Abstract
Ophraella communa is an effective bio-control agent of the invasive common weed. By now, the reference genes in O. communa have not yet been screened and validated. The aim of this study was to screen for the most stable reference genes in different backgrounds, such as different developmental stages, sexes, tissues, and male reproductive system with different body sizes. We selected 12 common housekeeping genes involved in different biological processes, including GAPDH, ACT1, ACT2, ARF1, ARF4, SDH, βTUBC, RPL4, RPL19, RPS18, EF1α, and COX as the candidate reference genes. To analyze the stability of the candidate reference genes, we first used three dedicated algorithms, GeNorm, NormFinder, and BestKeeper, and further comprehensive ranking was provided by ReFinder. The results showed that RPL19 and RPL4 exhibited the least variation in different developmental stages/sexes and in male reproductive systems with different body sizes. COX proved to be most suitable for normalizing the gene expression levels in different tissues, and coincidentally, RPL19 was also found to be second in terms of stability in this study. To the best of our knowledge, this is the first study to identify suitable reference genes for analyzing gene expression in O. communa; thus, this study would lay the foundation for future research on the molecular physiology and biochemistry of O. communa and other insects.
Keywords: Ophraella communa, RT-qPCR, reference gene, normalization, gene expression
Introduction
Ophraella communa LeSage (Coleoptera: Chrysomelidae) originated in North America (LeSage, 2012). It is a biological control agent of the invasive weed Ambrosia artemisiifolia L. (common ragweed) (Yamazaki et al., 2000; Zhou et al., 2010a, 2014; Gerber et al., 2011; Guo et al., 2011). For a biological control agent, reproductive regulation is a very important issue in order to facilitate its mass rearing. Gratifyingly, we have found that the O. communa males with a large body size can facilitate a female’s fecundity (unpublished data). Therefore, how to regulate the reproductive potential of the male beetle will be focused on in a further study. It is necessary to assess gene function in the beetle; quantification of gene expression is required as well.
In recent years, the ecology and physiology of O. communa has been studied extensively (Zhou et al., 2010a,b; Clark et al., 2014; Kim and Lee, 2018; Zhao et al., 2018). However, the molecular mechanisms of its biology remain largely unknown (Ma et al., 2019a,b). With the aim of investigating the genetic basis of its biology and physiology, the male transcriptome and proteome of O. communa have been sequenced (unpublished data). Remarkably, we found that the male’s body size is an important determinant in the evolutionary process and in the speciation of O. communa, as it affects female mate choice. Then, the large males could further improve the population fitness of their offspring (unpublished data). Currently, the mechanisms for higher fitness in large males are also being studied, so it is necessary to assess gene function in O. communa; quantification of gene expression is required as well.
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR), also referred to as qPCR, owing to its excellent repeatability, high sensitivity, accurate quantification, wide applicability, and easy operation, is the most common and sensitive technique for the detection and quantification of gene expression in different experimental samples, such as developmental stages, tissues, and other processing conditions (Bustin, 2002). It is especially useful to detect low-abundance mRNAs in limited samples (Bustin et al., 2005; Valasek and Repa, 2005; Chen et al., 2011; Hellemans and Vandesompele, 2014). However, the results of qPCR are often limited by factors such as the amount of the initial sample, cDNA synthesis efficiency, and primer amplification efficiency, which may result in inaccurate qPCR results (Bustin et al., 2005). Therefore, this approach requires normalization.
To date, internal control genes, also known as housekeeping genes or reference genes, are genes whose expression levels are stable regardless of the cell type and the experimental conditions. It is the most popular tool to normalize variability (Thellin et al., 1999; Vandesompele et al., 2002; Andersen et al., 2004; Huggett et al., 2005; Hellemans and Vandesompele, 2014). Many genes such as Actin, GAPDH, and rRNAs (18s or 28s) that are involved in fundamental biological processes are frequently used as reference genes (Keertan et al., 2004; de Kok et al., 2005). Nevertheless, it has become clear that no single reference gene retains a constant expression in all tissues, developmental stages, or experimental conditions (Silver et al., 2006; Lourenço et al., 2008; Lu et al., 2013, 2018a; Rodrigues et al., 2014; Tan et al., 2015; Dai et al., 2017; Luo et al., 2018). Therefore, the traditional genes are not applicable to all species and conditions (Shu et al., 2018; Zhou et al., 2018; Qu et al., 2019). Blindly choosing “classical” reference genes such as β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), or ribosomal 18S (18S) is unreliable and inaccurate. The reliability of qPCR largely depends on the stability of the reference genes. Previous studies have shown that it is crucial to select and validate the reference gene for data quantification (Lu et al., 2018b; Shakeel et al., 2018; Shu et al., 2018). Hence, it is extremely important to verify the stability of expression of the intended control gene under different experimental conditions. To our knowledge, research on the reference gene of O. communa is absent, which makes the study of the mechanisms of its physiological and biochemical behavior difficult.
In this study, we aim to identify the solidly expressed reference genes for RT-qPCR investigation under different conditions including developmental stages, sexes, tissues, and male reproductive system with different body sizes of O. communa. Based on prior experimental reports regarding reference genes in coleoptera and other insects, 12 candidate reference genes including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin 1 (ACT1), actin 2 (ACT2), succinate dehydrogenase (SDH), ADP-ribosylation factor 1 (ARF1), ADP-ribosylation factor 4 (ARF4), cytochrome oxidase (COX), β-tubulin C (βTUBC), elongation factor-1α (EF1α), ribosomal protein L4 (RPL4), ribosomal protein L19 (RPL19), and ribosomal protein S18 (RPS18), which are involved in various biological processes, were selected from the male transcriptome of O. communa. The results of the qPCR were analyzed using three statistical algorithms, GeNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and BestKeeper (Pfaffl et al., 2004). Finally, a comprehensive ranking of the stability of these candidates was given by ReFinder (Yang et al., 2015a). In addition, the expression profile of angiotensin-converting enzyme (ACE) was tested to verify our result. This study provides a valuable platform for gene expression and future functional research and serves as an example for similar studies in other insects.
Materials and Methods
Insect
Ophraella communa adults were collected from the fields in Laibin City in Guangxi Province in the summer of June 2017 and were maintained on common ragweed plants in the laboratory at the Chinese Academy of Agricultural Sciences (BJ, CAAS) for three generations. All the insects used for sampling were reared in an environmental chamber (PRX-450D, Ningbo Haishu Safe Experimental Equipment Co., Ltd., Zhejiang, China) at 27 ± 1°C, with 50 ± 10.0% relative humidity and a 14:10-hour photoperiod provided by artificial lights and with the robust common ragweed plants used as food.
Experimental Sample in This Study
Samples of Developmental Stages and Sexes
The adults deposited their eggs in batches of about 35 on the underside of common ragweed leaves. The number of sampled individuals for each replicate across the different developmental stages and sexes of O. communa included 150 eggs, 30 first instar nymphs, 20 second instar nymphs, eight third instar nymphs, eight pupa (third pupa), five sexually immature (1 day old) male and female adults, and five sexually mature (8 days old) male and female adults. Three independent biological replicates were collected for each sample.
Tissue
Male adults (8 days old) of O. communa were dissected for samples of five different tissues, including head, testis, male accessory glands (MAG), fat body, and gut. Three independent biological replicates were collected with each group of 12 insects.
Male Reproductive System With a Different Body Size
The testis and accessory glands of large and small males, respectively, were dissected for these samples. Three independent biological replicates were collected with each group of 12 insects.
All the above samples were collected in 1.5-ml tubes, immediately frozen in liquid nitrogen, and stored at −80°C until use.
RNA Extraction and cDNA Synthesis
Total RNA from all 48 samples was extracted using the Trizol reagent (Invitrogen, United States) according to the manufacturer’s protocol. The RNA integrity was tested by 1% agarose gel electrophoresis and quantified using NanoPhotometerTM P330 (Implen, Munich, Germany), with A260/A280 ratios ranging from 1.8 to 2.1 for all RNA samples. One microgram of total RNA was used to synthesize cDNAs using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech, Beijing, China) according to the manufacturer’s instructions.
Reference Gene Selection and Primer Design
A total of 12 candidate reference genes that are most frequently used in qPCR investigations were selected from the O. communa transcriptome data (Table 1). The primers were designed for gene cloning and qPCR using the Primer five programs. All the candidate reference genes were PCR-amplified from the bettle’s cDNA using the corresponding primers (Table 1) and the pEASY-T3 Simple Cloning Kit (TransGen, Beijing, China). Six independent subclones were sequenced and accurate nucleotide sequences were obtained after the sequence analysis. All the verified reference gene sequences were submitted to the GenBank database.
TABLE 1.
qPCR primers for candidate reference genes and characteristics of PCR amplification in O. communa.
| Gene name | Accession number | Primer sequence (5′–3′) | Product length (bp) | Tm (°C) | E (%) | R2 |
| RPL4 | MN641110 | F: TGTGGTAATGCTGTGGTAT | 104 | 60 | 99.14 | 0.998 |
| R: TCTAGCACTGCATGAACA | ||||||
| RPS18 | MN641111 | F: TCCGATGGCTGTATGTTAC | 126 | 60 | 98.145 | 0.998 |
| R: GCTGTTGGGAGGAGTTAT | ||||||
| ACT1 | MN641112 | F: CAGTGGTGATGGTGTTAC | 148 | 60 | 97.263 | 0.999 |
| R: CTCAGGAGGTATGCCAAT | ||||||
| ACT2 | MN641113 | F: GTTGCTCAGAGGTTATGC | 149 | 60 | 105.63 | 0.997 |
| R: GGTAAGGTGTAAGGTTCCA | ||||||
| ARF1 | MN641114 | F: GAGATGCCGTGTTGTTGA | 99 | 60 | 102.166 | 0.997 |
| R: TGCGTAGCGAATGAAGAC | ||||||
| ARF4 | MN641115 | F: GTGTAGATATGGAGCGAATG | 98 | 60 | 103.345 | 1 |
| R: GTTAGCGAGCACTAGAATC | ||||||
| SDH | MN641116 | F: ACTTCCGCTCATACTTGT | 149 | 60 | 98.306 | 0.999 |
| R: CCTCCTTCACCTCTACATC | ||||||
| GADPH | MN641117 | F: TGCTTCAACCGACACATT | 136 | 60 | 102.673 | 0.999 |
| R: CGACCAATACGACCGAAT | ||||||
| EF1α | MN641118 | F: AGAACAATCCACCAAGAGG | 98 | 60 | 101.81 | 0.996 |
| R: GTCCAACACAGGCGTATA | ||||||
| βTUBC | MN641119 | F: TCAGAGGTAGTCCTTCCA | 103 | 60 | 100.58 | 0.996 |
| R: CAATTATCGCAGTTCTCCG | ||||||
| COX | MN641121 | F: GCTGAACACAGTTATTCTGA | 150 | 60 | 98.533 | 0.998 |
| R: AGAGGCTCTATCTTGAAGT | ||||||
| RPL19 | MN641120 | F: AAGGAAGGCATTGTGGAT | 94 | 60 | 97.046 | 0.997 |
| R: GACGCAAATCTCGCATAC |
Quantitative Real-Time PCR Analysis
Quantitative PCR reactions were performed using an ABI 7500 PCR Detection System (Applied Biosystems, United States). The qPCR reactions consisted of 10 μl of 2 × TransStartTM Green qPCR SuperMix, 1 μl of the cDNA template, 0.4 μl of passive reference dye, and 0.4 μl each of the forward and reverse primers (10 μM) in a 20-μl final volume. The RT-qPCR procedure was as follows: an initial cycle at 94°C (30 s), followed by 40 cycles of 5 s at 94°C and 34 s at 60°C. Following the RT-qPCR procedure, the consistency and the specificity of the PCR-amplified products were subjected to melting curve analysis for all reactions. The amplification efficiencies (E) and correlation coefficients (R2) were determined for each gene using the standard curves with a 5-fold dilution series of the template (1/2, 1/10, 1/50, 1/250, and 1/1,250), where R2 is the slope of the standard curve. E was calculated according to the equation E = (10[−1/slope] −1) × 100 (Michael and Pfaffl, 2001). Each reaction was performed in triplicate.
Determination of Stability of Expression of the Candidate Reference Gene
The stability of the 12 candidate reference genes was evaluated using three different statistical programs, GeNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and BestKeeper (Pfaffl et al., 2004) under experimental conditions, and the instructions of all the three programs were consulted for the data arrangement and the subsequent analysis. Finally, the comprehensive ranking of each condition was obtained according to ReFinder. In addition, the number of reference genes for normalizing gene expression was decided by the pairwise variation (Vn/n+1) which was performed using the GeNorm program. Universally, Vn/n+1 less than the threshold value of 0.15 indicates that the most suitable number is n, and there is no need to introduce the n + 1 reference gene for normalization (Vandesompele et al., 2002).
Validation of Reference Genes
Angiotensin-converting enzyme (ACE), a zinc-dependent peptidase (Isaac et al., 1999, 2007; Macours and Biol, 2004), was reported as a seminal fluid protein in Drosophila melanogaster and Tribolium castaneum (Rylett et al., 2007; Xu et al., 2013). The ACE gene (GenBank number: MN641122) cloning primers used were as follows: forward (5′-AATCCATGCCTACGTACGGT AT-3′) and reverse (5′-CTTCCCATCTCCACAGGTCA-3′). The normalization of ACE expression in different tissues was conducted using the most stable gene combinations (COX/RPL19) as determined by geNorm, the most single stable gene (RPL19), and the least stable gene (SDH)as determined by ReFinder. The relative quantification of the ACE gene was calculated using the 2–ΔΔCT method (Schmittgen and Livak, 2008). One-way ANOVA followed by Tukey’s least significant difference test was used to determine the significance of the ACE expression levels between different tissues (SAS 9.4).
Results
Acquisition of Candidate Reference Gene Sequences
The sequences of the 12 candidate gene, including RPL4, RPS18, ACT1, ACT2, ARF1, ARF4, SDH, GAPDH, EF1α, βTUBC, COX, and RPL19, were obtained from the transcriptome database of the testis and the accessory glands of O. communa. The sequences were verified by qPCR. All amplicons were sequenced and confirmed to exhibit 99–100% identity with the corresponding transcriptome sequence. The sequence information is listed in Supplementary Table S1.
Specificity and Efficiency of RT-qPCR Primers
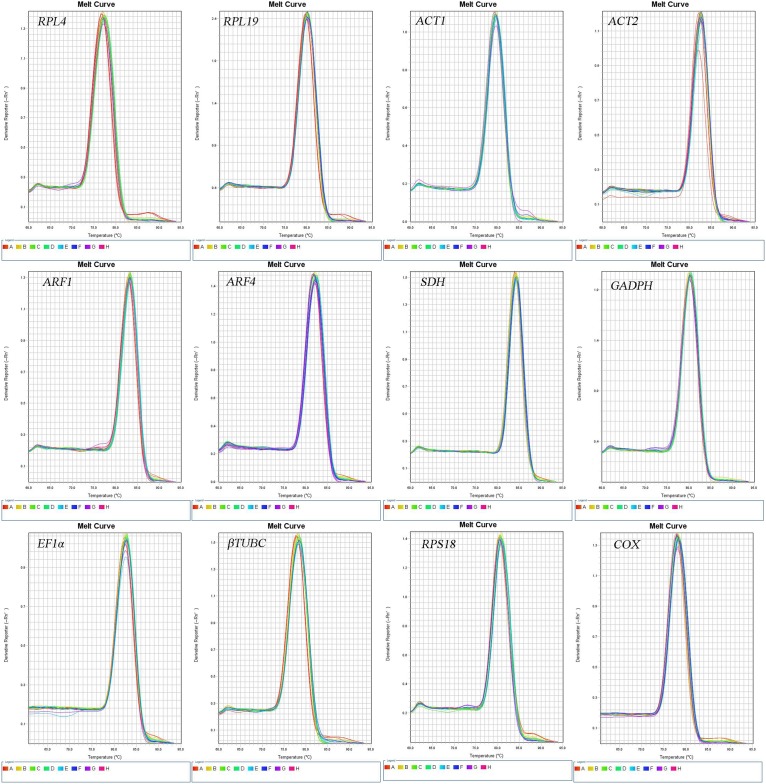
The accuracy of the RT-qPCR primers was confirmed by the presence of a single peak in the melting curve analyses and the absence of primer dimers, both of which also indicate the specificity of each primer pair (Figure 1). The amplification efficiency (E) of each primer (shown in Table 2) ranged from 97.05 to 105.63%, with associated R2 values of 0.996–1.000 (Table 1).
FIGURE 1.
Melting curves of the 12 candidate reference genes in O. communa.
TABLE 2.
Stability ranking of the 12 candidate reference genes’ expression in O. communa under different experimental conditions calculated by the three different analytical tools, geNorm, Normfinder, and BestKeeper, respectively.
| Experimental conditions | Rank | Bestkeeper |
NormFinder |
geNorm |
|||
| Gene | Stability | Gene | Stability | Gene | Stability | ||
| Developmental stage and sex | 1 | RPL19 | 0.179 | RPL19 | 0.221 | RPL19/RPL4 | 0.439 |
| 2 | RPL4 | 0.429 | RPL4 | 0.347 | – | – | |
| 3 | ACT2 | 0.514 | ACT1 | 0.369 | RPS18 | 0.525 | |
| 4 | ACT1 | 0.559 | ACT2 | 0.399 | ACT2 | 0.604 | |
| 5 | EF1α | 0.653 | EF1α | 0.428 | COX | 0.775 | |
| 6 | SDH | 0.66 | SDH | 0.446 | ARF1 | 0.818 | |
| 7 | RPS18 | 0.66 | RPS18 | 0.505 | ACT1 | 0.865 | |
| 8 | COX | 0.759 | βTUBC | 0.589 | SDH | 0.913 | |
| 9 | ARF1 | 0.793 | ARF1 | 0.59 | ARF4 | 0.971 | |
| 10 | ARF4 | 0.885 | COX | 0.657 | GADPH | 1.029 | |
| 11 | βTUBC | 0.919 | ARF4 | 0.695 | βTUBC | 1.095 | |
| 12 | GADPH | 1.15 | GADPH | 0.752 | EF1a | 1.206 | |
| Tissue | 1 | RPL19 | 0.24 | RPL4 | 0.162 | COX/ARF1 | 0.35 |
| 2 | RPS18 | 0.336 | COX | 0.294 | – | – | |
| 3 | COX | 0.361 | βTUBC | 0.301 | RPL19 | 0.373 | |
| 4 | ARF1 | 0.495 | RPS18 | 0.354 | RPL4 | 0.478 | |
| 5 | ACT2 | 0.535 | RPL19 | 0.385 | RPS18 | 0.533 | |
| 6 | RPL4 | 0.576 | ARF1 | 0.406 | βTUBC | 0.614 | |
| 7 | βTUBC | 0.77 | EF1Aα | 0.491 | ACT2 | 0.68 | |
| 8 | ARF4 | 0.86 | ARF4 | 0.562 | ARF4 | 0.751 | |
| 9 | EF1α | 1.026 | ACT2 | 0.656 | EF1α | 0.809 | |
| 10 | ACT1 | 1.047 | GADPH | 0.773 | ACT1 | 0.895 | |
| 11 | GADPH | 1.108 | ACT1 | 0.79 | GADPH | 0.97 | |
| 12 | SDH | 1.23 | SDH | 0.843 | SDH | 1.038 | |
| Male reproductive systems with different body sizes | 1 | RPL4 | 0.065 | ARF4 | 0.23 | RPL4/RPL19 | 0.053 |
| 2 | ACT2 | 0.089 | ACT1 | 0.271 | – | – | |
| 3 | RPL19 | 0.105 | GADPH | 0.326 | ACT2 | 0.169 | |
| 4 | GADPH | 0.397 | COX | 0.328 | ARF4 | 0.245 | |
| 5 | RPS18 | 0.552 | RPL19 | 0.355 | GADPH | 0.369 | |
| 6 | EF1a | 0.736 | RPL4 | 0.397 | ACT1 | 0.516 | |
| 7 | ACT1 | 0.736 | EF1a | 0.415 | EF1a | 0.584 | |
| 8 | COX | 0.809 | ARF1 | 0.451 | COX | 0.661 | |
| 9 | ARF4 | 0.846 | ACT2 | 0.557 | ARF1 | 0.708 | |
| 10 | ARF1 | 0.97 | RPS18 | 0.722 | RPS18 | 0.788 | |
| 11 | βTUBC | 1.22 | SDH | 0.845 | SDH | 0.878 | |
| 12 | SDH | 1.297 | βTUBC | 0.88 | βTUBC | 0.969 | |
Expression Profiles of Candidate Reference Genes
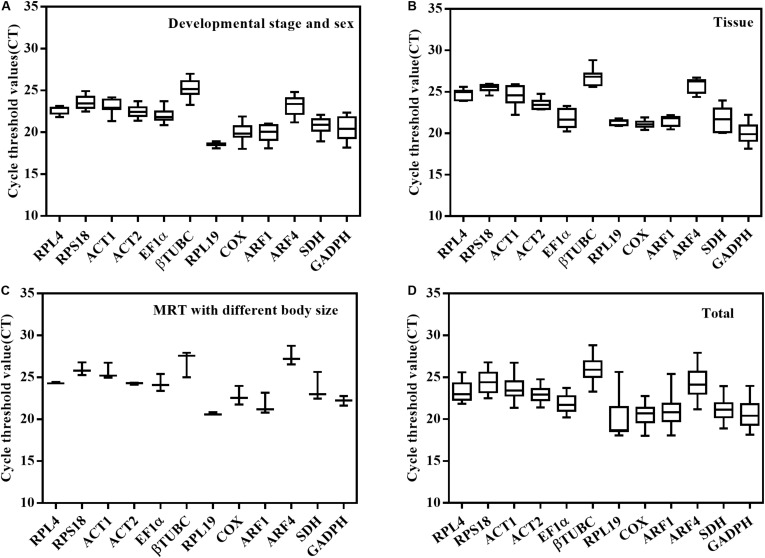
The expression profiles of the 12 candidate genes were analyzed by qPCR, and the Ct values exhibited different levels of transcript abundance (Figure 2). For the different developmental stages and sexes, the mean Ct values of the 12 candidate reference genes ranged from 18.01 to 26.98 cycles. In different tissues, the Ct values ranged from 18.13 to 28.83. Even in males of different sizes, it varied from 20.58 to 28.76.
FIGURE 2.
The expression profiles of the 12 candidate reference genes are shown as Ct values under three experimental conditions for O. communa. (A) Different developmental stages and sexes. (B) Different tissues. (C) Male reproductive systems with different body sizes. (D) Total Ct values exhibited different levels of transcript abundance in three biological samples.
Analysis of Stability of Candidate Reference Genes
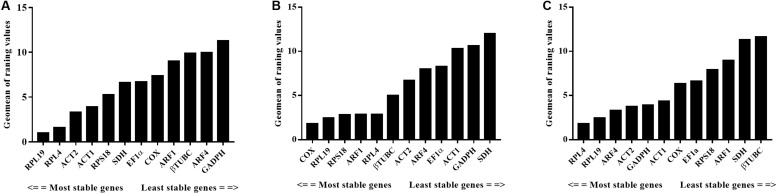
For different developmental stages and sexes—based on the analysis using GeNorm, NormFinder, and BestKeeper—the expression of RPL19 varied the least, while GAPDH tended to fluctuate the most. According to ReFinder parameters, the comprehensive ranking in these samples was as follows: RPL19 > RPL4 > ACT2 > ACT1 > RPS18 > SDH > EF1α> COX > ARF1 > βTUBC > ARF4 > GAPDH (Figure 3A). For the analysis of different tissue types, the stability of RPL4 and RPL19 were similarly excellent, while SDH and GAPDH were consistently recommended as the least stable by GeNorm, NormFinder, and BestKeeper. The comprehensive ranking was COX > RPL19 > RPL4 = ARF1 = RPS18 > βTUBC > ACT2 > ARF4 > EF1α> ACT1 > GAPDH > SDH (Figure 3B). For the male reproductive system with different body sizes, the overall variability of the reference genes was low except for βTUBC and SDH. The overall ReFinder stability ranking (from most to least stable) was RPL4 > RPL19 > ARF4 > ACT2 > GAPDH > ACT1 > COX > EF1α> RPS18 > ARF1 > SDH > βTUBC (Figure 3C). The stability of each sample’s stability was calculated using GeNorm, Normfinder, and BestKeeper, and all the specific values are shown in Table 2.
FIGURE 3.
Expression stability ranking of the 12 candidate reference genes analyzed using ReFinder under three experimental conditions. (A) Different developmental stages and sexes. (B) Different tissue. (C) Male accessory glands from large and small males. The most stable genes are on the right and the least stable genes are on the left.
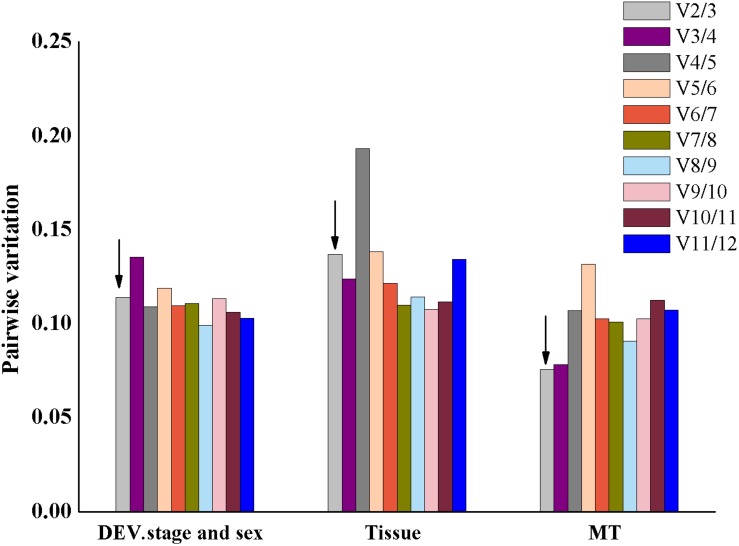
Selection of the Optimal Number of Reference Genes to Normalize the Gene Expression
We used GeNorm to compute the variation value to ensure the optimal number of reference genes. As shown in Figure 4, the V2/3 was below 0.15 across the different experimental conditions, which indicated that two reference genes were reliable for an accurate normalization of gene expression.
FIGURE 4.
Pairwise variation in the three groups calculated by geNorm is required for accurate normalization of gene expression. The threshold used was 0.15. The arrow indicates that the value of V2/V3 is less than 0.15.
Validation of Selected Reference Genes in O. communa
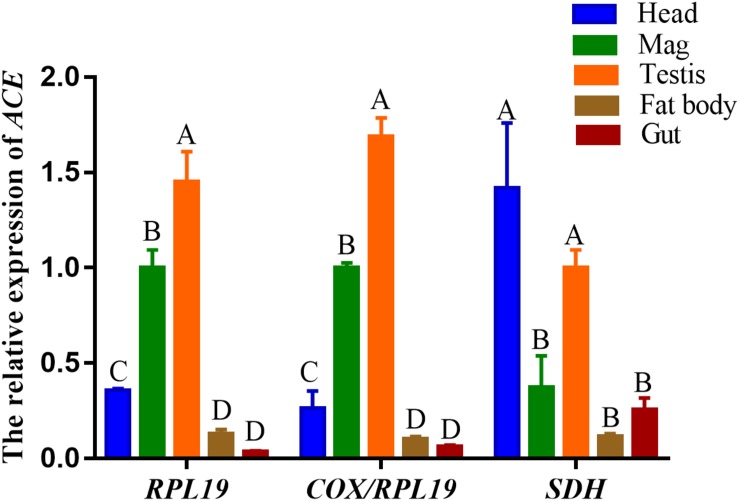
The ACE expression level was used to verify the validity of the reference genes between the different tissues. The best paired, best single, and the worst single reference genes—COX/RPL19, RPL19, and SDH, respectively, were used as internal controls. The ACE expression patterns were similar in that the highest expression of ACE was in the male reproductive tissues (testis and MAG) when normalized by the paired COX/RPL19 and single RPL19 (Figure 5). However, when we used SDH to normalize the expression levels, the highest expression level of ACE was in the heads and significantly higher than in the other tissues (Figure 5). This conclusion was completely inconsistent with the results seen through the normalization with RPL19 or COX/RPL19. This indicated that the accuracy of the RT-qPCR results may be influenced by the instability of the reference gene, which might even lead to an inaccurate or wrong conclusion.
FIGURE 5.
Relative quantification of ACE genes in different tissues. The expression levels of ACE genes were normalized using the best reference paired COX/RPL19, the single best stable gene RPL19, and the reference gene SDH that changed the most in tissues. Values are means ± SE. Different letters indicate significant differences in gene expression among the different tissues of O. communa (P < 0.05).
Discussion
Ophraella communa is a very valuable and effective biological control agent of Ambrosia artemisiifolia (Guo et al., 2011). Unfortunately, research on the molecular mechanisms of O. communa is scarce, making the study of its physiological and biochemical behavior difficult. Recently, the male transcriptome of O. communa was sequenced using Illumina high-throughput sequencing technologies (unpublished data), and further research on functional genomics and related gene expression studies will be initiated soon.
The quantitative accuracy of gene expression relies on the reference gene (Lu et al., 2018b). In our study, based on the study of reference genes in Coleoptera, 12 candidate reference genes, including traditional genes and novel genes, were selected. These genes have been evaluated as the most stable genes in different insects: RP18, RP4, AFR1, and ARF4 in Leptinotarsa decemlineata (Shi et al., 2013), β-actin and EF1a in Diabrotica virgifera virgifera (Rodrigues et al., 2014), Actin and RPS18 in T. castaneum (Lord et al., 2010; Toutges et al., 2010; Sang et al., 2015), EF1α, ACT1, and RPL19 in Colaphellus bowringi (Tan et al., 2015), SDHA, ACT and TUB in Galeruca daurica (Tan et al., 2017), GADPH in Hippodamia convergensin (Pan et al., 2015), and COX in Agasicles hygrophila. Besides that, these candidate genes have also been recommended for normalization gene expression in others (Lu et al., 2018b; Shakeel et al., 2018). Three different methods, GeNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), and BestKeeper (Pfaffl et al., 2004), were used to identify the stability of the 12 candidate genes across the condition of developmental stages and sexes, different tissues, and male reproductive system with different body sizes. However, because each method has its own strengths and appropriate application conditions, the expression stability obtained by different algorithms was different (Table 2). Therefore, another analysis tool, ReFinder (Yang et al., 2015a), was used to integrate the variation in the results between them and to define an overall final ranking (Xie et al., 2012).
Our current research focuses on the molecular mechanisms of the male fitness differences relative to different body sizes. The results of this study displayed that the reference gene transcript levels vary with the above experimental conditions (Figure 2). For the sample of different developmental stages and sexes, contrary to the results of other studies in Coleoptera insects, GAPDH, EF1α, and TUB performed the best stability under this condition (Rodrigues et al., 2014; Pan et al., 2015; Yang et al., 2018). Our results demonstrated that RPL19 and RPL4 showed the most stable gene expression, while GAPDH, EF1α, and βTUBC showed the least stability (Figure 3A). All four algorithms obtained the same two top-ranked genes with few differences in their respective orders. Similar to our results, in L. decemlineata, RPL4 and RPS18 exhibited excellent stability in different developmental stages and sexes. Ribosomal protein (RP) also presented superior stability in Coccinella septempunctata across this condition (Shi et al., 2013; Tan et al., 2015; Yang et al., 2016). For different tissues—consistent with the results in A. hygrophila—a novel reference gene, COX, that encodes for a key enzyme (Liu et al., 2017) and that responds to a variety of metabolic states showed the least variation (Figure 3B). In addition, like the ranking for developmental stages and sexes, the RPs, including RPL19, RPLl4, and RPS18, displayed high stability (Figure 3B), while SDH and GADPH were identified as the least stable genes. For the male reproductive system with different body sizes, the variation of each candidate gene is relatively small compared with the first two conditions (Figure 2C), and a similar ranking trend with previous condition was observed in this sample. RPL19 and RPL4 performed the least variation. To our knowledge, studies on the reference gene for body size are scarce. Our results provide a reference for the gene expression in other insects with different body sizes. Integrating the excellent stable reference genes under three conditions, RPL19 can be used to normalize the gene expression. Studies in C. bowringi similarly concluded that the RPL19 presented excellent stability across developmental stage, sex, and tissue (Tan et al., 2015). In D. virgifera virgifera, EF1a and Actin were respectively identified as the most stable gene among different tissues and developmental stages (Rodrigues et al., 2014). However, our results were almost the opposite. Therefore, the choice of the control gene for O. communa is of utmost importance.
Coincidentally, our results verified that RP gene expression was extremely stable under each experimental condition. Generally, ribosomal protein genes are expressed in all types of cells for the synthesis of new ribosomes, and RPs are among the most highly conserved proteins across all life forms (Hsiao et al., 2001). Recently, researchers found that even in T. castaneum, most RP gene expression was maintained stably following a fungal challenge (Lord et al., 2010; Lu et al., 2018b). In addition, besides Coleoptera insects, ribosomal genes showed high stability in diverse abiotic and biotic conditions in other organisms (Jonge et al., 2007; Popovici et al., 2009; Tong et al., 2009; Zhong et al., 2013; Yang et al., 2015b, 2016; Ma et al., 2016; Yan et al., 2016; Lu et al., 2018a; Luo et al., 2018). Our screening results also demonstrated that the RP gene is transcriptionally conserved under different physiological conditions. However, under some conditions, the expression levels of RP-encoding genes may be unstable. For example, RPS3 was the least stable gene in Spodoptera litura under different populations, exposed to different temperatures, or treated by different insecticides (Lu et al., 2013). Moreover, other classic references are GADPH, TUB, and Actin which have poor stability in O. communa (Figure 3). Therefore, it is very unwise to choose a reference gene blindly regardless of the conditions and species.
Furthermore, to certify the reliability of the reference gene in O. communa, the relative transcriptional level of the ACE gene was determined in different tissues using qPCR. GeNorm recommended that the use of two reference genes was optimal for normalization under each experimental condition (Figure 4). Therefore, ACE expression was normalized by the single stable RPL19, the pairwise stable RPL19 and COX, and the least stable SDH alone. Consistent with a previous study (Xu et al., 2013), when RPL19/COX and RPL19 alone were used as internal controls, ACE expression was found to be abundant in the reproductive organs, including the testis and the MAG. In contrast, the head and the testis showed a high ACE expression when normalized using SDH (Figure 5). This implies that different controls may lead to different conclusions. Therefore, we strongly recommend the validation of the stability of the reference gene expression prior to performing RT-qPCR experiments. Our study found that RPL19 and RPL4 showed excellent stability among the candidate reference genes for all three experimental conditions. This provides a fundamental premise to analyze qPCR data in O. communa and to further clarify the physiological and biochemical mechanisms in this important natural ragweed enemy.
Conclusion
In summary, we identified several stable housekeeping genes that are suitable as reference genes in O. communa. This is the first study to report the reference genes and to establish a standardized RT-qPCR analysis protocol in O. communa, which will contribute to the development of molecular research on this vital insect.
Data Availability Statement
The datasets generated for this study can be found in the NCBI using accession MN641110, MN641111, MN641112, MN641113, MN641114, MN641115, MN641116, MN641117, MN641118, MN641119, MN641121, MN641120, and MN641122.
Ethics Statement
The animal study was reviewed and approved by Ethics of Animal Experiment of Institute of Plant Protection, Chinese Academy of Agricultural Sciences.
Author Contributions
ZZ designed the study and revised the manuscript. YZ performed the research and wrote the manuscript. JC applied software for data analysis. GC searched for the sequence of candidate reference genes. CM, XG, ZQT, SC, and ZYT participated in the sample collection and data sorting. HC, JG, and FW guided in the data analysis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage for polishing the English.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Nos. 31672089 and 31972340).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00355/full#supplementary-material
References
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-pcr data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.Can-04-0496 [DOI] [PubMed] [Google Scholar]
- Bustin S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29 23–39. 10.1677/jme.0.0290023 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Nolan T., Pfaffl M. W. (2005). Quantitative real-time RT-PCR - a perspective. J. Mol. Endocrinol. 34 597–601. 10.1677/jme.1.01755 [DOI] [PubMed] [Google Scholar]
- Chen C., Tan R., Wong L., Fekete R., Halsey J. (2011). Quantitation of microRNAs by real-Time RT-qPCR. Methods Mol. Biol. 687 113–134. 10.1007/978-1-60761-944-4_8 [DOI] [PubMed] [Google Scholar]
- Clark S., Scavarda G., Chersi C., Massobrio V., Bosio G. (2014). Spread of the ragweed leaf beetle, Ophraella communa LeSage, 1986 (Coleoptera Chrysomelidae), in piedmont region (northwestern Italy). Boll. Soc. Ent. Ital. 146 17–30. 10.4081/BollettinoSEI.2014.17 [DOI] [Google Scholar]
- Dai T. M., Lu Z. C., Liu W. X., Wan F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS One 12:e0173821. 10.1371/journal.pone.0173821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok J. B., Roelofs R. W., Giesendorf B. A., Pennings J. L., Waas E. T., Feuth T., et al. (2005). Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab. Invest. 85 154–159. 10.1038/labinvest.3700208 [DOI] [PubMed] [Google Scholar]
- Gerber E., Schaffner U., Gassmann A., Hinz H. L., Seier M., Müller-Schärer H. (2011). Prospects for biological control of Ambrosia artemisiifolia in europe: learning from the past. Weed. Res. 51 559–573. 10.1111/j.1365-3180.2011.00879.x [DOI] [Google Scholar]
- Guo J. Y., Zhou Z. S., Zheng X. W., Chen H. S., Wan F. H., Luo Y. H. (2011). Control efficiency of leaf beetle, Ophraella communa, on the invasive common ragweed, Ambrosia artemisiifolia, at different growing stages. Biocontrol. Sci. Techn. 21 1049–1063. 10.1080/09583157.2011.603823 [DOI] [Google Scholar]
- Hellemans J., Vandesompele J. (2014). Selection of reliable reference genes for RT-qPCR analysis. Methods Mol. Biol. 1160 19–26. 10.1007/978-1-4939-0733-5_3 [DOI] [PubMed] [Google Scholar]
- Hsiao L. L., Dangond F., Yoshida T., Hong R., Jensen R. V., Misra J., et al. (2001). A compendium of gene expression in normal human tissues. Physiol. Genomics 7 97–104. 10.1152/physiolgenomics.00040.2001 [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6 279–284. 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- Isaac R. E., Ekbote U., Coates D., Shirras A. D. (1999). Insect angiotensin-converting enzyme: a processing enzyme with broad substrate specificity and a role in reproduction. Ann. N. Y. Acad. Sci. 897 342–347. 10.1111/j.1749-6632.1999.tb07904.x [DOI] [PubMed] [Google Scholar]
- Isaac R. E., Lamango N. S., Ekbote U., Taylor C. A., Hurst D., Weaver R. J., et al. (2007). Angiotensin-converting enzyme as a target for the development of novel insect growth regulators. Peptides 28 153–162. 10.1016/j.peptides.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Jonge H. J. M. D., Fehrmann R. S. N., Bont E. S. J. M., Hofstra R. M. W., Gerbens F., Kamps W. A., et al. (2007). Evidence based selection of housekeeping genes. PLoS One 2:e898. 10.1371/journal.pone.0000898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keertan D., Huggett J. F., Bustin S. A., Johnson M. A., Graham R., Alimuddin Z. (2004). Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37 118–119. 10.1016/j.biosystems.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Kim H. G., Lee D. H. (2018). Review of the biology and ecology of a ragweed leaf beetle, Ophraella communa (Coleoptera: Chrysomelidae), which is a biological control agent of an invasive common ragweed, Ambrosia artemisiifolia (Asterales: Asteraceae). Biocontrol. Sci. Techn. 29 185–200. 10.1080/09583157.2018.1540032 [DOI] [Google Scholar]
- LeSage L. (2012). A taxonomic monograph of the nearctic galerucine genus Ophraella Wilcox (Coleoptera: Chrysomelidae). Mem. Entomol. Soc. Can 118 3–75. 10.4039/entm118133fv [DOI] [Google Scholar]
- Liu J., Hiser C., Ferguson-Miller S. (2017). Role of conformational change and K-path ligands in controlling cytochrome coxidase activity. Biochem. Soc. Trans. 45:BST20160138. 10.1042/BST20160138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J. C., Hartzer K., Toutges M., Oppert B. (2010). Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 80 219–221. 10.1016/j.mimet.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Lourenço A. P., Mackert A., dos Santos Cristino A., Simões Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee. Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39 372–385. 10.1051/apido:2008015 [DOI] [Google Scholar]
- Lu J., Chen S., Guo M., Ye C., Qiu B., Wu J., et al. (2018a). Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9:1614. 10.3389/fphys.2018.01614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Yang C., Zhang Y., Pan H. (2018b). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560 10.3389/fphys.2018.01560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8:e68059. 10.1371/journal.pone.0068059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Ma C., Li Z., Zhu B. Q., Zhang J., Lei C. L., et al. (2018). Assessment of suitable reference genes for qRT-PCR analysis in Adelphocoris suturalis. J. Integr. Agr. 17 2745–2757. 10.1016/s2095-3119(18)61926-4 [DOI] [Google Scholar]
- Ma C., Cui S., Tian Z., Zhang Y., Chen G., Gao X., et al. (2019a). OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: Chrysomelidae). Front. Physiol. 10:1290 10.3389/fphys.2019.01290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Zhao C., Cui S., Zhang Y., Chen G., Chen H., et al. (2019b). Identification of candidate chemosensory genes of Ophraella communa LeSage (Coleoptera: Chrysomelidae) based on antennal transcriptome analysis. Sci. Rep. 9 1–14. 10.1038/s41598-019-52149-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K. S., Li F., Liang P. Z., Chen X. W., Liu Y., Gao X. W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect. Sci. 16:17. 10.1093/jisesa/iew003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macours N., Biol K. H. J. I. B. M. (2004). Zinc-metalloproteases in insects: ACE and ECE. Insect Biochem. Molec. 34 501–510. 10.1016/j.ibmb.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Michael W., Pfaffl A. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Yang X., Siegfried B. D., Zhou X. (2015). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10:e125868. 10.1371/journal.pone.0125868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-Based tool using pair-wise correlations. Biotechnol. Lett. 26 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Popovici V., Goldstein D. R., Antonov J., Jaggi R., Delorenzi M., Wirapati P. (2009). Selecting control genes for RT-qPCR using public microarray data. BMC Bioinformatics 10:42. 10.1186/1471-2105-10-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu R., Miao Y., Cui Y., Cao Y., Zhou Y., Tang X., et al. (2019). Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Mol. Biol. 20:9. 10.1186/s12867-019-0126-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues T. B., Khajuria C., Wang H., Matz N., Cunha Cardoso D., Valicente F. H., et al. (2014). Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS One 9:e109825. 10.1371/journal.pone.0109825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylett C. M., Walker M. J., Howell G. J., Shirras A. D., Isaac R. E. (2007). Male accessory glands of Drosophila melanogaster make a secreted angiotensin I-converting enzyme (ANCE), suggesting a role for the peptide-processing enzyme in seminal fluid. J. Exp. Biol. 210 3601–3606. 10.1242/jeb.009035 [DOI] [PubMed] [Google Scholar]
- Sang W., He L., Wang X. P., Zhu-Salzman K., Lei C. L. (2015). Evaluation of reference genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) under UVB stress. Environ. Entomol. 44 418–425. 10.1093/ee/nvv010 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shakeel M., Rodriguez A., Tahir U. B., Jin F. (2018). Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 40 227–236. 10.1007/s10529-017-2465-4 [DOI] [PubMed] [Google Scholar]
- Shi X. Q., Guo W. C., Wan P. J., Zhou L. T., Ren X. L., Ahmat T., et al. (2013). Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say). BMC Res. Notes 6:93. 10.1186/1756-0500-6-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Zhang J., Cui G., Sun R., Sethuraman V., Yi X., et al. (2018). Evaluation of reference genes for real-time quantitative pcr analysis in larvae of Spodoptera litura exposed to azadirachtin stress conditions. Front. Physiol. 9:372 10.3389/fphys.2018.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q. Q., Zhu L., Li Y., Liu W., Ma W. H., Lei C. L., et al. (2015). A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS One 10:e0118693. 10.1371/journal.pone.0118693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Zhou X.-R., Pang B.-P. (2017). Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). B Entomol. Res. 107 359–368. 10.1017/s0007485316000948 [DOI] [PubMed] [Google Scholar]
- Thellin O., Zorzi W., Lakaye B., De Borman B., Coumans B., Hennen G., et al. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75 291–295. 10.1016/S0168-1656(99)00163-7 [DOI] [PubMed] [Google Scholar]
- Tong Z., Gao Z., Wang F., Zhou J., Zhang Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10:1–13. 10.1186/1471-2199-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toutges M. J., Hartzer K., Lord J., Oppert B. (2010). Evaluation of reference genes for quantitative polymerase chain reaction across life cycle stages and tissue types of Tribolium castaneum. J. Agric. Food Chem. 58 8948–8951. 10.1021/jf101603j [DOI] [PubMed] [Google Scholar]
- Valasek M. A., Repa J. J. (2005). The power of real-time PCR. Adv. Physiol. Educ. 29 151–159. 10.1152/advan.00019.2005 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:7. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80 75–84. 10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- Xu J., Baulding J., Palli S. R. (2013). Proteomics of Tribolium castaneum seminal fluid proteins: identification of an angiotensin-converting enzyme as a key player in regulation of reproduction. J. Proteomics 78 83–93. 10.1016/j.jprot.2012.11.011 [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Imai C., Natuhara Y. (2000). Rapid population growth and food-plant exploitation pattern in an exotic leaf beetle, Ophraella communa LeSage (Coleoptera: Chrysomelidae), in western Japan. Appl. Entomol. Zool. 35 215–223. 10.1303/aez.2000.215 [DOI] [Google Scholar]
- Yan Z., Gao J., Lv X., Yang W., Wen S., Tong H., et al. (2016). Quantitative evaluation and selection of reference genes for quantitative RT-PCR in mouse Acute pancreatitis. Biomed. Res. Int. 2016:8367063. 10.1155/2016/8367063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Pan H., Liu Y., Zhou X. (2015a). Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite. Tetranychus Urticae 10:1. 10.1371/journal.pone.0120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Pan H., Liu Y., Zhou X. (2015b). Temperature and development impacts on housekeeping gene expression in Cowpea Aphid. Aphis craccivora (Hemiptera: Aphidiae). PLoS One 10:e0130593. 10.1371/journal.pone.0130593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Preisser E. L., Zhang H., Liu Y., Dai L., Pan H., et al. (2016). Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 7:1672. 10.3389/fpls.2016.01835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pan H., Yuan L., Zhou X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. 10.1038/s41598-018-20612-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Ma F., Chen H., Wan F., Guo J., Zhou Z. (2018). Heritability and evolutionary potential drive cold hardiness in the overwintering Ophraella communa beetles. Front. Physiol. 9:666 10.3389/fphys.2018.00666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Wang X., Wen J., Cai J., Wu C., Aly S. M. (2013). Selection of reference genes for quantitative gene expression studies in the house fly (Musca domestica L.) using reverse transcription quantitative real-time PCR. Acta Biochem. Biophys. Sin. 45 1069–1073. 10.1093/abbs/gmt111 [DOI] [PubMed] [Google Scholar]
- Zhou L., Chen F., Ye J., Pan H. (2018). Selection of reliable reference genes for RT-qPCR analysis of Bursaphelenchus mucronatus gene expression from different habitats and developmental stages. Front. Genet. 9:269 10.3389/fgene.2018.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. S., Chen H. S., Zheng X. W., Guo J. Y., Wei G., Min L., et al. (2014). Control of the invasive weed Ambrosia artemisiifolia with Ophraella communa and Epiblema strenuana. Biocontrol. Sci. Techn. 24 950–964. 10.1080/09583157.2014.897305 [DOI] [Google Scholar]
- Zhou Z. S., Guo J. Y., Chen H. S., Wan F. H. (2010a). Effects of temperature on survival, development, longevity, and fecundity of Ophraella communa (Coleoptera: Chrysomelidae), a potential biological control agent against Ambrosia artemisiifolia (Asterales: Asteraceae). Environ. Entomol. 39 1021–1027. 10.1603/EN09176 [DOI] [PubMed] [Google Scholar]
- Zhou Z. S., Guo J. Y., Michaud J. P., Li M., Wan F.-H. (2010b). Variation in cold hardiness among geographic populations of the ragweed beetle, Ophraella communa LeSage (Coleoptera: Chrysomelidae), a biological control agent of Ambrosia artemisiifolia L. (Asterales: Asteraceae), in China. Biol. Invas. 13 659–667. 10.1007/s10530-010-9857-x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI using accession MN641110, MN641111, MN641112, MN641113, MN641114, MN641115, MN641116, MN641117, MN641118, MN641119, MN641121, MN641120, and MN641122.