Abstract
Understanding the genetic quality of the gerbil, Meriones meridianus, plays an important role in the study of medical biology. However, no effective system has been established for evaluating a population’s genetic diversity to date. In the present study, we established a set of reasonable evaluative systems based on microsatellite markers of the Mongolian gerbil by using the method of cross-amplification of species. Following electrophoresis analysis, short tandem repeat (STR) scanning, and sequencing, 11 microsatellite loci were identified by matching the criteria characteristics and were used to evaluate the genetic diversity of two stocks of Meriones meridianus: Meriones meridianus jei Wang, 1964 (M. m. jei) and Meriones meridianus cryptorhinus Blanford, 1875 (M. m. cryptorhinus) from Xinjiang, China. The microsatellite loci screened were highly polymorphic and were suitable for genetic quality control of Meriones meridianus. In addition, the quality of the non-bred M. m. jei and M. m. cryptorhinus strains in our study is sufficient for them to be promising stocks in the future for the farmed animal industry.
Keywords: biomarker, genetic diversity, Meriones meridianus, microsatellite loci
Introduction
The gerbil, Meriones meridianus (Rodentia, Muridae, Gerbillinae), otherwise known as the midday gerbil, belongs to Rodentia, Cricetidae, Meriones, is native to the sandy deserts in Afghanistan, China, Iran, Azerbaijan, Kazakhstan, Kyrgyzstan, Mongolia, Russia, Tajikistan, Turkmenistan, and Uzbekistan [26]. It also widely distributed in the Inner Mongolia Xinjiang Plateau in China.
Meriones meridianus can serve as an animal model for biological and medical research in many aspects. For several decades, Meriones meridianus has been used as an animal model for research, human medicines, and animal anti-disease breeding, such as studies of echinococcosis [21], viral hepatitis [39], the sensibility of plague virus [31], a hantavirus infection model [38], a helicobacter infection model [9], and the duration and characteristics of the course of cutaneous leishmaniasis [27]. Meriones meridianus has also benefited a great deal of research on ethology [30].
So far, the major physiological and biochemical indices, including reproductive performance, reproductive biology [32], structure of parasympathetic ganglia [13], and thermogenesis parameters [2], have been systematically evaluated for Meriones meridianus populations. In order to breed better varieties of this species, two stocks, Meriones meridianus jei Wang, 1964 (M. m. jei) and Meriones meridianus cryptorhinus Blanford, 1875 (M. m. cryptorhinus), were captured in Turpan and Luntai areas of Xinjiang, china, in June 1992 [35]. Currently, forty-three generations of these stocks have been bred, and biochemical information of the stocks has been estimated from their urine [37]. The Meriones meridianus individuals used throughout this study were originated from the fortieth generations.
The genetic quality of Meriones meridianus has an important influence on the accuracy, repeatability, and scientificity of medical biological research results. If genetic quality standards have not been established, it is difficult to monitor the genetic quality within each stock and unify the quality among a population, and this has become one of the obstacles for widespread monitoring of genetic quality. While some basic research on the genetic diversity of Meriones meridianus has been conducted using molecular biology methods [10], to this day, there are still no suitable standards for genetic evaluation of Meriones meridianus.
Many methods are utilized to monitor genetic variation, such as biochemical marker gene detection, immunological trait marker gene detection, and DNA polymorphism. Among them, simple sequence repeats (SSRs) or microsatellites (MS) are extremely valuable tools for assessing population genetic diversity because of their distinctive features [29].
Microsatellites are characterized by short tandem repeats (STRs) of 1 to 6 nucleotide motifs, which widely spread in a random manner throughout the eukaryotic genome. They are commonly used in a wide range of applications, such as the evaluation of genetic diversity and population structure patterns [3, 16]. In previous studies, microsatellites have also been used as biomarkers for monitoring heritable traits in rodents [7, 33]. Because the whole genome of Meriones meridianus has not been sequenced completely, it is difficult to obtain suitable microsatellite markers by genome sequencing. It has been reported that cross-species amplification of species microsatellite loci in related species is an effective way to isolate microsatellite markers [14, 15, 22, 24]. Since the midday gerbils and Mongolian gerbils are the closely related species, the present study aimed to identify microsatellite loci for Meriones meridianus on the basis of a significant microsatellite marker system (28 loci markers) using the method of cross-amplification of species, which has already been established for the Mongolian gerbil [8]. On the basis of this research, we discuss the genetic structure of the Meriones meridianus population in the Xinjiang area of China.
Material and Methods
Ethics
All of the experimental procedures were conducted in accordance with the Guidelines of the Capital Medical University Animal Experiments and Experimental Animals Management Committee (No.AEEI-2017-032).
Sample background and collection
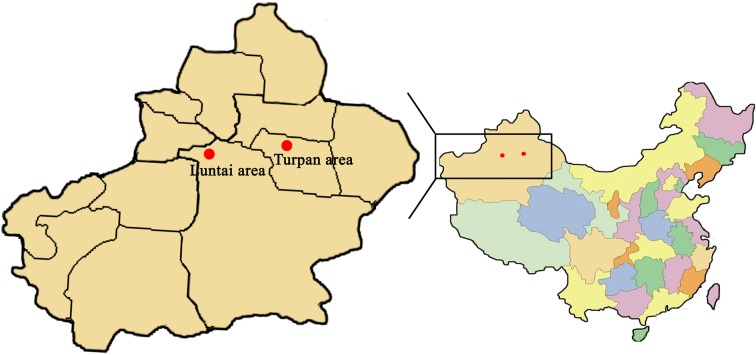
All the samples were available from the Center for Laboratory Animal Research of Xinjiang district in China. Forty Meriones meridianus subspecies (20 M. m. jei and 20 M. m. cryptorhinus; 1.5 months old; half male and half female; weight, 80–100 g and 60-80g respectively) were captured in the Turpan and Luntai areas of Xinjiang in June 1992. The locations of the sampled gerbil populations chosen as the original gerbils to be bred are shown in Fig. 1. Meriones meridianus individuals were bred at 17–26°C with a relative humidity of 30–65% and were placed in an experimental animal room with visible light and darkness for 12 h, respectively. Meriones meridianus individuals captured in the wild will likely fight if housed together, causing injury or death; therefore, one mature female and one mature male or two mature females and one mature male were selected for pairing. Pairings were determined by observation of several hours without fighting. The number of litters ranged from 1 to 8, and the average ratio of males to females was 1.4:1.0. To date, 43 generations of gerbils have been bred.. The Meriones meridianus individuals used in this study originated from the fortieth generations. In total, 63 DNA samples were extracted from 39 M. m. jei and 24 M. m. cryptorhinus individuals and preserved in sterile tubes at −80°C.
Fig. 1.
Maps of the Meriones meridianus populations showing the sample locations in the Turpan and Luntai areas in Xinjiang, China.
DNA extraction
In order to develop microsatellite loci for Meriones meridianus, total genomic DNA was extracted from 63 tail tissue samples following a previously described method [6]. The concentration and purity of DNA from each sample were determined by A260/280 measurement using a microplate absorbance reader system (Bio-Rad, Hercules, CA, USA), and the sample quality was further evaluated by 0.5% (w/v) agarose gel electrophoresis. DNA was then diluted to 100 ng/ µl and stored at −20°C for later use as polymerase chain reaction (PCR) templates.
PCR procedure
For each 20 µl PCR amplification, the following reagents were used: 2 µl 10X buffer, 1.0 µl each of 10 µM primer; 1.0 µl dNTPs (100 µM; Takara Co., Ltd., Tokyo, Japan), 0.2 µl Taq high-fidelity DNA polymerase (1.0U; Takara), and 1.0 µl template DNA (50–100 ng). Amplifications were performed in a gradient thermal cycler (ALS1296, Bio-Rad) using the following protocol: Pre-denaturation was performed at 94°C for 5 min, and then 35 cycles of denaturation were performed at 94°C for 30 s. The annealing temperature was set by a gradient PCR approach the temperature range of 45°C to 67°C. The optimum annealing temperature was selected, which allowed clear DNA bands to be distinguished in agarose gel. Extension was then performed at 72°C for 30 s, followed by a final extension at 72°C for 7 min. Amplified products were stored at 4°C for further analysis. The genotypes of 28 microsatellites loci for 63 Meriones meridianus individuals were detected using PCR with fluorescent primers, each labeled with a different commercially available dye label (FAM, HEX, TAMRA).
Agarose gel electrophoresis
Agarose was weighed and added into 0.5× TBE (Solarbio, Beijing, China) to make 2% gel, and ethidium bromide (EB) was added to a final concentration of 0.5 µg/ml. After the electrophoresis for 35 min at 120 V, images of the agarose gel were observed by ultraviolet imager and analyzed.
Microsatellite loci selection
All scanned data were analyzed with Gene Scan 3.7. The results revealed that there are two types of species, those that were homozygous, which mainly display a single waveform, there were both homozygous individuals, which mainly had no polymorphic positions, and those that were heterozygous, which had polymorphic positions, among the animals studied. The polymorphic microsatellite loci were selected for cloning and sequencing by Ruibiotech Co. (Beijing, China). Finally, by searching PubMed, we identified the appropriate target sequence.
Analysis of population genetic structure
The population genetic diversity and genetic structure were assessed using 11 microsatellite loci. Alleles of loci were classified as A, B, C or D according to amplified fragments. Genotypes of each microsatellite locus in all samples were input into POPGENE 3.2 in the form of AB and BB to observe significant genetic structure indices, including the observed and effective numbers of alleles, Shannon information index, observed and effective heterozygosity, and polymorphic loci (Poly-loci), were calculated for genetic variation analysis within the populations. In addition, the genetic distance between M. m. jei and M. m. cryptorhinus was measured according to the method for Nei’s standard distance [18].
Results
Selection of microsatellite loci
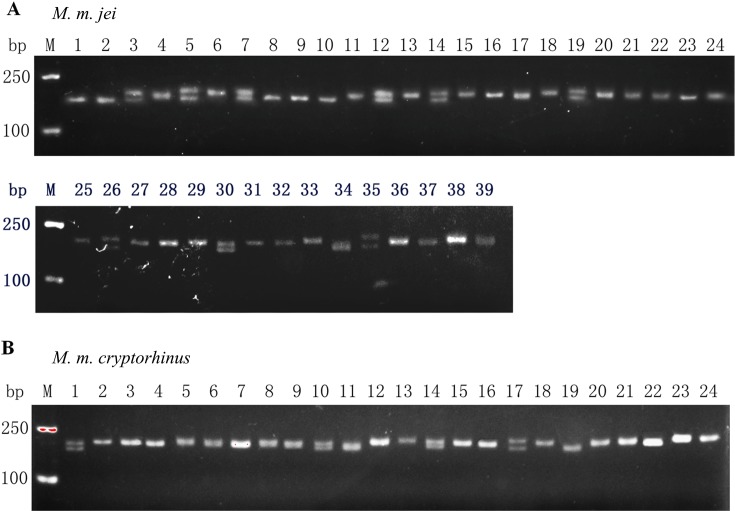
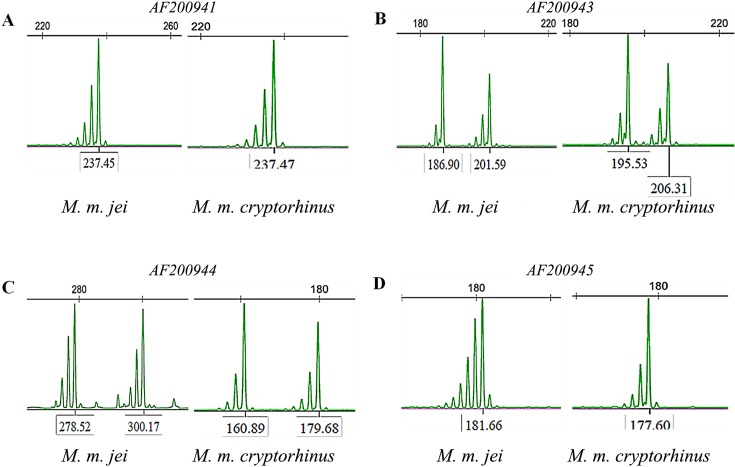
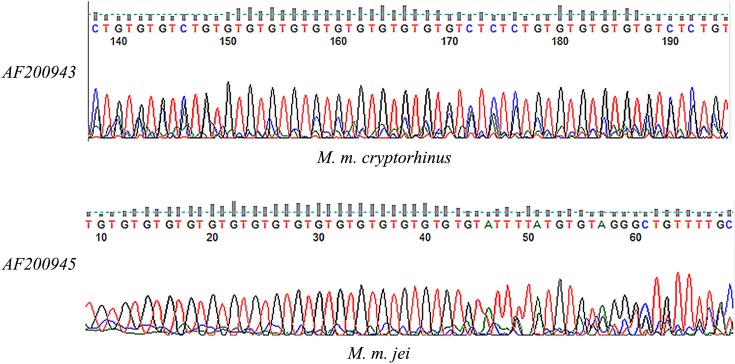
Previous studies have suggested that 28 microsatellite loci are optimal for identifying the Mongolian gerbil [8]. Since Meriones meridianus and the Mongolian gerbil both belong to the subfamily Gerbillinae, we can make use of the Mongolian gerbil microsatellite loci for filtering. The first round of selection was performed with 28 microsatellite loci of the Mongolian gerbil through PCR assays. After initial PCR assays, 19 primer sets generated the desired amplification products under modified conditions. Partial results of agarose gel electrophoresis are shown in Fig. 2, in which individual strips appear in pure individuals and two strips appear in hybrid individuals. Further STR screening, shown in Fig. 3, revealed that 13 loci matched microsatellite criteria. Primer sequences, annealing temperatures, range of allele genes, and GenBank accession numbers for the 13 microsatellites are shown in Table 1. To further verify the results obtained from STRscanning, we sent microsatellite loci to Ruibiotech Co. (Beijing, China) to confirm the sequencing. The sequencing was performed using an ABI3730XL auto-sequencing machine system. Partial results of cloning and sequencing are shown in Fig. 4. With the exception of D19Mit1 and DXMit17, all loci were polymorphic and showed different degrees of variability. D19Mit1 and DXMit17 were discarded, as they had one allele in M. m. jei and M. m. cryptorhinus population for further verification. As shown in Table 2, AF200941, AF200942, AF200943, AF200944, AF200945, and AF200947 loci were significantly polymorphic. Based on the results of PCR, STR scanning, and sequencing, an appropriate microsatellite loci system was established.
Fig. 2.
PCR amplification of the gerbil genome by primers for microsatellite loci: AF200943, which showed heterogeneity and homogeneity for the M. m. jei (A) and M. m. cryptorhinus (B) populations of Meriones meridianus gerbils in this study.
Fig. 3.
STR loci AF200941 (A), AF20043 (B), AF200944 (C), andAF200945 (D) showed microsatellite polymorphisms in Meriones meridianus gerbils.
Table 1. Characteristics of the 13 microsatellite loci isolated from Meriones meridianus gerbils.
| AN | Primer sequences(5’–3’) | M.m.jei | M.m.cryptorhinus | Mg2+(mM) | Chr | ||
|---|---|---|---|---|---|---|---|
| AT(°C) | Rng(bp) | AT(°C) | Rng(bp) | ||||
| AF200941 | TGGGTCCTTTGGAAGATGGCTTAAAATGAATCACTTA | 55°C | 170–230 | 53.8°C | 219–237 | 2 | |
| AF200942 | CAGGCACCCCCAGTTTGTCTACACAGGCTGAGGATGT | 54°C | 120–150 | 50.8°C | 137–143 | 2 | |
| AF200943 | GGCTCCTGATTCTACATTTCTCAACCATTGGCAACTCTC | 55°C | 180–200 | 53.8°C | 195–206 | 2 | |
| AF200944 | GCTGGGCTTTAATGTTTATTTGGTGGCTCACACTTTCTGT | 55°C | 200–300 | 57.5°C | 158–179 | 2 | |
| AF200945 | AGTCCCTATTACATCCACAAGTTATCCTGCAAAGCCTAAG | 58°C | 150–180 | 50.8°C | 160–177 | 2 | |
| AF200947 | GACAGAGTGGGAGGGGTATGTTGGCAAGTTTGGTTTGTTTGA | 55°C | 180–207 | 57.5°C | 197–199 | 2 | |
| GU562706 | CAGGAATAAAGTATAATGGGGTGCCCCATGATCAGTTGGGTTTT | 50°C | 216–228 | 50.8°C | 227 | 2 | D16Mit26 |
| GU562777 | GCTCCCTTTCCTCTTGAACCGGGCCCTTATTCTATCTCCC | 53°C | 110–130 | 50°C | 186–192 | 2 | D2Mit22 |
| GU562747 | AACACATGAAACGTGTGCGTTGATAGGCATGCTTAAGCCC | 50°C | 230–245 | 48.3°C | 243 | 2 | D3Mit130 |
| GU562700 | AATCCTTGTTCACTCTATCAAGGCCATGAAGAGTCCAGTAGAAACCTC | 50°C | 157 | 50°C | 157 | 2 | D19Mit1 |
| GU562704 | CCTCTGAGGAGTAACCAAGCCCACAGAGTTCTACCTCCAACCC | 50.8°C | 210–235 | 50.8°C | 228–236 | 2 | D17Mit38 |
| GU562695 | CCTGTTTGGGCACCTAGATTTAATAACCCATGTTTTCTGTGGG | 50°C | 215 | 45.4°C | 237 | 2 | DXMit17 |
| GU562725 | ACACTCAGAGACCATGAGTACACCGAGTTCACTACCCACAAGTCTCC | 53°C | 135 | 50.8°C | 111–127 | 2 | D8Mit56 |
AN, GenBank accession No.; AT, optimized annealing temperature; Mg2+ (mM), concentration of Mg2+ (mM) for PCR; Chr, position on mouse chromosome of the corresponding mouse locus; Rng, allele range.
Fig. 4.
Cloning sequences showing Meriones meridianus genomes generated at the loci AF200943 (up) and AF200945 (down) by Mongolian gerbils.
Table 2. Summary of the statistics for 11 microsatellite loci in the two Meriones meridianus gerbil populations.
| Loci | M. m. jei | M. m. cryptorhinus | FST | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obs-alle | Eff-alle | Shan- | Poly-loci% | Obs-Het | Exp-Het | Nei’s | Obs-alle | Eff-alle | Shan- | Poly-loci% | Obs-Het | Exp-Het | Nei’s | ||
| AF200941 | 7 | 4.8907 | 1.7 | – | 0.8462 | 0.8059 | 0.7955 | 5 | 3.1911 | 1.2995 | – | 0.2083 | 0.7012 | 0.6866 | 0.0865 |
| AF200942 | 7 | 2.5934 | 1.3315 | – | 0.6667 | 0.6224 | 0.6144 | 4 | 2.8444 | 1.1803 | – | 1 | 0.6622 | 0.6484 | 0.1355 |
| AF200943 | 10 | 4.8517 | 1.871 | – | 0.8462 | 0.8042 | 0.7939 | 2 | 1.0425 | 0.1013 | – | 0.0417 | 0.0417 | 0.0408 | 0.3832 |
| AF200944 | 12 | 5.5209 | 1.9866 | – | 0.7949 | 0.8295 | 0.8189 | 4 | 2.7961 | 1.1524 | – | 0.75 | 0.656 | 0.6424 | 0.0676 |
| AF200945 | 7 | 4.8057 | 1.7002 | – | 0.6154 | 0.8022 | 0.7919 | 4 | 2.8872 | 1.1386 | – | 0.7083 | 0.6676 | 0.6536 | 0.1017 |
| AF200947 | 8 | 6.6419 | 1.9467 | – | 0.8974 | 0.8605 | 0.8494 | 2 | 1.3318 | 0.4154 | – | 0.2917 | 0.2544 | 0.2491 | 0.2735 |
| Mit22 | 3 | 2.7531 | 1.0559 | – | 0.9474 | 0.6453 | 0.6368 | 2 | 1.9692 | 0.6853 | – | 0.875 | 0.5027 | 0.4922 | 0.0606 |
| D3Mit130 | 3 | 1.3011 | 0.4692 | – | 0.1026 | 0.2344 | 0.2314 | 2 | 1.0868 | 0.1732 | – | 0 | 0.0816 | 0.0799 | 0.7091 |
| D8Mit56 | 1 | 1 | 0 | – | 0 | 0 | 0 | 2 | 2 | 0.6931 | – | 1 | 0.5106 | 0.5 | 0.6 |
| D17Mit38 | 5 | 1.6751 | 0.8026 | – | 0.4615 | 0.4083 | 0.403 | 3 | 1.7916 | 0.6976 | – | 0.625 | 0.4512 | 0.4418 | 0.3641 |
| Mit26 | 5 | 4.1674 | 1.4669 | – | 0.5789 | 0.7702 | 0.76 | 1 | 1 | 0 | – | 0 | 0 | 0 | 0.3114 |
| Mean | 6.1818 | 3.6546 | 1.3028 | 90.91% | 0.6143 | 0.6166 | 0.6087 | 2.8182 | 1.9946 | 0.6852 | 90.91% | 0.5 | 0.4118 | 0.4032 | 0.2585 |
Obs-alle, observed number of alleles; Eff-alle, effcetive number of alleles; Obs-Het, observed heterozygosisty; Exp-Het, expected heterozygosisty; Nei’s, Nei’s genetic variation; FST, fixation index, Shan-, shannon’s information index analyzed for 11 loci.
Analysis of the genetic structure of Meriones meridianus
Samples of 39 M. m. jei and 24 M. m. cryptorhinus individuals were screened for the analysis of the genetic structures of 11 polymorphic MS loci. Supplementary Table 1 shows the genotypes of each microsatellite locus in all of the samples. The average effective number of alleles, heterozygosity, and average observation of heterozygosity as well as other factors, were analyzed in the determination of genetic variation in each population. As shown in Table 2, not all loci were equally variable. The average observed numbers of alleles in the two populations were 6.1818 and 2.8182, the average effective numbers of alleles were 3.6546 and 1.9946, the average Shannon information indices were 1.3028 and 0.6852, the average observed heterozygosity values were 0.6143 and 0.5000, and the average expected heterozygosity values were 0.6166 and 0.4118, respectively. The average Nei’s genetic distances were 0.6087 and 0.4032. The fixation index (FST) value, including all loci, was estimated to be 0.2585, which indicated a significant genetic differentiation between the M. m. jei and M. m. cryptorhinus populations.
Discussion
Meriones meridianus has been widely used as a valuable animal model in many medical fields [21, 31]. Its quality has an important influence on the accuracy, repeatability and scientificity of the research results. In the present study, we sought to construct a system for evaluating genetic quality using microsatellite markers for Meriones meridianus gerbil populations in China, which will be helpful for genetic quality control and monitoring of these populations. Molecular marker technology has brought with it the opportunity for the genetic breeding of experimental animals. For example, DNA sequencing [5], random amplified polymorphic DNA (RAPD) [17], amplified fragment length polymorphism (AFLP) [23], restriction fragment length polymorphisms (RFLP) [1], and other technologies have become powerful tools for genetic diversity analysis. Among these molecular biomarkers, the stability of the RAPD technology has been inadequate, while RFLP and other methods have required large amounts of high purity DNA. In the present study, microsatellite DNA polymorphism markers were selected as primers on both sides of the site, and PCR amplification was performed with high repeatability and comparability. Therefore, microsatellite DNA polymorphism markers represent an ideal tool for genetic diversity studies. Screening of microsatellite markers from near-source species is a feasible method [25, 33]. This approach is particularly useful for species that do not have sequence information available. Mongolian gerbil is a gerbil strain that has been widely used in a variety of studies [4, 11, 28]. In a previous study, we have screened 28 microsatellite marker systems to determine the genetic quality of a closed group of Mongolian gerbils through the use of cross-amplifying technology, using microsatellite sites in mice [6]. The genetic detection system was effectively applied to population quality monitoring for Mongolian gerbils in that study. Since midday gerbils and Mongolian gerbils are the closely related species, after further experiments, microsatellite loci suitable for Meriones meridianus were screened from 28 loci of Mongolian gerbils in the present study.
The results showed that 11 pairs (39%) could be stably amplified and had a single amplified stripe in agarose gel electrophoresis. After gene analysis of synthetic fluorescent primers, 11 microsatellite loci with good polymorphism and high heterozygosity were obtained. The results showed that it is feasible to obtain microsatellite sites by cross-amplification between near-source species (Meriones meridianus and Mongolian gerbils).
In our research, genetic variation between the two Meriones meridianus strains was studied using 11 microsatellite markers. Genetic variation in the group was determined on the basis of average effective heterozygosity, average effective allele number, average Shannon index, and other analytical indices used for estimating group genetic variation.
Nei (1987) stated that the mean number of alleles for different populations is a fair indicator of genetic variation and mutation drift balance. A higher mean effective number of alleles indicate that a population is able to maintain the original gene and avoids new variation in the case of pressures from genetic drift and artificial selection [20, 29]. In the M. m. jei group, the observed number of alleles varied from 1 to 12, with a mean of 6.1818, whereas the effective number of alleles varied from 1 to 6.6419, with a mean of 3.6546. The results indicate that the alleles were stable and evenly distributed in the population. In the M. m. cryptorhinus group, the observed number of alleles varied from 1 to 5, with a mean of 2.8182, and the effective number of alleles was 1 to 3, with a mean of 1.9946. The effective number of alleles was less than the actual observed number of alleles, which indicates that the allele distribution of the population was uneven; however, the fact that the population of this group was so small, only 24 animals, may have affected the number of effective alleles of this group.
Average effective heterozygosity is an important index that reflects the genetic diversity of a population. When the average effective heterozygosity of a group is less than 0.5, its genetic diversity is low; low genetic variability (heterozygote deficiency) coupled with inbreeding is the consequence of genetic bottleneck resulting from overexploitation and habitat-related events. The heterozygosity of an ideal closed group is between 0.5 and 0.7. In the M. m. jei stock, the average observed heterozygosity (HO) was 0.6143, and the average expected heterozygosity (HE) was 0.6166. This suggests that the population had high genetic diversity, which may be due to long-term natural selection for adaptation, the mixed nature of the breeds, or historic mixing of strains of different populations [12]. In addition, it was found that the mean observed heterozygosity was similar to the mean expected heterozygosity, indicating that there was no naked allele or inbreeding within this population. The results of this research suggested that the hybrid standard was strictly followed during the breading process for this population, thus obtaining good genetic variation and diversity.
The above indicates that the M. m. jei group will be able to adapt to changing environments and could be the ideal closed group in the future. According to the existing data, M. m. cryptorhinus stock (mean heterozygosity=0.4118; mean allelic number=1.9946) with less genetic variability will be unlikely to resist selection, mutation, and genetic drifting. This is likely the result of rapid population expansion from a small population. In addition, not many microsatellite loci (11) and breeds (24) were analyzed within the M. m. cryptorhinus stock; therefore, additional markers and samples are required to increase the accuracy and generality of the results. Thus, breeding strictly with non-inbred strains, which can increase genetic variation, is necessary in the future.
The (FST) is the correlation between two gametes randomly drawn from each subgroup and measures the degree of genetic differentiation and the heterogeneity of gene frequencies in sites studied [19]. The value of FST can range from 0 to 1. Wright had stated that values ranging from 0–0.05 indicate “little” genetic differentiation, those from 0.05–0.15 indicate “moderate” genetic differentiation, and those from 0.15–0.25 indicate “great” great genetic differentiation [36]. The higher the FST, the greater the implied different of a population from other populations. In this study, FST values (FST=0.2585) indicated a significant FST genetic differentiation between the M. m. jei and M. m. cryptorhinus groups. According to genetic data, M. m. jei [34] belongs to M. penicilliger [10], and M. m. cryptorhinus from the south of the Taklamakan Desert belongs to M. psammophilus [10, 40]. Based on the distance and barriers, there is a low genetic flow between the two subpopulations, which can be attributed to the differences between the two groups.
Evaluation of genetic variation and patterns of population/species genetic diversity provides insight into the breeding methods, which could to rapid changes in methods and conservation. In this study, we developed a microsatellite loci system for identifying the stock of Meriones meridianus. However, the ideal microsatellite tag would ideally be able to meet certain conditions. For instance, Barker proposed the analysis of at least 25 samples for each variety. The number of microsatellites should not be less than 25 (4 alleles per locus), and there should be no association between the position points. Currently, 11 microsatellites are not sufficient to establish a perfect marker system; further studies on the candidate sites of Meriones meridianus are required to determine the genetic structure of their populations.
Use of highly polymorphic microsatellite markers as genetic tags plays an important role in monitoring Meriones meridianus as a laboratory animal model; for instance, in our research, microsatellite loci system were shown to be powerful markers for quantifying genetic variations within and between populations of M. m. jei and M. m. cryptorhinus species. Population gene frequency is an important index of Meriones meridianus genetic variation sizes, which should be taken into account in the modeling period. It would be useful to keep the base genetic profiles of representative samples of Meriones meridianus in a holding facility.
In conclusion, there have been few genetic studies of microsatellite DNA markers in Meriones meridianus, since DNA markers are very rare. To date, no detailed information is available on the genetic diversity of cultured stocks of Meriones meridianus. In this study, we identified appropriate microsatellite markers for Meriones meridianus and elucidated ways to enrich their genetic database. Continuous monitoring of the genetic structure using microsatellite markers is essential for the successful propagation of Meriones meridianus.
Taken together, the combination of 11 loci selected in this experiment provides a good choice for genetic monitoring of the quality and the population genetic structure of outbred stocks.
Supplementary Material
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No. 31772545), Support Project of High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (No. IDHT20170516) and the Science and Technology Plan of Beijing Municipal Education Commission (No. KM201910025027).
References
- 1.Akhavan A.A., Yaghoobi-Ershadi M.R., Khamesipour A., Mirhendi H., Alimohammadian M.H., Rassi Y., Arandian M.H., Jafari R., Abdoli H., Shareghi N., Ghanei M., Jalali-zand N.2010. Dynamics of Leishmania infection rates in Rhombomys opimus (Rodentia: Gerbillinae) population of an endemic focus of zoonotic cutaneous leishmaniasis in Iran. Bull. Soc. Pathol. Exot. 103: 84–89. doi: 10.1007/s13149-010-0044-1 [DOI] [PubMed] [Google Scholar]
- 2.Bao W.D., Wang D.H., Wang Z.W.2002. Nonshivering thermogenesis in four rodent species from Kubuqi desert, Inner Mongolia, China. Folia Zool. (Brno) 511: 9–13. [Google Scholar]
- 3.Bilgiç H.B., Ünlü A.H., Aksulu A., Bakırcı S., Hacılarlıoğlu S., Eren H., Weir W., Karagenç T.2017. Selection of genetic markers to determine diversity in theileria annulata populations after recombination. Turkiye Parazitol. Derg. 41: 9–18. doi: 10.5152/tpd.2017.4970 [DOI] [PubMed] [Google Scholar]
- 4.Dai F., Zhuo X., Kong Q., Du J., Yu H., Zhou S., Song X., Tong Q., Lou D., Lou Q., Lu L., Lv Y., Sa X., Lu S.2019. Early detection of Toxoplasma gondii infection in mongolian gerbil by quantitative real-time PCR. J. Parasitol. 105: 52–57. doi: 10.1645/18-95 [DOI] [PubMed] [Google Scholar]
- 5.Dobigny G., Aniskin V., Granjon L., Cornette R., Volobouev V.2005. Recent radiation in West African Taterillus (Rodentia, Gerbillinae): the concerted role of chromosome and climatic changes. Heredity 95: 358–368. doi: 10.1038/sj.hdy.6800730 [DOI] [PubMed] [Google Scholar]
- 6.Du X., Chen Z., Li W., Tan Y., Lu J., Zhu X., Zhao T., Dong G., Zeng L.2010. Development of novel microsatellite DNA markers by cross-amplification and analysis of genetic variation in gerbils. J. Hered. 101: 710–716. doi: 10.1093/jhered/esq066 [DOI] [PubMed] [Google Scholar]
- 7.Du X., Cui J., Wang C., Huo X., Lu J., Li Y., Chen Z.2013. Detected microsatellite polymorphisms in genetically altered inbred mouse strains. Mol. Genet. Genomics 288: 309–316. doi: 10.1007/s00438-013-0751-y [DOI] [PubMed] [Google Scholar]
- 8.Du X.Y., Li W., Sa X.Y., Li C.L., Lu J., Wang Y.Z., Chen Z.W.2015. Selection of an effective microsatellite marker system for genetic control and analysis of gerbil populations in China. Genet. Mol. Res. 14: 11030–11042. doi: 10.4238/2015.September.21.16 [DOI] [PubMed] [Google Scholar]
- 9.Goto K., Jiang W., Zheng Q., Oku Y., Kamiya H., Itoh T., Ito M.2004. Epidemiology of Helicobacter infection in wild rodents in the Xinjiang-Uygur autonomous region of China. Curr. Microbiol. 49: 221–223. doi: 10.1007/s00284-004-4287-6 [DOI] [PubMed] [Google Scholar]
- 10.Ito M., Jiang W., Sato J.J., Zhen Q., Jiao W., Goto K., Sato H., Ishiwata K., Oku Y., Chai J.J., Kamiya H.2010. Molecular phylogeny of the subfamily Gerbillinae (Muridae, Rodentia) with emphasis on species living in the Xinjiang-Uygur Autonomous Region of China and based on the mitochondrial cytochrome b and cytochrome c oxidase subunit II genes. Zool. Sci. 27: 269–278. doi: 10.2108/zsj.27.269 [DOI] [PubMed] [Google Scholar]
- 11.Khakisahneh S., Zhang X.Y., Nouri Z., Hao S.Y., Chi Q.S., Wang D.H.2019. Thyroid hormones mediate metabolic rate and oxidative, anti-oxidative balance at different temperatures in Mongolian gerbils (Meriones unguiculatus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 216: 101–109. doi: 10.1016/j.cbpc.2018.11.016 [DOI] [PubMed] [Google Scholar]
- 12.Kotzé A. M.G. Genetic relationship in South African cattle breeds. in Proceedings of the 5th world congress on genetics applied to livestock production. 1994. Guelph, Canada.
- 13.Kuder T. 1985. Topography and macroscopic structure of parasympathetic ganglia connected with the trigeminal nerve in midday gerbil (Meriones meridianus-Mammalia: Rodentia). Acta Biologica Cracoviensia. Ser. Zoologia 27: 61. [Google Scholar]
- 14.Lei D.J., Zhao G., Xie P., Li Y., Yuan H., Zou M., Niu J.G., Ma X.F.2017. Analysis of genetic diversity of Leuciscus leuciscus baicalensis using novel microsatellite markers with cross-species transferability. Genet. Mol. Res. 16: 16. doi: 10.4238/gmr16029376 [DOI] [PubMed] [Google Scholar]
- 15.Maduna S.N., Rossouw C., Roodt-Wilding R., Bester-van der Merwe A.E.2014. Microsatellite cross-species amplification and utility in southern African elasmobranchs: A valuable resource for fisheries management and conservation. BMC Res. Notes 7: 352. doi: 10.1186/1756-0500-7-352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mun J.H., Kim D.J., Choi H.K., Gish J., Debellé F., Mudge J., Denny R., Endré G., Saurat O., Dudez A.M., Kiss G.B., Roe B., Young N.D., Cook D.R.2006. Distribution of microsatellites in the genome of Medicago truncatula: a resource of genetic markers that integrate genetic and physical maps. Genetics 172: 2541–2555. doi: 10.1534/genetics.105.054791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naseri Z., Jalal R., Darvish J.2007. Determination of Meriones Species (Rodentia, Gerbillinae) by RAPD-PCR. Iran. J. Anim. Biosyst. 2: 35–40. [Google Scholar]
- 18.Nei M.1972. Genetic distance between populations. Am. Nat. 106: 283–292. doi: 10.1086/282771 [DOI] [Google Scholar]
- 19.Nei M.1977. F-statistics and analysis of gene diversity in subdivided populations. Ann. Hum. Genet. 41: 225–233. doi: 10.1111/j.1469-1809.1977.tb01918.x [DOI] [PubMed] [Google Scholar]
- 20.Ojango J.M., Mpofu N.,, Marshall K., Andersson-Eklund L. 2011. Quantitative methods to improve the understanding and utilization of animal genetic resources. In: Animal genetics training resource. Version. 3. (Ojango, J.M., Malmfors, B. and Okeyo, A.M. eds), Swedish University of Agricultural Sciences, Nairobi, Kenya, ILRI and Uppsala, Sweden. [Google Scholar]
- 21.Osman I., Jiao W., Liao L., Chai J.1998. [Comparative observation on experimental infection with Echinococcus multilocularis in Cricetulus Migratorius and Meriones meridianus]. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 16: 130–132.(in Chinese) [PubMed] [Google Scholar]
- 22.Pirog A., Jaquemet S., Blaison A., Soria M., Magalon H.2016. Isolation and characterization of eight microsatellite loci from Galeocerdo cuvier (tiger shark) and cross-amplification in Carcharhinus leucas, Carcharhinus brevipinna, Carcharhinus plumbeus and Sphyrna lewini. PeerJ 4: e2041. doi: 10.7717/peerj.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razzoli M., Papa R., Valsecchi P., Nonnis Marzano F.2003. AFLP to assess genetic variation in laboratory gerbils (Meriones unguiculatus). J. Hered. 94: 507–511. doi: 10.1093/jhered/esg097 [DOI] [PubMed] [Google Scholar]
- 24.Reid K., Hoareau T.B., Bloomer P.2012. High-throughput microsatellite marker development in two sparid species and verification of their transferability in the family Sparidae. Mol. Ecol. Resour. 12: 740–752. doi: 10.1111/j.1755-0998.2012.03138.x [DOI] [PubMed] [Google Scholar]
- 25.Ruggeri P., Splendiani A., Giovannotti M., Nisi Cerioni P., Caputo V.2012. Isolation of novel microsatellite loci in the black goby Gobius niger and cross-amplification in other gobiid species (Perciformes, Gobiidae). J. Fish Biol. 81: 2044–2052. doi: 10.1111/j.1095-8649.2012.03444.x [DOI] [PubMed] [Google Scholar]
- 26.Smith A.T., Yan X.2008. A guide to the mammals of China. pp. 1–487, 495–525. (Smith, A.T. and Xie, Y. eds.), Princeton University Press, Princeton. [Google Scholar]
- 27.Strelkova M.V.1975. [Duration and character of the course of cutaneous leishmaniasis in the midday gerbil (Meriones meridianus Pall.)] ]. Parazitologia 9: 532–534.(in Russian) [PubMed] [Google Scholar]
- 28.Suzuki R., Satou K., Shiroma A., Shimoji M., Teruya K., Matsumoto T., Akada J., Hirano T., Yamaoka Y.2019. Genome-wide mutation analysis of Helicobacter pylori after inoculation to Mongolian gerbils. Gut Pathog. 11: 45. doi: 10.1186/s13099-019-0326-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takezaki N., Nei M.1996. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tchabovsky A.V., Savinetskaya L.E., Ovchinnikova N.L., Safonova A., Ilchenko O.N., Sapozhnikova S.R., Vasilieva N.A.2019. Sociability and pair-bonding in gerbils: a comparative experimental study. Curr. Zool. 65: 363–373. doi: 10.1093/cz/zoy078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tikhomirova M.J.1934. Meriones meridianus Pall, a Reservoir of Plague Virus in sandy Regions of the Volga-Ural Steppes. Revue demicrobiologie (et) d’epidemiologie (et de parasitologie). 13: 89–101.
- 32.Verevkin M.V.1985. Reproductive biology of the midday jird (Meriones meridianus). Zool. Zh. 64: 276–281. [Google Scholar]
- 33.Wang C., Zhang S.J., Du X.Y., Xu Y.M., Huo X.Y., Liao L.F., Chen Z.W.2015. Development of novel DNA markers for genetic analysis of grey hamsters by cross-species amplification of microsatellites. Genet. Mol. Res. 14: 14339–14347. doi: 10.4238/2015.November.13.19 [DOI] [PubMed] [Google Scholar]
- 34.Wang S.1964. [New species and subspecies of mammals from Sinkiang,China]. Acta Zootaxonomica Sinica 1: 6–15.(in Chinese) [Google Scholar]
- 35.Wang Y., Zhao L., Fang F., Liao J., Liu N.2013. Intraspecific molecular phylogeny and phylogeography of the Meriones meridianus (Rodentia: Cricetidae) complex in northern China reflect the processes of desertification and the Tianshan Mountains uplift. Biol. J. Linn. Soc. Lond. 110: 362–383. doi: 10.1111/bij.12123 [DOI] [Google Scholar]
- 36.Wright S.1968, Evolution and the genetics of populations, Vol. 4:Variability within and among natural populations. University of Chicago Press, Chicago. [Google Scholar]
- 37.Yi-mei X., Shen S., Yun L., Shun-sheng Y., Hong-qiong Z., Li-fu L.2017. Establishment of urine collection method and determination of some biochemical indexes for meriones meridianus in Xinjiang. Lab. Anim. Comp. Med. 37: 55–58. [Google Scholar]
- 38.Zhang Y.Z., Zou Y., Fu Z.F., Plyusnin A.2010. Hantavirus infections in humans and animals, China. Emerg. Infect. Dis. 16: 1195–1203. doi: 10.3201/eid1608.090470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao S, Liao L, Zou L. 2001. Study of Experiment on HEV infection in Gerbil (Meriones meridianus) from generation to generation. Zhongguo Mei Jie Sheng Wu Xue Ji Kong Zhi Za Zhi 12: 215–217. [Google Scholar]
- 40.Zou G., Zhou L., Zha X.2008. Geographical pattern and historical demography of Midday gerbil Meriones meridianus (Gerbillidae, Rodentia) inferred from the sequences of the mitochondrial DNA control region. Russ. J. Theriology 7: 25–32. doi: 10.15298/rusjtheriol.07.1.04 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.