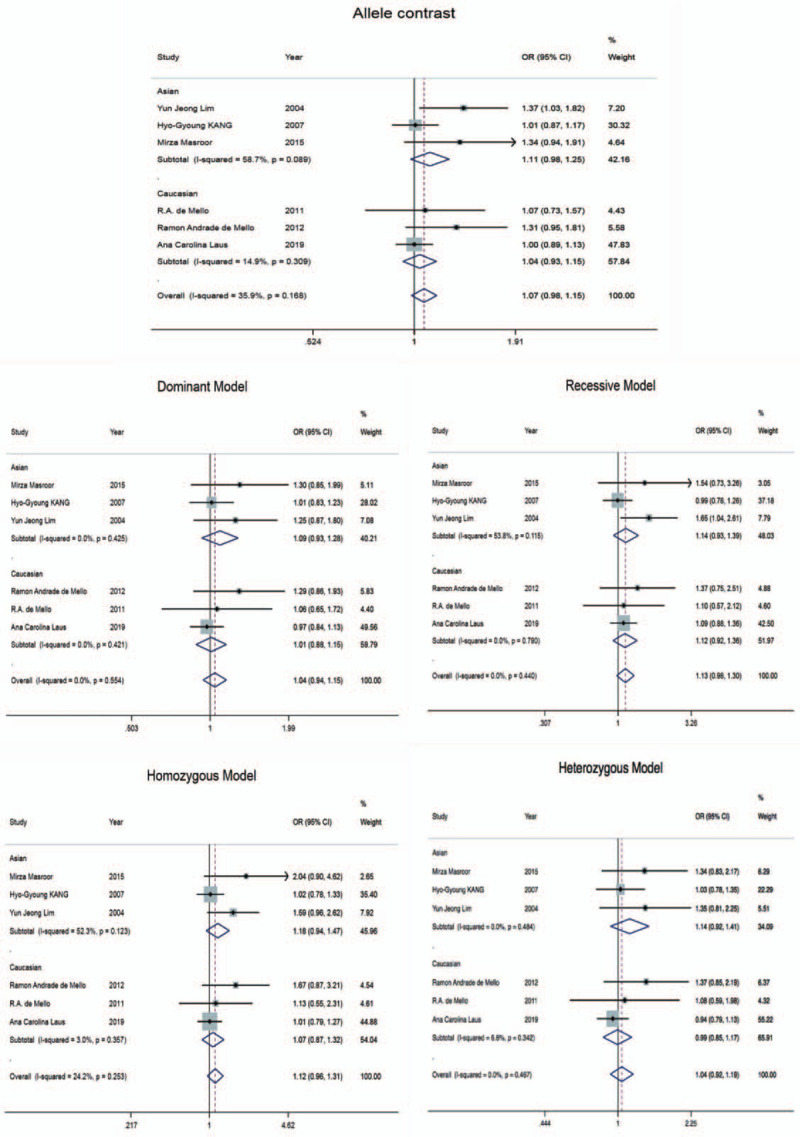
Forest plot for the association between EGF 61A/G polymorphism and lung cancer risk in 5 gene models. EGF = epidermal growth factor.
3.4. Publication bias
The assessment results of publication bias are shown in Funnel Figure 3, and the results of Egger’ test are showed in Table 2. The results showed that among the 5 genotypes, except the P value in the recessive gene model was .132, the rest were all less than .05, which was not very symmetrical on funnel map, indicating that there was publication bias. However, from the specific value of P value, the allele model P = .043, approaching .05, indicating that publication bias is almost not exist.
Figure 3.

Funnel plot for the assessment of publication bias.
3.5. Sensitivity analysis
The 6 papers included in this study were published in high-quality academic journals. The results of sensitivity analysis was showed in Figure 4. It showed that, in the allele model, after removing the large proportion of the study of Laus et al, [16] the difference was statistically significant. In the recessive gene model, after the removal of the study of Kang et al, [12] the difference was statistically significant. The results of meta-analysis of the other 3 genotypes showed that the combined effects of genetic models did not change significantly. Therefore, the conclusions of allele model and recessive gene model should be made carefully.
Figure 4.
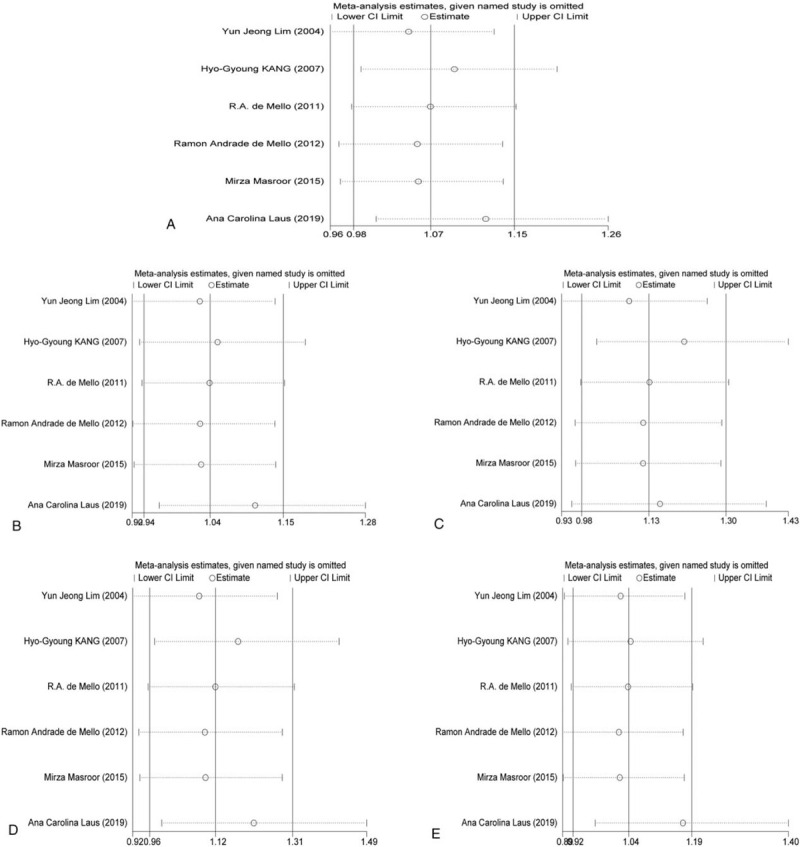
Sensitivity analysis results.
4. Discussion
Since the past decade, the EGF 61A/G polymorphism has been studied and considered as a risk factor for cancer, [17] such as liver cancer, gastric cancer, malignant melanoma,[18,19] and so on. In the study of EGF 61A/G polymorphism and malignant melanoma, [17] the authors pointed out that the EGF 61G/A locus was close to the regulatory region of EGF gene, which can also explain the high expression of serum EGF biologically. In September, 2011, at the European Multidisciplinary Conference held in Stockholm, the association between EGF A/G polymorphism and non-small cell lung cancer[13,14] was demonstrated for the first time in the Portuguese population. This is inconsistent with previous studies by Kang et al[11,12] and other people. The reason for this inconsistency may be ethnic differences. Current studies have indicated that the presence of the EGF 61G allele is indeed considered as a key point in the carcinogenic process, because it can increase serum EGF, thus stimulating proliferation, angiogenesis, and cancer cell metastasis. This interaction between serum EGF and its receptor is very important in the framework of non-small cell lung cancer. It induces tumor invasion through 4 main pathways:
phospholipase C γ;
phosphatidylinositol 3-kinase;
signal transduction and activation factor;
Ras, Raf, MEK, extracellular regulated protein kinases, mitogen-activated protein kinase. [20]
However, another Korean study conducted by Lim et al[11,12] and other people showed that EGF + 61A/G polymorphism was associated with lung cancer. Therefore, the conclusions are inconsistent between different studies. Thus, to evaluate the relation between EGF 61A/G locus and susceptibility of lung cancer, this study collected case-control studies on the relation between EGF 61A/G polymorphism and lung cancer in recent years, and conducted a Meta-analysis to comprehensively evaluate the correlation between EGF 61A/G polymorphism and lung cancer risk.
Different from the studies on EGF 61A/G polymorphism and malignant tumors like gastric cancer, liver cancer, and melanoma, the studies on the association between EGF 61A/G polymorphism and lung cancer are not enough. At present, there are only 6 literature that can extract data and meet the research requirements. Only Asians and Caucasians have had some study on this topic, while Africans have not. From the overall results, the EGF 61A/G allele G related to allele A, OR = 1.07, 95% CI is 0.98 to 1.05, the interval contained 1. In dominant gene model, recessive gene model, homozygote model, and heterozygote model, 95% CI contained 1, and the difference is not statistically significant.
Of course, this study also has limitations.
The number of papers included is not enough, and there are few studies on the association between EGF 61 polymorphism and lung cancer. From the first study in 2004, up to now, there are only 6 papers. From the test results of publication bias, there is significant bias;
the number of cases and the control group is only 1487 and 2044, and the sample size is relatively small;
The research scope is narrow, only Asian and Caucasian studies, not Africans;
The sensitivity of allele model and recessive gene model is poor;
The influence of gender, diet and other factors is not included.
Although the current results of meta-analysis shows that there is no association between the polymorphism of EGF 61A/G and the risk of lung cancer, in term of the limitations of the study, this conclusion still need more researches to support, and more studies are needed to be conducted. Up to now, no meta-analysis is conducted on the association between EGF 6A/G polymorphism and lung cancer susceptibility, so it is meaningless to compare with the results of other researchers. Of course, the occurrence of cancer is caused by multiple factors, especially the influence of genes and environment. Therefore, in the future, a lot of researches still should be done to study the interaction between genes and genes, genes and environment, environment and environment, and explore the association between EGF polymorphism and susceptibility of lung cancer from a broader and deeper perspective.
Author contributions
Study concept and design: Quan Chen, Pengcheng Wang.
Acquisition of data: Quan Chen, Yiming Zheng, Bingbing Wu, Xia Chen, Pengfei Ge, Pengcheng Wang.
Analysis and interpretation of data: Quan Chen, Pengcheng Wang.
Drafting of the manuscript: Quan Chen, Pengcheng Wang; Critical revision of the manuscript for important intellectual content: Quan Chen, Pengcheng Wang.
Statistical analysis: Quan Chen, Yiming Zheng, Bingbing Wu, Xia Chen, Pengfei Ge, Pengcheng Wang.
Administrative, technical, and material support: Quan Chen, Yiming Zheng, Bingbing Wu, Xia Chen, Pengfei Ge, Pengcheng Wang.
Study supervision: Quan Chen, Pengcheng Wang.
All authors have read and approved the manuscript.
Correction
In the original version, R.A. de Mello et al appeared incorrectly in Table 1 and is now appearing correctly as Ataman et al.
Footnotes
Abbreviations: CBM = China Biology Medicine, CI = confidence interval, CJFD = China Academic Journals Full-text Database, \ = China National Knowledge Infrastructure, EGF = epidermal growth factor, Embase = The excerpta medica database, ERK = extracellular regulated protein kinases, Mek = mitogen-activated protein kinase, OR = odds ratio, VIP = China Science and Technology Journal Database.
How to cite this article: Chen Q, Zheng Y, Wu B, Chen X, Ge P, Wang P. Association between polymorphisms of epidermal growth factor 61 and susceptibility of lung cancer: a meta-analysis. Medicine. 2020;99:17(e19456).
This study was funded by the Research Fund Project of Taizhou People's Hospital (GLZL201904).
The authors have no conflicts of interest to disclose.
References
- [1]. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [2]. Yang K, Zhang C, Sun L, et al. Cigarette smoke condensate could promote human bronchial epithelial BEAS-2B cell migration through shifting neprilysin trafficking. J Cancer Res Ther 2018;14:S680–7. [DOI] [PubMed] [Google Scholar]
- [3]. Wang L, Wang LL, Shang D, et al. Gene polymorphism of DNA repair gene X-ray repair cross complementing group 1 and xeroderma pigmentosum group D and environment interaction in non-small-cell lung cancer for Chinese nonsmoking female patients. Kaohsiung J Med Sci 2019;35:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Codony-Servat J, Garcia-Roman S, Molina-Vila MA, et al. Anti-epidermal growth factor vaccine antibodies enhance the efficacy of tyrosine kinase inhibitors and delay the emergence of resistance in EGFR mutant lung cancer cells. J Thorac Oncol 2018;13:1324–37. [DOI] [PubMed] [Google Scholar]
- [5]. Westover D, Zugazagoitia J, Cho BC, et al. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 2018;29: Suppl_1: i10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Peng Q, Li S, Qin X, et al. EGF +61A/G polymorphism contributes to increased gastric cancer risk: evidence from a meta-analysis. Cancer Cell Int 2014;14:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Jiang G, Yu K, Shao L, et al. Association between epidermal growth factor gene +61A/G polymorphism and the risk of hepatocellular carcinoma: a meta-analysis based on 16 studies. BMC Cancer 2015;15:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Zhu Y, Chen Z, Jiang H, et al. The genetic association between EGF A61G polymorphism (rs4444903) and risk of colorectal cancer: an update meta-analysis and trial sequential analysis. Medicine (Baltimore) 2019;98:e14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [10]. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [11]. Lim YJ, Kim JW, Song JY, et al. Epidermal growth factor gene polymorphism is different between schizophrenia and lung cancer patients in Korean population. Neurosci Lett 2005;374:157–60. [DOI] [PubMed] [Google Scholar]
- [12]. Kang HG, Choi JE, Lee WK, et al. +61A>G polymorphism in the EGF gene does not increase the risk of lung cancer. Respirology 2007;12:902–5. [DOI] [PubMed] [Google Scholar]
- [13]. Ataman OU, Bentzen SM, Saunders MI, et al. Failure-specific prognostic factors after continuous hyperfractionated accelerated radiotherapy (CHART) or conventional radiotherapy in locally advanced nonsmall-cell lung cancer: a competing risks analysis. Brit J Cancer 2001;85:1113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. de Mello RA, Ferreira M, Costa S, et al. Association between EGF +61 genetic polymorphisms and non-small cell lung cancer increased risk in a Portuguese population: a case-control study. Tumour Biol 2012;33:1341–8. [DOI] [PubMed] [Google Scholar]
- [15]. Masroor M, Amit J, Javid J, et al. Clinical implication of EGF A61G polymorphism in the risk of non small cell lung adenocarcinoma patients: a case control study. Asian Pac J Cancer Prev 2015;16:7529–34. [DOI] [PubMed] [Google Scholar]
- [16]. Laus Ana Carolina, de Paula Flavia Escremim, de Lima Marcos Alves, et al. EGF+61 A>G polymorphism is not associated with lung cancer risk in the Brazilian population. Mol Biol Rep 2019;46:2417–25. [DOI] [PubMed] [Google Scholar]
- [17]. Zhang Y, Cao C, Liang K. Genetic polymorphism of epidermal growth factor 61A>G and cancer risk: a meta-analysis. Cancer Epidemiol 2010;34:150–6. [DOI] [PubMed] [Google Scholar]
- [18]. Wu SJ, Jiang SY, Wu J, et al. Association between EGF +61 A>G polymorphism and gastric cancer risk: a meta-analysis. J Huazhong Univ Sci Technolog Med Sci 2015;35:327–32. [DOI] [PubMed] [Google Scholar]
- [19]. Zhong JH, You XM, Gong WF, et al. Epidermal growth factor gene polymorphism and risk of hepatocellular carcinoma: a meta-analysis. Plos One 2012;7:e32159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Hu-Lieskovan S, Vallbohmer D, Zhang W, et al. EGF61 polymorphism predicts complete pathologic response to cetuximab-based chemoradiation independent of KRAS status in locally advanced rectal cancer patients. Clin Cancer Res 2011;17:5161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


