Abstract
Genetic diversity is of great importance and a prerequisite for genetic improvement and conservation programs in pigs and other livestock populations. The present study provides a genome wide analysis of the genetic variability and population structure of pig populations from different production systems in South Africa relative to global populations. A total of 234 pigs sampled in South Africa and consisting of village (n = 91), commercial (n = 60), indigenous (n = 40), Asian (n = 5) and wild (n = 38) populations were genotyped using Porcine SNP60K BeadChip. In addition, 389 genotypes representing village and commercial pigs from America, Europe, and Asia were accessed from a previous study and used to compare population clustering and relationships of South African pigs with global populations. Moderate heterozygosity levels, ranging from 0.204 for Warthogs to 0.371 for village pigs sampled from Capricorn municipality in Eastern Cape province of South Africa were observed. Principal Component Analysis of the South African pigs resulted in four distinct clusters of (i) Duroc; (ii) Vietnamese; (iii) Bush pig and Warthog and (iv) a cluster with the rest of the commercial (SA Large White and Landrace), village, Wild Boar and indigenous breeds of Koelbroek and Windsnyer. The clustering demonstrated alignment with genetic similarities, geographic location and production systems. The PCA with the global populations also resulted in four clusters that where populated with (i) all the village populations, wild boars, SA indigenous and the large white and landraces; (ii) Durocs (iii) Chinese and Vietnamese pigs and (iv) Warthog and Bush pig. K = 10 (The number of population units) was the most probable ADMIXTURE based clustering, which grouped animals according to their populations with the exception of the village pigs that showed presence of admixture. AMOVA reported 19.92%–98.62% of the genetic variation to be within populations. Sub structuring was observed between South African commercial populations as well as between Indigenous and commercial breeds. Population pairwise FST analysis showed genetic differentiation (P ≤ 0.05) between the village, commercial and wild populations. A per marker per population pairwise FST analysis revealed SNPs associated with QTLs for traits such as meat quality, cytoskeletal and muscle development, glucose metabolism processes and growth factors between both domestic populations as well as between wild and domestic breeds. Overall, the study provided a baseline understanding of porcine diversity and an important foundation for porcine genomics of South African populations.
Keywords: pigs, diversity, population structure, genetic characterization, SNP60K
Introduction
Pigs were domesticated over 5,000 years ago, leading to the gradual and cumulative development of modern pig breeds with very distinctive phenotypes and production abilities (Zeder et al., 2006; Rothschild and Ruvinsky, 2010). Domesticated pig (Sus Scrofa domesticus) originated from the Sus scrofa, which is commonly known as the wild boar belonging to the Suidae family (Jones, 1998). This family includes species of wild pigs such as Phacochoerus africanus (Common warthog), Potamochoerus larvatus (Bush pig) and Hylochoerus meinertzhageni (Giant Forest hog) some that are indigenous to Africa (Jones, 1998). The Wild Boars are widely distributed covering areas such as Europe, Asia, and North Africa and were introduced as game species in all other continents including Africa (Jones, 1998; Scandura et al., 2011).
Pig breeds worldwide are either of well-defined ancestry or in certain instances crossbreds from populations of diverse origins (Amills et al., 2010). South African pig production consists of a commercial intensive sector with defined breeds and an extensive sector that is mainly associated with small-scale farmers in the rural areas. Village production system is characterized by non-descript populations raised under extensive low-input management. Commercial breeds such as the Large White, Landrace and Duroc have worldwide distribution in modern commercial farming systems including South Africa and are widely used (Amills et al., 2010). Indigenous breeds classified under Sus indica such as Kolbroek and Windsnyer are geographically restricted to Southern Africa (Nicholas, 1999). The Kolbroek, which is of Chinese origin, is speculated to have pigs that ended up in the hands of South African farmers when a sailing ship wrecked at the Cape Hangklip (Ramsay et al., 1994). Although the origin of the Windsnyer is unknown, there are observed similarities to Chinese breeds (Nicholas, 1999) thereby suggesting that it is of Chinese origin. Regardless of their origins and domestication routes, pig breeds in South Africa have become closed genetic pools restricted to specific farming systems and molded by artificial selection and possibly genetic drift (Amills et al., 2010). In addition to these domesticated breeds are the Warthog, Bush pig and Red River Hog wild pigs that are native to Africa and are found roaming in forests or in the zoos (Porter, 1993). The common Warthog (Phacochoerus Africanus) which was first discovered at Cape Verde, Senegal is one of the three species found in Africa. The Cape Warthog (Phacochoerus aethiopicus) is now extinct due to the rinderpest epizootic of the 1860s (Pallas, 1766; Gmelin, 1788; D’Huart and Grubb, 2003). Another Warthog (Phacochoerus delamerei) species was described in Somalia and later renamed Phacochoerus aethiopicus delamerei as it is similar to the Cape Warthog (Lönnberg, 1908, 1912; Roosenvelt and Heller, 1915). Muwanika et al. (2003) studied the phylogeography of the common Warthog in Africa and found three clades representing West, South and East African Warthogs. There is no enough evidence to support the origin of the Bush pig, which was assumed to have originated from Asia (White and Harris, 1977). There are recordings of the Bush pig in the Swellendam and Outeniqualand in the Western Cape provinces of South Africa (Rookmaaker, 1989). Hybrids between the domestic and Bush pigs have been recorded with the introduction of Bush pigs to South Africa being as far as 1400 years ago (Linnaeus, 1758; Mujibi et al., 2018). The existence of hybrids is a concern, as they could become asymptomatic carriers of diseases such African swine fever (Jori and Bastos, 2009).
Indigenous breeds are often geographically restricted and harbor unique genetic variants that may provide future breeds with the flexibility to change in response to product market preferences and production environments. While low-input and indigenous breeds may not compete with exotic breeds in terms of production performance, they are considered hosts to unique genetic diversity that should be protected as sources of variation. Local pigs are important because of their hardiness and ability to survive in extreme conditions (Taverner and Dunkin, 1996; Zadik, 2005). Most indigenous breeds are, however, threatened by small and fragmented flock sizes, which predispose them to lose genetic diversity as a result of genetic drift and indiscriminate crossbreeding with exotic germplasm that can lead to genetic erosion and the eradication of the local genetic pool. Globally, 35% of pig breeds are classified as at risk or already extinct (FAO, 2009) demonstrating the threat to local biodiversity.
Genomics have emerged as an effective tool for assessing diversity within and amongst populations. Swart et al. (2010) observed low differentiation among pig populations in Southern Africa using microsatellites. Heterozygosity levels ranged from 0.531 to 0.692 for commercial and indigenous breeds. The availability of the Porcine SNP60K BeadChip has opened new avenues of examining genetic diversity (Ramos et al., 2009) at a genome wide scale relative to that using microsatellite and other low-coverage markers. Mujibi et al. (2018) observed close clustering of Warthogs and Bush pigs using the Porcine SNP60K BeadChip. The Porcine SNP60K BeadChip has been used to infer on population structure and selection signatures in Chinese and European pig populations (Ai et al., 2013). Using this SNP panel in South African pig populations will provide comprehensive information on the genomic architecture of local, exotic and wild pig populations, which will guide future management and conservation. The objective of the present study was to provide a large-scale analysis of the genetic diversity and structure of South African local pig populations using the Porcine SNP 60K BeadChip. The study investigated diversity of South African pigs relative to global populations of 389 pigs consisting of villages and out-group pigs from South America, Europe, United States, and China amongst other countries.
Materials and Methods
Breeds/Populations Sampled
South African specimens were collected from a total of 234 samples from different production systems, representing village, intensively farmed populations in conservation units and free ranging populations. Village and non-descript pig populations were sampled from Alfred Nzo (ALN; n = 17) and Oliver Reginald Tambo (ORT; n = 22) districts in Eastern Cape province and Mopani (MOP; n = 27) and Capricorn (CAP; n = 25) districts in Limpopo province. Commercial pig breeds of Large White (LWT; n = 20), South African Landrace (SAL; n = 20) and Duroc (DUR; n = 20) were sampled from commercial farmers in Limpopo province. Indigenous populations Kolbroek (KOL; n = 20.) and Windsnyer (WIN; n = 20) were sampled from the Agricultural Research Council-Animal Production Institute in Pretoria, South Africa (Table 1). Vietnamese Potbelly breed (VIT; n = 5) was sampled from the Johannesburg Zoo and represents a breed that is endangered in Vietnam, its country of origin but has been raised in a conservation zoo in South Africa. European Wild Boar (n = 4), Warthogs (n = 31), and Bush pigs (n = 3) were sampled as representatives of the wild pig populations. The European Wild Boar and Bush pigs were sampled from the surrounding villages in the North-West whilst the Warthog samples were collected from geographically separated National Parks from North-West (n = 4), Eastern Cape (n = 3), and Limpopo (n = 24). The distribution of the sampled individuals is illustrated in Figure 1. Ear tissue samples were collected using the tissue sampling applicator gun while pliers were used to collect the hair samples according to standard procedures and ethical approval from ARC-Irene Animal Ethics committee (APIEC16/028).
TABLE 1.
Population category and sample size of the 13 pig populations.
| Category | Population | Code | N |
| Village | Mopani | MOP | 27 |
| Village | Capricorn | CAP | 25 |
| Village | Oliver Reginald Tambo | ORT | 22 |
| Village | Alfred Nzo | ALN | 17 |
| Commercial | Large White | LWT | 20 |
| Commercial | SA Landrace | SAL | 20 |
| Commercial | Duroc | DUR | 20 |
| Indigenous | Kolbroek | KOL | 20 |
| Indigenous | Windsnyer Type | WIN | 20 |
| Asian | Vietnamese Potbelly | VIT | 5 |
| Wild | Wild Boar | WBO | 4 |
| Wild | Warthog | WAT | 31 |
| Wild | Bush Pig | BSP | 3 |
FIGURE 1.
Map showing geographic locations of the 13 pig populations in the present study.
Genotyping and Quality Control
DNA was extracted at the Agricultural Research Council-Biotechnology Platform from the ear tissue and hair samples using a commercially available Perkin Elmer Genomic DNA kit according to the manufacturer’s protocol. DNA concentration was quantified using the Qubit® 2.0 Fluorometer. Gel electrophoresis (5%) was used to assess the quality and integrity of the DNA.
All 234 animals were genotyped using PorcineSNP60 v2 genotyping BeadChip (Illumina, United States) containing 62,163 SNPs with an average gap of 43.4 kb. Genotyping was done using the standard infinium assay at the ARC-Biotechnology Platform in South Africa. GenomeStudio version 2.0 (Illumina, United States) was used to process the genotype data, including raw data normalization, clustering and genotype calling. A final custom report was created to be able to generate a Plink Ped (Pedigree file) and Map (SNP panel file) for use in downstream analysis.
Golden Helix SNP Variation Suite (SVS) version 8.5 was used to update the SNPs marker file (Golden Helix Inc., 2016) based on the pig genome assembly (Sus Scrofa v10.2). Markers were then filtered to exclude SNPs located on the sex chromosomes. From this data set, Minor allele frequency (MAF) and deviation from Hardy–Weinberg equilibrium (HWE) were estimated per population for the 10 populations that excluded BSP, VIT, and WBO, which were left out due to small sample sizes. Additional quality control (QC) was also performed per population to remove SNPs with less than 85% call rate, MAF < 0.02 and HWE < 0.0001. The resultant filtered dataset was used to calculate observed (HO), and expected (HE) heterozygosities, inbreeding (FIS) and effective population size (Ne).
Quality control was then performed overall population to remove SNPs with less than 85% call rate, MAF < 0.02 and HWE < 0.0001 and generate a dataset used for analysis of molecular variance (AMOVA) and FST analysis. Using this dataset, further QC filtered for SNPs in high LD (r2 = 0.2) and closely related individual [Identity By Descent (IBD) ≥ 0.45] to produce a filtered dataset used for population structure analysis using ADMIXTURE and Principle Component Analysis (PCA).
Genetic Diversity Within Population
The MAF, HE and HO were calculated as measures of within population genetic variation using PLINK 1.07 (Purcell et al., 2007). In addition, inbreeding coefficient (FIS) was calculated on Golden Helix SNP Variation Suite (SVS) version 8.5 (Golden Helix Inc., 2016). Effective population size (Ne) trends across generations were estimated based on a relationship between r2 (expected LD), Ne and C (recombination rate). SNeP software (Version 1.1) tool was used based on the following formula suggested by Corbin et al. (2012) using the equation:
where:
NT(t): Effective population size estimated t generations ago
Ct: Recombination rate t generations ago
r2adj: Linkage disequilibrium estimation adjusted for sampling biasness
α: a constant.
The recombination rate was estimated by using the following formula proposed by Sved (1971):
The Bush pig, Vietnamese Potbelly and Wild Boar were excluded from the diversity within population analysis due to their small sample sizes. The few available samples were sampled from zoos and game reserves in the country where only few animals are often rescued and kept in conservation.
Population Differentiation and Structure
Analysis of Molecular Variance (AMOVA) was used to determine the genetic variance within populations (FIS), among populations within group (FSC) and among groups (FCT) using ARLEQUIN v3.5 (Excoffier et al., 2005). The populations were categorized into villages, commercial, indigenous and wild populations and consisted of animals sampled in South Africa as well global populations from Burgos-Paz et al. (2013) which consisted of 389 genotypes of villages and out-group pigs from 24 countries of America (United States), South America (Mexico, Cuba, Guadeloupe, Guatemala, Costa Rica, Columbia, Ecuador, Peru, Brazil, Bolivia, Paraguay, Argentina, and Uruguay), Europe (Spain, Portugal, Italy, Poland, Hungary, Tunisia, Denmark, Holland, United Kingdom) and China. Variance components were also estimated for groups consisting of different categories, i.e., village and indigenous; indigenous and commercial; South African village and global villages; South African commercial and global commercial etc.
Principal Component Analysis (PCA) using SVS version 8.5 (Golden Helix Inc., 2016) and the eigenvector method was used to determine population clustering. ADMIXTURE version 1.20 (Alexander and Lange, 2011) was used to detect the most likely clusters (K) for the population. ADMIXTURE was run from K = 2 to K = 15. The number of potential genetic clusters (K) was tested from 1–15 to reassign each sample to its population of origin. The optimum K-value was that with the lowest cross-validation error value. Initially, all the 13 populations sampled from South Africa were included in the population structure analysis. After this the South African data set was merged to Porcine SNP60K genotype data from Burgos-Paz et al. (2013) described above.
Population pairwise FST values were estimated according to the formula of Weir and Cockerham (1984) implemented in the Golden Helix SNP Variation Suite (SVS) version 8.5 (Golden Helix Inc., 2016). Based on population pairwise FST values, PCA and ADMIXTURE based clustering, FST analysis per marker was estimated between pairs of highly differentiated populations of the village populations, indigenous populations and commercial breeds as well as amongst highly differentiated commercial breeds and wild populations. To reduce noise, an FST averaged smooth value was used to identify genomic regions differentiating pairs of populations. Manhattan plots of per marker FST values between pairs of populations were plotted against chromosomal coordinates using the porcine assembly (Sus Scrofa 10.2). Highly differentiating SNPs (FST ≥ 0.8) were subsampled and genes associated with these SNPs searched using genome browse including their associations with known QTLs in the pig genome based on the Sus Scrofa 10.2 on Ensembl1.
Results
Genotypes and Quality Control
The percentage of polymorphic and number of SNPs (NSNP) remaining after QC per population and overall is presented in Table 2. Two hundred and eleven individuals with a genotyping rate of 85% remained after QC. Windsnyer pigs had the highest percentage of informative markers (95%) after QC, whilst Warthog had the lowest at 82%. About 31,705 SNPs were removed leaving 30,458 polymorphic SNPs of the loci distributed over 18 autosomal chromosomes, which were used for AMOVA and FST analysis. After LD and IBD pruning, 23,345 SNPs and 176 individuals were used for the population structure analysis.
TABLE 2.
Summary of the genetic diversity measures across South African Pig populations.
| POP | N | %SNP | MAF ± SD | NSNP | HO ± SD | HE ± SD | FIS ± SD | P-value |
| MOP | 27 | 92 | 0.262 ± 0.149 | 52,925 | 0.299 ± 0.129 | 0.369 ± 0.131 | 0.198 ± 0.134 | 0.495 |
| CAP | 24 | 94 | 0.264 ± 0.147 | 54,078 | 0.332 ± 0.140 | 0.371 ± 0.126 | 0.117 ± 0.155 | 0.582 |
| ORT | 22 | 93 | 0.259 ± 0.153 | 52,238 | 0.315 ± 0.145 | 0.370 ± 0.130 | 0.163 ± 0.113 | 0.553 |
| ALN | 15 | 94 | 0.238 ± 0.157 | 53,580 | 0.336 ± 0.160 | 0.359 ± 0.134 | 0.056 ± 0.168 | 0.695 |
| LWT | 18 | 93 | 0.227 ± 0.161 | 49,773 | 0.358 ± 0.177 | 0.348 ± 0.144 | 0.023 ± 0.009 | 0.721 |
| SAL | 19 | 94 | 0.221 ± 0.162 | 49,191 | 0.372 ± 0.186 | 0.345 ± 0.144 | 0.052 ± 0.085 | 0.704 |
| DUR | 19 | 94 | 0.177 ± 0.168 | 40,632 | 0.359 ± 0.182 | 0.337 ± 0.147 | 0.067 ± 0.153 | 0.764 |
| KOL | 20 | 94 | 0.173 ± 0.167 | 39,560 | 0.364 ± 0.182 | 0.339 ± 0.144 | 0.051 ± 0.087 | 0.727 |
| WIN | 19 | 95 | 0.220 ± 0.164 | 47,402 | 0.385 ± 0.171 | 0.360 ± 0.134 | 0.056 ± 0.158 | 0.733 |
| WAT | 28 | 82 | 0.076 ± 0.109 | 3,967 | 0.188 ± 0.155 | 0.204 ± 0.151 | 0.398 ± 0.475 | 0.710 |
%SNP used to calculate MAF analysis; NSNP, the number of SNPs in the subset 62,163 SNP; HO, observed heterozygosity; HE, expected heterozygosity; SD, standard deviation; FIS, inbreeding co-efficient; MAF, minor allele frequency, P < 0.05.
Genetic Diversity Across Populations
Genetic diversity parameters among the 10 populations are summarized in Table 2. Warthog pigs had the lowest HO (0.188 ± 0.155) and Windsnyer the highest (0.385 ± 0.171). Expected heterozygosity values ranged from 0.204 ± 0.151 from Warthog to 0.371 ± 0.126 for Capricorn. The highest inbreeding coefficient (FIS) was for Warthog at 0.398 ± 0.475 while the Duroc had the lowest and slightly negative value of −0.067 ± 0.153. FIS values were positive for all village populations as well as Warthog suggesting some level of inbreeding within these populations. MAF was the highest in village population from Capricorn (0.264 ± 0.147) and the least in Warthog pigs (0.076 ± 0.109).
Effective Population Size
Figure 2 shows trends in effective population size across all of the studied populations. The Warthog was excluded in this analysis because the number of polymorphic SNPs was not enough to generate results. Effective population size values are presented in Supplementary Table S1. There was a general decline in Ne across all the populations across generations. The indigenous and commercial populations had higher effective population size compared to the village populations. The Kolbroek had the lowest effective population size 12 generations prior.
FIGURE 2.
Average effective population size plotted against generation in the past.
AMOVA
Genetic differentiation between populations is presented in Supplementary Table S2. The major proportion of the genetic variance was attributed to variation within South African populations with FIS values ranging from 76.41 to 98.62%. Diversity within populations (FIS) in village populations from this study and those from Burgos-Paz et al. (2013) was 35.52% while variation among groups (FCT) was 62.35%. Diversity of South African commercial pigs was 76.41% within populations, 18.17% among populations within group and 5.42% among groups. When including the commercial breeds from Burgos-Paz et al. (2013), the diversity parameters changed to FIS = 30.97%, FSC = 8.31% and FCT = 60.72%. High FCT (>60%) were observed in the category consisting of South African indigenous and Chinese indigenous (FCT = 70.08%) as well as that consisting of the South African Wild Boar and the worldwide Wild Boar (FCT = 73.58%).
Population Structure
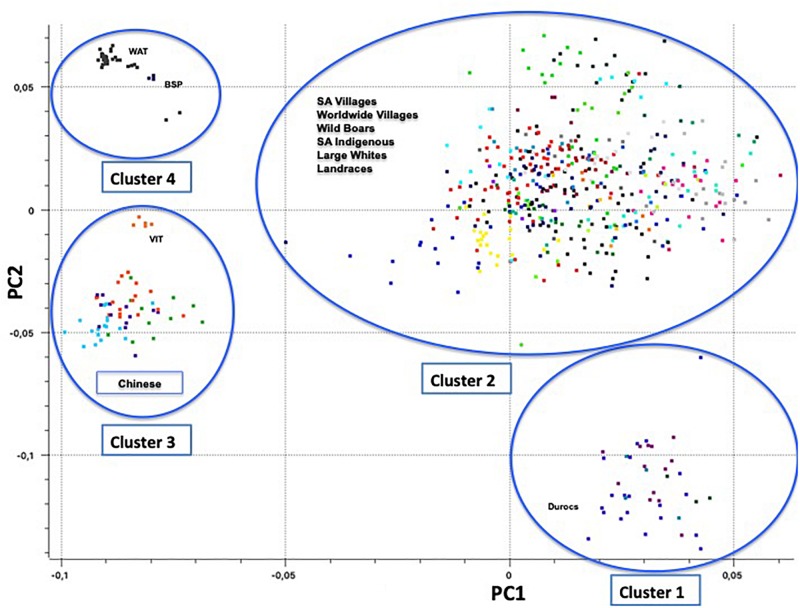
Principal component one (PC1) and principal component two (PC2) explained approximately 30.7% and 11.8% of the total variation, respectively. The PCA of South African breeds yielded four main genetic clusters (Figure 3). The Duroc clearly separated from the Large White and South African Landrace that clustered together with the wild boar and village populations. The Warthog and the Bush pig clustered together as a third cluster whilst the fourth cluster consisted of Vietnamese potbelly sampled from the zoo. The PCA analysis using South African samples and those from Burgos-Paz et al. (2013) demonstrated the same clustering with all the village pigs grouping together with the Large White and Landraces separated from clusters of (i) Warthog and Bush pig, (ii) Chinese and Vietnamese breeds and (iii) Duroc (Figure 4).
FIGURE 3.
Principal Component Analysis based population clustering.
FIGURE 4.
Principal Component Analysis based population clustering including Burgos-Paz et al. (2013) genotypes (22,430 SNPs).
Genetic structure of the South African breeds was further investigated using ADMIXTURE. The results presented in Figure 5 show the Warthog and Bush pig populations clustering together and clearly separated from the rest of the other populations at K = 2. Duroc separated from the rest of the populations at K = 3 followed by Vietnamese at K = 4. K = 4 clustered animals in the same way observed with PCA based clustering. Beyond K = 8, the genetic clusters of the commercial, indigenous, Asian and wild breeds are maintained whilst the added K is distributed within the village populations. K = 10 which was the optimal K (Supplementary Figure S1) with lowest CV (0.551) resulted in the eight distinct genetic clusters of commercial, indigenous, Asian and wild breeds plus highly admixed clusters consisting of all village pig populations from Limpopo and Eastern Cape provinces of South Africa.
FIGURE 5.
ADMIXTURE based clustering K = 2 – K = 10. Each individual is represented by a single column divided into K colored segments, where K is the number of clusters assumed with lengths proportional to each of the K inferred cluster.
Population Differentiation
Population pairwise FST values are shown in Table 3. Low FST were observed between village populations with values ranging from 0.022–0.060 (P < 0.05) within South Africa and in global populations. The highest differentiation was found between Warthog and Duroc at FST = 0.481. Warthog and Kolbroek pigs showed the high differentiation at 0.468. All other populations had FST values above 0.282. The extent of differentiation between Warthog and all the other populations was high ranging from 0.312 (Warthog and Creole from Columbia) to 0.589 (Warthog and Vietnamese). Highest FST observed was between Vietnamese and Bush pig populations at 0.700 (Supplementary Table S3).
TABLE 3.
Pairwise genetic differentiation (FST values) between 10 pig populations.
| MOP | CAP | ORT | ALN | LWT | DUR | SAL | KOL | WIN | WAT | ||
| Villages | MOP | ||||||||||
| CAP | 0.022* | ||||||||||
| ORT | 0.031* | 0.026* | |||||||||
| ALN | 0.059 | 0.060 | 0.040* | ||||||||
| Commercial | LWT | 0.091 | 0.073 | 0.096 | 0.130 | ||||||
| DUR | 0.134 | 0.126 | 0.143 | 0.174 | 0.183 | ||||||
| SAL | 0.094 | 0.073 | 0.099 | 0.132 | 0.120 | 0.194 | |||||
| Indigenous | KOL | 0.120 | 0.116 | 0.129 | 0.162 | 0.189 | 0.237 | 0.194 | |||
| WIN | 0.061 | 0.064 | 0.077 | 0.106 | 0.143 | 0.189 | 0.144 | 0.173 | |||
| Wild | WAT | 0.282 | 0.306 | 0.314 | 0.350 | 0.433 | 0.481 | 0.435 | 0.468 | 0.410 |
Significant levels: *P < 0.05.
Per Marker Pairwise FST, SNP Annotation and Association With Porcine QTLs
Per population, per marker pairwise FST values were computed for highly differentiated populations and are illustrated in Table 4, Supplementary Figure S2. SNPs. High FST values (≥0.8) where considered breed differentiating and the associated SNPs were functionally annotated for genes within a 1 MB region. Fixed SNPs (FST = 1.0) where observed on chromosome 9 between Duroc and Warthog, on chromosome 12 between Koelbroek and Warthog and on chromosome 18 between Windsnyer and Warthog. For all the pairwise comparisons, 281 SNPs (FST ≥ 0.8) were detected (Supplementary Figure S2) with only 123 candidate genes within 1 MB of those SNPs. Pairwise comparison of village pigs from Alfred NZO, South Africa and Warthog yielded genes related to acute heat stress (RPL18) and inflammatory response (IL17B and ARHGAP23) as illustrated in Table 4 and Supplementary Figure S2a. Gene ADGRB3 was in close proximity of SNPs rs81353971, rs81353988, rs81353991, rs81297001, and rs81333295 that were of significant between Duroc and Warthog. Inflammatory response genes such as ARHGAP23 were associated with the significant SNPs observed between Koelbroek, Large White and Windsnyer populations. For reproduction traits, genes CD28, TCP11L2, TLK1, ATPB2, GPR137C, ZNF609, ARHGAP22, EPSTI1, GPR63, TCTE3, PTP4A2, ZSCAN20, CLU, and CACNA2D3 were observed within 14 significant SNPs on chromosomes 1, 2, 5, 6, 11, 14, and 15. Genes that had association with meat traits such as DLX1, BRPF1, CLPTM1, FANCD2, SEC13, FHL3, FSTL5, CEP135, EXOC1, FOXO1, ASTN2, MYO18B, PLXNA1, DNAH2, HECTD2, TMEM39B, TXLNA, CSMD2, COL16A1, SCARA3, ZFAND3, and PTPRD were also reported. Comparison with indigenous pigs showed genes that were associated with mastitis resistance (ARHGAP39, ARPC4, PHC2, and BCL2L15) and hair follicle development (FOXN1). A total of eight SNPs associated with growth traits (ADGRB3, TSPAN, and ZFAND3) were detected. PTPN3 gene associated with immune response was observed between indigenous and Wild Boar. Wild Boar and Duroc comparison resulted in genes associated with adaptation (HDAC1 and GNAI3).
TABLE 4.
Most significant SNPs detected with FST analysis and the associated genes.
| Population | SNP | Chr | Position | Genes | Function |
| ALN and WAT | rs81355030 | 1 | 84,376,735 | RPL18 | Acute heat stress (Newton et al., 2012) |
| rs81367521 | 2 | 150,546,025 | IL17B | Embryonic development, tissue regeneration and inflammation (Bie et al., 2017) | |
| rs81285672 | 12 | 23,638,629 | ARHGAP23 | Inflammatory response (Liu, 2015) | |
| DUR and | rs81353971 | 1 | 49,024,494 | ADGRB3 | Growth traits (Emrani et al., 2017) |
| WAT | rs81353988 | 1 | 49,350,539 | ADGRB3 | |
| rs81353991 | 1 | 49,392,902 | ADGRB3 | ||
| rs81297001 | 1 | 49,458,254 | ADGRB3 | ||
| rs81333295 | 1 | 49,592,586 | ADGRB3 | ||
| rs80946298 | 13 | 33,531,504 | DOCK3 | Induces axonal growth (Kimura et al., 2016) | |
| rs81444796 | 13 | 33,481,604 | DOCK3 | ||
| rs81478683 | 13 | 34,024,632 | IQCF3 | Conjunctival UV to auto fluorescence (Yazar et al., 2015) | |
| rs81478482 | 13 | 34,117,528 | ACY1 | Amino acid and heat shock protein (Martínez-Montemayor et al., 2008) | |
| rs81454214 | 15 | 107,134,695 | CD28 | Endometrial gene expression (Gu et al., 2014) | |
| KOL and WAT | rs81341610 | 3 | 4,508,681 | LOC102160627 | Uncharacterized |
| rs80993200 | 4 | 234,605 | ARHGAP39 | Milk production related and mastitis resistance (Wang et al., 2015) | |
| rs80851822 | 5 | 13,913,761 | POLR3B | Residual feed intake (Gondret et al., 2017) | |
| rs80873063 | 5 | 13,940,475 | TCP11L2 | Regulated in small atretic follicles for healthy follicles (Hatzirodos et al., 2014a) | |
| rs80999600 | 5 | 66,998,856 | TSPAN9 | ADG (Fontanesi et al., 2014) | |
| rs80929588 | 5 | 67,092,749 | TSPAN9 | ||
| rs80883075 | 5 | 67,132,255 | TEAD4 | Regulation in organ size control and cell proliferation (Frankenberg et al., 2016) | |
| rs81385003 | 5 | 67,297,728 | ITFG2 | Disease resistance (Moioli et al., 2016) | |
| rs81285672 | 12 | 23,638,629 | ARHGAP23 | Inflammatory response (Liu, 2015) | |
| rs81325261 | 12 | 44,771,203 | FOXN1 | Regulation of hair follicle development (Song et al., 2017) | |
| rs80801871 | 13 | 33,170,033 | DOCK3 | Induces axonal growth (Kimura et al., 2016) | |
| rs80802886 | 13 | 33,202,454 | DOCK3 | ||
| rs81444784 | 13 | 33,306,071 | DOCK3 | ||
| rs81444796 | 13 | 33,481,604 | DOCK3 | ||
| rs80946298 | 13 | 33,531,504 | DOCK3 | ||
| rs81478683 | 13 | 34,024,632 | IQCF3 | Conjuctival UV to auto fluorescence (Yazar et al., 2015) | |
| rs335091311 | 15 | 148,461 | STAM2 | Residual feed intake (Gondret et al., 2017) | |
| rs80852223 | 15 | 77,232,829 | TLK1 | Decrease expression in the endometrium (Gray et al., 2006) | |
| rs80999734 | 15 | 77,318,065 | TLK1 | ||
| rs81453662 | 15 | 78,190,260 | DLX1 | Muscling and meat availability (Li et al., 2010) | |
| LWT and WAT | rs81349766 | 1 | 182,224,202 | GPR137C | Litter size (Sosa-Madrid et al., 2018) |
| rs81296498 | 1 | 182,722,677 | DDHD1 | Lipid metabolism (Parker Gaddis et al., 2018) | |
| rs81349773 | 1 | 182,756,343 | DDHD1 | ||
| rs332395415 | 1 | 246,195,557 | ABCA1 | Mediates the transport of excess cholesterol (Schwartz et al., 2000) | |
| rs321979518 | 1 | 246,199,966 | ABCA1 | ||
| rs81383185 | 5 | 21,606,108 | RNF41 | Lipid rafts in immune signalling (McGraw and List, 2017) | |
| rs80820161 | 5 | 21,745,636 | STAT2 | Milk production (Salehi et al., 2015) | |
| rs80894897 | 5 | 21,727,701 | PAN2 | Fat yield (Suchocki et al., 2016) | |
| rs80940129 | 5 | 21,970,939 | BAZ2A | Nutrition related (Cornelis and Hu, 2013) | |
| rs325229936 | 5 | 22,338,939 | MYO1A | Coat color and pigmentation (Gutiérrez-Gil et al., 2007) | |
| rs81285672 | 12 | 23,6386,29 | ARHGAP23 | Inflammatory response (Liu, 2015) | |
| rs80854565 | 14 | 89,185,576 | ARHGAP22 | Fertility (Browett et al., 2018) | |
| rs80833618 | 14 | 89,227,581 | ARHGAP22 | ||
| rs80957034 | 14 | 89,255,703 | ARHGAP22 | ||
| rs80962102 | 14 | 89,309,115 | ARHGAP22 | ||
| SAL and WAT | rs81395957 | 6 | 51,328,753 | NECTIN2 | Cell recognition and adhesion (Wang et al., 2010) |
| rs81395929 | 6 | 51,427,663 | CLPTM1 | Marbling score (Lim et al., 2013) | |
| WIN and WAT | rs81381252 | 4 | 65,339 | ZNF609 | Fertility (Hatzirodos et al., 2014b) |
| rs81285672 | 12 | 23,638,629 | ARHGAP23 | Inflammatory response (Liu, 2015) | |
| rs81325261 | 12 | 44,771,203 | FOXN1 | Regulation of hair follicle development (Song et al., 2017) | |
| rs331955329 | 13 | 66,004,327 | MTMR14 | Reduced with age accelerates skeletal muscle aging (Romero-Suarez et al., 2010) | |
| rs80971430 | 13 | 66,026,240 | BRPF1 | Intramuscular fatty acid (Puig-Oliveras et al., 2016) | |
| rs80945527 | 13 | 66,104,857 | ARPC4 | Mastitis resistance (Grossi et al., 2014) | |
| rs80885182 | 13 | 66,270,725 | FANCD2 | Muscle weight (Lionikas et al., 2010) | |
| rs45430493 | 13 | 66,515,894 | SEC13 | Muscle weight (Lionikas et al., 2012) | |
| rs81248260 | 13 | 66,583,753 | ATPB2 | Heat stress on reproductive performance (Dash et al., 2016) | |
| rs81446451 | 13 | 66,668,301 | ATPB2 | ||
| rs81446497 | 13 | 66,691,206 | ATPB2 | ||
| rs81446475 | 13 | 66,725,741 | ATPB2 | ||
| rs81446484 | 13 | 66,777,686 | ATPB2 | ||
| rs81478601 | 13 | 66,795,578 | ATPB2 | ||
| IND and DUR | rs80866460 | 4 | 106,698,421 | PTPN22 | Immune response (Lamsyah et al., 2009) |
| rs81413279 | 9 | 79,010,742 | NXPH1 | DMI (Olivieri et al., 2016) | |
| rs81413279 | 9 | 79,010,742 | ABCB5 | Immune function (Lee et al., 2017) | |
| rs81306790 | 6 | 89,661,963 | PHC2 | Mastitis (Chen et al., 2015) | |
| rs80854994 | 4 | 106,719,032 | PTPN22 | Immune response (Lamsyah et al., 2009) | |
| rs80854994 | 4 | 106,719,032 | BCL2L15 | Mastitis (Chen et al., 2015) | |
| Villages and DUR | rs81282695 | 6 | 94,442,844 | POU3F1 | Neurobehavioral functioning (Eusebi et al., 2018) |
| rs81282695 | 6 | 94442844 | FHL3 | Carcass traits (Zuo et al., 2004, 2007) | |
| Villages and KOL | rs81430450 | 11 | 24,063,007 | DNAJC15 | Feeding efficiency (Reyer et al., 2017a) |
| rs81430450 | 11 | 24,063,007 | EPSTI1 | Fertility traits (Gaddis et al., 2016), fat deposition (Zhang et al., 2018) | |
| SAL&LWT and IND | rs81232179 | 8 | 51,070,662 | FSTL5 | Meat quality (Ryu and Lee, 2016); skeletal muscle (Novianti et al., 2010) |
| rs45431508 | 8 | 69,912,174 | CXCL8 | Pig disease (Wang et al., 2019) | |
| rs81400554 | 8 | 55,181,102 | CEP135 | Intramuscular fat (Hamill et al., 2012); milk production (Rui et al., 2013) | |
| rs81400554 | 8 | 55,181,102 | EXOC1 | Marbling score (Wu et al., 2016) | |
| rs81400740 | 8 | 63,119,376 | EPHA5 | Feed efficiency (Reyer et al., 2017b) | |
| rs81400500 | 8 | 52,213,568 | NPY5R | Feed efficiency and fat deposition (Chen et al., 2018) | |
| rs81400500 | 8 | 52,213,568 | NPY1R | Feed efficiency and fat deposition (Chen et al., 2018) | |
| rs81302014 | 8 | 69,950,857 | RASSF6 | Body conformation (Fang and Pausch, 2019) | |
| rs80904678 | 11 | 15,274,089 | FOXO1 | Meat quality and carcass traits (Ropka-Molik et al., 2018) | |
| rs81400500 | 8 | 52,213,568 | SLC7A11 | Feed efficiency (Vigors et al., 2016) | |
| rs81300083 | 9 | 78,940,661 | NXPH1 | DMI (Olivieri et al., 2016) | |
| IND and VIT | rs81350922 | 1 | 257,096,974 | ASTN2 | Carcass weight in cattle (Júnior et al., 2016) |
| rs80970078 | 14 | 43,524,181 | MYO18B | Meat quality and carcass traits (Ropka-Molik et al., 2018) | |
| DRGA0006738 | 6 | 117,857,953 | NOL4 | Fatness (Li et al., 2011) | |
| rs80860919 | 1 | 64,018,444 | GPR63 | Fertility traits (Moran et al., 2017) | |
| rs80921694 | 13 | 73,023,057 | PLXNA1 | Meat quality (Martínez-Montes et al., 2016) | |
| rs81327396 | 12 | 53,063,765 | DNAH2 | Intramuscular fat (Luo et al., 2012); carcass weight (Kang et al., 2013) | |
| Villages and WBO | rs81244815 | 2 | 50,167,007 | SWAP70 | Disease resistance (Ma et al., 2011; Zhang et al., 2018) |
| rs81244815 | 2 | 50,167,007 | SBF2 | Fertility (Zhang et al., 2014); immune function (Ibeagha-Awemu et al., 2016) | |
| rs81401075 | 8 | 73,841,435 | FRAS1 | Sow reproductive traits (Fischer et al., 2015), feed efficiency (Messad et al., 2019) | |
| rs81401075 | 8 | 73,841,435 | NPY2R | Obesity (Siddiq et al., 2007; Hunt et al., 2011) | |
| Villages and VIT | INRA0003181 | 1 | 95,198,598 | SLC14A2 | Conformation traits (Le et al., 2017) |
| rs81332040 | 6 | 45,777,816 | ZNF382 | Conformation traits (Le et al., 2017) | |
| INRA0045852 | 14 | 10,3086,988 | HECTD2 | Fat and meat quality traits (Piórkowska et al., 2018) | |
| rs80980839 | 4 | 93,722,493 | RHBG | Ammonia transporter (Xiang et al., 2016) | |
| rs80971176 | 5 | 49,876,132 | SOX5 | Ear morphology (Edea et al., 2017) | |
| WBO and DUR | rs80837120 | 1 | 565,627 | TCTE3 | Involved in spermatogenesis (Du et al., 2016) |
| rs81389959 | 6 | 88,334,239 | PTP4A2 | Reproductive traits (Verardo et al., 2016); intramuscular fat (Martínez-Montes et al., 2016) | |
| rs81390106 | 6 | 88,751,010 | TMEM39B | Intramuscular Fat (Cesar et al., 2018) | |
| rs81390106 | 6 | 88,751,010 | TXLNA | Meat quality (Ropka-Molik et al., 2018) | |
| rs81390106 | 6 | 88,751,010 | HDAC1 | Altitude (Ban et al., 2015) | |
| rs81390106 | 6 | 88,751,010 | MARCKSL1 | Feed intake (Lindholm-Perry et al., 2016) | |
| rs81317489 | 6 | 89,640,457 | ZSCAN20 | Scrotal circumference (Sweett et al., 2018) | |
| rs81317489 | 6 | 89,640,457 | CSMD2 | Meat pH trait (Dong et al., 2014); Body weight (Yoshida et al., 2017) | |
| rs80894853 | 9 | 78,663,586 | NXPH1 | DMI (Olivieri et al., 2016) | |
| rs81389936 | 6 | 88,264,983 | COL16A1 | Carcass and meat quality traits (Choi et al., 2012) | |
| rs80790807 | 4 | 106,750,789 | PTPN22 | Immune response (Lamsyah et al., 2009) | |
| rs80790807 | 4 | 106,750,789 | BCL2L15 | Mastitis (Chen et al., 2015) | |
| rs80911350 | 14 | 11,345,116 | SCARA3 | Meat quality traits (Tizioto et al., 2015) | |
| rs80911350 | 14 | 11,345,116 | CLU | Fertility (Kumar et al., 2015), intramuscular fat (de Jager et al., 2013) | |
| rs343528814 | 13 | 36,608,977 | CACNA2D3 | Reproductive traits (Smith et al., 2019); body width in gilts and sows (Rothschild, 2010), body weight traits (Borowska et al., 2017), altitude (Zhang et al., 2014) | |
| rs81478390 | 13 | 53,707,241 | RYBP | Body conformation traits - body weight, body length, body height, and chest circumference (Zhou et al., 2016) | |
| rs81330369 | 9 | 7,449,894 | FCHSD2 | Milk production traits (Kemper et al., 2015) | |
| rs80975991 | 7 | 33,481,446 | ZFAND3 | Growth and carcass quality traits (Li and Kim, 2015) | |
| rs80855522 | 4 | 11,0552,282 | GNAI3 | Heat tolerance (Berihulay et al., 2019) | |
| rs80988392 | 1 | 213,780,848 | PTPRD | Meat quality (Raschetti et al., 2013) | |
Discussion
The Porcine SNP60K BeadChip was developed in 2009 (Ramos et al., 2009) and has been used to analyze genetic diversity and population structure in several pig populations (Ai et al., 2013; Burgos-Paz et al., 2013; Yang et al., 2017; Mujibi et al., 2018). This is the first report using the Porcine SNP60K BeadChip to explore diversity of domestic and wild pig populations covering the commercial, village, wild and conserved pigs farmed and reared in Africa. Pigs are possibly known to have reached Sub-Saharan Africa through the Nile corridor and later dispersed to the West-Central Africa (Blench, 2000). There are 541 pig breeds worldwide (Rischkowsky and Pilling, 2007) but the dominating commercial breeds in the pork industry are the Large White, Landrace, Duroc, Hampshire, Berkshire and Piétrain (Rothschild and Ruvinsky, 2010). The source of the improved breeds found in Southern Africa is believed to be the European settlers in 1600s (Krige, 1950; Blench and MacDonald, 2000; Swart et al., 2010). This was when Jan van Riebeeck brought some pigs to the Cape of Good Hope (Naude and Visser, 1994). The Large White, South African Landrace and the Duroc are the breeds mostly found and used in the commercial sector while the Kolbroek and Windsnyer are considered as indigenous and are mostly found in rural areas (Kem, 1993; Ramsay et al., 2000). The Vietnamese, Bush pig and Wild Boar populations constitute a small component of the genetic pool of pigs in the country often restricted to the game reserves and zoos.
The Porcine SNP60K BeadChip was designed using genomic resources from Western pig genomes (Ramos et al., 2009) and hence the number of SNPs after QC for the commercial population was higher (Table 2). The village populations had a higher number of polymorphic SNPs and moderate-high MAF compared to that of commercial pigs. Non-descript livestock populations including pigs are often observed to be highly diverse probably due to open mating systems and gene flow between populations. In South Africa similar observations of highly diverse and polymorphic populations were observed in village chicken populations (Khanyile et al., 2015), cattle (Makina et al., 2014), and village goats (Mdladla et al., 2016). The Warthog and other indigenous pigs were observed to be the least polymorphic and diverse which could be attributed to ascertainment bias as the Kolbroek, Windsnyer, Vietnamese Potbelly, Warthog and Bush pigs were not used in the development of the Porcine SNP60K BeadChip. Overall, the porcine SNP panel showed moderate MAF for the village, commercial and indigenous purebred pig populations such as the Windsnyer implying utility of the chip in the prevalent farmed pig populations of South Africa.
A study conducted by Swart et al. (2010) using microsatellite markers in various Southern African pig breeds revealed higher levels of diversity within population than was observed in this study for the same breeds (Table 2). High heterozygosity levels (0.61–0.75) were also reported by Halimani et al. (2012). In contrast to Swart et al. (2010) the Large White had the lowest diversity (Ho = 0.358) compared to the South African Landrace (Ho = 0.372) and other breeds of the Duroc and Kolbroek. It must be noted that these previous studies used microsatellite markers that are highly polymorphic markers and cannot be compared to SNPs that are biallelic in nature. High gene diversity is therefore expected in microsatellites markers. However, results on genetic diversity from this study were comparable to other studies that used the Porcine SNP60K BeadChip in Chinese and Western pig populations (Ai et al., 2013).
The heterozygosity values for the indigenous pigs were relatively similar to those of the commercial pigs (Table 2). A lower diversity was expected for the commercial pigs as they are under selection while the indigenous pigs are known to be rich reservoirs of distinct alleles, coupled with presence of gene flow (Amills et al., 2012). However, the indigenous pig populations are also of very small flock sizes and often fragmented and restricted to specific farming communities and conservation units hence diversity was low. Small and fragmented populations and the possibility of natural selection due to disease and unfavorable climatic conditions could explain the genetic diversity observed in the village populations. The high inbreeding levels observed in the Warthog populations might have been promoted by its family structuring where pigs are organized into fragmented breeding and social units (Table 2). Somers et al. (1995) noted that a group of Warthogs consist of about 40% of adults with changes seasonally. The number of mature individuals is estimated to be between 2000 and 5000 in the Kruger National Park (Ferreira et al., 2013). The geographical separation of the three national parks from which the warthogs were sampled, could have created small and fragmented subpopulations leading to escalated FIS values due to Wahlund effect. As expected, we found that the village pig populations of South Africa had high inbreeding values compared with other populations. The negative FIS values for commercial and indigenous populations are reflective of their intensive production environment as individuals are outbred to avoid mating to close relatives.
The low levels of effective population size (Ne) in the recent 12–22 generations for both commercial and indigenous populations are of concern (Supplementary Table S1). More so in the indigenous breeds since low levels of genetic diversity are likely to diminish overtime and increase the risk of extinction. The effective population of the Kolbroek of 34 at 12 generations ago is even lower than the minimum threshold Ne of 50 set by the FAO (2000). Franklin (1980) recommended a Ne of at least more than 500 while Willi et al. (2006) suggested Ne of more than 1,000 to maintain the evolutionary potential of any population. The genetic diversity of these populations will likely continue to be negatively impacted by the small number of founders and them being farmed in fragmented populations. Small effective population size of the Kolbroek might be due to pigs being raised in a research facility with limited boars and sows. Large White, Duroc and South African Landrace are commercial pigs that have undergone strong selection for meat and carcass traits thus resulting in small effective population sizes. Long-term sustainability of the populations might be compromised due to the small population size as it increases the effects of genetic drift and reduction in fitness traits (Frankham et al., 1998).
The high FIS values observed within populations across breeds are similar to previous studies (SanCristobal et al., 2006; Swart et al., 2010; Gama et al., 2013; Edea et al., 2014). An overall AMOVA FIS value of 93.95% was comparable to Halimani et al. (2012) value of 92.90% in indigenous pigs of Southern Africa. Diversity amongst South African populations that ranged from FCT = 0.92 (village pigs) to FCT = 5.42 (Commercial populations) might be due to gene flow between different populations within a sub-populations. Moderate diversity within population (i.e., FIS ranging from 19.92 in the category consisting of South African Wild Boar and worldwide Wild Boar to FIS = 35.52 in the categories consisting on South African villages and Worldwide villages) relative to elevated FCT in the same categories implies a higher genetic variation distributed among groups from different geographic locations. This genetic variation observed amongst groups of the South African and Burgos-Paz et al. (2013) pig populations (i.e., FCT = 62.35–73.58) is higher than the variation reported amongst Angora goats from South Africa, France and Argentina using 50K SNP BeadChip (Visser et al., 2016), which could be explained by limited exchange of breeding animals across geographic boundaries in the studied pig populations. The amongst population within groups diversity values ranging from FSC = 0.46 for South African villages to FSC = 18.17 for South African commercial demonstrates evidence of population sub-structure and genetic differentiation between the well-defined commercial and indigenous breeds relative to non-descript village populations that are characterized by weak population boundaries.
The PCA demonstrates the impact of domestication and geographic history on the clustering of populations. European populations as represented by Wild Boar, South African Landrace, and Large White, clustered together as expected (Figure 3). Considering the history that the Wild Boar is an ancestor to the domestic pigs of today, some gene flow may have remained from the Wild Boar in the domestic pigs (Giuffra et al., 2000). The clustering of the Wild Boars reflects a European ancestry of those populations within that cluster. The slight difference between the Wild Boar and domestic populations might have been due to geographic isolation and artificial selection. Geographic structures were evident amongst most of the pig populations that were aligned to production systems and their founder effects. The clustering of the Windsnyer and the village populations could be due to gene flow between indigenous breeds and village populations. Limpopo populations had a closer proximity to Large White and South African Landrace, and farmers in this region are more likely to buy pigs from commercial herds. The Large White and South African Landrace are also closer together as these are both European breeds. It was interesting that generally the village populations were closer to the Windsnyer and Kolbroek as these are both indigenous breeds in South Africa. Although not much is known about our indigenous breeds, different theories suggest that the Kolbroek might have far Eastern alleles while the Windsnyer is known to be dominant in other parts of Southern Africa like Mozambique, Zambia and Zimbabwe (Holness, 1973, 1991). The village populations and other Large Whites and Landraces from the global data set clustered together with the South African village, commercial and indigenous pigs demonstrating genetic similarities that could be aligned to founder effects and similarities in production systems.
The clustering of Duroc away from other commercial populations (Large White and South African Landrace) was expected. The Duroc breed was created in the United States with pigs of several ancestries, including African pigs (Porter, 1993). Studies conducted by Kotze and Visser (1996) and Swart et al. (2010) using the microsatellite markers on the Large White, South African Landrace and Duroc also reported similar results. The Large White and South African Landrace were more genetically similar when compared to the Duroc. The inclusion of global populations did not alter this clustering (Figure 4).
The distance of Vietnamese Potbelly population from the rest of the domestic pigs is clear evidence of independent domestication that took place between the European and Asian subspecies of the wild boar (Giuffra et al., 2000). The PCA including pigs genotyped from all over the world clearly shows the geographical effect of the populations as the Vietnamese Potbelly clustered in close proximity to the Chinese population.
ADMIXTURE K = 2 presented the first level of ancestry of the Suidae family representing Phacochoerus africanus (Warthog) and Potamochoerus larvatus (Bush pig) versus Sus scrofa (domesticated pigs including the Wild Boar) species (Figure 5). The presence of the Wild Boar genomic signature in the domestic pigs from K = 2 to K = 7 is not surprising (Figure 5). It is well documented that the domestic pigs diverged from each other and originated from the ancestral wild boars around 8,000–10,000 years ago (Giuffra et al., 2000; Laval et al., 2000; Larson et al., 2005). The Asian and European ancestral wild boars also originated from different subspecies thus the Vietnamese Potbelly diverged early (K = 2) from the rest of the domestic pig population. The results for the village populations showed high levels of admixture and weak between population sub-structuring. As opposed to pigs from the commercial sector that practices the intensive production systems, pigs in the villages are farmed under semi-intensive of free-range production systems, which might explain the admixture observed in this study. There is considerable indiscriminate crossbreeding that is taking place in village populations (Rege and Gibson, 2003). European and Asian pigs were used to improve the South African pig breeds but the actual contribution is unknown. Although phenotypically distinct from each other, the Bush pigs and warthogs clustered together which is suggestive of either common founder effect or selection pressures in the natural environments.
According to Wright (1978), FST estimation with values of less than 0.05 represents low differentiation while values between 0.05 and 0.15 represent a moderate genetic differentiation and those between 0.15 and 0.25 and beyond reflect highly differentiated populations. The low levels of genetic differentiation of the village populations from this study (Table 3) is consistent to pairwise FST values of Halimani et al. (2012) of village populations from Zimbabwe and South Africa. Most pig farmers from the villages practice free ranging or semi-controlled farming where there is continuous gene flow between populations within villages thereby explaining the low levels of population sub-structuring observed. Moderate FST values implies closer relationship between the South African Landrace and Large White and agrees with their breeding history, whereby the Landrace was developed from crossing the Large White from England and a Denmark indigenous. Greater genetic differentiation between the Warthog and the other pig populations (FST = 0.36–0.53) might be attributed to the (i) pressures of natural selection (ii) the separate histories of domestic and wild populations and (iii) the unique population dynamics of Warthogs that are known to live in clans of adult females, males and their offspring while maintaining minimal contacts with other clans (Cumming, 1975; Somers et al., 1994). In South Africa, Warthog populations are restricted to nature reserves thus creating a physical barrier and huge genetic differentiation between them and other pig populations. This will be in contrast to the greater interaction between village, commercial and indigenous populations. Low FST values between the villages in South African and village populations from South America (Supplementary Table S3) from Burgos-Paz et al. (2013) study, might be an indication that either common founder populations or similarities in production systems leading to common selection pressures. Ramírez et al. (2009) demonstrated that the African and South American pigs were derived from Europe and Far Eastern pigs. The very high genetic differentiation between the Vietnamese Potbelly and Bush pig agrees with the PCA and Admixture clustering.
Per marker pairwise FST were estimated between pairs highly differentiated populations which were from villages, commercial, indigenous, Asian and wild populations (Table 3). From the pairwise FST, Warthog was found to be genetically different from the rest of the populations. The per marker pairwise FST analysis used a threshold of 0.8 and above to plot Manhattan graphs of the Warthog against the rest of the populations. From the SNPs showing a threshold of FST ≥ 0.8, we looked at candidate genes and QTLs that can be associated with those SNPs to infer on traits that might have genetically differentiated the Warthog from Alfred Nzo, Duroc, Kolbroek, Large White, South African Landrace, and Windsnyer populations (Supplementary Figure S2).
Majority of the SNPs that were above the threshold between the Warthog and the rest of the populations were from chromosomes 1, 4, 5, 12, 13, and 15 (Table 4). Chromosomes 2 (Warthog vs. Alfred Nzo), 3 (Warthog vs. Kolbroek), 6 (Warthog vs. South African Landrace) and 14 (Warthog vs. Large White) seemed to be less common. Chromosome 1 with a total number of 12 SNPs was associated with reproduction and growth traits while the indigenous populations of Kolbroek and Windsnyer were differentiated on chromosome 4 that was also linked to reproduction and growth traits.
Warthog vs. Alfred Nzo had three SNPS (FST ≥ 0.8) that are associated with reproduction (RPL18, IL17B) and growth (IL17B, ARHGAP23) characteristics (Table 4). It is known that good nutrition is vital to be able to maximize growth performance. Genes IL17B and ARHGAP23 are linked to inflammatory response (Liu, 2015; Bie et al., 2017) and the gastrointestinal tract where they play a role in the digestion and absorption of the nutrients. Inflammatory responses lead to reduction of feed intake, which in turn affects the growth of the animal (Liu, 2015). Selection on genes associated with inflammation in the populations of Warthog vs. Alfred Nzo might be an effect of the different diets these populations scavenge on. Medzhitov (2008) noted the inflammation response to be a protective mechanism from the stress and harmful environment.
Growth linked genes ADGRB3, and ACY1 were dominant in differentiating Warthog vs. Duroc populations with an overall total of 10 SNPs. Emrani et al. (2017) associated ADGRB3 to body weight traits in the broiler chickens. The association of ADGRB3 gene to Duroc rather than Large White or South African Landrace breeds might be linked to the higher percentage of intramuscular fat in Duroc compared to the other two commercial breeds (De Vries et al., 2000). Mature males of Warthog can also reach up to 100 kg and possesses good meat and carcass qualities (Hoffman and Sales, 2007).
A total number of 20 significant SNPs (FST ≥ 0.8) were linked to the Warthog vs. Kolbroek populations. Growth traits were associated with five of the SNPs between Warthog vs. Kolbroek. Indigenous Kolbroek are reported to be smaller in size when compared to commercial breeds such as Large White (Chimonyo et al., 2005). Kutwana et al. (2015) reported no significant difference (P > 0.05) between the Kolbroek and Large White populations that had higher fat percentages when compared to the other commercial breeds (Nicholas, 1999).
Chromosome 13 was also highly notable with significant SNPs differentiating Warthog vs. Kolbroek and Warthog vs. Windsnyer. Only two SNPs appeared for Warthog vs. South African Landrace and were on chromosome 6. The Warthog vs. Windsnyer had a total of fourteen SNPs differentiating them. The identification of BRPF1 gene in the Warthog vs. Windsnyer populations is an important observation as this gene is associated with the intramuscular fat (IMF). When it comes to the value and taste of the pork meat, intramuscular fat is an important characteristic because meat that is high in IMF tends to be juicy and tender (Eikelenboom et al., 1996; de Koning et al., 1999). The gene ATPB2 associated with six significant SNPs is linked to heat stress and reproductive performance (Dash et al., 2016). Heat stress might result in poor reproduction for both sows and boars. Pigs cannot sweat and this makes them sensitive to high environmental temperatures making and of concern particularly to commercial pig farmers (Ross et al., 2015).
Genes linked to immune response and mastitis were observed in Indigenous vs. Duroc comparisons. PTPN22 gene on chromosome 4 has a regulatory effect on T- and B- cell activation in immune response (Lamsyah et al., 2009). PTPN22 plays a role in susceptibility to tuberculosis. Pigs are generally natural hosts of mycobacterial infections (de Lisle, 1994). Porcine TB has been reported in South Africa where infections are commonly via infected cattle fecal matter fed to piglets as well as interactions with wild pigs (Muwonge et al., 2012). NXPH1 gene is associated with DMI (dry matter intake) in cattle (Olivieri et al., 2016). Both PTPN and NXPH1 genes were fixed in the Duroc implying natural selection of the Duroc when compared to both indigenous and Wild Boars. Breeds in the commercial sector are mainly selected for growth, carcass and meat quality traits. The indigenous and village population on the other hand has not been systematically selected for such traits.
The NPY5R located on chromosome 8, was associated with feed efficiency and fat deposition. This gene was also reported in Jinhua and Rongchang pigs that belong to Chinese breeds (Chen et al., 2018). Fat deposition genes observed in Indigenous vs. Vietnamese, Villages vs. Kolbroek and South African Landrace with Large White vs. Indigenous are evidence in agreement with suggestions that Kolbroek and other indigenous pigs tend to carry their weight in their bellies and backs (Hoffman et al., 2005). Hoffman et al. (2005) also reported breed type and diet to have an influence on the composition of the meat. This study therefore presented a diverse genomic architecture of South African pigs with differentiating selection pressures for meat and carcass quality traits in the different pigs raised in diverse production systems.
Conclusion
Overall, the study demonstrated the utility of the Porcine SNP60K BeadChip in elucidating genetic diversity and population genomic structure of South African pig populations relative to other global populations. Village pigs demonstrated distinctiveness from other domestic and commercial populations within South Africa and when compared to global populations. The study provided baseline knowledge with regards to the genetic diversity of the domestic and wild pig populations of South Africa, which is a prerequisite for population/breed characterization, utilization and conservation. A more in-depth analysis of patterns of genetic variations is required to get more insight into factors shaping genetic diversity of these populations.
Data Availability Statement
The datasets generated for this study can be found in Dryadhttps://doi.org/10.5061/dryad.b0t10b0.
Ethics Statement
Ear tissue samples were collected from pigs using the Tissue Sampling Applicator Gun while pliers were used to collect the hair samples according to standard procedures and ethical approval from ARC-Irene Animal Ethics committee (APIEC16/028).
Author Contributions
NH collected samples, analyzed the data, and wrote the draft manuscript. FM, PS, and ED designed the experiment and sourced funding. KH analyzed the genomic data for the experiment. FM, PS, and ED coordinated the conduct of the study and writing of manuscript and revisions. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Agricultural Research Council-Biotechnology Platform (ARC-BTP) for funding the genotyping of samples. We express our gratitude to all the pig farmers, the Department of Agriculture (Eastern Cape and Limpopo) and various stakeholders who allowed us to use their animals in this study. NH holds fellowships from the National Research Foundation and ARC-Professional Development Program.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00344/full#supplementary-material
Cross validation plot for inferring the number of K populations in the analysis of population structure.
Genome-wide Manhattan plot of FST among the pig populations of (a) Alfred Nzo and Warthog, (b) Duroc and Warthog, (c) Kolbroek and Warthog, (d) Large White and Warthog, (e) South African landrace and Warthog, (f) Windsnyer and Warthog, (g) Indigenous and Duroc, (h) Villages and Duroc, (i) Villages and Kolbroek, (j) South African Landrace with Large White and Indigenous, (k) Indigenous and Vietnamese, (l) Villages and Wild Boar, (m) Villages and Vietnamese, (n) Wild Boar and Duroc. The solid lines indicate the FST ≥ 0.8 thresholds.
Average effective population size estimates across generations for the different populations analyzed.
Partitioning of genetic variance for the different populations analyzed.
FST between SA populations and genotypes from Burgos-Paz et al. (2013).
References
- Ai H., Huang L., Ren J. (2013). Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western pigs revealed by genome-wide SNP markers. PLoS One 8:e56001. 10.1371/journal.pone.0056001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. H., Lange K. (2011). Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12:246. 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amills M., Clop A., Ramírez O., Pérez-Enciso M. (2010). “Origin and diversity of pig breeds,” in Encyclopedia of Life Sciences ed. Kehrer-Sawatzki H. (Chichester: John Wiley & Sons; ), 1–7. 10.1002/9780470015902.a002288 [DOI] [Google Scholar]
- Amills M., Ramírez O., Galman-Omitogun O., Clop A. (2012). Domestic pigs in Africa. Afr. Archaeol. Rev. 30 73–82. 10.1007/s10437-012-9111-2 [DOI] [Google Scholar]
- Ban D. M., Zhang B., Wang Z. X., Zhang H., Wu C. X. (2015). Differential gene expression of epigenetic modifying enzymes between Tibet pig and Yorkshire in high and low altitudes. Genet. Mol. Res. 14 3274–3280. 10.4238/2015.April.13.6 [DOI] [PubMed] [Google Scholar]
- Berihulay H., Abied A., He X., Jang L., Ma Y. (2019). Adaptation mechanisms of small ruminants to environmental heat stress. Animals 9:75. 10.3390/ani9030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bie Q., Jin C., Zhang B., Dong H. (2017). IL-17B: a new area of study in the IL-17 family. Mol. Immunol. 90 50–56. 10.1016/j.molimm.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Blench R. M. (2000). “A history of pigs in Africa,” in Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography, eds Blench R. M., Mac Donald K. (Abingdon: Routledge Books; ). [Google Scholar]
- Blench R. M., MacDonald K. C. (eds) (2000). “The origins and development of the African livestock,”in Archaelogy, Genetics, Linguistics and Ethnography. London: UCL Press [Google Scholar]
- Borowska A., Reyer H., Wimmers K., Varley P., Szwackowski T. (2017). Detection of pig genome regions determining production traits using an information theory approach. Livest. Sci. 205 31–35. 10.1016/j.livsci.2017.09.012 [DOI] [Google Scholar]
- Browett S., McHugo G., Richardson I. W., Magee D. A., Park S. D. E., Fahey A. G., et al. (2018). Genomic characterisation of the indigenous Irish Kerry cattle breed. Front. Genet. 9:51. 10.3389/fgene.2018.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Paz W., Souza C. A., Megens H. J., Ramayo-Caldas Y., Melo M., Lemús- Flores C., et al. (2013). Porcine colonization of the Americas: a 60k SNP story. Hereditary 110 321–330. 10.1038/hdy.2012.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesar A. S., Regitano L. C., Reecy J. M., Poleti M. D., Oliveira P. S. N., De Oliveira G. B., et al. (2018). Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits. BMC Genomics 19:499. 10.1186/s12864-018-4871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang J., Wang Y., Wu Y., Fu J., Liu J. F. (2018). Genome-wide detection of selection signatures in Chinese indigenous Laiwu pigs revealed candidate genes regulating fat deposition in muscle. BMC Genet. 19:31. 10.1186/s12863-018-0622-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cheng Z., Zhang S., Werling D., Wathes D. C. (2015). Combining genome wide association studies and differential gene expression data analyses identifies candidate genes affecting mastitis caused by two different pathogens in the dairy cow. Open J. Anim. Sci. 5 358–393. 10.4236/ojas.2015.54040 [DOI] [Google Scholar]
- Chimonyo M., Bhebhe E., Dzama K., Halimani T. E., Kanengoni A. (2005). Improving smallholder pig production for food security and livelihoods of the poor in Southern Africa. Proc. Afr. Crop Sci. Conf. 7 569–573. [Google Scholar]
- Choi I., Bates R. O., Raney N. E., Steibel J. P., Ernst C. W. (2012). Evaluation of QTL for carcass merit and meat quality traits in a US commercial Duroc population. Meat Sci. 92 132–138. 10.1016/j.meatsci.2012.04.023 [DOI] [PubMed] [Google Scholar]
- Corbin L. J., Liu A. Y., Bishop S. C., Woolliams J. A. (2012). Estimation of historical effective population size using linkage disequilibrium with marker data. J. Anim. Breed. Genet. 129 257–270. 10.1111/j.1439-0388.2012.01003.x [DOI] [PubMed] [Google Scholar]
- Cornelis M. C., Hu F. B. (2013). Systems epidemiology: a new direction in nutrition and metabolic disease research. Curr. Nutr. Rep. 2 225–235. 10.1007/s13668-013-0052-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming D. H. M. (1975). A Field of Study of the Ecology and Behaviour of Warthog. Harare: National Museums and Monuments of Rhodesia, [Google Scholar]
- D’Huart J., Grubb P. (2003). Distribution of the common warthog (Phacochoerus africanus) and the desert warthog (Phacochoerus aethiopicus) in the Horn of Africa. Afr. J. Ecol. 39 156–169. 10.1046/j.0141-6707.2000.00298.x [DOI] [Google Scholar]
- Dash S., Chakravarty A. K., Singh A., Upadhyay A., Singh M., Yousef S. (2016). Effect of heat stress on reproductive performances of dairy cattle and buffaloes: a review. Vet. World 9 235–244. 10.14202/vetworld.2016.235-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager N., Hudson N. J., Reverter A., Barnard R., Café L. M., Greenwood P. L., et al. (2013). Gene expression phenotypes for lipid metabolism and intramuscular fat in skeletal muscle of cattle. J. Anim. Sci. 91 1112–1128. 10.2527/jas.2012-5409 [DOI] [PubMed] [Google Scholar]
- de Koning D. J., Janss L. L., Rattubk P., van Oers P. A., de Vries B. J., Groenen M. A., et al. (1999). Detection of quantitative traits loci for backfat thickness and intramuscular fat content in pig. Genetics 152 1679–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lisle G. W. (1994). Mycobacterial infections in pigs. Surveillance 21 23–25. [Google Scholar]
- De Vries A. G., Faucitano L., Sosnicki A., Plastow G. H. (2000). The use of gene technology for optimal development of pork meat quality. Food Chem. 69 397–405. 10.1016/S0308-8146(00)00049-2 [DOI] [Google Scholar]
- Dong Q., Liu H., Li X., Wei W., Zhao S., Cao J. (2014). A genome-wide association study of five meat quality traits in Yorkshire pigs. Front. Agr. Sci. Eng. 1 137–143. 10.15302/J-FASE-2014014 24740293 [DOI] [Google Scholar]
- Du Y., Li M., Chen J., Duan Y., Wang X., Qiu Y., et al. (2016). Promoter targeted bisulfite sequencing reveals DNA methylation profiles associated with low sperm motility in asthenozoospermia. Hum. Reprod. 31 24–33. 10.1093/humrep/dev283 [DOI] [PubMed] [Google Scholar]
- Edea Z., Bhuiyan M. S. A., Dessie T., Rothschild M. F., Dadi H., Kim K. S. (2014). Genome-wide genetic diversity, population structure and admixture analysis in African and Asian cattle breeds. Animal 9 218–226. 10.1017/S1751731114002560 [DOI] [PubMed] [Google Scholar]
- Edea Z., Hong J. K., Jung J. H., Kim D. W., Kim E. S., Shin S. S., et al. (2017). Detecting selection signatures between Duroc and Duroc synthetic pig populations using high-density SNP chip. Anim. Genet. 48 473–477. 10.1111/age.12559 [DOI] [PubMed] [Google Scholar]
- Eikelenboom G., Hoving-Bolink A. H., Van Der Wal P. G. (1996). The eating quality of pork: 2. The influence of intramuscular fat. Fleischwirtschaft 76 517–518 [Google Scholar]
- Emrani H., Torshizi R.V., Masoudi A.A., Ehsani A. (2017). Identification of new loci for body weight traits in F2 chicken population using genome-wide association study. Livest. Sci. 206 125–131. 10.1016/j.livsci.2017.10.016 [DOI] [Google Scholar]
- Eusebi P. G., Cortés O., Carleos C., Dunner S., Cañon J. (2018). Detection of selection signatures for agonistic behaviour in cattle. J. Anim. Breed. Genet. 135 170–177. 10.1111/jbg.12325 [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. (2005). ARLEQUIN ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fang Z. H., Pausch H. (2019). Multi-trait meta-analyses reveal 25 quantitative trait loci for economically important traits in Brown Swiss cattle. bioRxiv [Preprint]. 10.1101/517276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2000). Secondary Guidelines of Farm Animal Genetic Resources Management Plans. Management of Small Populations at Risk. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- FAO (2009). Status and Trends Report on Animal Genetic Resources–2008. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Ferreira S., Gaylard A., Greaver C., Hayes J., Cowell C., Ellis G. (2013). Summary Report: Animal abundances in Parks 2012/2013. Skukuza: SANParks. [Google Scholar]
- Fischer D., Laiho A., Gyenesei A., Sironen A. (2015). Identification of reproduction-related gene polymorphisms using whole transcriptome sequencing in the Large White pig population. G3 5 1351–1360. 10.1534/g3.115.018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L., Schiavo G., Galimberti G., Calò D. G., Russo V. (2014). A genomewide association study for average daily gain in Italian Large White pigs. J. Anim. Sci. 92 1385–1394. 10.2527/jas2013-7059 [DOI] [PubMed] [Google Scholar]
- Frankenberg S. R., De Barros F. R. O., Rossant J., Renfree M. B. (2016). The mammalian blastocyst. J. Dev. Biol. 5 210–232. 10.1002/wdev.220 [DOI] [PubMed] [Google Scholar]
- Frankham R., Ballou J. D., Briscoe D. A. (1998). An Introduction to Conservation Genetics. Cambridge: Cambridge University Press. [Google Scholar]
- Franklin I. R. (1980). “Evolutionary change in small populations,” in Conservation Biology: An Evolutionary-Ecological Perspective, eds Soulé M., Wilcox B. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Gaddis K. P., Null D. J., Cole J. B. (2016). Explorations in genome-wide association studies and network analyses with dairy cattle fertility traits. J. Dairy Sci. 99 6420–6435. 10.3168/jds.2015-10444 [DOI] [PubMed] [Google Scholar]
- Gama L. T., Martínez A. M., Carolino I., Landi V., Delgado J. V., Vicente A. A. (2013). Genetic structure, relationships and admixture with wild relatives in native pig breeds from Iberia and its island. Genet. Sel. Evol. 45:18. 10.1186/1297-9686-45-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E., Kijas J. M., Amarger V., Carlborg O., Andersson L. (2000). The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmelin J. F. (1788). Systema Naturae per Regna Tria Naturae Secundum Classes, Ordines, Genera, Species Cum Caracteribus, Differentiis, Synonmis, Locis, Vol. 1 Leipzig: CRC Press. [Google Scholar]
- Gondret F., Vincent A., Houée-Bigot M., Siegel A., Lagarrigue S., Causeur D., et al. (2017). A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genomics 18:244. 10.1186/s12864-017-3639-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. A., Abbey C. A., Beremand P. D., Choi Y., Farmer J. L., Adelson D. L., et al. (2006). Identification of endometrial genes regulated by early pregnancy, progesterone, and interferon tau in the ovine uterus. Biol. Reprod. 74 383–394. 10.1095/biolreprod.105.046656 [DOI] [PubMed] [Google Scholar]
- Grossi D. A., Abo-Ismail M. K., Koeck A., Miller S. P., Stothard P., Plastow G., et al. (2014). “Genome-wide association analyses for mastitis in Canadians Holsteins,” in Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Wageningen. [Google Scholar]
- Gu T., Zhu M., Schroyen M., Qu L., Nettleton D., Kuhar D., et al. (2014). Endometrial gene expression profiling in pregnant Meishan and Yorkshire pigs on day 12 of gestation. BMC Genomics 15:156. 10.1186/1471-2164-15-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Gil B., Wiener P., Williams J. L. (2007). Genetic effects on coat colour in cattle: dilution of eumelanin and phaeomelanin pigments in an F2-backcross Charolais × Holstein population. BMC Genomics 8:56. 10.1186/1471-2156-8-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halimani T. E., Muchadeyi F. C., Chimonyo M., Dzama K. (2012). Some insights into the phenotypic and genetic diversity of indigenous pigs in Southern Africa. S. Afr. J. Anim. Sci. 42 507–510. 10.4314/sajas.v42i5.1 [DOI] [Google Scholar]
- Hamill R. M., McBryan J., McGee C., Mullen A. M., Sweeney T., Talbot A., et al. (2012). Functional analysis of muscle gene expression profiles associated with tenderness and intramuscular fat content in pork. Meat Sci. 92 440–450. 10.1016/j.meatsci.2012.05.007 [DOI] [PubMed] [Google Scholar]
- Hatzirodos N., Hummitzsch K., Irving-Rodgers H. F., Harland M. L., Morris S. E., Rodgers R. J. (2014a). Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genomics 15:40. 10.1186/1471-2164-15-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzirodos N., Irving-Rodgers H. F., Hummitsch K., Harland M. L., Morris S. E., Rodgers R. J. (2014b). Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics 15:24. 10.1186/1471-2164-15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S. C., Hasstedt S. J., Xin Y., Dalley B. K., Milash B. A., Yakobson E., et al. (2011). Polymorphisms in the NPY2R Gene Show Significant Associations with BMI that are Additive to FTO, MC4R, and NPFFR2 Gene Effects. Obesity 19 2241–2247. 10.1038/oby.2011.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mdladla K., Dzomba E. F., Huson H., Muchadeyi F. C. (2016). Population genomic structure and linkage disequilibrium analysis of South African goat breeds using genome-wide SNP data. Anim. Genet. 47 471–482. 10.1111/age.12442 [DOI] [PubMed] [Google Scholar]
- Hoffman L. C., Sales J. (2007). Physical and chemical quality characteristics of warthog (Phacochoerus africanus) meat. Livest. Res. Rural Dev. 19:153 10.1016/j.meatsci.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Hoffman L. C., Styger W. F., Brand T. S., Muller M. (2005). The growth, carcass yield, physical and chemical characteristic of two South African indigenous pig breeds. S. Afr. J. Anim. Sci. 6 25–35 [Google Scholar]
- Holness D. H. (1973). The role of indigenous pigs as source of protein in Africa: a review. Rhode. Agric. J, 73 59–63. [Google Scholar]
- Holness D. H. (1991). The Tropical Agriculturalist. London: Macmillan Education Ltd. [Google Scholar]
- Ibeagha-Awemu E. M., Peters S. O., Akwanji K. A., Imumorin I. G., Zhao X. (2016). High density genome wide genotyping-by-sequencing and association identifies common and low frequency SNPs, and novel candidate genes influencing cow milk traits. Sci. Rep. 6:31109. 10.1038/srep31109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. F. (1998). “Genetic aspects of domestication, common breeds and their origin,” in The Genetics of the Pig, eds Rothschild M. F., Ruvinsky A. (Wallingford: CABI Publishing; ). [Google Scholar]
- Jori F., Bastos D. S. (2009). Role of wild suids in the epidemiology of African swine fever. Ecohealth 6 296–310. 10.1007/s10393-009-0248-7 [DOI] [PubMed] [Google Scholar]
- Júnior G. F., Costa R. B., De Camargo G. M., Carvalheiro R., Rosa G. J., Baldi F., et al. (2016). Genome scan for postmortem carcass traits in Nellore cattle. J. Anim. Sci. 94 4087–4095. 10.2527/jas2016-0632 [DOI] [PubMed] [Google Scholar]
- Kang K., Seo D. W., Lee J. B., Jiung E., Park H., Cho I., et al. (2013). Identification of SNPs affecting porcine carcass weight with the 60K SNP chip. J. Anim. Sci. Technol. 55 231–235. 10.5187/JAST.2013.55.4.231 [DOI] [Google Scholar]
- Kem E. H. (ed.) (1993). Pig Production in South Africa. Irene: Agricultural Research Council. [Google Scholar]
- Kemper K. E., Reich C. M., Bowman P. J., van der Jagt C. J., Chamberlain A. J., Mason B. A., et al. (2015). Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions. Genet. Sel. Evol. 47:29. 10.1186/s12711-014-0074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanyile K. S., Dzomba E. F., Muchadeyi F. C. (2015). Population genetic structure, linkage disequilibrium and effective population size of conserved and extensively raised chicken populations of Southern Africa. Front. Genet. 6:13. 10.3389/fgene.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Namekata K., Guo X., Harada C., Harada T. (2016). Neuroprotection, growth factors and BDNF-TrkB signalling in retinal degeneration. Int. J. Mol. Sci. 17:1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotze A., Visser D. P. (1996). “Status of genetic variation in purebred pig breeds in South Africa,” in Proceedings of the 7th All Africa Conference on Animal Agriculture, Pretoria. [Google Scholar]
- Krige J. E. (1950). The Social Systems of the Zulus, 2nd Edn Pietermaritzburg: Shuter and Shooter. [Google Scholar]
- Kumar S., Deb R., Singh U., Ganguly I., Mandal D. K., Tyagi S., et al. (2015). Bovine circadian locomotor output cycles kaput (CLOCK) and clusterin (CLU) mRNA quantitation in ejaculated crossbred bull spermatozoa. Reprod. Domest. Anim 50 505–509. 10.1111/rda.12522 [DOI] [PubMed] [Google Scholar]
- Kutwana H. W., Gxasheka M., Tyasi T. L. (2015). Body weight and morphological traits of Large White and Kolbroek pig breeds. Int. J. Adv. Res. 3 105–109 [Google Scholar]
- Lamsyah H., Rueda B., Baassi L., Elaouad R., Bottini N., Sadki K., et al. (2009). Association of PTPN22 gene functional variants with development of pulmonary tuberculosis in Moroccan population. Tissue Antigens 74 228–232. 10.1111/j.1399-0039.2009.01304.x [DOI] [PubMed] [Google Scholar]
- Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., Robins J., et al. (2005). Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307 1618–1621. 10.1126/science.1106927 [DOI] [PubMed] [Google Scholar]
- Laval G., Iannuccelli N., Legault C., Milan D., Groenen M. A., Giuffra E., et al. (2000). Genetic diversity of eleven European pig breeds. Genet. Sel. Evol. 32 187–203. 10.1186/1297-9686-32-2-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. H., Christensen O. F., Nielsen B., Sahana G. (2017). Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 49:12. 10.1186/s12711-017-0289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Taye M., Kwon T., Yoon J., Jang D., Suzuki S., et al. (2017). Identifying candidate positive selection genes in Korean imported pig breeds. Genes Genomics 39 557–565. 10.1007/s13258-017-0529-4 [DOI] [Google Scholar]
- Li X., Kim S. W., Choi J. S., Lee Y. M., Lee C. K., Choi B. H., et al. (2010). Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content. Mol. Biol. Rep. 37 3931–3939. 10.1007/s11033-010-0050-1 [DOI] [PubMed] [Google Scholar]
- Li X., Kim S.-W., Do K.-T., Ha Y.-K., Lee Y.-M., Yoon S.-H., et al. (2011). Analyses of porcine public SNPs in coding-gene regions by re-sequencing and phenotypic association studies. Mol. Biol. Rep. 38 3805–3820. 10.1007/s11033-010-0496-1 [DOI] [PubMed] [Google Scholar]
- Li Y., Kim J. J. (2015). Multiple linkage disequilibrium mapping methods to validate additive quantitative trait loci in Korean native cattle (Hanwoo). Asian Australas J. Anim. Sci. 28 926–935. 10.5713/ajas.15.0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Lee S., Kim N., Cho Y., Chai H., Seong H., et al. (2013). Gene co-expression analysis to characterize genes related to marbling trait in Hanwoo (Korean) cattle. Asian-Australas J. Anim. Sci. 26 19–29. 10.5713/ajas.2012.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnaeus C. (1758). Systema Naturae per Regna Tria Naturae, secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis. Tomus I. Editio Decima, Reformata. Stockholm: Laurentii Salvii. [Google Scholar]
- Lindholm-Perry A. K., Butler A. R., Kern R. J., Hill R., Kuehn L. A., Wells J. E., et al. (2016). Differential gene expression in the duodenum, jejunum and ileum among crossbred beef steers with divergent gain and feed intake phenotypes. Anim. Genet. 47 408–427. 10.1111/age.12440 [DOI] [PubMed] [Google Scholar]
- Lionikas A., Cheng R., Lim J. E., Palmer A. A., Blizard D. A. (2010). Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol. Genomics 42A 33–38. 10.1152/physiolgenomics.00100.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionikas A., Meharg C., Derry J. M. J., Ratkevicius A., Carroll A. M., Vandenbergh D. J., et al. (2012). Resolving candidate genes of mouse skeletal muscle QTL via RNA-Seq and expression network analyses. BMC Genomics 13:592. 10.1186/1471-2164-13-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. (2015). Fatty acids, inflammation and intestinal health in pigs. J. Anim. Sci. Biotechnol. 6:41. 10.1186/s40104-015-0040-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnberg E. (1908). Remarks on some wart-hog skulls in the British Museum. Proc. Zool. Soc. London 78 936–940. 10.1111/j.1469-7998.1908.00936.x [DOI] [Google Scholar]
- Lönnberg E. (1912). Mammals collected by the Swedish zoological expedition to British East Africa 1911. Kungliga Svenska Vetenskapakademiens Handligar 48 1–188. [Google Scholar]
- Luo W., Cheng D., Chen S., Wang L., Li Y., Ma X. (2012). Genome-wide association analysis of meat quality traits in a porcine Large White × Minzhu intercross population. Int. J. Biol. Sci. 8 580–595. 10.7150/ijbs.3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. J., Zhang X. L., Wang L. X., Liu Z. (2011). Studies on difference of immune and production indexes between Songliao black pig and large white pig. China Anim. Husbandry Vet. Med. 38 52–55. [Google Scholar]
- Makina S. O., Muchadeyi F. C., van Marle-Koöster E., Macneil M. D., Maiwashe A. (2014). Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Front. Genet. 5:333 10.3389/fgene.2014.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Montemayor M. M., Hill G. M., Raney N. E., Rilington V. D., Tempelman R. J., Link J. E., et al. (2008). Gene expression profiling in hepatic tissue of newly weaned pigs fed pharmacological zinc and phytase supplemented diets. BMC Genomics 9:412. 10.1186/1471-2164-9-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Montes A. M., Muiños-Bühl A., Fernández A., Folch J. M., Ibáñez-Escriche N., Fernández A. (2016). Deciphering the regulation of porcine genes influencing growth, fatness and yield-related traits through genetical genomics. Mamm. Genome 28 130–142. [DOI] [PubMed] [Google Scholar]
- McGraw K., List A. (2017). Chapter five-Erythropoietin receptor signalling and lipid rafts. Vitam. Horm. 105 79–100. 10.1016/bs.vh.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2008). Origin and physiological roles of inflammation. Nature 454 428–435. 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- Messad F., Louveau I., Koffi B., Gilbert H., Gondret F. (2019). Investigation of muscle transcriptomes using gradient boosting machine learning identifies molecular predictors of feed efficiency in growing pigs. BMC Genomics 20:659. 10.1186/s12864-019-6010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moioli B., D’Andrea S., De Grossi L., Sezzi E., de Sanctis B., Catillo G., et al. (2016). Genomic scan for identifying candidate genes for paratuberculosis resistance in sheep. Anim. Prod. Sci. 56 1046–1055. 10.1071/ANI14826 [DOI] [Google Scholar]
- Moran B., Butler S., Moore S., MacHugh D. E., Creevey C. J. (2017). Differential gene expression in the endometrium reveals cytoskeletal and immunological genes in lactating dairy cows genetically divergent for fertility traits. Reprod. Fertil. Dev. 29 274–282. 10.1071/RD15128 [DOI] [PubMed] [Google Scholar]
- Mujibi F. D., Okoth E., Cheruiyot E. K., Onzere C., Bishop R. P., Fèvre E. M., et al. (2018). Genetic diversity, breed composition and admixture of Kenyan domestic pigs. PLoS One 13:e0190080. 10.1371/journal.pone.0190080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muwanika V. B., Nyakaana S., Siegismund H. R., Arctander P. (2003). Phylogeography and population structure of the common warthog (Phacochoerus africanus) inferred from variation in mitochondrial DNA sequences and microsatellite loci. J. Hered. 91 361–372. 10.1038/sj.hdy.6800341 [DOI] [PubMed] [Google Scholar]
- Muwonge A., Johansen T. B., Vigdis E., Godfroid J., Olea-Popelka F., Demelash B., et al. (2012). Mycobacterium bovis infections in slaughter pigs in Mubende district, Uganda: a public health concern. BMC Vet. Res. 8:168. 10.1186/1746-6148-8-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naude R. T., Visser D. P. (1994). “ń Generiese kwalitatiewe benadering ten einde die doeltreffende produksie van verbruikersaanneemlike varkvleis te verseker,” in Proceedings of the 6de Nasionale SAVPO-Kongres te Elangeni Hotel, KwaZulu-Natal. [Google Scholar]
- Newton J. R., De Santis C., Jerry D. R. (2012). The gene exptression response of the catadromous perciform barramundi Lates calcarifer to an acute heat stress. J. Fish. Biol. 81 81–93. 10.1111/j.1095-8649.2012.03310 [DOI] [PubMed] [Google Scholar]
- Nicholas G. (1999). Kolbroek-the Unique Local Breed. Farmer’s Weekly, August. [Google Scholar]
- Novianti I., Pitchford W. S., Bottema C. D. (2010). Beef cattle muscularity candidate genes. J. Ilmu Ilmu Peternakan 20 1–10. [Google Scholar]
- Olivieri B. F., Mercadante M. E., Cyrillo J. N., Branco R. H., Bonilha S. M., Albuquerque L. G., et al. (2016). Genomic regions associated with feed efficiency indicator traits in an experimental Nellore cattle population. PLoS One 11:e0164390. 10.1371/journal.pone.0164390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas P. S. (1766). Miscellanea Zoological Quibus Novae Imprimis Atque Obscurae Animalium Species Describuntur et Observationibus Iconibusque Illustrantur. The Hague: Nabu Press. [Google Scholar]
- Parker Gaddis K. L., Megonigal J. H., Jr., Clay J. S., Wolfe C. W. (2018). Genome-wide association study for ketosis in US jerseys using producer-recorded data. J. Dairy Sci. 101 413–424. 10.3168/jds.2017-13383 [DOI] [PubMed] [Google Scholar]
- Piórkowska K., Żukowski K., Ropka-Molik K., Tyra M., Gurgul A. (2018). A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 41 125–136. 10.1590/1678-4685-GMB-2016-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter V. (1993). Pigs: A Handbook to the Breeds of the World. Mountfield, HK: Helm Information Ltd. [Google Scholar]
- Puig-Oliveras A., Revilla M., Castelló A., Fernández A. I., Folch J. M., Ballester M. (2016). Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular fatty acid content and composition in porcine meat. Sci. Rep. 6:31803. 10.1038/srep31803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas l, Ferreira M. A., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez O., Ojeda A., Tomas A., Gallardo D., Huang L. S., Folch J. M., et al. (2009). Integrating Y-chromosome, mitochondrial, and autosomal data to analyse the origin of pig breeds. Mol. Biol. Evol. 26 2061–2072. 10.1093/molbev/msp118 [DOI] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P., Affara N. A., Amaral A. J., Archibald A. L., Beever J. E., et al. (2009). Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4:e6524. 10.1371/journal.pone.0006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay K., Smuts M., Els H. C. (2000). Adding value to South African landrace breeds conservation through utilisation. Anim. Genet. Resour. Inf. 27 9–15. [Google Scholar]
- Ramsay K. A., Reed D. S., Bothma A. J., Lepen J. M. (1994). Profitable and Environmentally Effective Farming with Early-Domesticated Livestock in Southern Africa. Pretoria: Department of Agriculture. [Google Scholar]
- Raschetti M., Castiglioni B., Caroli A., Guiatti D., Pagnacco G., Chessa S. (2013). SNP identification in swine candidate genes for meat quality. Livest. Sci. 155 165–171. 10.1111/age.12388 [DOI] [PubMed] [Google Scholar]
- Rege J. E. O., Gibson J. P. (2003). Animal genetic resources and economic development: issues in relation to economic valuation. Ecol. Econ. 45 319–330. 10.1016/S0921-8009(03)00087-9 [DOI] [Google Scholar]
- Reyer H., Oster M., Magowan E., Dannenberger D., Ponsuksili S., Wimmers K. (2017a). Strategies towards improved feed efficiency in pigs comprise molecular shifts in hepatic lipid and carbohydrate metabolism. Int. J. Mol. Sci. 18:1674. 10.3390/ijms18081674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyer H., Shirali M., Ponsuksili S., Murani E., Varley P. F., Jensen J., et al. (2017b). Exploring the genetics of feed efficiency and feeding behaviour traits in a pig line highly selected for performance characteristics. Mol. Genet. Genomics 292 1001–1011. 10.1007/s00438-017-1325-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischkowsky B., Pilling D. (eds) (2007). The State of the World’s Animal Genetic Resources for Food and Agriculture. Rome: Food and Agriculture Organization. [Google Scholar]
- Rookmaaker L. C. (1989). The Zoological Exploration of Southern Africa 1650- 1790. Rotterdam: A.A Balkema. [Google Scholar]
- Romero-Suarez S., Shen J., Brotto L., Hall T., Mo C., Valdivia H. H., et al. (2010). Muscle-specific inositide phosphatase (MIP/MTMR14) is reduced with age and its loss accelerates skeletal muscle aging process by altering calcium homeostasis. J. Aging 2 504–513. 10.18632/aging.100190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropka-Molik K., Bereta A., Żukowski K., Tyra M., Piórkowska K., Żak G., et al. (2018). Screening for candidate genes related with histological microstructure, meat quality and carcass characteristic in pig based on RNA-seq data. Asian Australas J. Anim. Sci. 31 1565–1574. 10.5713/ajas.17.0714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosenvelt T., Heller E. (1915). Life-histories of African game animals. London: John Murray. [Google Scholar]
- Ross J. W., Hale B. J., Gabler N. K., Rhoads R. P., Keating A. F., Baumgard L. H. (2015). Physiological consequences of heat stress in pigs. Anim. Prod. Sci. 55 1381–1390. 10.1071/AN15267 [DOI] [Google Scholar]
- Rothschild M. F. (2010). Association of Genetic Markers with Structural Soundness and Its Relationship to Gilt Development and Sow Longevity Available online at: https://www.pork.org/wp-content/uploads/2009/06/06-019-ROTHSCHILD-ISU.pdf (accessed February 20, 2017). [Google Scholar]
- Rothschild M. F., Ruvinsky A. (2010). The Genetics of the Pig. Wallingford: CABI. [Google Scholar]
- Rui L., Sun D. X., Wang Y., Yu Y., Zhang Y., Chen H., et al. (2013). Fine mapping QTLs affecting milk production traits on BTA6 in Chinese Holstein with SNP markers. J. Integr. Agric. 12 110–117. 10.1016/S2095-3119(13)60211-7 [DOI] [Google Scholar]
- Ryu J., Lee C. (2016). Genetic association of marbling score with intragenic nucleotide variants at selection signals of the bovine genome. Animal 10 566–570. 10.1017/S1751731115002633animal [DOI] [PubMed] [Google Scholar]
- Salehi A., Sobhani R., Aminafshar M., Sayyadnejhad M. B., Nasiri K. (2015). Single nucleotide of FGF2 gene in Iranian Holstein proven bulls. Mol. Biol. Res. Commun. 4 57–62. [PMC free article] [PubMed] [Google Scholar]
- SanCristobal M., Chevalet C., Haley C. S., Joosten R., Rattink A. P., Harlizius M. A. M., et al. (2006). Genetic diversity within and between European pig breeds using microsatellite markers. Anim. Genet. 37 189–198. 10.1111/j.1365-2052.2005.01385.x [DOI] [PubMed] [Google Scholar]
- Scandura M., Iacolina L., Apollonio M. (2011). Genetic diversity in the European wild boar Sus scrofa: phylogeography, population structure and wild × domestic hybridization. Mamm. Rev. 41 125–137. 10.1111/j.1365-2907.2010.00182.x [DOI] [Google Scholar]
- Schwartz K., Lawn R. M., Wade D. P. (2000). ABC1 Gene expression and ApoA-I-Mediated cholesterol efflux regulated by LXR. Biochem. Biophys. Res. Commun. 274 794–802. 10.1006/bbrc.2000.3243 [DOI] [PubMed] [Google Scholar]
- Siddiq A., Gueorguiev M., Samson C., Hercberg S., Heude B., Levy-Marchal C., et al. (2007). Single nucleotide polymorphisms in the neuropeptide Y2 receptor (NPY2R) gene and assosciation with severe obesity in French white subjects. Diabetologia 50 574–584. 10.1007/s00125-006-0555-2 [DOI] [PubMed] [Google Scholar]
- Smith S. P., Phillips J. B., Johnson M. L., Abbot P., Capra J. A., Rokas A. (2019). Genome-wide association analysis uncovers variants for reproductive variation across dog breeds and links to domestication. Evol. Med. Public Health 2019 93–103. 10.1093/emph/eoz015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers M. J., Penzhorn B. L., Rasa A. E. (1994). Home range size use and dispersal of warthogs in the Eastern Cape, South Africa. J. Afr. Zool. 108 361–373. [Google Scholar]
- Somers M. J., Rasa O. A., Penzhorn B. L. (1995). Group structure and social behaviour of warthogs Phacochoerus aethiopicus. Acta Theriol. 40 257–281. [Google Scholar]
- Song J., Yang D., Ruan J., Zhang J., Chen Y. E., Xu J. (2017). Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci. Rep. 7:12202. 10.1038/s41598-017-12201-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Madrid B. S., Santacreu M. A., Fontanesi L., Blasco A., Ibañez-Escriche N. (2018). A genomic region on chromosome 17 has a major impact on litter size traits in rabbits. Proc. World Congr. Genet. Appl. Livestock Product. 11:163. [Google Scholar]
- Suchocki T., Wojdak-Maksymiec K., Szyda J. (2016). Using gene networks to identify genes and pathways involved in milk production traits in Polish Holstein dairy cattle. Czech J. Anim. Sci. 61 526–538. 10.17221/43/2015-CJAS [DOI] [Google Scholar]
- Sved J. A. (1971). Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2 125–141. 10.1016/0040-5809(71)90011-6 [DOI] [PubMed] [Google Scholar]
- Swart H., Kotze A., Olivier P. A. S., Grobler J. P. (2010). Microsatellite-based characterization of Southern African domestic pigs (Sus scrofa domestica). S. Afr. J. Anim. Sci. 40 121–132. 10.4314/sajas.v40i2.57280 [DOI] [Google Scholar]
- Sweett H., Miglior F., Livernois A., Fonseca P., Id-Lahoucine S., Troya E., et al. (2018). Genome-wide association study to identify genomic regions and single nucleotide polymorphisms functionally associated with bull fertility. J. Anim. Sci. 96 138–139. 10.1093/jas/sky404.303 [DOI] [Google Scholar]
- Taverner M. R., Dunkin A. C. (eds) (1996). Introduction to Pig Production. Pig Production. New York, NY: Elsevier Science. [Google Scholar]
- Tizioto P. C., Taylor J. F., Decker J. E., Gromboni C. F., Mudadu M. A., Schnabel R. D. (2015). Detection of quantitative trait loci for mineral content of Nelore longissimus dorsi muscle. Genet. Sel. Evol. 47:15. 10.1186/s12711-014-0083-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardo L. L., Silva F. F., Lopes M. S., Madsen O., Bastiaansen J. W. M., Knol E. F., et al. (2016). Revealing new candidate genes for reproductive traits in pigs: combining Bayesian GWAS and functional pathways. Genet. Sel. Evol. 48:9. 10.1186/s12711-016-0189-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigors S., Sweeney T., O’Shea C. J., Kelly A. K., O’Doherty J. V. (2016). Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal 10 1848–1855. 10.1017/S1751731116000847 [DOI] [PubMed] [Google Scholar]
- Visser C., Lashmar S. F., Van Marle-Köster E., Poli M. A., Allain D. (2016). Genetic diversity and population structure in South African, French and Argentinian angora goats from genome-wide SNP Data. PLoS One 11:e0154353. 10.1371/journal.pone.0154353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. T., Zhang Z., Fan M., Wang L., Dong G. L. (2010). Expression of CD112 in colon carcinoma tissues and cell lines and their clinical significance. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 26 477–479. [PubMed] [Google Scholar]
- Wang S., Li D., Zhu M., Xie R., Duan S., Yin Z., et al. (2019). A single nucleotide polymorphism of the porcine CXCL8 gene is associated with serum CXCL8 level. Ital. J. Anim. Sci. 18 474–479. 10.1080/1828051X.2018.1539349 [DOI] [Google Scholar]
- Wang X., Ma P., Liu J., Zhang Q., Zhang Y., Ding X., et al. (2015). Genome-wide association study in Chinese Holstein cows reveal two candidate gene for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 16:111. 10.1186/s12863-015-0263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. D., Harris J. M. (1977). Suid evolution and correlation of African hominid localities. Science 198 13–21. 10.1126/science.331477 [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Willi Y., Van Buskirk J., Hoffman A. A. (2006). Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37 433–458. 10.1146/annurev.ecolsys.37.091305.110145 [DOI] [Google Scholar]
- Wright S. (1978). Evolution and the Genetics of Population, Variability Within and Among Natural Populations. Chicago, IL: The University of Chicago Press. [Google Scholar]
- Wu X., Zhang Q., Xu S., Jin P., Luan P., Li Y., et al. (2016). Differential expression of six chicken genes associated with fatness traits in a divergently selected broiler population. Mol. Cell. Probe 30 1–5. [DOI] [PubMed] [Google Scholar]
- Xiang R., Oddy V. H., Archibald A. L., Vercoe P. E., Dalrymple B. P. (2016). Epithelial, metabolic and innate immunity transcriptomic signatures differentiating the rumen from other sheep and mammalian gastrointestinal tract tissues. PeerJ 4:e1762. 10.7717/peerj.1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Cui L., Perez-Enciso M., Traspov A., Crooijmans R. P. M. A., Zinovieva N., et al. (2017). Genome-wide SNP data unveils the globalization of domestication pigs. Genet. Sel. Evol. 49:71. 10.1186/s12711-017-0345-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar S., Cuellar-Partida G., McKnight C. M., Quach-Thanissorn P., Mountain J. A., Coroneo M. T. (2015). Genetic and environmental factors in conjunctival UV autofluorescence. JAMA Opthalmol. 133 406–412. 10.1001/jamaophthalmol.2014.5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G. M., Lhorente J. P., Carvalheiro R., Yáñez J. M. (2017). Bayesian genome-wide association analysis for body weight in farmed Atlantic salmon (Salmo salar L.). Anim. Genet. 48 698–703. 10.1111/age.12621 [DOI] [PubMed] [Google Scholar]
- Zadik B. J. (2005). The Iberian Pig in Spain and the Americas at the Time of Columbus. Master’s thesis, University of California, Berkeley, 36–51. [Google Scholar]
- Zeder M. A., Emshwiller E., Smith B. D., Bradley D. G. (2006). Documenting domestication: the intersection of genetics and archaeology. Trends Genet. 22 139–155. 10.1016/j.tig.2006.01.007 [DOI] [PubMed] [Google Scholar]
- Zhang W., Fan Z., Han E., Hou R., Zhang L., Galaverni M. (2014). Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet Plateau. PLoS Genet. 10:e1004466. 10.1371/journal.pgen.1004466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xiao Q., Zhang Q. Q., Sun H., Chen J. C., Li Z. C., et al. (2018). Genomic analysis reveals genes affecting distinct phenotypes among different Chinese and western pig breeds. Sci. Rep. 8:13352. 10.1038/s41598-018-31802-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Ji J., Peng S., Zhang Z., Fang S., Li L., et al. (2016). A GWA study reveals genetic loci for body conformation traits in Chinese Laiwu pigs and its implications for human BMI. Mamm. Genome 27 610–621. 10.1007/s00335-016-9657-4 [DOI] [PubMed] [Google Scholar]
- Zuo B., Xiong Y., Yang H., Wang J. (2007). Full-length cDNA, expression pattern and association analysis of the porcine FHL3 gene. Asian Australas J. Anim. Sci. 20 1473–1477. 10.5713/ajas.2007.1473 [DOI] [Google Scholar]
- Zuo B., Xiong Y. Z., Deng C. Y., Su Y. H., Wang M. G., Lei M. G., et al. (2004). cDNA cloning, genomic structure and polymorphism of the porcine FHL3 gene. Anim. Genet. 35 230–233. 10.1111/j.1365-2052.2004.01119.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cross validation plot for inferring the number of K populations in the analysis of population structure.
Genome-wide Manhattan plot of FST among the pig populations of (a) Alfred Nzo and Warthog, (b) Duroc and Warthog, (c) Kolbroek and Warthog, (d) Large White and Warthog, (e) South African landrace and Warthog, (f) Windsnyer and Warthog, (g) Indigenous and Duroc, (h) Villages and Duroc, (i) Villages and Kolbroek, (j) South African Landrace with Large White and Indigenous, (k) Indigenous and Vietnamese, (l) Villages and Wild Boar, (m) Villages and Vietnamese, (n) Wild Boar and Duroc. The solid lines indicate the FST ≥ 0.8 thresholds.
Average effective population size estimates across generations for the different populations analyzed.
Partitioning of genetic variance for the different populations analyzed.
FST between SA populations and genotypes from Burgos-Paz et al. (2013).
Data Availability Statement
The datasets generated for this study can be found in Dryadhttps://doi.org/10.5061/dryad.b0t10b0.