Abstract
Objectives This study aimed to describe an alternative approach for accessing electronic medical records (EMRs) from clinical decision support (CDS) functions based on Arden Syntax Medical Logic Modules, which can be paraphrased as “map the entire record.”
Methods Based on an experimental Arden Syntax processor, we implemented a method to transform patient data from a commercial patient data management system (PDMS) to tree-structured documents termed CDS EMRs. They are encoded in a specific XML format that can be directly transformed to Arden Syntax data types by a mapper natively integrated into the processor. The internal structure of a CDS EMR reflects the tabbed view of an EMR in the graphical user interface of the PDMS.
Results The study resulted in an architecture that provides CDS EMRs in the form of a network service. The approach enables uniform data access from all Medical Logic Modules and requires no mapping parameters except a case number. Measurements within a CDS EMR can be addressed with straightforward path expressions. The approach is in routine use at a German university hospital for more than 2 years.
Conclusion This practical approach facilitates the use of CDS functions in the clinical routine at our local hospital. It is transferrable to standard-compliant Arden Syntax processors with moderate effort. Its comprehensibility can also facilitate teaching and development. Moreover, it may lower the entry barrier for the application of the Arden Syntax standard and could therefore promote its dissemination.
Keywords: electronic medical record, Arden Syntax, Medical Logic Modules, curly braces problem
Background and Significance
The Arden Syntax is a Health Level Seven (HL7) standard for clinical decision support (CDS) functions in the form of Medical Logic Modules (MLMs). 1 2 3 4 An MLM can be compared with a condition-action rule and typically reports a noteworthy clinical situation. A common example would be an MLM that sends an alert notification to the attending physicians in case of a critical laboratory value. The internal decision logic of an MLM is built from programming language constructs (for an introduction see Hripcsak 5 ). Integrating CDS based on the Arden Syntax into a hospital environment can be difficult, since vendors rarely permit third party database access to electronic medical records (EMRs). Moreover, there may be problems regarding data formats due to the heterogeneity of data models (syntactic interoperability), or the meaning of data items, which may require the use of terminologies (semantic interoperability). If these challenges can be overcome, EMR access from MLMs can be enabled by providing a data mapper to transform patient data items to Arden Syntax data types. Since the Arden Syntax encloses any institution-specific query parameters in a pair of curly braces (see specification, 7.1.8 6 ), the term “curly braces problem” has evolved. The standard stipulates that any institution must implement their specific solution to the curly braces problem. 1
Universitätsklinikum Erlangen (UKER), Germany, is a tertiary care hospital with approximately 1,400 beds. In 2006, UKER introduced a commercial patient data management system (PDMS) at a surgical intensive care unit (ICU). 7 8 9 Soon after the introduction, local clinicians demanded various CDS functions such as monitoring lung-protective ventilation parameters 10 or efficient testing of inflammation markers. 11 Thus, we integrated a commercial Arden Syntax processor with the PDMS. 12 With respect to EMR access from MLMs, we applied the conventional approach where each query retrieves a specific measurement. 13 In 2017, the availability of a new data interface for our PDMS prompted a redesign of our CDS integration. Simultaneously, we migrated to an experimental Arden Syntax version that provides multiple convenient features that facilitate the operation of MLMs. For example, it natively supports the mapping of XML-encoded patient data of arbitrary complexity. We discontinued the conventional data access approach where measurements are queried individually, and designed an alternative solution to the curly braces problem that can be paraphrased as “map the entire record.” The objective of this paper is to describe this practical approach that has proven beneficial in clinical routine at UKER ever since its introduction.
Mapping Time Series Data in Medical Logic Modules
A time series is “a sequence of observations registered at consecutive time instants.” 14 An electronic medical record (EMR) usually contains various measurements in the form of time series. For example, ICU patients are closely monitored with point-of-care testing by means of a blood gas analyzer (BGA), determining a fixed set of observations from blood samples.
Fig. 1 shows a detail of a screenshot from the PDMS on the left. It displays the results from point-of-care testing, including four blood samples. The glucose measurement includes the values 130, 147, 137, and 137 mg/dL. Clinical users can view the corresponding timestamps via tooltip. Clinical databases usually employ the Entity-Attribute-Value (EAV) model, 15 where time series are stored as illustrated in Fig. 1 on the right; the entity is a case number. The highlighted table rows correspond to the glucose values in the screenshot.
Fig. 1.
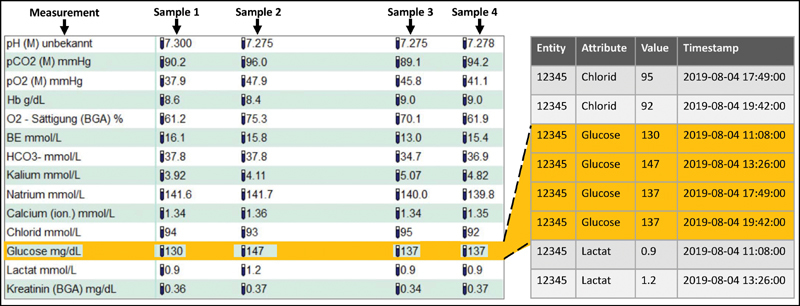
EMR data from point-of-care testing. On the left is a detail of a screenshot from the PDMS where the glucose values are highlighted. On the right is a detail from an EAV-modeled database in which the corresponding values are highlighted. BGA, blood gas analyzer; EAV, Entity-Attribute-Value; EMR, electronic medical record; PDMS, patient data management system.
In the Arden Syntax standard, all elementary data types, such as NUMBER, are timestamped. This means that they are pairs of value and timestamp. The Arden Syntax LIST data type is an ordered sequence of other types. Consequently, an Arden Syntax list of timestamped elementary types represents a time series. To access data items from EMRs, the Arden Syntax provides a READ statement, which typically queries a specific measurement in the form of a list. In other words, a READ statement reads a line from an EMR as shown in Fig. 1 on the left. For example, the following Arden Syntax code reads the glucose values from an EMR and performs a trend calculation based on the three most recent values:
glucose:= READ {Glucose};
WRITE SLOPE OF LATEST 3 FROM glucose;
Whenever the processor executes a READ statement, it invokes the institution-specific data mapper. Using the parameters in the curly braces expression, the mapper constructs a database query and transforms the query result to an Arden Syntax list, which is then assigned to a variable name. Thus, after executing the above example, the variable “glucose” contains a time series in the form of a list that can be processed with the rich set of operators provided by the Arden Syntax standard, such as SLOPE, INTERVAL, and INCREASE.
Methods
We performed a complete reengineering of the Arden Syntax integration at UKER. As the technical platform for MLM execution, we employed an experimental Arden Syntax version, termed Programming Language, Arden-INspired (PLAIN), 16 which is based on an alternative Arden Syntax grammar. 17 It provides multiple extended features, including a natively integrated mapper for XML-encoded patient data. The specific XML format is called PLAIN Data Markup Language (PDML); it supports data structures of arbitrary complexity, up to entire EMRs, or even large sequences of EMRs. The PDML format is explained in the Supplementary Material (available in the online version). PLAIN natively supports network and file access to PDML-encoded patient data. Fig. 2 includes a PDML-encoded time series.
Fig. 2.
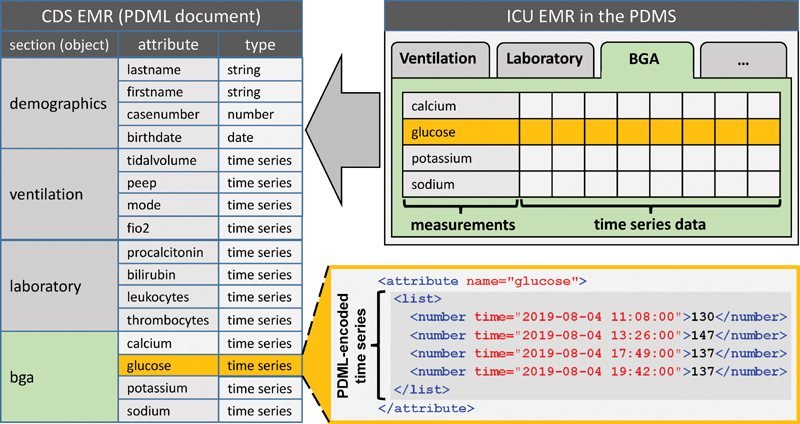
Basic structure of the PDML-encoded CDS EMR, which corresponds to the tabbed view in the PDMS. The code on the lower right shows a PDML-encoded time series. BGA, blood gas analyzer; CDS, clinical decision support; EMR, electronic medical record; ICU, intensive care unit; PDML, PLAIN Data Markup Language; PDMS, patient data management system.
Recent versions of our local PDMS (Integrated Care Manager, Dräger, Germany) provide a clinical application interface (CAI) for third party data access, which supports a proprietary protocol to query measurements from an EMR. For interoperability of the CAI with the MLMs, we applied a lightweight communication server that has been developed in a doctoral thesis and is not yet published. It has a layered architecture and supports the integration of adapters for the connection to various data sources and communication endpoints. We implemented a CAI adapter that transforms query results to PDML.
Instead of querying each measurement individually, we designed a strategy to access entire EMRs at once. To avoid confusion regarding the term EMR, we need to distinguish between an ICU EMR and a CDS EMR. Upon patient admission, the PDMS creates an ICU EMR in a proprietary database where MLMs have no access. Instead, we provide MLMs with a PDML document containing the superset of all measurements possibly required. We denote this document as the CDS EMR; it constitutes a partial copy of the ICU EMR. The Supplementary Material (available in the online version) includes a CDS EMR as an example.
The fundamental idea of our approach was to compose CDS EMRs with an internal structure that reflects the tabbed view of the ICU EMR in the graphical user interface of the PDMS. Fig. 2 illustrates this tabbed view on the upper right. The corresponding CDS EMR, shown on the left, constitutes a PDML document with sections and subsections built from objects and their attributes (see specification, annex A6 6 ). The first section contains demographic data; the other sections correspond to the tabs and contain measurements in the form of time series. The CAI adapter creates a section for each tab that includes at least one measurement required by any MLM, and executes a sequence of queries to the CAI to retrieve the respective patient data.
We customized the communication server to provide the CDS EMRs created by the CAI adapter in the form of a network service. One service endpoint creates a CDS EMR just at the moment it is requested (“fresh CDS EMR”). For MLMs, where a fast response time must be guaranteed, a CDS EMR buffer has been integrated. We implemented a continuously running copy process that creates a fresh CDS EMR every 2 minutes for each admitted patient and stores it into the buffer (“buffered CDS EMR”). CDS EMRs are generally timestamped on creation to indicate their age. Finally, we measured performance on a normal workday with 100 random accesses each to fresh and buffered CDS EMRs.
Results
The approach described in this paper enables our MLMs to access patient data in the form of CDS EMRs in a uniform way. With our solution, any MLM retrieves an entire CDS EMR in the form of a PDML-encoded document, which always contains the superset of data items possibly required. Whenever new MLMs require additional measurements, the CAI will be configured to include them. From that moment on, they will be contained in any CDS EMR. At present, a CDS EMR at UKER contains 22 time series in five sections, with a tendency toward growth due to new MLMs that require additional measurements.
Fig. 3 shows a detail from an MLM authoring environment. Line 1 reads a CDS EMR from the network service (“phb” is the name of the communication server). In PLAIN, the argument of READ is a string that refers either to a network resource or to a file. A specific EMR is addressed with a case number, whereas “$case” is a placeholder for the actual case number. PLAIN substitutes placeholders whenever strings are processed, as described elsewhere. 18 Once the CDS EMR is retrieved, the PDML mapper transforms it to an Arden Syntax object; the result is then assigned to the variable name “emr.” Individual measurements can be addressed with path expressions. Line 2 shows a WRITE statement that uses such a path expression to address bilirubin from the laboratory section, and outputs it in the form of a table via a user-defined function (“@table,” such functions are another extended feature of PLAIN). In clinical routine, we generally access CDS EMRs from network resources. In development, education, and research, we usually access them from files.
Fig. 3.
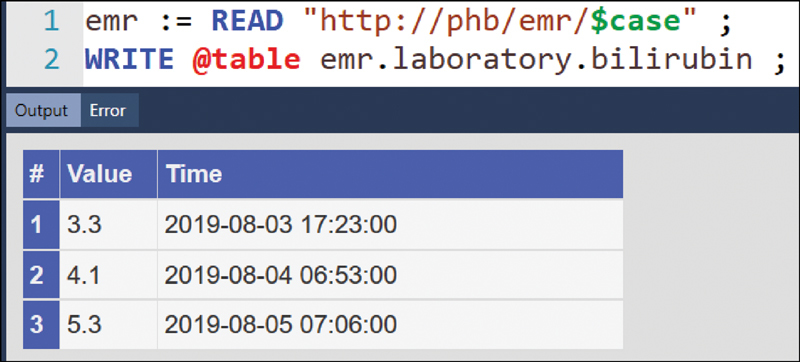
Detail from an authoring environment where an MLM reads a CDS EMR from a network resource and displays the bilirubin time series from the laboratory section in the form of a table. CDS, clinical decision support; EMR, electronic medical record; MLM, Medical Logic Module.
Fig. 4 shows the resulting architecture which has been adopted into routine at UKER. Whenever a READ statement is executed, the processor calls the integrated data mapper (A). If the MLM requests a fresh CDS EMR (B), the service interface of the communication server passes the request to the CAI adapter (C). The CAI adapter performs a sequence of queries from the CAI (D). This query sequence uses a single-network connection and is thus considerably faster than a sequence of independent queries that use distinct connections. The CAI adapter composes a CDS EMR from the query results and returns it to the mapper (H). If the MLM requests a buffered CDS EMR (E), the service interface takes it from the CDS EMR buffer (F), which is kept up to date by the copy process (G), and returns it to the mapper (H). Due to the interval of the copy process, buffered CDS EMRs may be up to two minutes old. It is possible to complement a returned CDS EMR with precalculated data items, as described elsewhere. 19
Fig. 4.
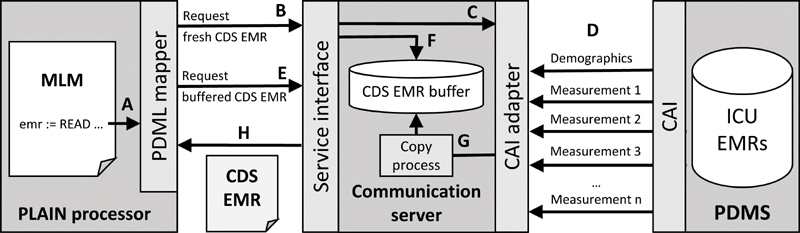
Basic architecture of the implemented system. The service interface of the communication server returns a CDS EMR. CDS, clinical decision support; EMR, electronic medical record.
The performance measurement on a normal workday showed that retrieving a fresh CDS EMR took 1.6 ± 0.7 s (average and standard deviation). In approximately 17% of the queries, however, a substantial additional communication delay of up to 3 seconds occurred, which can be attributed to concurrent queries to the CAI from different computers. Accessing a buffered CDS EMR took 68 ± 46 ms and no additional delay, since in this case the CAI is not directly involved. The actual transformation to Arden Syntax types by the PDML mapper took less than 10 ms, the remaining time was consumed by network communication. For comparison, a conventional query to the CAI took 250 ± 104 ms, with additional delays similar to those above, whereas the actual database query took 61 ms, while the remaining time was consumed for network communication.
In manually invoked MLMs, like for information retrieval at the point of care, we usually request buffered CDS EMRs, so the results can be displayed immediately. Data-driven MLMs, in contrast, usually request fresh CDS EMRs, since they typically require the very latest values of time series, which may not yet be contained in the buffered version.
Discussion
Enabling MLMs to access clinical databases is often a challenge, and multiple publications concentrate on this matter. 13 20 21 22 23 24 25 26 If access is possible, MLMs typically query only those measurements required for the actual decision making. Solving the curly braces problem by mapping XML documents that contain large proportions of the EMR, in contrast, appears undescribed in the literature. The only related work is an earlier investigation at UKER on the mapping of tree-structured microbiology data. 26 Other approaches to abstract from clinical databases by providing XML-encoded EMRs, such as HL7 virtual medical records, 27 do not directly reflect the Arden Syntax type system and thus cannot be mapped like CDS EMRs.
If direct access to a clinical database is not possible, a successful solution to the curly braces problem depends on an alternative data access option, such as a service interface. Before the CAI became available, we were limited to an export interface for the creation of reports and doctors' letters, which could not be invoked from outside the PDMS. Thus, the only possible solution was to create text documents at evenly spaced intervals, to parse them after creation, and to store the extracted data into another database. This approach was successful, but also laborious and caused a substantial delay in data provision. 12 The CAI, in contrast, can be invoked from outside the PDMS. Our approach does, however, not fully eliminate data replication, as the CDS EMR buffer stores copies of patient data. Nevertheless, this replication process is considerably less complicated than the former one. It does not even require a database, as XML documents can be stored in the file system. From a technical perspective, the buffer would not be necessary, but local clinicians demand fast response times in case of manually invoked MLMs, thus corroborating that “speed is everything,” the first commandment for effective CDS. 28 Retrieving a buffered CDS EMR is so fast that clinicians will rarely perceive a delay at all. Therefore, it does not matter that CDS EMRs include multiple data items that an MLM might not even process, which constitutes a certain waste of resources. There is even room for further acceleration if buffer and processor were on the same machine, thus avoiding network communication.
Generalizability of Our Approach
Our implementation employs an experimental Arden Syntax processor with extended features. This raises the question whether our approach is transferable to standard-compliant processors without these features. One requirement is Arden Syntax version 2.5 or higher, since the object data type is used to encode CDS EMRs. Another requirement is a PDML mapper, which is relatively easy to create, because the designers of the Arden Syntax intentionally kept the type system simple. The tutorial in the supplementary material should demonstrate that PDML-encoded CDS EMRs are not a technical challenge. Provided that access is possible at all, transforming patient data to PDML is a relatively straightforward task, regardless of the source data format. Enabling access to files and network resources is a task that also requires only moderate programming effort. In summary, our approach is fully compliant with the Arden Syntax, but presupposes a certain initial effort to incorporate the PDML mapper and the support of file and network access, which are not natively provided by standard-compliant processors. Thus, a limitation of our approach exists for those Arden Syntax processors that are firmly integrated into commercial systems and for which the customers are therefore unable to make technical extensions on their own.
We believe that native support for PDML-encoded patient data would be beneficial for the Arden Syntax standard. First, one could even create CDS EMRs by hand and access them without requiring a database connection beforehand. Second, one is no longer limited to regular data structures, but can work on tree-structured patient data of arbitrary complexity. In version 2.10, ArdenML has been included into the standard as an XML-based representation of the programming language constructs. 29 A comprehensible XML format for patient data, however, is still missing today, although it might be even more important, since it could facilitate the often tedious task of enabling EMR access from MLMs. Consequently, we would suggest integrating a format like PDML into the standard, as well as native support for access to files and network resources.
Comprehensibility of Our Approach
During everyday use, we observed advantages of CDS EMRs with respect to comprehensibility. Their simplicity makes them human-readable to some degree, and many clinicians intuitively understand them. Since the internal structure of CDS EMRs corresponds to the tabbed view in the PDMS, our clinicians understand the path expressions to address specific measurements. Moreover, MLM authors can easily view the contents of CDS EMRs during the authoring process if necessary. We made similar experiences with respect to teaching. For most students, CDS EMRs are comprehensible without in-depth computer knowledge. In our lectures, we let them manually create CDS EMRs to make the data mapping understandable. As a first exercise, they encode a single-time series as a list, like included in Fig. 2 . Thereafter, they manually compose CDS EMRs from multiple time series, and when they process them in MLMs, they usually understand the transformation of patient data from PDML-encoded documents to Arden Syntax data structures. For informatics students, this understanding is normally already present before our lectures, but students of medicine and medical technology, who are less familiar with information science, can gain new insights. Therefore, CDS EMRs might be advantageous for obtaining basic Arden Syntax skills.
Semantic Interoperability
MLMs have been designed as a unit of knowledge sharing between institutions 1 where semantic interoperability is an issue. This raises the question of whether it would make sense to use terminologies like Logical Observation Identifiers Names and Codes (LOINC) 30 instead of institution-specific path expressions. As another experimental feature, PLAIN provides native support of LOINC annotations. This feature is provided in the form of an extension to the type system, complemented with an optional LOINC attribute in PDML-encoded patient data, and additional language constructs to process the LOINC annotations. For example, the following statement extracts all those items from a CDS EMR that are annotated with the LOINC code “2345-7,” which refers to “Glucose [mass/volume] in Serum or Plasma.”
glucose:= EXTRACT LOINC “2345-7” FROM emr;
LOINC annotations can enable semantic interoperability in case of sharing MLMs. Our local clinicians, however, prefer addressing data items in a way that corresponds to what they know from routine documentation. For example, glucose is included in the BGA tab ( Fig. 2 ), thus they want to address this measurement as “emr.bga.glucose,” rather than using a code such as “2345-7” that is not immediately understandable. Hussain et al also observed that physicians prefer localized concepts to terminologies. 31 Therefore, whether to address measurements with path expressions or terminologies mostly depends on whether knowledge sharing actually happens and institutional boundaries are crossed.
Relationship to FHIR and EAV/CR
Accessing patient data in the form of CDS EMRs bears some similarities with the Fast Healthcare Interoperability Resources (FHIR) standard, 32 insofar, as both approaches access complex patient data structures from network resources. We did not use FHIR because the CAI returns a proprietary format instead of FHIR resources, and transforming query results to FHIR resources only to immediately transform them to PDML would yield no benefit in our local setting. Nevertheless, FHIR is a promising technology, so it is worth considering how the integrated PDML mapper can handle FHIR resources. In FHIR, a time series can be represented as a “Bundle” resource, which contains the individual values and their timestamps as an array of “Observation” resources. In case of a direct transformation of a glucose time series to Arden Syntax types, values can be accessed as follows:
glucose:= bundle.entry.resource.valueQuantity.value;
Since the DOT operator of the Arden Syntax (9.18.1 6 ) automatically loops through the Observation resources within the Bundle resource, the above path expression returns a list of values, but without the corresponding timestamps, which are provided in a different attribute. In other words, the result is not a time series. Consequently, those operators that require timestamped values, such as SLOPE, cannot calculate a result. This problem can be overcome by implementing a customized FHIR mapper that transforms a Bundle resource to an Arden Syntax time series, but different processors have different application programming interfaces, thus FHIR mappers are likely limited to a specific processor. It seems more reasonable to build a processor-independent FHIR-to-PDML converter, which preserves the inherent structure of resources, but automatically provides the values of elementary Arden Syntax types with the corresponding timestamps. We already implemented a prototypic converter as a proof of concept. Therefore, a conversion from FHIR to PDML is not a superfluous intermediary step, but has advantages, and PDML should not be seen as a competitor to FHIR and other data formats but as a complement at a lower level.
Our approach also has similarities with an extension of the EAV data model, called EAV with classes and relationships (EAV/CR). 33 34 In EAV/CR, an EAV-modeled database is complemented with a metadata infrastructure that enables the definition of complex objects. In this respect, CDS EMRs are compliant with the EAV/CR model. Thus, a clinical information system that incorporates this model could enable the convenient creation of CDS EMRs from the object definitions in the metadata.
Potential Use in Clinical Research Projects
The German Medical Informatics Initiative is a large-scale project that aims at providing clinical data for medical research. 35 36 37 UKER is part of one of the research consortia within this project, termed Medical Informatics in Research and Care in University Medicine (MIRACUM). 38 The hospitals within the consortia are currently building data integration centers to enable patient cohort identification for clinical trials across institutions. Currently, we investigate the suitability of MLMs as sharable patient cohort identification algorithms within MIRACUM. For this purpose, we provided our research database with a service interface to generate CDS EMRs, in analogy to the approach described in this paper. This leverages the strengths of the Arden Syntax with respect to the convenient processing of time series data, enabling sophisticated but comprehensible algorithms that, for example, consider the chronological progression of a patient's condition by analyzing time series data and performing calculations at query runtime.
Conclusion
For our local hospital, CDS EMRs are an appropriate solution to the curly braces problem. Since they enable data access from MLMs in a uniform manner, they simplify the application of the Arden Syntax at UKER. Our approach is transferrable to standard-compliant Arden Syntax processors with moderate effort. It may lower the entry barrier for the application of the Arden Syntax and thus promote its dissemination.
Clinical Relevance Statement
This study presents an uncommon but practical solution to the curly braces problem that is in routine use at a university hospital for more than 2 years. We believe that other institutions could also benefit from this approach. We also assume that the Arden Syntax itself may benefit from the integration of this approach into the standard.
Multiple Choice Questions
-
Which measurements are included in a CDS EMR at UKER?
All measurements documented for a specific patient.
The superset of measurements required by any MLM.
All measurements within a specific tab of the PDMS.
The measurements denoted in a curly braces expression.
Correct Answer: The correct answer is option b. A CDS EMR always contains the superset of measurements that might be required by any MLM. If additional measurements will be required by a new MLM, they will be included in any CDS EMR.
-
How are CDS EMRs represented technically?
As a sequence of entries in an EAV-modeled database.
In a proprietary format provided by the application interface.
As PDML documents built from objects and their attributes.
As FHIR Bundle resources composed from Observation resources.
Correct Answer: The correct answer is option c. A CDS EMR is a PDML-encoded document with an internal structure built from objects and their attributes, which reflects the tabbed view of an EMR in the PDMS.
Conflict of Interest None declared.
Note
To make our approach more understandable for the reader, the Supplementary Material (available in the online version) provides a tutorial on PDML with commented examples, including a CDS EMR.
Protection of Human and Animal Subjects
Ethical approval was not required.
Supplementary Material
References
- 1.Hripcsak G, Ludemann P, Pryor T A, Wigertz O B, Clayton P D. Rationale for the Arden Syntax. Comput Biomed Res. 1994;27(04):291–324. doi: 10.1006/cbmr.1994.1023. [DOI] [PubMed] [Google Scholar]
- 2.Jenders R A, Adlassnig K-P, Fehre K, Haug P. Evolution of the Arden syntax: key technical issues from the standards development organization perspective. Artif Intell Med. 2018;92:10–14. doi: 10.1016/j.artmed.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adlassnig K-P, Haug P, Jenders R A. Arden Syntax: then, now, and in the future. Artif Intell Med. 2018;92:1–6. doi: 10.1016/j.artmed.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Samwald M, Fehre K, de Bruin J, Adlassnig K-P. The Arden Syntax standard for clinical decision support: experiences and directions. J Biomed Inform. 2012;45(04):711–718. doi: 10.1016/j.jbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Hripcsak G. Writing Arden Syntax Medical Logic Modules. Comput Biol Med. 1994;24(05):331–363. doi: 10.1016/0010-4825(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 6.Health Level Seven International. The Arden Syntax for Medical Logic Systems, Version 2.8. 2012. Available at:https://www.hl7.org/implement/standards/product_brief.cfm?product_id=268. Accessed March 30, 2020
- 7.Castellanos I, Schüttler J, Prokosch H-U, Bürkle T. Does introduction of a Patient Data Management System (PDMS) improve the financial situation of an intensive care unit? BMC Med Inform Decis Mak. 2013;13:107. doi: 10.1186/1472-6947-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellanos I, Ganslandt T, Prokosch H U, Schüttler J, Bürkle T. [Implementation of a patient data management system. Effects on intensive care documentation] (in German) Anaesthesist. 2013;62(11):887–890. doi: 10.1007/s00101-013-2239-x. [DOI] [PubMed] [Google Scholar]
- 9.Bürkle T, Castellanos I, Tech H, Prokosch H-U.Implementation of a patient data management system - an evaluation study of workflow alterations Stud Health Technol Inform 2010160(Pt 2):1256–1260. [PubMed] [Google Scholar]
- 10.Castellanos I, Martin M, Kraus S et al. Effects of staff training and electronic event monitoring on long-term adherence to lung-protective ventilation recommendations. J Crit Care. 2018;43:13–20. doi: 10.1016/j.jcrc.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Castellanos I, Kraus S, Toddenroth D, Prokosch H-U, Bürkle T. Using Arden syntax medical logic modules to reduce overutilization of laboratory tests for detection of bacterial infections-success or failure? Artif Intell Med. 2018;92:43–50. doi: 10.1016/j.artmed.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Kraus S, Castellanos I, Toddenroth D, Prokosch H-U, Bürkle T. Integrating Arden-Syntax-based clinical decision support with extended presentation formats into a commercial patient data management system. J Clin Monit Comput. 2014;28(05):465–473. doi: 10.1007/s10877-013-9430-0. [DOI] [PubMed] [Google Scholar]
- 13.Johansson B, Shahsavar N, Ahlfeldt H, Wigertz O.Database and knowledge base integration--a data mapping method for Arden Syntax knowledge modules Methods Inf Med 199635(4,5):302–308. [PubMed] [Google Scholar]
- 14.Lamigueiro O P. Boca Raton, FL: CRC Press; 2018. Displaying Time Series, Spatial, and Space-Time Data with R. 2nd ed. [Google Scholar]
- 15.Dinu V, Nadkarni P.Guidelines for the effective use of entity-attribute-value modeling for biomedical databases Int J Med Inform 200776(11-12):769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraus S. Generalizing the Arden Syntax to a common clinical application language. Stud Health Technol Inform. 2018;247:675–679. [PubMed] [Google Scholar]
- 17.Kraus S, Rosenbauer M, Schröder L, Bürkle T, Adlassnig K-P, Toddenroth D. A detailed analysis of the Arden Syntax expression grammar. J Biomed Inform. 2018;83:196–203. doi: 10.1016/j.jbi.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Kraus S, Toddenroth D, Unberath P, Prokosch H-U, Hueske-Kraus D. An extension of the Arden Syntax to facilitate clinical document generation. Stud Health Technol Inform. 2019;259:65–70. [PubMed] [Google Scholar]
- 19.Kraus S, Prokosch H-U. Complementing medical records with precalculated data items to facilitate decision support and phenotyping. Stud Health Technol Inform. 2019;258:36–40. [PubMed] [Google Scholar]
- 20.Hripcsak G, Johnson S B, Clayton P D. Desperately seeking data: knowledge base-database links. Proc Annu Symp Comput Appl Med Care. 1993:639–643. [PMC free article] [PubMed] [Google Scholar]
- 21.Sujansky W, Altman R. Towards a standard query model for sharing decision-support applications. Proc Annu Symp Comput Appl Med Care. 1994:325–331. [PMC free article] [PubMed] [Google Scholar]
- 22.Wilcox A, Hripcsak G, Chen C. Creating an environment for linking knowledge-based systems to a clinical database: a suite of tools. Proc AMIA Annu Fall Symp. 1997:303–307. [PMC free article] [PubMed] [Google Scholar]
- 23.Arkad K, Gao X M, Ahlfeldt H. Query-handling in MLM-based decision support systems. Med Inform (Lond) 1995;20(03):229–240. doi: 10.3109/14639239508995007. [DOI] [PubMed] [Google Scholar]
- 24.Johansson B, Shahsavar N, Ahlfeldt H, Wigertz O. Database and knowledge base integration in decision support systems. Proc AMIA Annu Fall Symp. 1996:249–253. [PMC free article] [PubMed] [Google Scholar]
- 25.Jenders R A, Corman R, Dasgupta B. Making the standard more standard: a data and query model for knowledge representation in the Arden syntax. AMIA Annu Symp Proc. 2003;2003:323–330. [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus S, Enders M, Prokosch H-U, Castellanos I, Lenz R, Sedlmayr M. Accessing complex patient data from Arden Syntax Medical Logic Modules. Artif Intell Med. 2018;92:95. doi: 10.1016/j.artmed.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Marcos C, González-Ferrer A, Peleg M, Cavero C. Solving the interoperability challenge of a distributed complex patient guidance system: a data integrator based on HL7's Virtual Medical Record standard. J Am Med Inform Assoc. 2015;22(03):587–599. doi: 10.1093/jamia/ocv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bates D W, Kuperman G J, Wang S et al. Ten commandments for effective clinical decision support: making the practice of evidence-based medicine a reality. J Am Med Inform Assoc. 2003;10(06):523–530. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Haug P J, Rocha R A, Choi I. Modeling the Arden Syntax for medical decisions in XML. Int J Med Inform. 2008;77(10):650–656. doi: 10.1016/j.ijmedinf.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 30.McDonald C J, Huff S M, Suico J G et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(04):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 31.Hussain M, Afzal M, Ali T et al. Data-driven knowledge acquisition, validation, and transformation into HL7 Arden Syntax. Artif Intell Med. 2018;92:51–70. doi: 10.1016/j.artmed.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Benson T, Grieve G. London, United KIngdom: Springer International Publishing and Imprint and Springer; 2016. Principles of Health Interoperability: SNOMED CT, HL7 and FHIR. 3rd ed. 2016. [Google Scholar]
- 33.Nadkarni P M, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999;6(06):478–493. doi: 10.1136/jamia.1999.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadkarni P M, Nadkarni P. London; United Kingdom: Springer; 2011. Metadata-Driven Software Systems in Biomedicine: Designing Systems That Can Adapt to Changing Knowledge (Health Informatics) [Google Scholar]
- 35.Semler S C, Wissing F, Heyder R.German medical informatics initiative Methods Inf Med 201857(S 01):e50–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haux R.Health information systems - from present to future? Methods Inf Med 201857(S 01, Suppl 1):e43–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehring S, Eulenfeld R.German medical informatics initiative: unlocking data for research and health care Methods Inf Med 201857(S 01, Suppl 1):e46–e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prokosch H-U, Acker T, Bernarding Jet al. MIRACUM: medical informatics in research and care in university medicine Methods Inf Med 201857(S 01):e82–e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


