Abstract
X-linked severe combined immunodeficiency (X-SCID) is an inherited genetic disorder. A majority of X-SCID subjects carries point mutations in the Interleukin-2 receptor gamma chain (IL2RG) gene. In contrast, Il2rg-knockout mice recapitulating X-SCID phenotype lack a large part of Il2rg instead of point mutations. In this study, we generated novel X-SCID mouse strains with small insertion and deletion (InDel) mutations in Il2rg by using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9. To this end, we injected Streptococcus pyogenes Cas9 (SpCas9) mRNA and single guide RNA targeting the exon 2, 3 or 4 of Il2rg into mouse zygotes. In the F0 generation, we obtained 35 pups and 25 out of them were positive for Surveyor assay, and most of mutants displayed dramatic reductions of T and B lymphocytes in the peripheral blood. By amplicon sequencing, 15 out of 31 founder mice were determined as monoallelic mutants with possible minor mosaicisms while 10 mice were mosaic. Finally, we established new strains with 7-nucleotide deletion and 1-nucleotide insertions in the exon 2 and the exons 3 and 4, respectively. Although no IL2RG protein was detected on T cells of exons 3 and 4 mutants, IL2RG protein was unexpectedly detected in the exon 2 mutants. These data indicated that CRISPR/Cas9 targeting Il2rg causes InDel mutations effectively and generates genetically X-SCID mice. Genetic mutations, however, did not necessarily grant phenotypical alteration, which requires an intensive analysis after establishing a strain to confirm their phenotypes.
Keywords: animal model, clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, genome-editing, II2rg, X-linked severe combined immunodeficiency (X-SCID)
Introduction
The interleukin-2 receptor γ chain (IL2RG), encoded from chromosome X, is an essential common receptor for IL-2, 4, 7, 9, 15 and 21 [3, 21] and plays an vital role in lymphoid-lineage cell development through activation of Janus kinase 3 [11, 20]. Mutations in IL2RG gene, causing the truncation or altered amino sequence of the protein, impair the interaction of the receptor complexes to cytokines, and as a result, the differentiation of T and natural killer (NK) cells is blocked [4]. Allogeneic transplantation of hematopoietic stem cells (HSCs) is the first-line treatment of X-SCID [4]. Gene therapy, autologous transplantation of HSCs introduced with IL2RG gene by retro- or lentiviral vectors, is another therapy in clinical trials [13]. However, due to limited availability of HLA-matched donors and leukemia caused by the insertional mutagenesis of the viral vector [14], another alternative therapy might be required. With recent advances in targeted gene correction using designed nucleases [2], autologous transplantation of gene-corrected HSCs is one alternative and promising method for X-SCID treatment.
To develop genome-editing therapy, a better murine model of X-SCID is preferred. Nearly 200 types of mutations causing X-SCID have been reported and they occur in any of eight exons, mainly as point or frameshift mutations [29]. In contrast, current murine models of X-SCID are either deletion of exon 3 and entire exon 4–8 [3] or two-third of the exon 7, the intron 7 and the half of exon 8 of Il2rg [27]. The mice with the loss-of-function mutations of Il2rg are without functional T, B and NK cells and are good models for gene therapies. However, as the nature of the mutations, they are not suitable for the disease model of genome-editing therapies. Therefore, Il2rg-knockout mice with small insertion or deletion (InDel) mutations are awaited.
Genetically engineered mouse models with site-specific transgene insertion, gene replacement or conditional knock-out alleles were mostly generated through a homologous recombination in embryonic stem (ES) cells. In contrast, clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) allows direct generation of targeted genome-edited animals due to high efficiency of genome editing [8, 10, 35]. More importantly, CRISPR/Cas9 enable to generate an animal model with small mutations efficiently [18].
In this study, we developed novel Il2rg-knockout mouse strains that have short InDel mutations in exons 2, 3 or 4 by CRISPR/Cas9.
Materials and Methods
Animals
C57BL/6J and Multi Cross Hybrid ICR (MCH [ICR]) mice were purchased from SLC Japan (Shizuoka, Japan) and CLEA Japan (Tokyo, Japan), respectively. All mice were housed at Center for Experimental Medicine, Jichi Medical University. All animal experiments reported here were approved by the Institutional Animal Care and Concern Committee at Jichi Medical University. Animal care and all experiments were performed in accordance with the committee’s guidelines.
Designing and preparing guide RNA
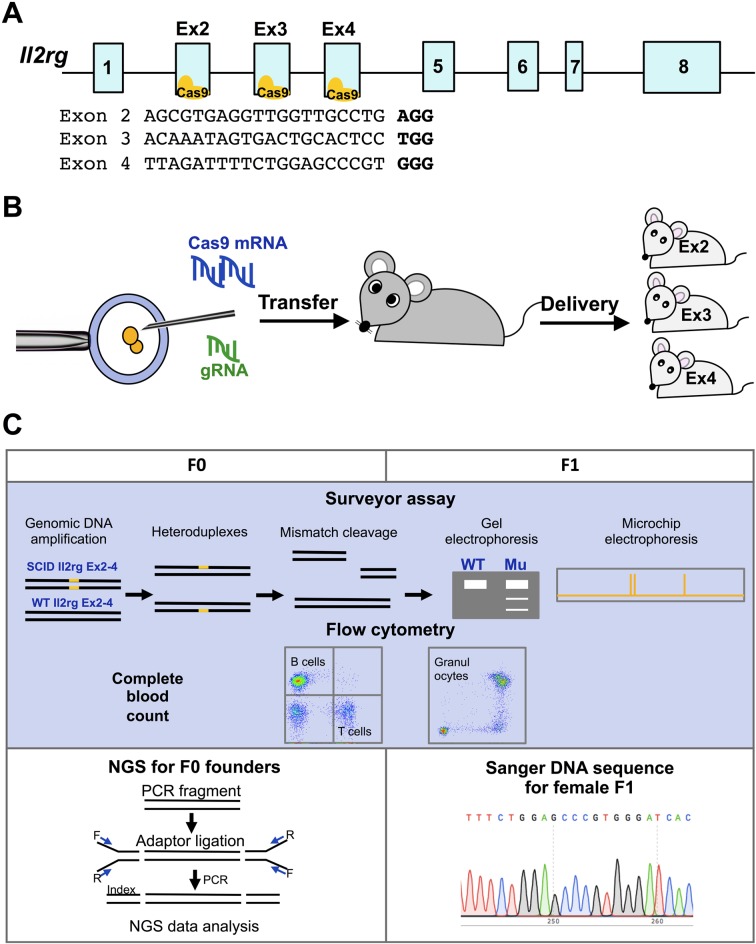
The guide sequences of guide RNAs (gRNAs) for generating Il2rg-knockout mice were designed in exon 2 (AGCGTGAGGTTGGTTGCCTG), exon 3 (ACAAATAGTGACTGCACTCC) or exon 4 (TTAGATTTTCTGGAGCCCGT) of Il2rg. All gRNA sequences were designed using online software, Benchling (https://benchling.com). Their specificities (off-target scores and potential off-target sites) and potential on-target efficiencies (on-target scores) were also examined with Geneious 11.1.5 (GRCm38.p4 for mouse chromosome dataset) (Biomatters Ltd., Auckland, New Zealand), CCTop, Cas-OFFinder and CRISPRdirect [1, 25, 32]. We calculated off-target scores using Geneious R11. For the calculation, we used mouse genomic DNA sequences, GRCm38.p4, with allowing maximum three mismatches and one indel to the off-target sites[17]. To generate single guide RNAs (sgRNAs) for Il2rg exon 2, 3 and 4, GeneArt Precision gRNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) was used, followed by in vitro transcription (IVT) with CUGA3/CUGA7 (Nippon Gene Tech., Tokyo, Japan) as following manufactures’ instructions. Briefly, we assembled T7 promoter, the target-specific sequences and trans-activating CRISPR RNA (tracrRNA) fragment to generate the DNA template for IVT. The oligonucleotides for the T7 promoter and the target-specific sequences were synthesized as follows; Il2rg exon2: fwd; TAATACGACTCACTATAG-AGCGTGAGGTTGGTTG, rev; TTCTAGCTCTAAAAC-CAGGCAACCAACCTCACGC; Il2rg exon3: fwd; TAATACGACTCACTATAG-ACAAATAGTGACTGCA, rev; TTCTAGCTCTAAAAC GGAGTGCAGTCACTATTTG, Il2rg exon4: fwd; TAATACGACTCACTATAG-TTAGATTTTCTGGAGC; rev; TTCTAGCTCTAAAAC-ACGGGCTCCAGAAAATCTA. Then, IVT reactions were performed using CUGA3/CUGA7. After IVT, the template DNA were removed by 1U/µl DNase I digestion. Finally, sgRNAs were purified with phenol/chloroform extraction, and the quality of the RNA was assessed by gel electrophoresis using 2% agarose gel with 3-(N-morpholino) propanesulfonic acid (MOPS) buffer and 37% formaldehyde.
Co-injection of Cas9 mRNA and gRNA into pronucleus in zygotes
20 ng/µl Streptococcus pyogenesis Cas9 (SpCas9) mRNA (Thermo Fisher Scientific) and 5 ng/µl sgRNAs targeting exon 2, 3 or 4 of mouse Il2rg were microinjected into pronucleus of zygotes. Microinjected zygotes were transferred into pseudopregnant MCH (ICR) mice at the one- or two-cell stage and founders were delivered by Caesarian section.
Surveyor assay and DNA sequencing for target sites
The genomic DNA was extracted from the peripheral blood or tail-cut tissues with the DNA extraction kit (Qiagen, Venlo, Netherlands) and CRISPR/Cas9-mediated genomic mutations were detected with the Surveyor mutation detection kit (Integrated DNA Technologies, Skokie, IL, USA). The target site of Il2rg exon 2 to 4 was amplified with ExTaq DNA polymerase (Takara Bio, Otsu, Japan) and the primer set 5’-AGATCCTTCTTAGTCCTTCAGCTGCT-3’ and 3’-GCTTGGCCTTAGCCACTGC-5’. Heteroduplex was formed under the denaturing and re-annealing protocol with a thermal cycler as indicated in the manufacturer’s instructions. Then, the samples were treated with Surveyor nuclease and the DNA fragments were analyzed by agarose gel electrophoresis and by MultiNA, a microchip electrophoresis system (Shimadzu, Tokyo, Japan) with DNA-12000 Reagent Kit (Shimadzu, Tokyo, Japan).
To analyze the introduced mutations, Sanger DNA sequencing was performed in all mutated F1 female mice. The peripheral blood was obtained from all F1 mice, and DNA was extracted using the DNA extraction kit. PCR fragments of the targeted sites were amplified and then cloned to the pTAC-2 vector (Biodynamics, Tokyo, Japan). After transformation to One Shot Stbl3 Chemically Competent E. coli(Thermo Fisher Scientific), four clones were collected for each mouse. Plasmids were extracted by the Qiaprep Spin Miniprep kit (Qiagen, Hilden, Germany) for DNA sequencing.
Amplicon sequence
To precisely examine the mosaicisms in the founder lines, we performed amplicon sequencing. We first amplified exon 2–4 of Il2rg using the primer set described above, then performed nested PCR to amplify each exon using primer pairs (Il2rg Ex2: 5’-TCTACAGCCCCTGAACACCT-3’, 3’-CAGTCTCTCCCAGCTAACCTCCCT-5’; Il2rg Ex3: 5’-TGGCCCATCATTCTTTTGCCTTG-3’, 5’-CTGTACAGCTCGCCTCTGG-3’; Il2rg Ex4: 5’-CTCCAAGATCCTGACTTGTCTAGGC-3’, 3’-GTCCAGCTTCGATCTCTGTTGCT-5’). Illumina sequence adapters and indices (SeqCap Adapter Kit A, Roche, Basel, Switzerland) were ligated using KAPA LTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA, USA). Pooled library was sequenced with MiSeq v2 Nano, 150-bp, paired-end (Illumina, San Diego, CA, USA). As we pooled 33 samples with only 12 indices (ones from each exon line were assigned to one index), we demultiplex based on the sequence results using R package ShortRead [24]. Low coverage mutants were excluded from the analysis (Ex3 #7 and 9). The mutations in each founder line were examined and mosaicisms were calculated for each mutation using R package amplican[22] with the demultiplexed, full-length and paired-end sequences. In some cases, amplican counted reads separately if reads contain some PCR or sequence errors. Therefore, we further examined the mosaicisms only at the target site sequences (Figs. 3B and C) after merging the paired-end sequences with Paired-End reAd mergeR (PEAR) [34].
Fig. 3.
Mutation and mosaicism analysis of F0 founder lines. (A) Schematic representation of amplicon sequencing for detection of mutations and mosaicisms in the founder lines. (B) Allele frequencies in 30 founders. Blue to light blue indicate allele frequencies of up to top three and the alleles correspond to at least 10% of reads; yellow indicates the wild-type allele; gray indicates the rest of reads. (C) Detected mutation types at the each exon. Blue to light blue indicate mutation types; yellow indicates the wild-type. (D) Sequences around the target sites. The wild-type alleles of the corresponding exons were shown on the top (blue, PAM; green, gRNA; underline, potential microhomology). Sequences with at least 10% of allele frequencies in each mouse were shown (red, mutations). Freq indicates the allele frequencies of the sequence in the corresponding mouse.
Flow cytometry
The peripheral blood was obtained through the jugular vein of mice and mononuclear cells were isolated through erythrocyte lysis. The cells were stained with anti-mCD45-FITC, anti-CD45.2-BV711 (BD Bioscience, East Rutherford, NJ, USA), anti-CD3-PE, anti-CD3-APC, anti-CD19-PE/Cy7, anti-CD11b-BV421 and anti-Gr-1-BV510 (BioLegend, San Diego, CA, USA) to identify lymphoid and myeloid lineages of cells. Anti-CD132-PE (clone TUGm2, BioLegend) was used to detect IL2RG protein. Flow cytometry was performed with BD LSR Fortessa II, The BD FACS Aria Fusion (BD Bioscience).
Results
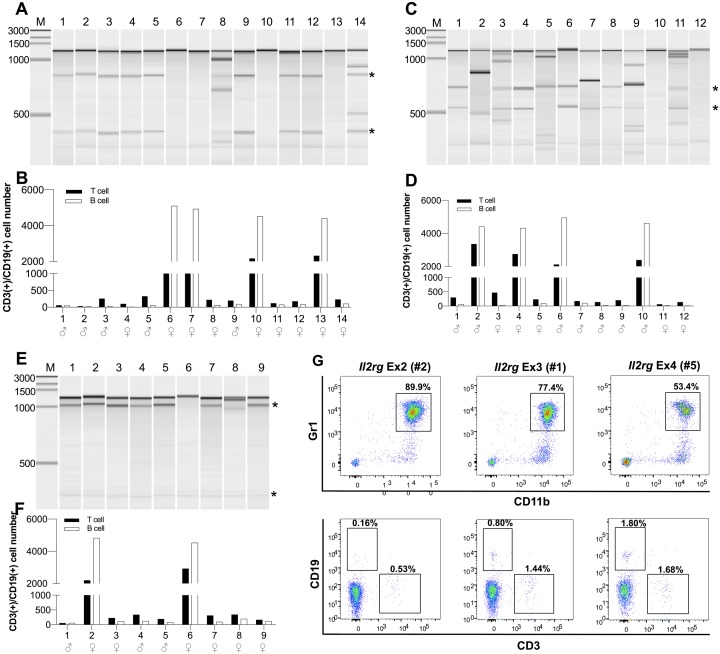
In this study, we selected gRNA for exons 2, 3 and 4 of Il2rg with at least 90% off-target score and higher on-target score (Fig. 1A). SpCas9 mRNA and gRNA targeting exons 2, 3 or 4 of Il2rg were injected into mouse zygotes (Fig. 1B, Table 1). To identify mutated founders, we performed genotyping and phenotyping for all F0 founders by Surveyor assay and flow cytometry, respectively (Figs. 1C and 2A–G). In the F0 generation, we obtained 35 offspring (14, 12 and 9 mice for exons 2, 3 and 4, respectively) (Table 1). Among them, 29 were positive for the Surveyor assay (71–92%) (Figs. 2A, C and E). We confirmed dramatic reductions of T and B lymphocytes (CD3+ and CD19+, respectively) and the existence of granulocytes (Gr1+/CD11b+) in the peripheral blood of most mutated founders (Figs. 2B, D and F) and representative flow-cytometric results are shown in Fig. 2G (Il2rg Ex2 #2, Ex3 #1, Ex4 #5). Of note, some female mutants positive for Surveyor assay also displayed the reductions of T and B cells, indicating bi-allelic mutations (e.g. Il2rg Ex2 #4, #8, #11, #12 and #14), while other mutated males and females retained normal counts of T and B cells, suggesting in-frame mutations, heterozygous mutants or mosaicism (e.g. Il2rg Ex3 #2, #4 and #6).
Fig. 1.
Experimental design for the generation of CRISPR/Cas9-mediated X-SCID mice. (A) A schematic diagram showing the target sites of gRNAs in the Il2rg gene exon 2, 3 and 4. (B) A schematic presentation for generation of Il2rg-mutant mice by CRIPSR/Cas9. (C) Experimental design of Il2rg-knockout mice. For F0 and F1 generation, all of offspring were subjected to tail cut at 4 weeks after delivery for sampling and Surveyor assays were performed for genotyping. Amplicon sequencing was also performed for founder mice to determine mosaicisms. With F1 generation, Sanger DNA sequencing was performed to confirm mutations in the strains.
Table 1. Generation of founders.
| Target | Numbers of zygote transferred |
Numbers of offspring with mutations (%)a | ||
|---|---|---|---|---|
| Total | Male | Female | ||
| Il2rg Ex2 | 42 | 10/14 (71.4%) | 5/5 (100%) | 5/9 (55.5%) |
| Il2rg Ex3 | 42 | 11/12 (91.6%) | 6/7 (85.7%) | 5/5 (100%) |
| Il2rg Ex4 | 16 | 8/9 (88.9%) | 3/3 (100%) | 5/6 (83.3%) |
aAnalyzed by Surveyor assay.
Fig. 2.
Phenotypical and genotypical analysis of Il2rg-targeted F0 founders. Phenotyping with flow cytometry and genotyping with Surveyor assays of founder mice. (A, C, E) Electrophoresis results of Surveyor assays. Expected mutated bands are indicated with black asterisks. (B, D, F) Flow cytometry results: Black bars, the numbers of CD3+ T cells; white bars, the numbers of CD19+ B cells in the peripheral blood of the mice. (G) Representative flow cytometry results showing the reductions of T and B cells and the existence of granulocytes (Gr+/CD11b+ cells) in the peripheral blood of Il2rg Ex2 #2, Ex3 #1 and Ex4 #5 mice.
To further examine mosaicisms of founder lines, we performed amplicon sequencing (Fig. 3A). Among 33 examined (Ex2 #6 and #7 were not examined), two were not sequenced well (Ex3 #7 and #9, <1,000 reads/sample) and excluded from the analysis. Ex3 #5 had large deletion and insertion (90% of the reads were the identical sequence, data not shown). In six mice, the wild-type sequences were identified at 96.4% in Ex2 (#10, 13), 94.5% in Ex3 (#10, 12) and 95.1% in Ex4 (#6, 8) of the reads (Fig. 3B-D), confirming the Surveyor assays (Fig. 2). Fifteen mutants (9 males and 6 females) were monoallelic with mutated sequences with minor possible mosaicisms (at least 90% of the reads were identical at the target sites) while 10 mutants (4 males and 6 females) were more heterogeneous with two to four different sequences detected (Figs. 3B–D). Of note, 3-nucleotide deletion in Ex3 (Ex3 #4, 6) did not alter T and B cell counts, whereas 9-nucleotide deletion in Ex4 (Ex4 #4, 7) were enough to abolish T and B cell development. Overall, mutations were in most part deletions between 2- to 14-nucleotide, 1-nucleotide substitution or 1-nucleotide insertion except one mutant with larger insertion and deletion (Figs. 3C and D, Supplementary Fig. 1). Specifically, 14-nucleotide deletion in the Ex2 was the most frequent mutation found in the Ex2 founder mice (45.5%, Fig. 3C). The target site at the Ex3 was mainly unedited (33.1%) while 1-nucleotide insertion was the most frequent mutation (24.2%). At the Ex4, 9-nucleotide deletion was the majority of editing (32.0%). We also performed Surveyor assay for detecting possible off-target mutations. Total 24 sites were selected using CCTop and Cas-OFFinder for Il2rg Ex 2, 3 and 4 (Supplementary Table 1). Based on Surveyor assay, we did not observe any specific mutations in 8 potential off-target sites for Ex2 ; 9 for Ex3; and 7 for Ex4 (Supplementary Fig. 3). Taken together, the injection of Cas9 mRNA and gRNA caused mutations effectively.
To test germ-line transmission and to determine mosaicism in germ cells, we crossed one male founder from each target with the wild-type C57BL/6J female mice (Fig. 4A). In the female offspring of F1 generation, we found that total 13 out of 15 mice (87.7%) had mutated alleles in the corresponding exons; that is, Ex2, 4/4 (100%); Ex3, 5/6 (83.3%); and Ex4, 4/5 (80%) (Figs. 4A and B, Supplementary Fig. 2), suggesting germline mosaicisms seems to be greater than somatic mosaicisms in the founder mice. We also performed DNA sequencing to confirm mutations in all mutated female offspring. We detected the 7-nucleotide deletion in Il2rg Ex2 strain and the 1-nucleotide insertion in the strains of Il2rg Ex3 and Ex4 (Fig. 4C), thus, we named each strain as follows: Il2rg Ex2 Del (7), Il2rg Ex3 Ins (1) and Il2rg Ex4 Ins (1). From F2 generation, we genotyped only male offspring. In the F2 generation, we also confirmed the dramatic reduction of T and B lymphocytes in the peripheral blood in male mice by flow cytometry (Fig. 4D). Furthermore, we obtained mutant male and heterozygote female mice in generation F2 to F9. Overall, the mutations were carried in approximately 45.3% of male offspring by following Mendelian law (Fig. 4A).
Fig. 4.
Germline transmission and the expression of Il2rg on T cells of the three strains. (A) A diagram of germ-line transmission. One male founder from each target was crossed with wild-type C57BL/6J mice. For F2 to F9 generations, total numbers of Il2rg-mutated male mice were indicated. (B) Surveyor assays for detecting mutations in Il2rg Exon 2, 3 or 4 in F1 female mice. The expected mutated bands were indicated with white asterisks. (C) Sequencing results of the new strains: a 7-nucleotide deletion in the Il2rg Ex2 Del (7) strain; and 1-nucleotide insertions in the strains Ex3 Ins (1) and Ex4 Ins (1) (red dash and text indicate mutations). (D) Flow cytometry of CD3, CD19 and IL2RG of the peripheral blood from C57BL/6J (wild-type), NOG and the three strains of Il2rg-mutated mice. Top panels show CD3 (T cells) and CD19 (B cells) of the peripheral blood. Bottom panels show IL2RG in CD3+ fractions from top panel. To note, almost all wild-type T cells express IL2RG, while 76.5% and less than 1% in Ex2 Del (7) and Ex 3 Ins (1) and Ex 4 (1), respectively. No T cells were detected in NOG mice.
Finally, we examined IL2RG protein expression on CD3+ T cells by flow cytometry (between F5 to F7 generation). Among the three strains of mice, T cells in Il2rg Ex2 Del (7) were positive for IL2RG protein (Fig. 4D), while T cells in two other strains (Il2rg Ex3 Ins (1) and Il2rg Ex4 Ins (1)) did not express IL2RG protein (Fig. 4D). The results show that even CRISPR/Cas9-mediated approach is highly efficient to induce targeted mutations like small InDels, some of mutations may remain inefficient to fully impair gene function.
Discussion
In this study, we generated novel strains of Il2rg-knockout mice by CRISPR/Cas9 mRNA injection. We proved that targeted gene disruption with CRISPR/Cas9 works effectively and provides several advantages. First, CRISPR/Cas9 can induce small mutations like small frameshift insertions or deletions that are enough to disrupt Il2rg. Second, Il2rg-knockout mice were generated within three months with high efficiency (mutation efficiencies were more than 71%). Finally, the new Il2rg-knockout mice were C57BL/6J background, thus no backcross was required to establish the strains.
We found CRISPR/Cas9-mediated mutations varied from 14-nucleotide deletion to one-nucleotide insertion except one with a larger InDel mutation and also found mosaicisms (15 mutants with minor mosaicisms or potentially due to PCR/sequencing error while 10 mutants with major mosaicisms) confirmed by the amplicon sequencing. Mosaicism is one of the remaining issues with CRISPR/Cas9-mediated mutagenesis. Injection and electroporation of Cas9 mRNA and gRNA are commonly used as a delivery method of CRISPR/Cas9 to zygotes [15, 28]. However, these methods induce mosaicism because the delivery is carried out at E0.5 or later while mouse first genome replication starts around E0.4 [16]. The time lag of the translation from mRNA to protein is another factor to delay the genome editing in a zygote. Recently, electroporation of Cas9 protein and gRNA into zygotes becomes a more popular method as higher quality Cas9 protein and gRNA became available. More importantly, non-mosaic animals were established by this method [7, 8, 16].
Interestingly, there were predominant types of mutations in two targets (Ex2, 14-nucleotide del; Ex4, 8- or 9-nucleotide del and 1-nucleotide ins). CRISPR/Cas9-mediated double strand breaks are mostly repaired by non-homologous end joining (NHEJ). However, in some cases, alternative NHEJ takes place called microhomology-mediated end-joining (MMEJ) [26]. At the sites of the targets, we identified potential microhomology sequences (shown as underbars in Fig. 3D). For example, around the target site of Ex2, two CCTCA were spanned by Cas9 cleavage site and the nine nucleotides in between the microhomology and one CCTCA seemed to be excised to form 14-nucleotide deletion.
One-nucleotide insertion at the exon 3 or 4 of Il2rg and even 9-nucleotide and in-frame deletion at the exon 4 were enough to abolish IL2RG protein expression on T cells. In contrast, the mutant mice with 7-nucleotide deletion in exon 2 carried T cells expressing IL2RG protein, which highlights the importance of an extensive analysis for newly generated mutants. Similar results were recently reported [31]. Among 193 frameshifts generated by CRISPR-Cas9, one third of induced frameshift mutations resulted in complete loss of protein expression, however one third of them still remained expressed, mostly at very low level. Moreover, choosing target exons is more important for generating phenotypically correct knockout animals. Triple-targeting CRISPR would be an option for this purpose [33]. The nature of IL2RG protein expression from the mutated Il2rg allele remains to be characterized. The reduction of T-cell number is obvious in the exon 2 mutants. Therefore, the IL2RG function or activity is supposed to be lower. To detect IL2RG protein, we used anti-CD132-PE (TUGm2) monoclonal antibody. The exact epitope of this antibody is unknown, but it is predicted around Lys158 of IL2RG [12]. Because all of the gRNAs used in this study are located in 5’ side of Lys158, all indels equally disrupt the epitope sequence if splicing occurs in the same manner as wild-type sequence. Therefore, there might be exon skipping, alternative splicing or alternative start codon as proposed in a previous study [30]. As a signal peptide is coded in exon 1, exon skipping or alternative splicing are more likely for the expression of the altered IL2RG protein. Several studies reported that CRISPR/Cas9-mediated mutations can cause exon skipping by boundary deletion of intron and exon (3’ splice site), generating new splicing donor sites or larger exon removal [30].
As noted in the introduction, the Il2rg-knockout mice established previously have large deletions of Il2rg [11, 19, 27]. In contrast, our new Il2rg-knockout strains have short InDel mutations, which more faithfully reflect mutations seen in X-SCID patients and thus should be preferable for developing genome-editing therapies. With the C57Bl/6J background, our strains have CD45.2+ lymphocytes while NODShi.Cg-PrkdcscidIl2rgtm1Sug (NOG) mice have CD45.1, which allows us to perform congenic transplantation of HSCs from the new strains to NOG, or vice versa. In addition, due to their contract restrictions, it is not allowed to mate NOG and NOD.Cg-PrkdcscidIl2rgtm1Wjl (NSG) mice with other strain. These features also give advantage to our new strains to develop therapeutics. Patient-derived induced pluripotent stem (iPS) cells or bone marrow samples from X-SCID patient can also be used for the models of X-SCID [5, 23]. However, it is still challenging to generate HSCs from iPS cells, and the bone marrow samples are insufficient for the demands. Needless to say, generation of disease model animals with the exactly same mutations as patients is helpful to develop clinical treatments and mouse models with patient-specific mutations such as X-linked retinoschisis or Borjeson-Forssman-Lehmann syndrome were reported [6, 9]. However, as there are more than 200 mutations identified to cause X-SCID, it would be more complicated to select and generate each mutation to model a patient unless one specific mutation is the focus of a research. To note, though the homologous recombination (HR)-mediated targeted knock-in with single-stranded oligodeoxynucleotides (ssODNs) became more efficient, it is still less efficient than NHEJ-mediated knock-out [36]. Thus, NHEJ-mediated knock-out might be the first choice to generate a new mouse model to recapitulate disease phenotypes thus far.
In summary, we have generated three strains of Il2rg-knockout mice with small InDel mutations by using CRISPR/Cas9 mRNA and gRNA. The novel strains established by CRISPR/Cas9 should provide a valuable model for evaluating genome-editing therapy. It was highly effective to produce such mutations with minor mosaicisms, although a frameshift InDel mutation may not completely abolish the protein expression but just alter the function. As genome-editing therapy has gained more interest, more animal models with small mutations mimic to patients are awaited.
Author Contributions
S.B. designed experiments, performed experiments, analyzed data, prepared figures, wrote and revised a manuscript; H.U. designed experiments, performed experiments, analyzed data, wrote and revised a manuscript; H.H. designed experiments and wrote a manuscript; H.S. designed experiments and performed experiments; T.A. designed experiments; Y.N. performed experiments; O.N. generated reagents; T.O. designed experiments, performed experiments, analyzed data and wrote a manuscript, and Y.H designed experiments, analyzed data, wrote and revised a manuscript.
Conflict of Interests
All authors declare no competing financial interests in this study.
Supplementary Material
Acknowledgments
We appreciate Hiroko Hayakawa, Tamaki Aoki, Mika Kishimoto, Yaeko Sutoh, Eri Noguchi, Nawin Chanthra, Takahiro Ohnuki, Rie Ishihara and Mari Karube for their technical assistance. This study was supported in part by the Japan Agency for Medical Research and Development (AMED) Basic Science and Platform Technology Program for Innovative Biological Medicine (18am0301002s0105 to O.N.) and the Ministry of Education, Culture, Sports, Science and Technology for the Supported Program for the Strategic Research Foundation at Private Universities (S1311030 to Y.H.). This publication was also subsidized by JKA through its promotion funds from KEIRIN RACE (for the purchase of BD LSR Fortessa II).
References
- 1.Bae S., Park J., Kim J.S.2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. doi: 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birling M.C., Herault Y., Pavlovic G.2017. Modeling human disease in rodents by CRISPR/Cas9 genome editing. Mamm. Genome 28: 291–301. doi: 10.1007/s00335-017-9703-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao X., Shores E.W., Hu-Li J., Anver M.R., Kelsall B.L., Russell S.M., Drago J., Noguchi M., Grinberg A., Bloom E.T., Pau L.W.E., Katz S.I., Love P.E., Leonard W.J.1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2: 223–238. doi: 10.1016/1074-7613(95)90047-0 [DOI] [PubMed] [Google Scholar]
- 4.Cavazzana-Calvo M., Hacein-Bey S., de Saint Basile G., Gross F., Yvon E., Nusbaum P., Selz F., Hue C., Certain S., Casanova J.L., Bousso P., Deist F.L., Fischer A.2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288: 669–672. doi: 10.1126/science.288.5466.669 [DOI] [PubMed] [Google Scholar]
- 5.Chang C.W., Lai Y.S., Westin E., Khodadadi-Jamayran A., Pawlik K.M., Lamb L.S., Jr, Goldman F.D., Townes T.M.2015. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 12: 1668–1677. doi: 10.1016/j.celrep.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 6.Chen D., Xu T., Tu M., Xu J., Zhou C., Cheng L., Yang R., Yang T., Zheng W., He X., Deng R., Ge X., Li J., Song Z., Zhao J., Gu F.2018. Recapitulating X-Linked juvenile retinoschisis in mouse model by knock-in patient-specific novel mutation. Front. Mol. Neurosci. 10: 453. doi: 10.3389/fnmol.2017.00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S., Lee B., Lee A.Y., Modzelewski A.J., He L.2016. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 291: 14457–14467. doi: 10.1074/jbc.M116.733154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S., Sun S., Moonen D., Lee C., Lee A.Y., Schaffer D.V., He L.2019. CRISPR-READI: Efficient generation of knockin mice by CRISPR RNP electroporation and AAV donor infection. Cell Rep. 27: 3780–3789.e4. doi: 10.1016/j.celrep.2019.05.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng C., Deng P. Y., Ikeuchi Y., Yuede C., Li D., Rensing N., Huang J., Baldridge D., Maloney S. E., Dougherty J. D., Constantino J., Jahani-Asl A., Wong M., Wozniak D. F., Wang T., Klyachko V. A., Bonni A.2018. Characterization of a mouse model of Borjeson-Forssman-Lehmann syndrome. Cell Rep 25: 1404–1414 e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwish M., Nishizono H., Uosaki H., Sawada H., Sadahiro T., Ieda M., Takao K.2019. Rapid and high-efficient generation of mutant mice using freeze-thawed embryos of the C57BL/6J strain. J. Neurosci. Methods 317: 149–156. doi: 10.1016/j.jneumeth.2019.01.010 [DOI] [PubMed] [Google Scholar]
- 11.DiSanto J.P., Müller W., Guy-Grand D., Fischer A., Rajewsky K.1995. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. USA 92: 377–381. doi: 10.1073/pnas.92.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh T.S., Jo Y., Lee B., Kim G., Hwang H., Ko E., Kang S.W., Oh S.O., Baek S.Y., Yoon S., Lee J.S., Hong C.2017. IL-7 induces an epitope masking of gammac protein in IL-7 receptor signaling complex. Mediators Inflamm. 2017: 9096829. doi: 10.1155/2017/9096829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina S., Hauer J., Lim A., Picard C., Wang G.P., Berry C.C., Martinache C., Rieux-Laucat F., Latour S., Belohradsky B.H., Leiva L., Sorensen R., Debré M., Casanova J.L., Blanche S., Durandy A., Bushman F.D., Fischer A., Cavazzana-Calvo M.2010. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363: 355–364. doi: 10.1056/NEJMoa1000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., Sorensen R., Forster A., Fraser P., Cohen J.I., de Saint Basile G., Alexander I., Wintergerst U., Frebourg T., Aurias A., Stoppa-Lyonnet D., Romana S., Radford-Weiss I., Gross F., Valensi F., Delabesse E., Macintyre E., Sigaux F., Soulier J., Leiva L.E., Wissler M., Prinz C., Rabbitts T.H., Le Deist F., Fischer A., Cavazzana-Calvo M.2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302: 415–419. doi: 10.1126/science.1088547 [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto M., Takemoto T.2015. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 5: 11315. doi: 10.1038/srep11315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto M., Yamashita Y., Takemoto T.2016. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 418: 1–9. doi: 10.1016/j.ydbio.2016.07.017 [DOI] [PubMed] [Google Scholar]
- 17.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F.2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. doi: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S., Asahara H., Fukami M., Takada S.2014. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 4: 5396. doi: 10.1038/srep05396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M., Hiramatsu H., Kobayashi K., Suzue K., Kawahata M., Hioki K., Ueyama Y., Koyanagi Y., Sugamura K., Tsuji K., Heike T., Nakahata T.2002. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100: 3175–3182. doi: 10.1182/blood-2001-12-0207 [DOI] [PubMed] [Google Scholar]
- 20.Kawahara A., Minami Y., Miyazaki T., Ihle J.N., Taniguchi T.1995. Critical role of the interleukin 2 (IL-2) receptor gamma-chain-associated Jak3 in the IL-2-induced c-fos and c-myc, but not bcl-2, gene induction. Proc. Natl. Acad. Sci. USA 92: 8724–8728. doi: 10.1073/pnas.92.19.8724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovanen P.E., Leonard W.J.2004. Cytokines and immunodeficiency diseases: critical roles of the gamma(c)-dependent cytokines interleukins 2, 4, 7, 9, 15, and 21, and their signaling pathways. Immunol. Rev. 202: 67–83. doi: 10.1111/j.0105-2896.2004.00203.x [DOI] [PubMed] [Google Scholar]
- 22.Labun K., Guo X., Chavez A., Church G., Gagnon J.A., Valen E.2019. Accurate analysis of genuine CRISPR editing events with ampliCan. Genome Res. 29: 843–847. doi: 10.1101/gr.244293.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menon T., Firth A.L., Scripture-Adams D.D., Galic Z., Qualls S.J., Gilmore W.B., Ke E., Singer O., Anderson L.S., Bornzin A.R., Alexander I.E., Zack J.A., Verma I.M.2015. Lymphoid regeneration from gene-corrected SCID-X1 subject-derived iPSCs. Cell Stem Cell 16: 367–372. doi: 10.1016/j.stem.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgulis A., Gertz E.M., Schäffer A.A., Agarwala R.2006. WindowMasker: window-based masker for sequenced genomes. Bioinformatics 22: 134–141. doi: 10.1093/bioinformatics/bti774 [DOI] [PubMed] [Google Scholar]
- 25.Naito Y., Hino K., Bono H., Ui-Tei K.2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123. doi: 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T., Suzuki K.T.2014. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5: 5560. doi: 10.1038/ncomms6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohbo K., Suda T., Hashiyama M., Mantani A., Ikebe M., Miyakawa K., Moriyama M., Nakamura M., Katsuki M., Takahashi K., Yamamura K., Sugamura K.1996. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor gamma chain. Blood 87: 956–967. doi: 10.1182/blood.V87.3.956.bloodjournal873956 [DOI] [PubMed] [Google Scholar]
- 28.Oliver D., Yuan S., McSwiggin H., Yan W.2015. Pervasive genotypic mosaicism in founder mice derived from genome editing through pronuclear injection. PLoS One 10: e0129457. doi: 10.1371/journal.pone.0129457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puck J.M., Pepper A.E., Henthorn P.S., Candotti F., Isakov J., Whitwam T., Conley M.E., Fischer R.E., Rosenblatt H.M., Small T.N., Buckley R.H.1997. Mutation analysis of IL2RG in human X-linked severe combined immunodeficiency. Blood 89: 1968–1977. [PubMed] [Google Scholar]
- 30.Sharpe J.J., Cooper T.A.2017. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 18: 109. doi: 10.1186/s13059-017-1240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smits A.H., Ziebell F., Joberty G., Zinn N., Mueller W.F., Clauder-Münster S., Eberhard D., Fälth Savitski M., Grandi P., Jakob P., Michon A.M., Sun H., Tessmer K., Bürckstümmer T., Bantscheff M., Steinmetz L.M., Drewes G., Huber W.2019. Biological plasticity rescues target activity in CRISPR knock outs. Nat. Methods 16: 1087–1093. doi: 10.1038/s41592-019-0614-5 [DOI] [PubMed] [Google Scholar]
- 32.Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L.2015. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10: e0124633. doi: 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunagawa G.A., Sumiyama K., Ukai-Tadenuma M., Perrin D., Fujishima H., Ukai H., Nishimura O., Shi S., Ohno R.I., Narumi R., Shimizu Y., Tone D., Ode K.L., Kuraku S., Ueda H.R.2016. Mammalian reverse genetics without crossing reveals Nr3a as a short-sleeper gene. Cell Rep. 14: 662–677. doi: 10.1016/j.celrep.2015.12.052 [DOI] [PubMed] [Google Scholar]
- 34.Xiao A., Cheng Z., Kong L., Zhu Z., Lin S., Gao G., Zhang B.2014. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30: 1180–1182. doi: 10.1093/bioinformatics/btt764 [DOI] [PubMed] [Google Scholar]
- 35.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R.2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. doi: 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimi K., Kunihiro Y., Kaneko T., Nagahora H., Voigt B., Mashimo T.2016. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 7: 10431. doi: 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.