Summary
Recently, a unique form of X chromosome dosage compensation has been demonstrated in human preimplantation embryos, which happens through the dampening of X-linked gene expression from both X chromosomes. Subsequently, X chromosome dampening has also been demonstrated in female human pluripotent stem cells (hPSCs) during the transition from primed to naive state. However, the existence of dampened X chromosomes in both embryos and hPSCs remains controversial. Specifically, in preimplantation embryos it has been shown that there is inactivation of X chromosome instead of dampening. Here, we performed allelic analysis of X-linked genes at the single-cell level in hPSCs and found that there is partial reactivation of the inactive X chromosome instead of chromosome-wide dampening upon conversion from primed to naive state. In addition, our analysis suggests that the reduced X-linked gene expression in naive hPSCs might be the consequence of erasure of active X chromosome upregulation.
Keywords: X chromosome dampening, X chromosome inactivation, X chromosome upregulation, human pluripotent stem cells, naive and primed pluripotency, human embryos, XIST, single-cell RNA-seq
Highlights
-
•
Partial reactivation of inactive X, not X-dampening upon conversion of primed hPSCs to naive state
-
•
Reduction in X-linked gene expression in naive cells is independent of XIST
-
•
Presence of upregulated active X chromosome in primed hPSCs
-
•
X-upregulation erasure can cause X-linked expression reduction in naive hPSCs
In this article, Gayen and colleagues show that conversion of primed human pluripotent stem cells (hPSCs) to naive state is associated with partial reactivation of inactive X, not X dampening, and erasure of active X upregulation can lead to the reduction in X-linked gene expression in naive hPSCs.
Introduction
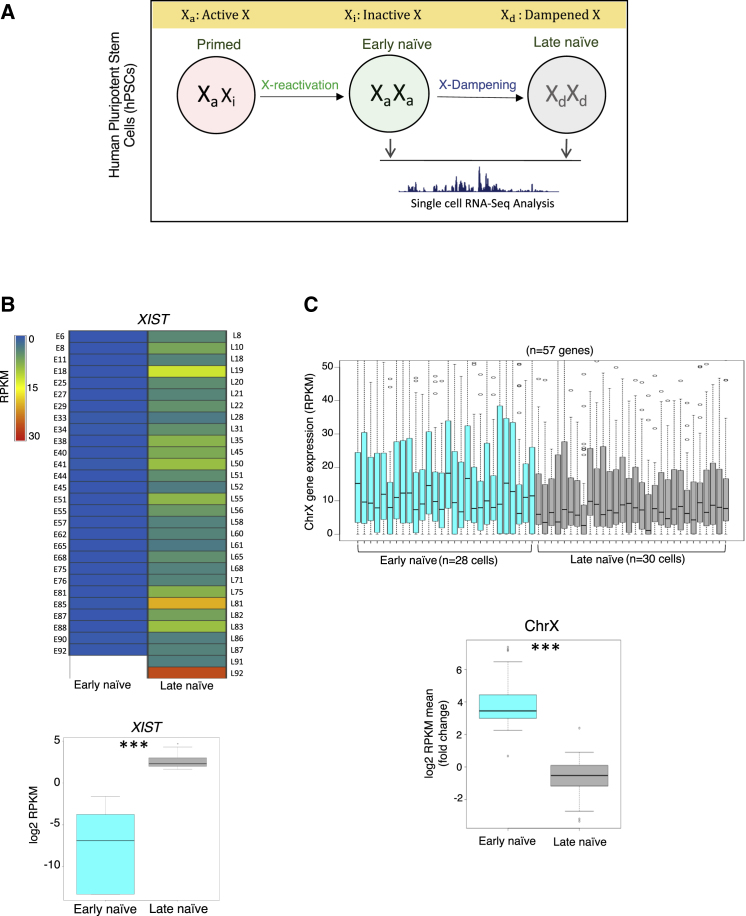
In therian mammals, to balance the X chromosome dosage between males and females, one X chromosome becomes inactivated in female cells (Lyon, 1961). The dosage imbalance between a single active X chromosome and two copies of autosomes (AA) is compensated through upregulation of the active X chromosome in both males and females (Deng et al., 2011, Larsson et al., 2019, Ohno, 1967). Recently, another form of X chromosome dosage compensation has been demonstrated in human preimplantation embryos, termed X chromosome dampening. Based on single-cell transcriptome analysis of human preimplantation embryos, Petropoulos et al. (2016) found that X-linked gene expression gradually decreased from morula to blastocyst stage while both X chromosomes were maintaining active state (Petropoulos et al., 2016). Based on these results, they proposed that dampening of X-linked gene expression from both X chromosome as a likely dosage compensation mechanism during human preimplantation development. However, the dampening phenomenon in human embryos remains controversial (De Mello et al., 2017, Saiba et al., 2018). De Mello et al. (2017) found evidence of inactivation of the X chromosome instead of dampening in preimplantation embryos when they reanalyzed the same transcriptome dataset of Petropoulos et al. (2016) with more stringency. In addition, Sahakyan et al., 2017a showed that naive human pluripotent stem cells (hPSCs) also exhibit the X chromosome dampening found in embryos (Sahakyan et al., 2017a). However, similar to the human embryos, X chromosome states in hPSCs remains unclear (Kaur et al., 2019, De Mello et al., 2017). Conventional hPSCs derived from blastocysts represent a primed state instead of naive state and are therefore unable to recapitulate the preimplantation X chromosome states (Nichols and Smith, 2009, Sahakyan et al., 2017b). To model the preimplantation X chromosome state, Sahakyan et al., 2017a converted primed hPSCs to the naive state using 5iLAF culture conditions (Figure 1A) (Theunissen et al., 2014). The primed cell line used for their study, UCLA1, harbored one active X chromosome and one inactive X chromosome. The transition of primed to naive state happened through an intermediate early naive state (Figure 1A). Primarily based on RNA-sequencing (RNA-seq) analysis using a bulk cell population, they suggested that the inactive X chromosome was reactivated upon transition from primed to the early naive state and that was followed by X chromosome dampening in late naive cells (Figure 1A). However, considering the heterogeneity of cell states during the conversion process, in this study we have analyzed an available single-cell RNA-seq (scRNA-seq) dataset of early and late naive cells from Sahakyan et al. (2017a) to gain better insight into X chromosomal states (Figure 1A).
Figure 1.
Transition of Early to Late Naive State is Associated with Increased XIST Expression and Reduction in X-Linked Gene Expression
(A) Schematic representation of different stages and corresponding X chromosome states of conversion of primed hPSCs to naive state as described in Sahakyan et al., 2017a. In this study, we performed analysis of the scRNA-seq dataset acquired from the early and late naive state.
(B) Comparison of XIST expression (RPKM) between early naive (n = 28) cells and late naive (n = 30) cells. p < 0.00001 (Mann-Whitney U test).
(C) Comparison of X-linked gene expression (n = 57 genes) between early naïve (n = 28) cells and late naive (n = 30) cells. p < 0.00001 (Mann-Whitney U test).
Results
Increased XIST Expression and Reduction in X-Linked Gene Expression upon Conversion of Early to Late Naive State
First, in early and late naive cells, we quantified the expression of XIST, a master regulator of X inactivation. We found that a majority of the early naive cells had very low level of XIST expression, whereas late naive cells mostly showed a higher level of XIST expression (Figure S1A). Overall our analysis revealed that transition of early to late naive state was associated with a significant increase of XIST expression (Figure S1B). Next, based on quality (reads per kilobase per million mapped reads [RPKM] sum and mean) and XIST expression level, we selected the top 28 early cells having a low level of XIST and the top 30 late cells having a higher level of XIST expression for further analysis (Figure 1B). Again, comparison of XIST expression in these cells (28 early versus 30 late) showed a significant increase in late naive cells compared with early naive cells (Figure 1B). In addition, we found that there was a significant reduction in X-linked gene expression in late naive cells compared with early naive cells (Figure 1C; Table S1). Altogether, our analysis of scRNA-seq data showed increased XIST expression and reduction in X-linked gene expression upon transition from the early to late naive state.
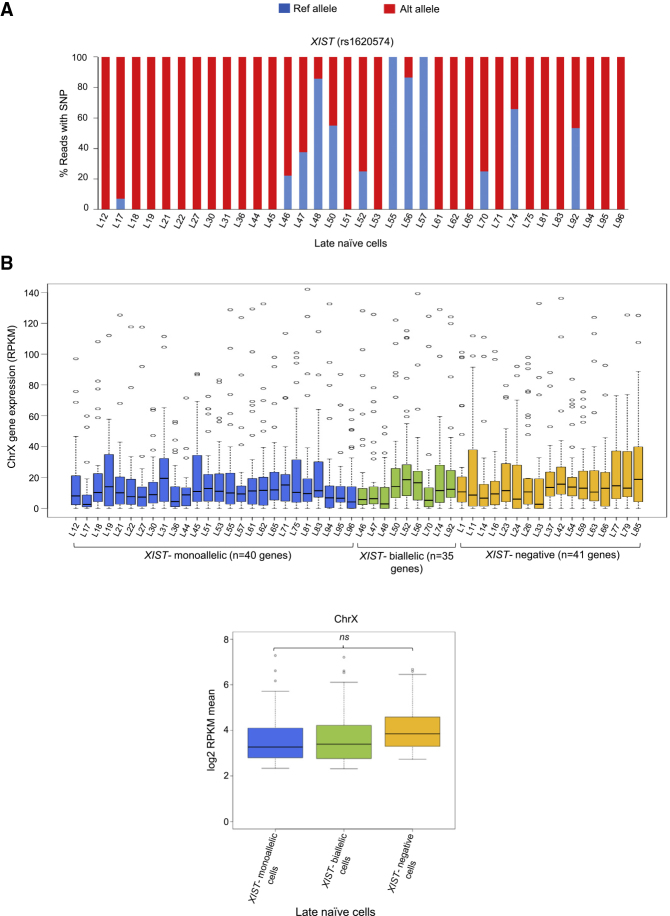
Reduction in X-Linked Gene Expression in Late Naive Cells Is Independent of XIST
It has been shown that X chromosome dampening in preimplantation embryos is associated with the expression of XIST from both X chromosomes (Petropoulos et al., 2016). In fact, X dampening initiates concomitantly with the initiation of XIST expression; therefore, it is thought that XIST might have an important role in the dampening process. To test this, we examined what fraction of cells shows XIST expression from both X chromosomes in late naive cells and whether the reduction in X-linked gene expression is restricted to the XIST biallelically expressed cells. We found that about 26% cells (9 of 35) expressed XIST from both X chromosomes (Figure 2A and Table S2). Interestingly, comparison of the global X-linked gene expression level of XIST-biallelic versus -monoallelic cells did not show any significant difference (Figure 2B and Table S2). Moreover, we observed that there was no significant difference in X-linked gene expression when we compared it against the XIST-negative cells (Figure 2B and Table S2). Based on these data, we concluded that reduction in X-linked gene expression in late naive cells was independent of XIST.
Figure 2.
Reduction in X-Linked Gene Expression in Late Naive Cells Is Independent of XIST
(A) Allelic expression of XIST in late naive cells (n = 35 cells).
(B) Comparison of X-linked gene expression among XIST-monoallelic (n = 26 cells and 40 genes), XIST-biallelic (n = 9 cells and 35 genes), and XIST-negative (n = 17 cells and 41 genes) cells. Non-significant at p < 0.05 (Mann-Whitney U test).
No Evidence of X Chromosome Inactivation or Dampening
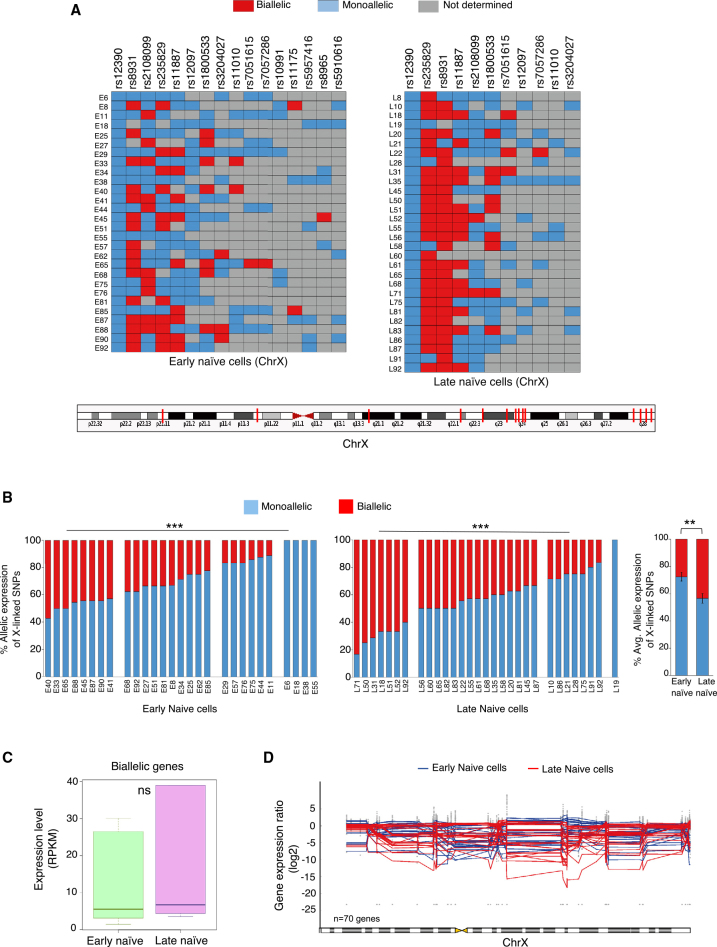
We then looked into possible mechanisms behind the reduction in X-linked gene expression in late naive hPSCs. First, we postulated that it could be due to X chromosome inactivation or dampening. To test for X inactivation, we profiled allelic expression of multiple X-linked genes distributed across the X chromosome based on single-nucleotide polymorphisms (SNPs) in early and late cells (Figure 3 and Table S3). We found that the majority of early cells showed monoallelic expression of many genes along with biallelically expressed X-linked genes, which indicated incomplete reactivation of X-linked genes (Figures 3A and 3B). Moreover, we found significant variation in the proportion of biallelic versus monoallelic genes between these cells (Figure 3B). Interestingly, late naive cells showed a pattern of allelic expression similar to that of as early cells. However, there was a significant increase in the fraction of SNPs showing biallelic expression in late naive cells compared with early naive cells (Figure 3B). Altogether, these data indicated that late naive cells do not harbor inactive X chromosome but rather have a partially reactivated X chromosome. If the late naive cells underwent X inactivation, monoallelic expression of most of the X-linked genes would be expected. In contrast, we observed increased biallelic expression upon transition from early to late stage. Next, we examined whether X dampening was causing reduction of X-linked gene expression in late naive cells. From the allelic analysis of X-linked gene expression it was clear that late naive cells harbor partially reactivated X chromosome. If these cells harbored dampened X chromosomes, we would have expected biallelic expression of most of the genes chromosome wide, which was not observed. In addition, we compared median expression of biallelically expressed genes of early cells with that of the late naive cells. If X dampening was occurring, we would have expected a significant decrease in median expression of biallelically expressed genes in late cells. However, significant differences were not observed (Figure 3C). Taken together, we concluded that there was neither X inactivation nor dampening upon conversion of early to late naive cells. Second, to determine whether loss of an X chromosome in late cells is causing the reduction in X-linked gene expression in late naive cells, we explored X chromosome ploidy of these cells. It was clear that cells harbored two X chromosomes as evidenced by the biallelic expression of some X-linked genes (Figure 3A). However, the possibility existed that these cells may lose part(s) of the X chromosome. To test for this, we analyzed the gene expression ratio of X-linked genes across the X chromosome, but significant differences between early and late naive cells were not identified (Figure 3D and Table S4). Therefore, we confirmed that loss of a portion of the X chromosome is not the cause of reduction in X-linked gene expression in late naive cells.
Figure 3.
Partial Reactivation of Inactive X Chromosome upon Conversion of Primed to Naive State
(A) Allelic expression analysis of X-linked genes in early and late naive cells at single-cell level. Bottom: genomic position on the X chromosome of the X-linked genes analyzed.
(B) Histogram showing the percentage of SNPs showing monoallelic and biallelic in each cell of early (n = 28) cells and late naive (n = 30) cells. p < 0.00001, p < 0.01 (average plot) (Student's t test).
(C) Comparison of median of expression level of biallelically expressed genes between early naive (n = 28) cells and late naive (n = 30) cells. Non-significant at p < 0.05 (Mann-Whitney U test).
(D) Ploidy analysis of the cells of early naive (n = 28) cells and late naive (n = 30) cells through the analysis of gene expression ratio across the X chromosome.
Erasure of Active X Upregulation Might Be the Cause of Reduced X-Linked Gene Expression in Late Naive Cells
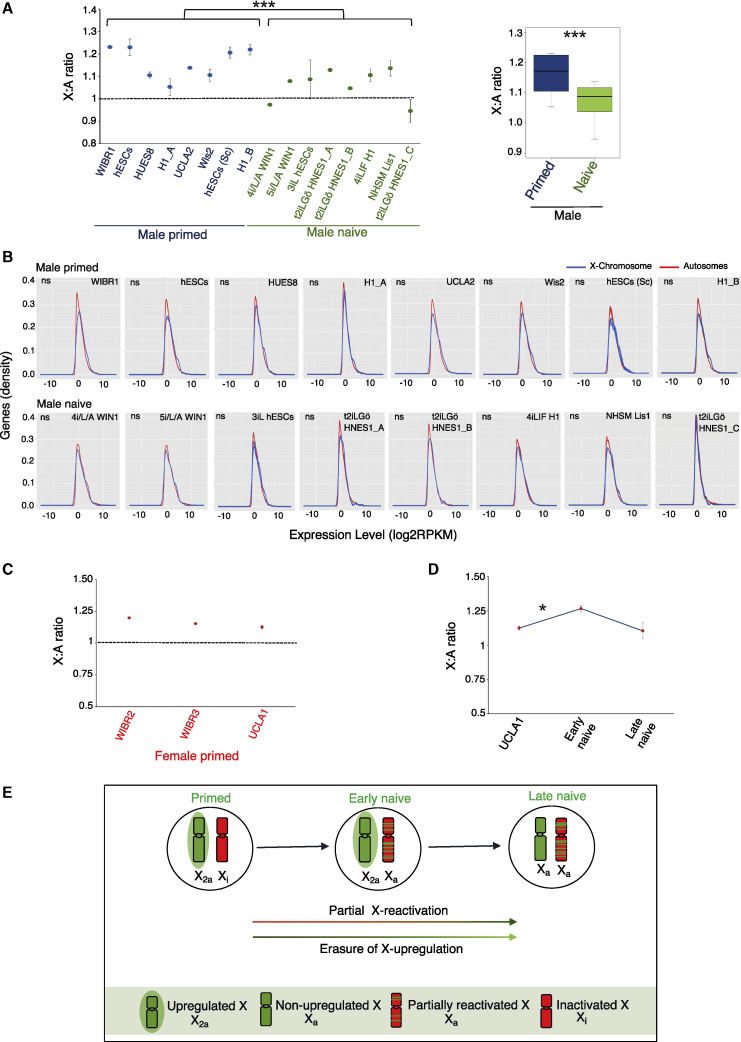
Next, we investigated whether the erasure of active X chromosome upregulation might be causing the reduction in X-linked gene expression in late naive cells as proposed by De Mello et al. (2017). In recent years, the existence of upregulated active X chromosome has been extensively demonstrated in mammals as hypothesized by Ohno(De Mello et al., 2017, Deng et al., 2013, Deng et al., 2011, Li et al., 2017, Lin et al., 2011, Sangrithi et al., 2017), although some studies have found lack of active X upregulation (Chen and Zhang, 2016, Xiong et al., 2010). To probe this further, we analyzed the X-to-autosomal (X:A) gene expression ratio of different male primed hPSC lines. If a diploid male cell has upregulated active X, the X:A ratio should be more than 0.5 and closer to 1. Indeed, we found that the X:A ratio of all male primed cells was greater than 1, indicating that primed hPSCs harbor an upregulated active X chromosome (Figure 4A and Table S5). We then asked whether the active X upregulation becomes erased in naive hPSCs. To test this, we compared the X:A ratio of male primed cells against different male naive cell lines. Interestingly, a significant reduction of X:A ratio in naive cells compared with the primed cells was observed, suggesting erasure of active X chromosome upregulation in naive cells (Figure 4A). We made sure that the X:A ratio was not affected by the difference between X-linked and autosomal gene expression distribution for each dataset (Figure 4B). We focused on male cells for analysis of active X upregulation, as in female cells X-linked gene expression is often confounded with X chromosome inactivation/reactivation/erosion. However, we profiled the X:A ratio in three different primed female cells (including UCLA1), which are known to harbor one inactive X chromosome (Figure 4C). We found that the X:A ratio of female primed cells was greater than 1, which indicated that these cells harbor an upregulated active X chromosome (Figures 4C and S2B). Next, we examined the X:A ratio dynamics during the primed-to-naive conversion of UCLA1 female cells. A significant increase in the X:A ratio upon transition from primed to early naive state was observed (Figures 4D and S2B). Considering the partial X reactivation upon transition of primed to early naive cells, it is obvious that the X:A ratio should increase provided that the active X upregulation is not completely erased. If there was complete erasure of active X upregulation, the X:A ratio should not have an observable increase. Therefore, these data indicated that erasure of active X upregulation was incomplete in the early naive state. Conversely, a decrease in the X:A ratio was observed in the late naive state compared with the early naive state, which suggested that erasure of active X upregulation was occurring (Figures 4D and S2B). In this scenario, it is possible that the erasure of active X upregulation led to the overall reduction in X-linked gene expression in late naive cells. However, the decrease in X:A ratio from early to late naive state was not significant as expected. We think that a significant increase in biallelic X-linked gene expression from early to late naive state was masking this to some extent. In summary, our data suggest that X dampening is not the determining factor for the reduction in X-linked gene expression in late naive cells, and erasure of active X upregulation lead to the decrease in X-linked gene expression (Figure 4E).
Figure 4.
Reduction in X-Linked Gene Expression in Naive hPSCs Might Be Due to the Erasure of Active X Upregulation
(A) Comparison of X:A ratio between male primed and naive hPSCs. p < 0.001(Student's t test). Number of independent replicates of RNA-seq dataset used were as follows. Male primed: WIBR1 (n = 1), hESCs (n = 3), HUES8 (n = 2), H1_A (n = 2), UCLA2 (n = 1), Wis2 (n = 2), hESCs (Sc) (n = 8 single cells), H1_B (n = 3). Male naive: 4i/L/A WIN1 (n = 1), 5i/L/A WIN1 (n = 1), 3iL hESCs (n = 3), t2iLGo HNES1_A (n = 3), t2iLGo HNES1_B (n = 1), 4iLIF H1 (n = 2), NHSM Lis1 (n = 2), t2iLGo HNES1_C (n = 2).
(B) Histograms representing the distribution of X-linked and autosomal gene expression for male primed and naive hPSCs with their different replicates (p > 0.05 by Kolmogorov-Smirnov test).
(C) Analysis of X:A ratio in different primed female hPSCs. Replicates of RNA-seq dataset used: WIBR2 and WIBR3 (n = 1); UCLA1 (n = 2).
(D) Comparison of X:A ratio in UCLA1 primed, early naive cells (cl4), and late naive cells (cl9 and cl12) based on two replicates of RNA-seq dataset for each. p < 0.05.
(E) Proposed model representing the X chromosome states during the conversion of primed hPSCs to the naive state.
Discussion
X chromosome states in female human preimplantation embryos have remained elusive to date. While it has been reported that preimplantation embryos carry dampened X chromosomes, other studies have provided evidence of inactivation of one of the X chromosomes. One of the major challenges in resolving this issue is the lack of availability of a surplus number of human embryos for experimentation. Therefore, hPSCs derived from human embryos serve as an alternative system. However, conventional hPSCs represent the primed state instead of the naive state of preimplantation embryos (Davidson et al., 2015, Sahakyan et al., 2017b). Recently, Sahakyan et al., 2017a converted primed hPSCs to naive state to model the X chromosome states of preimplantation embryos and suggested that naive hPSCs also carry dampened X chromosomes. However, our analysis indicates that the conversion of primed hPSCs to naive state is associated with the partial reactivation of the inactive X chromosome instead of the chromosome-wide dampening (Figure 4E). The main reason behind the dissimilar outcomes between our study and that of Sahakyan et al., 2017a is likely that their conclusion is primarily based on analysis of bulk RNA-seq of the cell population, whereas our conclusion is based on analysis of an scRNA-seq dataset. Since scRNA-seq provides better clarity regarding cellular state and gene expression to distinguish the heterogeneity among cells in a population, we believe that our analysis provides better insight into X chromosome states of hPSCs. In addition, our study suggests that erasure of active X upregulation might be causing the reduction in X-linked gene expression in late naive cells (Figure 4E).
Although these two studies led to dissimilar outcomes, several observations in our analyses were consistent with findings by Sahakyan et al., 2017a. For example, we also found that early naive cells were mostly XIST negative and transitioned to XIST positive in the late naive state, which was accompanied by a reduction in X-linked gene expression (Figure 1). Moreover, about 26% of cells in late naive state showed XIST expression from both X chromosomes, which was similar to what was reported by Sahakyan et al., 2017a. We should point out that in preimplantation embryos the majority of the cells (~85%) express XIST from both X chromosomes (Okamoto et al., 2011) and it is believed that XIST might have an important role in X dampening. However, we found that reduction in X-linked gene expression was independent of XIST as XIST-biallelic, -monoallelic, and -negative cells showed almost similar levels of gene expression in late naive cells (Figure 2B).
On the other hand, allelic analysis of X-linked gene expression at the single-cell level revealed striking differences between the two studies. We found that early and late naive cells harbored partially reactivated X chromosome as indicated by monoallelic expression of many genes in each cell of both cell states (Figure 3). This was contrary to the findings of Sahakyan et al., 2017a whereby they found that most of the genes had biallelic expression. Importantly, we found significant variations in allelic patterns of gene expression in different cells within a population, such as when the same genes, which were monoallelic in some cells, showed biallelic expression in other cells. In this scenario, bulk cell population analysis will always show biallelic gene expression, which might be the case for Sahakyan's observation. Interestingly, while Sahakyan et al., 2017a interpreted the reduction in X-linked gene expression in late naive cells as dampening, we found a lack of dampening, since there was no significant difference in the median expression of biallelically expressed genes between early and late naive cells (Figure 3C). In fact, Theunissen et al. (2016) found that female naive cells had significantly higher X-linked gene expression compared with male naive cells, which also suggested that naive female cells harbored active X chromosomes instead of dampened X chromosomes (Theunissen et al., 2016). In addition, we observed that the proportion of biallelically expressed genes increased significantly in late naive cells compared with early naive cells, which indicated that the cells were still undergoing the reactivation process. Some observations by Sahakyan et al., 2017a also indicated that there was an incomplete erasure of epigenetic memory on the inactive X in the naive state. They found, upon differentiation, that naive hPSCs underwent non-random X inactivation dissimilar to the normal development, and the same X chromosome that was inactive in the primed hPSCs was again inactivated. Even naive cells showed an accumulation of H3K27me3 repressive marks on one of the X chromosomes, which had not been observed in preimplantation blastocysts (Okamoto et al., 2011). We think that the presence of H3k27me3 is the result of incomplete erasure of inactive X marks, which is consistent with our observation of a partially reactivated X chromosome. Taken together, these findings suggest that naive cells were still in the process of removing inactive X epigenetic marks.
Our study suggests that erasure of active X upregulation might be causing the reduction in X-linked gene expression upon transition from early to late naive state. Many recent studies have demonstrated the existence of upregulated active X chromosomes in mammals (Deng et al., 2013, Larsson et al., 2019). We also found evidence for upregulated active X in different male primed hPSC lines (Figure 4A). Importantly, our analysis of male primed and naive cells strongly indicates that there is erasure of active X upregulation upon conversion of primed to naive state (Figure 4A). Erasure of upregulation has been demonstrated previously in spermatids during oogenesis and germ cell reprogramming of both sexes (Di and Disteche, 2006, De Mello et al., 2017, Sangrithi et al., 2017). Moreover, our observation is consistent with some other studies in mouse, which showed that naive embryonic stem cells (ESCs) have lower X:A ratio compared with the differentiated cells (Lin et al., 2007, Marks et al., 2015). In particular, during female ESC differentiation, upregulation increases concomitantly with the initiation of X inactivation (Larsson et al., 2019). In fact, Theunissen et al. (2016) found a reduction in X-linked gene expression in male naive hPSCs compared with that in primed hPSCs, which may be due to the erasure of active X chromosome upregulation (Theunissen et al., 2016). Furthermore, it has been shown that male blastocysts had significantly lower X:A ratio compared with the primed hPSCs (De Mello et al., 2017). Taken together, we believe that primed-to-naive conversion of hPSCs is accompanied by erasure of active X upregulation. In addition, our data indicate that the erasure of upregulation for female UCLA1 might still be ongoing in the early naive state and only reaches near completion in the late naive state, thereby leading to a reduction in X-linked gene expression upon transition from early to late naive state (Figure 4D). In fact, during germ cell reprogramming it has been shown that erasure of upregulation and reactivation of inactive X does not occur simultaneously; rather, loss of upregulation occurs later than loss of X inactivation (Sangrithi et al., 2017).
Collectively, our study indicates that the conversion of primed to naive state is associated with the incomplete reactivation of X chromosome rather than X inactivation or X dampening. Importantly, our data also indicates that erasure of active X upregulation lead a reduction of X-linked gene expression in naive hPSCs. Although our results argue against dampening and propose that erasure of active X upregulation leads to reduced X-linked gene expression in late naive cells, further work must be done using better scRNA-seq data. Finally, better culture conditions are necessary to establish naive hPSCs that recapitulate the X chromosome states of preimplantation embryos.
Experimental Procedures
Data Acquisition
RNA-seq datasets were acquired from the Gene Expression Omnibus under the accession number GEO: GSE87239 (Sahakyan et al., 2017a). For additional datasets, see Supplemental Experimental Procedures.
Variant Calling
First, reads were mapped to the human genome (hg38) using STAR. To mark the duplicate reads from the aligned reads of single cells, we used Picard tools v2.18.11 (https://broadinstitute.github.io/picard/). Next, we retrieved the allelic read counts for SNPs by using GATK (v3.8) “HaplotypeCaller.” We considered those SNPs for our analysis that were present in the UCLA1 cell line database (GSM2420529). Furthermore, we annotated those SNPs using dbSNP Build 152 (GRCh38.p12).
Allelic Expression Analyses
For allelic expression analysis, we considered the SNPs having ≥3 reads per SNP site in a cell. We proceeded with those SNPs having informative reads in at least five different cells of each category: early and late naive. The allelic expression was calculated by directly counting the allele-specific reads covering a SNP position mapped to the reference or the alternative allele and then dividing it by the total number of reads covering that position. A SNP was considered monoallelic if at least 90% of the allelic reads was coming from only one allele. We only considered allelic ratios of SNPs for those genes that had RPKM ≥1. Finally, we considered only those SNPs for which the allelic data were available in at least four cells for early and late naive cells. We validated the allele-specific expression pipeline through analysis of genes of an autosome (Chr17), which showed mostly biallelic expression of SNPs (Figure S2A). Moreover, SNPs belonging to the same gene showed almost similar allelic expression pattern in most of the cells with few exceptions.
Author Contributions
S.G., S. Mandal, D.C., and H.K. conceptualized the study. S.G. supervised the study. Bioinformatics analyses were done by S. Mandal and D.C. S.G., S. Mandal, D.C., M.A., and S. Majumdar wrote, edited, and proofread the manuscript. The final manuscript was edited and approved by all the authors.
Acknowledgments
We thank R.V. Pavithra, M. Karunyaa, and Amitesh Panda for their help in art work and discussion. This study is supported by Department of Biotechnology (DBT), India (BT/PR30399/BRB/10/1746/2018), DBT-Ramalingaswamy fellowship (BT/RLF/Re-entry/05/2016), and IISC-MHRD, India Startup grant awarded to S.G. We also thank DST-FIST [SR/FST/LS11-036/2014(C)], UGC-SAP [F.4.13/2018/DRS-III (SAP-II)], and DBT-IISc Partnership Phase II (BT/PR27952-INF/22/212/2018) for financial support.
Published: April 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.03.027.
Supplemental Information
References
- Chen X., Zhang J. The X to autosome expression ratio in haploid and diploid human embryonic stem cells. Mol. Biol. Evol. 2016;33:3104–3107. doi: 10.1093/molbev/msw187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K.C., Mason E.A., Pera M.F. The pluripotent state in mouse and human. Development. 2015;142:3090–3099. doi: 10.1242/dev.116061. [DOI] [PubMed] [Google Scholar]
- Deng X., Hiatt J.B., Nguyen D.K., Ercan S., Sturgill D., Hillier L.W., Schlesinger F., Davis C.A., Reinke V.J., Gingeras T.R. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Berletch J.B., Ma W., Nguyen D.K., Hiatt J.B., Noble W.S., Shendure J., Disteche C.M. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di K.N., Disteche C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Kaur H., Rv P., Gayen S. Dampened X-chromosomes in human pluripotent stem cells: dampening or erasure of X-upregulation? Chromosoma. 2019 doi: 10.1007/s00412-019-00717-5. [DOI] [PubMed] [Google Scholar]
- Larsson A.J.M., Coucoravas C., Sandberg R., Reinius B. X-chromosome upregulation is driven by increased burst frequency. Nat. Struct. Mol. Biol. 2019;26:963–969. doi: 10.1038/s41594-019-0306-y. [DOI] [PubMed] [Google Scholar]
- Li X., Hu Z., Yu X., Zhang C., Ma B., He L., Wei C., Wu J. Dosage compensation in the process of inactivation/reactivation during both germ cell development and early embryogenesis in mouse. Sci. Rep. 2017;7:3729. doi: 10.1038/s41598-017-03829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Gupta V., Vermilyea M.D., Falciani F., Lee J.T., O’Neill L.P., Turner B.M. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:2809–2820. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Halsall J.A., Antczak P., O’Neill L.P., Falciani F., Turner B.M. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nat. Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marks H., Kerstens H.H.D., Barakat T.S., Splinter E., Dirks R.A.M., van Mierlo G., Joshi O., Wang S.Y., Babak T., Albers C.A. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16:149. doi: 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mello J.C.M., Fernandes G.R., Vibranovski M.D., Pereira L.V. Early X chromosome inactivation during human preimplantation development revealed by single-cell RNA-sequencing. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-11044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Perspective naive and primed pluripotent states. Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Ohno S. Vol. 68. Springer-Verlag; 1967. p. 1375. (Sex Chromosomes and Sex-Linked Genes). [Google Scholar]
- Okamoto I., Patrat C., Thépot D., Peynot N., Fauque P., Daniel N., Diabangouaya P., Wolf J.P., Renard J.P., Duranthon V. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Petropoulos S., Deng Q., Panula S.P., Codeluppi S., Reyes A.P., Linnarsson S., Sandberg R., Lanner F. Single-cell RNA-seq reveals lineage and X-chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–1026. doi: 10.1016/j.cell.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Plath K., Rougeulle C. Regulation of X-chromosome dosage compensation in human: mechanisms and model systems. Philos. Trans. R. Soc. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0363. 20160363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiba R., Arava M., Gayen S. Dosage compensation in human pre-implantation embryos: X-chromosome inactivation or dampening? EMBO Rep. 2018;19:e46294. doi: 10.15252/embr.201846294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi M.N., Royo H., Mahadevaiah S.K., Ojarikre O., Bhaw L., Sesay A., Peters A.H.F.M., Stadler M., Turner J.M.A. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev. Cell. 2017;40:289–301.e3. doi: 10.1016/j.devcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O’Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Chen X., Chen Z., Wang X., Shi S., Wang X., Zhang J., He X. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat. Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.