Abstract
There is a growing interest in breeding and production of hexaploid triticale (× Triticosecale Wittmack ex A. Camus) in European Union and in the world. It is reported that triticale can be an alternative to wheat (Triticum aestivum L.) for livestock feed production and has a potential to become preferred industrial energy crop. Fungal diseases, mainly leaf and stripe rusts, are the limiting factors of triticale growth and yield. Geneticists and breeders are now focusing on accumulation of the major genes for durability of rust resistance. Slow-rusting genes Lr34/Yr18 and Lr46/Yr19 are being exploited in many wheat breeding programs. This type of horizontal resistance is reported to be effective over space and time. Classical breeding techniques supported by marker-assisted selection (MAS) are the main tools in breeding programs. The aim of this study was to assess the possibility of transfer of slow-rusting genes from resistant genotypes of wheat into hexaploid triticale through cross-hybridizations. A total of 5,094 manual pollinations were conducted between two triticale cultivars Fredro and Twingo and 33 accessions of common wheat, which were reported as sources of slow-rusting resistance genes. The investigation of the slow-rusting gene transmission was performed using both molecular markers analyses and genomic in situ hybridization (GISH). In total, 34 F1 hybrid plants were obtained, and 29 of them carried both slow-rusting loci. Therefore, these hybrids may be used for triticale prebreeding program.
Keywords: genomic in situ hybridization, molecular markers, resistance genes, slow rust, triticale, wheat
Introduction
Triticale (× Triticosecale Wittmack ex A. Camus) is an artificial manmade grain, created through hybridization of wheat derivatives (Triticum sp.) with rye (Secale cereale L.). Initially, triticale was expected to combine the high robustness of rye with the great yield potential (Kwiatek and Nawracała, 2018). At present, this crop is widely used as an alternative for other cereals, mostly for wheat, as a valuable source of livestock feed (Ayalew et al., 2018). In recent years, the interest in triticale production has increased because of its potential to become an industrial energy crop (Sanaei and Stuart, 2018). The economic importance of triticale is reflected by a significant acreage in Europe, which accounts for 90% of the world production (FAO, 2018). Triticale, as an artificial crop, is characterized by low genetic variation. Moreover, the increasing harvesting area of this crop is associated with the rapid development of fungal pathogens, which are continuously adapting to triticale. Hence, the need to develop more basic and applied research on this crop connected with resistance breeding.
Leaf rust caused by Puccinia triticina Eriks. and stripe rust caused by Puccinia striiformis Westend f. sp. tritici are the most important foliar diseases of triticale, as well as other major cereal species. It is reported that rye genome in triticale improves the resistance for leaf rust (Mikhailova et al., 2009). Genetic resistance is the most economical and preferable method of reducing yield losses due to leaf rust (Kolmer, 1996). More than 70 leaf rust resistance genes (Lr) have been mapped to specific chromosomes and described in the Catalog of Gene Symbols for Wheat (McIntosh, 1995; McIntosh et al., 2016). Many of the leaf and stripe rust resistance genes are race specific and have been overcome by the new races of Puccinia spp. (Periyannan et al., 2017). However, in the wheat genepool, three genes were identified that confer durable adult plant resistance against multiple fungal diseases (Ellis et al., 2014). These genes were named Lr34 (= Yr18/Sr57/Pm38; chromosome 7D), Lr46 (= Yr29/Sr58/Pm39; 1B), and Lr67 (= Yr46/Sr55/Pm46; 4D) (Dyck, 1977; Dyck and Samborski, 1979; Singh et al., 1998). Their expression results in partial resistance against all races of the fungal wheat pathogens causing leaf rust, stripe rust, and powdery mildew (Blumeria graminis f. sp. tritici).
New plant-breeding techniques (NPBTs) developed over the past two decades have provided comprehensive opportunities for efficient trait development in crops (Eriksson et al., 2018). For example, transgenic barley (Hordeum vulgare L.) lines expressing slow-rusting allele Ta-Lr34res of wheat showed enhanced resistance against leaf rust and powdery mildew of barley (Boni et al., 2018). Unfortunately, diverging opinions and politically motivated arguments are hampering the political progress to decide on the regulation of NPBTs in the European Union. Hence, classical breeding techniques, such as wide cross-hybridization supported by marker-assisted selection (MAS), are still the main tool for breeding programs.
The aim of this work was to assess the possibility of transfer of two main slow-rusting genes Lr34 and Lr46 from wheat-resistant genotypes derived from the United States Department of Agriculture (USDA)/Agricultural Research Service (ARS) Small Grains Laboratory, Aberdeen (ID, United States) gene bank into two elite cultivars of hexaploid triticale through cross-hybridizations. The evaluation of the chromosome transmission was performed using both molecular marker analyses and genomic in situ hybridization (GISH). This initial study is to obtain the starting pool triticale–wheat hybrids, carrying slow-rusting genes, which will be applied in the triticale breeding programs.
Materials and Methods
Plant Material
Seeds of 33 accessions of wheat, which were reported as sources of slow-rusting genes (Table 1), and two cultivars of winter triticale were germinated on Petri dishes. The plantlets were transferred to soil and cultivated for 6–8 weeks under short-day conditions (8 h light/16 h dark, 20/18°C). Finally, the winter genotypes were transferred for 6 weeks to vernalizing conditions (10 h light/14 h dark, 4°C) and then returned to long-day conditions (13 h light/11 h dark, 20/16°C).
TABLE 1.
Pollen viability of wheat accessions and seed production ability of triticale after pollination with pollen grains from wheat components.
| No. | Pollinators (Triticum aestivum L.) | Maternal components (× Triticosecale Wittmack) | ||||||||||||
| Cultivar/genotype | Plant ID | Growth habit | Source | csLV34 linked to Lr34 | Xgwm44 linked to Lr46 | csLV46G22 linked to Lr46 | Pollen viability (%) | Triticale cv. “Fredro” (Danko | Triticale cv. “Twingo” (Danko | |||||
| Breeding Company, Poland) | Breeding Company, Poland) | |||||||||||||
| Number of pollinated flowers | Number of hybrid seeds | Crossing efficiency (%) | Number of pollinated flowers | Number of hybrid seeds | Crossing efficiency (%) | |||||||||
| 1. | Frontana | Cltr 12470 | Spring | USDA | + | + | + | 90.29 | 62 | 0 | 0.00 | 124 | 0 | 0.00 |
| 2. | Chris | Cltr 13751 | Spring | USDA | + | + | − | 99.36 | 68 | 0 | 0.00 | 350 | 5 | 2.86 |
| 3. | H_N_ROD6_13751 | PI 191772 | Spring | USDA | − | − | − | 89.33 | not crossed | not crossed | ||||
| 4. | Frontana 3671 | PI 193932 | Spring | USDA | + | + | + | 92.23 | 56 | 0 | 0.00 | 56 | 12 | 21.43 |
| 5. | Frontana LF 320 | PI193933 | Spring | USDA | + | + | + | 88.29 | 58 | 0 | 0.00 | 116 | 0 | 0.00 |
| 6. | Frontana LF 321 | PI193934 | Spring | USDA | + | + | 88.74 | 52 | 2 | 3.85 | 100 | 2 | 2.00 | |
| 7. | Fronthatch-1 | PI 290745 | Spring | USDA | + | + | + | 89.36 | 60 | 0 | 0.00 | 64 | 0 | 0.00 |
| 8. | Fronthatch-2 | PI 297014 | Spring | USDA | + | + | + | 89.09 | 56 | 0 | 0.00 | 66 | 0 | 0.00 |
| 9. | Fronthatch-3 | PI 299419 | Spring | USDA | + | + | − | 88.92 | 62 | 0 | 0.00 | 58 | 0 | 0.00 |
| 10. | Toropi | PI 344200 | Spring | USDA | − | − | + | 90.34 | not crossed | not crossed | ||||
| 11. | Fortaleza | PI 351779 | Spring | USDA | + | + | − | 89.74 | 64 | 0 | 0.00 | 56 | 0 | 0.00 |
| 12. | Sparrow | PI 519725 | Spring | USDA | + | + | + | 91.24 | 62 | 0 | 0.00 | 58 | 1 | 1.72 |
| 13. | Pavon F76 | PI 519847 | Spring | USDA | − | + | + | 89.60 | 88 | 0 | 0.00 | 100 | 0 | 0.00 |
| 14. | Pavon 76 | PI 520003 | Spring | USDA | − | + | − | 89.36 | 64 | 0 | 0.00 | 64 | 0 | 0.00 |
| 15. | Pavon-1 | PI 520054 | Spring | USDA | + | + | + | 89.81 | 68 | 0 | 0.00 | 48 | 0 | 0.00 |
| 16. | Pavon-2 | PI 520172 | Spring | USDA | − | + | + | 88.24 | 62 | 0 | 0.00 | 64 | 0 | 0.00 |
| 17. | Myna | PI 520340 | Spring | USDA | + | + | − | 90.46 | 58 | 0 | 0.00 | 68 | 0 | 0.00 |
| 18. | Junco | PI 519947 | Spring | USDA | − | − | − | 88.10 | not crossed | not crossed | ||||
| 19. | Tanager | PI 519878 | Spring | USDA | − | + | + | 90.13 | 62 | 0 | 0.00 | 56 | 0 | 0.00 |
| 20. | Parula | PI 520340 | Spring | USDA | + | − | − | 89.36 | 68 | 0 | 0.00 | 170 | 0 | 0.00 |
| 21. | Rayon 89 | PI 591786 | Spring | USDA | − | − | − | 95.81 | not crossed | not crossed | ||||
| 22. | Cumpas 88 | PI 591786 | Spring | USDA | + | − | + | 91.20 | 58 | 0 | 0.00 | 62 | 0 | 0.00 |
| 23. | Mochis 88 | PI 591791 | Spring | USDA | + | − | − | 94.83 | 54 | 0 | 0.00 | 160 | 9 | 13.13 |
| 24. | P8901-AP | PI 613175 | Spring | USDA | + | − | − | 89.19 | 56 | 0 | 0.00 | 64 | 0 | 0.00 |
| 25. | P8901-AQ | PI 613176 | Spring | USDA | + | − | − | 91.08 | 64 | 0 | 0.00 | 64 | 0 | 0.00 |
| 26. | Tlaxcala F2000 | PI 619634 | Spring | USDA | + | + | + | 89.34 | 54 | 0 | 0.00 | 68 | 0 | 0.00 |
| 27. | Lr34 | GSTR 433 | Spring | USDA | + | − | − | 89.41 | 62 | 0 | 0.00 | 58 | 0 | 0.00 |
| 28. | IWA8608696 | PI 624623 | Spring | USDA | + | + | − | 88.34 | 58 | 0 | 0.00 | 56 | 0 | 0.00 |
| 29. | Anza | PI 638742 | Spring | USDA | + | + | − | 90.91 | 56 | 0 | 0.00 | 62 | 0 | 0.00 |
| 30. | UC1110 | PI 671999 | Spring | USDA | + | + | − | 89.63 | 64 | 0 | 0.00 | 128 | 0 | 0.00 |
| 31. | Kern | PI 672001 | Spring | USDA | + | + | − | 88.59 | 68 | 0 | 0.00 | 164 | 0 | 0.61 |
| 32. | TX89D6435 | PI 584759 | Winter | USDA | + | + | − | 93.70 | 62 | 0 | 0.00 | 658 | 3 | 0.46 |
| 33. | Purdue | Cltr 13227 | Winter | USDA | + | + | + | 91.13 | 68 | 0 | 0.00 | 138 | 0 | 0.00 |
| Mean | 90.46 | 61.86 | 0.07 | 0.001 | 113.79 | 1.10 | 0.01 | |||||||
| Total | n/a | 1,794 | 2 | n/a | 3,300 | 32 | n/a | |||||||
Pollen Viability Evaluation and Cross-Hybridizations
F1 hybrid plants were obtained through cross-hybridization between triticale (female parent) and wheat (pollinator) performed in the glasshouse chambers of the Department of Genetics and Plant Breeding at the Poznań University of Life Sciences (PULS), Poland. In this purpose, 10 seeds from each accession were sown in plastic pots in three replications. Evaluation of pollen vitality was made on the wheat genotypes. Pollen grains were stained with 2% aceto-carmine in glycerine (vol. 1:1) for the presence of cytoplasm. The evaluation of pollen vitality was made using a Delta Genetic Pro microscope (Delta Optical, Poland).
Florets of maternal components (triticale plants) were emasculated to avoid self-fertilization in order to cross with the pollen of wheat. The emasculated florets were counted and pollinated with freshly collected pollen of wheat within a period of 3 months (April–June 2019). The percentage ratio of the total amount of seeds from each plant with the total amount of pollinated flowers of each plant was calculated [crossing efficiency (CE)].
Identification of Molecular Markers Linked to Lr34 and Lr46 Genes
The following molecular markers csLV34 (Lagudah et al., 2006) linked to Lr34 and Xgwm44 (William et al., 2003) and csLV46G22 (Lagudah pers. comm.) linked to Lr46 were used to confirm the presence of alleles connected with slow-rusting resistance in wheat genotypes and F1 hybrids. Genomic DNA was extracted from seedling leaves using the Plant and Funghi DNA Purification Kit (EURx, Poland). The PCR reaction volume was 20 μl, consisting of 100 nM each of the two primers, 2 × TaqNova-DNA hot-start polymerase buffer (Blirt, Poland), and 50 ng of genomic DNA as template. A typical PCR procedure was as follows: 5 min at 95°C, then 35 cycles of 30 s at 94°C, 30 s at 50–60°C (Skowrońska et al., 2019), 1 min at 72°C, and 5 min at 72°C. PCR products were run on 2% agarose gel (Lab Empire, Poland) with 1% Tris–borate–ethylenediaminetetraacetic acid (TBE) buffer. DNA was visualized via Midori Green Direct (Nippon Genetics Europe, Germany) that was added to the samples.
Genomic in situ Hybridization
Chromosome sets of F1 hybrids were analyzed using GISH. This approach was to calculate the cross-hybridization efficiency (CE). It was performed on mitotic chromosomes of root meristem cells collected from hybrid plants. Mitotic metaphase accumulation and fixation procedures were carried out according to Kwiatek et al. (2016). Total genomic DNA was isolated using the DNeasy Plant Maxi Kit 24 (Qiagen, Germany). DNA of Aegilops tauschii Coss. (2n = 2x = 14 chromosomes; DD; PI 603226; US National Plant Germplasm System), a progenitor of the D-genome of wheat, was labeled by nick translation with Atto-488 dye (Atto-488NT kit; Jena Bioscience, Germany) for the investigation of D-genome chromosomes. Total genomic DNA of rye (Imperial; PI 323382; US National Plant Germplasm System) was labeled in the same way by Atto-550 dye. Blocking DNA from T. durum Desf. (2n = 4x = 28 chromosomes; AABB; Ceres; HR Smolice; Poland) was sheared by boiling for 30–45 min and used at a ratio of 1:50 (probe/block). GISH was performed according to Kwiatek et al. (2016). Slides were analyzed with the use of an Axio Observer 7 (Carl Zeiss, Oberkochen, Germany) fluorescence microscope. Image processing was done using ZEN Pro software (Carl Zeiss, Oberkochen, Germany). Each plant was evaluated by an analysis of chromosome sets of 10 cells.
Results
Identification of Molecular Markers Linked to Loci of Slow-Rusting Genes in Wheat Accessions
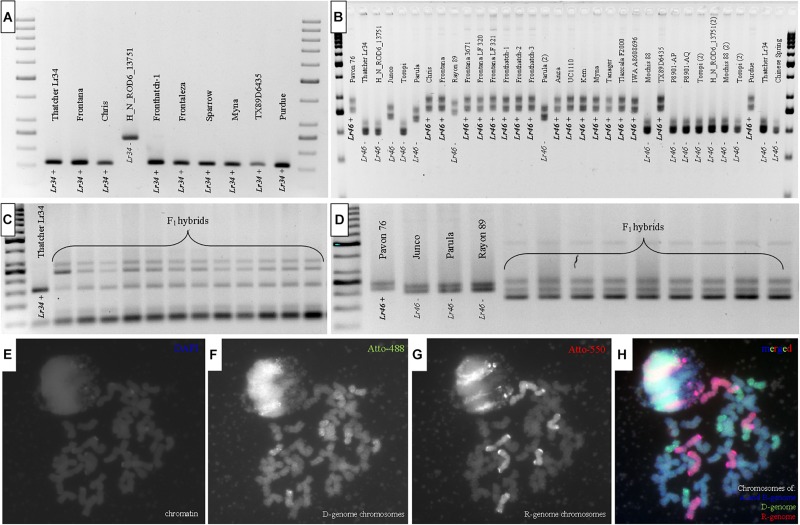
The molecular identification of Lr34 and Lr46 alleles was performed in all wheat and triticale cultivars and genotypes using csLV34 and Xgwm44 markers, respectively. The Thatcher near-isogenic line Lr34 (Lr34; GSTR 433; USDA) and Pavon 76 (Lr46; PI 520003; USDA) lines were used as a positive control (R; Figures 1A,B). The PCR reactions using csLV34 resulted in the amplification products of 160 bp in size and were found in DNA extracts of Lr34-positive control (GSTR 433) and 24 wheat cultivars and genotypes (Table 1). A PCR product of 240 bp in size was identified for eight wheat genotypes, indicating susceptible (S) allele (Figure 1A and Table 1). PCR reaction with Xgwm44 marker yielded two amplicons: 260 and 280 bp for a product for Pavon76 and 17 wheat accessions (Figure 1B). Eleven wheat genotypes were characterized by 200 and 230 bp bands. Two wheat accessions showed 240- and 270-bp bands. “Toropi” wheat showed 235- and 265-bp band pattern (S-allele). The second Lr46 marker, csLV46G22, gave different results, compared to Xgwm44. R-allele was identified in 16 wheat accessions. The results of Xgwm44 and csLV46G22 markers were similar for 21 of 33 wheat accessions. Ten wheat accessions carried all three resistant allele markers for Lr34 (csLV34) and Lr46 (both Xgwm44 and csLV46G22) (Table 1).
FIGURE 1.
Amplification products of PCR reactions of (A) wheat accessions (pollen donors) and (C) F1 hybrids with csLV34 marker linked to Lr34 (= Yr18/Sr57/Pm38) locus and (B) wheat accessions and (D) F1 hybrids with Xgwm44 marker linked to Lr46 (= Yr29/Sr58/Pm39). Lr34 or Lr46+ indicate resistant alleles; Lr34- or Lr46– indicate susceptible alleles. Genomic in situ hybridization preformed on mitotic chromosomes of plants derived through cross-hybridization between triticale and wheat: (E) chromatin (DAPI), (F) D-genome chromosmes (Atto-488, green), (G) R-genome chromosomes (Atto-550, red), (H) merged.
Pollen Viability of Wheat Accessions With Slow-Rusting Genes
The viability of pollen grains of wheat accessions was high and ranged between 88.29 and 99.36% (Table 1). The highest pollen viability was observed for “Chris” wheat cultivar. The lowest viability of pollen grains was evaluated in “Junco” cultivar. The mean pollen viability evaluated for all 33 wheat accession used for this experiment amounts to 90.45%. Following wheat accessions: Frontana, Frontana 3671, Toropi, Sparrow, Myna, Tanager, Rayon 89, Mochis 88, P8901-AQ, Anza, TX89D6435, and Purdue showed high viability of pollen grains (in excess of 90%).
Evaluation of Triticale × Wheat Cross-Hybridizations
Cross-hybridization of winter triticale with wheat slow-rusting pollinators was performed in order to evaluate the CE through triticale × wheat crossing combinations (Table 1). Total amount of 5,094 flowers of triticale were pollinated with wheat pollen grains. The crossing evaluation showed, that in general, triticale cv. Twingo produced more hybrid seeds (32 seeds) than cv. Fredro (two seeds). Overall, hybrid seeds were provided by only seven crossing combinations. The total CE for both triticale cultivars was surprisingly low and amounts to 0.1% for Twingo × wheat combinations and 0.001% for Fredro × wheat combinations. Twingo × Frontana 3671 cross-hybridization was the most effective and yielded 12 seeds (CE, 21.43%).
Identification of Molecular Markers Linked to Loci of Slow-Rusting Genes in Triticale × Wheat F1 Hybrids
Similar set of three molecular markers, csLV34, Xgwm44, and csLV46G22 was used to screen the presence of Lr34 and Lr46 loci in F1 hybrids (H) (Figure 1C and Table 2). The PCR reactions for csLV34 marker yielded three types of amplicons: 160 bp from resistant wheat (chromosome 7D) and 250 and 300 bp from R-genome of triticale (Figure 1C). The evaluation of electropherograms generated after separation of PCR products, which were amplified using Xgwm44 marker, was more difficult. Three products were amplified using DNA samples from F1 hybrids (H): 150, 200, and 240 bp, which were characteristics for both 1B chromosome that originated from resistant wheat accessions and 1B chromosome from triticale (Figure 1D). The resistant allele of csLV46G22 marker was identified in plants of four crossing combinations. Moreover, those combinations (Fredro × Frontana LF 321, Twingo × Frontana LF 321, Twingo × Frontana 3671, and Twingo × Sparrow) carried all three resistant allele markers linked to Lr34 and Lr46 slow-rusting genes (Table 2).
TABLE 2.
Chromosome sets of triticale × wheat hybrids and the identification of marker loci linked to slow-rusting genes.
| No. | Crossing combination | F1 plant ID | Number of chromosomes | Number of A- and B-genome chromosomes | Number of R-genome chromosomes | Number of D-genome chromosomes | csLV34 linked to Lr34 | Xgwm44 linked to Lr46 | csLV46G22 linked to Lr46 |
| 1. | Fredro × Frontana LF 321 | 1 | 42 | 28 | 7 | 7 | + | + | + |
| 2. | 2 | 42 | 28 | 7 | 7 | + | + | + | |
| 3. | Twingo × Chris | 1 | 42 | 28 | 7 | 7 | + | + | − |
| 4. | 2 | 42 | 28 | 7 | 7 | + | + | − | |
| 5. | 3 | 42 | 28 | 7 | 7 | + | + | − | |
| 6. | 4 | 42 | 28 | 7 | 7 | + | + | − | |
| 7. | 5 | 42 | 28 | 7 | 7 | + | + | − | |
| 8. | Twingo × Frontana 3671 | 1 | 42 | 28 | 7 | 7 | + | + | + |
| 9. | 2 | 42 | 28 | 7 | 7 | + | + | + | |
| 10. | 3 | 42 | 28 | 7 | 7 | + | + | + | |
| 11. | 4 | 42 | 28 | 7 | 7 | + | + | + | |
| 12. | 5 | 42 | 28 | 7 | 7 | + | + | + | |
| 13. | 6 | 42 | 28 | 7 | 7 | + | + | + | |
| 14. | 7 | 42 | 28 | 7 | 7 | + | + | + | |
| 15. | 8 | 42 | 28 | 7 | 7 | + | + | + | |
| 16. | 9 | 42 | 28 | 7 | 7 | + | + | + | |
| 17. | 10 | 42 | 28 | 7 | 7 | + | + | + | |
| 18. | 11 | 42 | 28 | 7 | 7 | + | + | + | |
| 19. | 12 | 42 | 28 | 7 | 7 | + | + | + | |
| 20. | Twingo × Frontana LF 321 | 1 | 42 | 28 | 7 | 7 | + | + | + |
| 21. | 2 | 42 | 28 | 7 | 7 | + | + | + | |
| 22. | Twingo × Sparrow | 1 | 42 | 28 | 7 | 7 | + | + | + |
| 23. | Twingo × Mochis 88 | 1 | 42 | 28 | 7 | 7 | + | − | − |
| 24. | 2 | 42 | 28 | 7 | 7 | + | − | − | |
| 25. | 3 | 42 | 28 | 7 | 7 | + | − | − | |
| 26. | 4 | 42 | 28 | 7 | 7 | + | − | − | |
| 27. | 5 | 42 | 28 | 7 | 7 | + | − | − | |
| 28. | 6 | 42 | 28 | 7 | 7 | + | − | − | |
| 29. | 7 | 42 | 28 | 7 | 7 | + | − | − | |
| 30. | 8 | 42 | 28 | 7 | 7 | + | − | − | |
| 31. | 9 | 42 | 28 | 7 | 7 | + | − | − | |
| 32. | Twingo × TX89D6435 | 1 | 42 | 28 | 7 | 7 | + | + | − |
| 33. | 2 | 42 | 28 | 7 | 7 | + | + | − | |
| 34. | 3 | 42 | 28 | 7 | 7 | + | + | − |
Chromosome Constitution of F1 Hybrids Revealed by Genomic in situ Hybridization
Evaluation of chromosome constitution was made for all 34 F1 seeds in order to confirm their hybrid origin (Figures 1E–H and Table 2). The karyotype for triticale × wheat F1 hybrid contained 14 chromosomes (seven pairs) of A-genome, the same amount of B-genome chromosomes, seven chromosomes (monosomic) belonging to D-genome (Figure 1F), which were derived from wheat parent, and seven monosomic chromosomes of R-genome derived from triticale parent (Figure 1G).
Discussion
Development of transgenic triticale as a source of bioindustrial products is currently one of the main challenges for breeders. For now, pollen-mediated gene flow from related species into conventional triticale varieties is the only pathway for transgene movement in large-scale breeding programs leading in the European Union. Currently, most breeders lean toward the use of slow-rusting genes for durable and race non-specific resistance of cereals. The main aim of this study was to obtain the starting pool triticale–wheat hybrids, which will be further used for backcrossing program, evaluation of slow-rusting gene expression, and inoculation tests.
The Lr34 gene has been widely used in breeding of wheat cultivars worldwide, and its resistance has remained effective over many years despite large-scale agricultural use. The gene was identified in several distinct groups of genetic material, e.g., CIMMYT lines, Chinese landraces, and European winter wheat germplasm, for example Frontana, which was developed in Brazil or the Italian wheat Mentana (Kolmer et al., 2013). Lagudah et al. (2009) developed a sequence-tagged site (STS) marker, csLV34, that maps 0.4 cM from Lr34 locus and was validated in many lines and cultivars from different breeding programs worldwide. The Lr34 allele yielded a 150-bp product, and a 229-bp band was amplified in non-Lr34 germplasm (Lagudah et al., 2009). In this study, the product for resistant genotypes of wheat was the same. A 230-bp band was amplified for wheat accessions with the lack of Lr34 allele (Rod, Toropi, Junco, etc.). However, different bands were amplified for susceptible triticale cultivars and F1 hybrids (250 and 350 bp). Hence, it could be assumed that the csLV34 ortholog is placed on R-genome chromosomes (probably on 7R chromosome) but yields different amplicons, which are useful for identification of Lr34 locus in triticale × wheat hybrids.
The second major slow-rusting gene, Lr46, was first described in 1998 by Singh et al. (1998) in Mexican cultivar Pavon 76 and located on chromosome 1B (William et al., 2003). Suenaga et al. (2003) determined that the microsatellite locus Xwmc44 is located 5.6 cM proximal to the putative quantitative trait locus (QTL) for Lr46. However, it is reported that the Xwmc44 resistance allele in some cases is not diagnostic of Lr46, since numerous varieties without Lr46 have Xwmc44 products of similar sizes (Dubcovsky and Soria, 2017). In this study, Xwmc44 marker yielded four different amplification products for F1 hybrids. The size differences between amplicons were difficult to evaluate by standard electrophoresis using 2% agarose gel. In comparison, we have used csLV46G22 marker (primer sequences and protocols were kindly provided by Prof. E. Lagudah, CSIRO, Australia), which is highly reliable and close to 100% diagnostic marker for the Lr46 gene (Lagudah, personal communication). This cleaved amplified polymorphic sequence (CAPS) marker is codominant and appeared to be useful for genotyping on the F1 hybrids. Moreover, we showed the poor diagnostic ability of the Xwmc44 marker used to trace the Lr46 gene. Moreover, we identified mismatch results between Xgwm44 and csLV46G22 in 12 of 33 wheat accessions.
The GISH experiment confirmed the presence of seven monosomic chromosomes of D-genome derived from wheat accessions in most of the seeds obtained from cross-hybridizations of triticale and wheat in this study. This method has direct applications on the fundamental research, as well as in detecting the amount of introgressed chromatin during the production of prebreeding germplasm (Schwarzacher et al., 1992; Kwiatek et al., 2019). A number of reports showed the ability of cytogenetic methods to determine the introgression of D-genome chromosomes of wheat or A. tauschii Coss. into triticale genetic background (Lukaszewski et al., 1987; Salmanowicz et al., 2013; Kwiatek et al., 2015). The Lr34 locus is located on 7D chromosome of slow-rusting wheat accessions; hence, GISH was an additional method to prove the hybrid origin of F1 plants and to confirm the transfer of D-genome chromosomes with Lr34 locus into hybrid plants, per se. Unfortunately, Lr46 locus identification cannot be supported by GISH because this method is not a applicable to discriminate B-genome chromosomes of F1 hybrids, which were originated from both wheat and triticale parental forms.
A combination of MAS and GISH enabled to select triticale × wheat hybrid plants with loci slow-rusting genes. Twingo × Frontana 3671 hybrids seems to be the most promising prebreeding forms, considering the accumulation of both Lr34- and Lr46-resistant alleles. Those combinations will be used for seed propagation, further backcrossing, and resistance evaluation. The crucial approach for this research is to improve the crossability of triticale cultivars with chosen wheat pollinators. What is more, field and greenhouse resistance tests will be essential to confirm whether the transferred slow-rusting genes will provide resistance in the triticale background.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
MK initiated the project and designed the study. MK and MM obtained the hybrid plants. RS and MM performed the identification of molecular markers and made the mitotic chromosome preparations. RS performed the GISH analysis. RS and WU analyzed the microscope images. MK and RS wrote the manuscript. AT supervised the methodology of molecular marker analyses. JN supervised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge and thank Prof. Evans Lagudah for providing protocol instructions and primer sequences for csLV46G22 marker and his invaluable help with data interpretation, Dr. Harrold Bockelman at the USDA/ARS Small Grains Laboratory, Aberdeen (ID, United States) for providing the seeds of wheat accessions, Dr. Zofia Banaszak at the Danko Breeding Station, for providing seeds of triticale cultivars. In addition, we would also like to thank all of the reviewers and manuscript editor for their careful review of the manuscript and for their excellent suggestions for improving our initial work.
Footnotes
Funding. This publication is being cofinanced by the framework of Ministry of Science and Higher Education program as “Regional Initiative Excellence” in years 2019–2022, project No. 005/RID/2018/19.
References
- Ayalew H., Kumssa T. T., Butler T. J., Ma X.-F. (2018). Triticale improvement for forage and cover crop uses in the southern great plains of the United States. Front. Plant Sci. 9:1130. 10.3389/fpls.2018.01130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni R., Chauhan H., Hensel G., Roulin A., Sucher J., Kumlehn J., et al. (2018). Pathogen-inducible Ta-Lr34res expression in heterologous barley confers disease resistance without negative pleiotropic effects. Plant Biotechnol. J. 16 245–253. 10.1111/pbi.12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J., Soria M. A. (2017). MAS Wheat. Univ. California, Davis. Available online at: https://maswheat.ucdavis.edu/ (accessed 2019). [Google Scholar]
- Dyck P. L. (1977). Genetics of leaf rust reaction in three introductions of common wheat. Can. J. Genet. Cytol. 19 711–716. 10.1139/g77-077 [DOI] [Google Scholar]
- Dyck P. L., Samborski D. J. (1979). Ault-plant leaf rust resistance in PI 250413, an introduction of common wheat. Can. J. Plant Sci. 59 329–332. 10.4141/cjps79-053 [DOI] [Google Scholar]
- Ellis J. G., Lagudah E. S., Spielmeyer W., Dodds P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5:641. 10.3389/fpls.2014.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson D., Harwood W., Hofvander P., Jones H., Rogowsky P., Stöger E., et al. (2018). A welcome proposal to amend the GMO legislation of the EU. Trends Biotechnol. 36 1100–1103. 10.1016/j.tibtech.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2018). The State of Food and Agriculture 2018. Migration, Agriculture and Rural Developement. Rome: FAO. [Google Scholar]
- Kolmer J. A. (1996). Genetics of resistance to wheat leaf rust. Annu. Rev. Phytopathol. 34 435–455. 10.1146/annurev.phyto.34.1.435 [DOI] [PubMed] [Google Scholar]
- Kolmer J. A., Hanzalova A., Goyeau H., Bayles R., Morgounov A. (2013). Genetic differentiation of the wheat leaf rust fungus Puccinia triticina in Europe. Plant Pathol. 62 21–31. 10.1111/j.1365-3059.2012.02626.x 28398164 [DOI] [Google Scholar]
- Kwiatek M., Belter J., Majka M., Wiśniewska H. (2016). Allocation of the S-genome chromosomes of Aegilops variabilis Eig. carrying powdery mildew resistance in triticale (× Triticosecale Wittmack). Protoplasma 253 329–343. 10.1007/s00709-015-0813-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek M., Majka M., Wiśniewska H., Apolinarska B., Belter J. (2015). Effective transfer of chromosomes carrying leaf rust resistance genes from Aegilops tauschii Coss. into hexaploid triticale (X Triticosecale Witt.) using Ae. tauschii × Secale cereale amphiploid forms. J. Appl. Genet. 56 163–168. 10.1007/s13353-014-0264-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek M. T., Kurasiak-Popowska D., Mikołajczyk S., Niemann J., Tomkowiak A., Weigt D., et al. (2019). Cytological markers used for identification and transfer of Aegilops spp. chromatin carrying valuable genes into cultivated forms of Triticum. Comp. Cytogenet. 13 2–19. 10.3897/CompCytogen.v13i1.30673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek M. T., Nawracała J. (2018). Chromosome manipulations for progress of triticale (×Triticosecale) breeding. Plant Breed. 137 823–831. 10.1111/pbr.12652 [DOI] [Google Scholar]
- Lagudah E. S., Krattinger S. G., Herrera-Foessel S., Singh R. P., Huerta-Espino J., Spielmeyer W., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 119 889–898. 10.1007/s00122-009-1097-z [DOI] [PubMed] [Google Scholar]
- Lagudah E. S., McFadden H., Singh R. P., Huerta-Espino J., Bariana H. S., Spielmeyer W. (2006). Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor. Appl. Genet. 114 21–30. 10.1007/s00122-006-0406-z [DOI] [PubMed] [Google Scholar]
- Lukaszewski A. J., Apolinarska B., Gustafson J. P. (1987). Introduction of the D-genome chromosomes from bread wheat into hexaploid triticale with a complete rye genome. Genome 29 425–430. 10.1139/g87-074 [DOI] [Google Scholar]
- McIntosh R., Dubcovsky J., Rogers J., Morris C., Appels R., Xia X. (2016). Catalogue of Gene Symbols for Wheat: 2015-2016 Supplement. Available at: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf (accessed April 2, 2019). [Google Scholar]
- McIntosh R. A. (1995). “Catalogue of gene symbols for wheat,” in Proc. 8th Int. Wheat Genet. Symp, eds Li Z. S., Xin Z. Y. (Beijing: China Agricultural Scientech; ). [Google Scholar]
- Mikhailova L. A., Merezhko A. F., Funtikova E. Y. (2009). Triticale diversity in leaf rust resistance. Russ. Agric. Sci. 35:320 10.3103/S1068367409050097 [DOI] [Google Scholar]
- Periyannan S., Milne R. J., Figueroa M., Lagudah E. S., Dodds P. N. (2017). An overview of genetic rust resistance: from broad to specific mechanisms. PLoS Pathog. 13:e1006380. 10.1371/journal.ppat.1006380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmanowicz B. P., Langner M., Wiśniewska H., Apolinarska B., Kwiatek M., Błaszczyk L. (2013). Molecular, physicochemical and rheological characteristics of introgressive Triticale/Triticum monococcum ssp. monococcum lines with wheat 1D/1A chromosome substitution. Int. J. Mol. Sci. 14 15595–15614. 10.3390/ijms140815595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanaei S., Stuart P. (2018). Systematic assessment of triticale-based biorefinery strategies: techno-economic analysis to identify investment opportunities. Biofuels Bioprod. Bioref. 12 S46–S59. 10.1002/bbb.1499 [DOI] [Google Scholar]
- Schwarzacher T., Anamthawat-Jónsson K., Harrison G. E., Islam A. K., Jia J. Z., King I. P., et al. (1992). Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor. Appl. Genet. 84 778–786. 10.1007/BF00227384 [DOI] [PubMed] [Google Scholar]
- Singh R. P., Mujeeb-Kazi A., Huerta-Espino J. (1998). Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88 890–894. 10.1094/PHYTO.1998.88.9.890 [DOI] [PubMed] [Google Scholar]
- Skowrońska R., Kwiatek M., Tomkowiak A., Nawracała J. (2019). Development of multiplex PCR to detect slow rust resistance genes Lr34 and Lr46 in wheat. J. Appl. Genetics 60, 301–304. 10.1007/s13353-019-00520-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga K., Singh R. P., Huerta-Espino J., William H. M. (2003). Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 93 881–890. 10.1094/PHYTO.2003.93.7.881 [DOI] [PubMed] [Google Scholar]
- William M., Singh R. P., Huerta-Espino J., Islas S. O., Hoisington D. (2003). Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93 153–159. 10.1094/PHYTO.2003.93.2.153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.