Abstract
The maintenance of tissue homeostasis and health relies on the efficient removal of damaged or otherwise suboptimal cells. One way this is achieved is through cell competition, a fitness quality control mechanism that eliminates cells that are less fit than their neighbours. Through this process, cell competition has been shown to play diverse roles in development and in the adult, including in homeostasis and tumour suppression. However, over the last few years it has also become apparent that certain oncogenic mutations can provide cells with a competitive advantage that promotes their expansion via the elimination of surrounding wild-type cells. Thus, understanding how this process is initiated and regulated will provide important insights with relevance to a number of different research areas. A key question in cell competition is what determines the competitive fitness of a cell. Here, we will review what is known about this question by focussing on two non-mutually exclusive possibilities; first, that the activity of a subset of transcription factors determines competitive fitness, and second, that the outcome of cell competition is determined by the relative cellular metabolic status.
Keywords: Cell competition, Cell fitness, Metabolism, Apoptosis, Growth regulation, P53, Myc, Hippo, NF-κB, Stat
1. General introduction
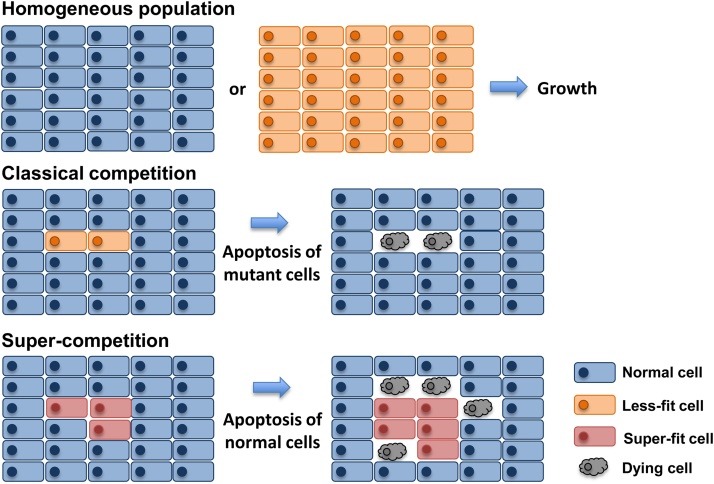
In a multicellular organism, mechanisms to ensure the removal of damaged or suboptimal cells are essential for maintaining tissue health. It has been proposed that in addition to sensing their own intrinsic fitness, cells also determine their fitness relative to surrounding cells in the tissue. Through this mechanism, those cells that are comparatively weaker than their neighbours (generically called losers) can be eliminated, even though their defects are not sufficiently severe to trigger the intrinsic apoptotic pathway (summarised in Fig. 1). This process has been termed cell competition and accumulating evidence indicates that it has diverse functions in development, homeostasis and tumour suppression. However, in addition to its beneficial role in eliminating defective and potentially harmful cells, competition has also been implicated in cancer progression. This has been primarily observed through a process termed super-competition, by which cells acquire mutations that give them a competitive advantage and allow them to induce the elimination of surrounding wild-type cells. Given this behaviour, these mutant cells are generically termed super-competitors.
Fig. 1.
Competition versus Super-competition.
Cell competition occurs between cells which are viable and proliferate in a heterogeneous environment. During competition cells which are defective or sub-optimal are eliminated when surrounded by wild-type cells. In contrast to this, during super-competition, cells which have acquired mutations that make them more competitive, induce apoptosis in the surrounding wild-type cells, highlighting the relative nature of cell competition.
Much remains to be understood about the mechanisms by which cells are eliminated through competitive interactions. In particular, the factors which determine cell fitness and the process by which cells are able to recognise their relative fitness levels remain elusive. Here we will approach the first of these questions by contrasting what is known about the transcriptional versus the metabolic regulation of cell competition. For example, while a number of transcription factors have been shown to be critical for cell competition, most notably Myc, p53, NF-κB and TEAD, it is often unclear which of their transcriptional targets are acting downstream to regulate the outcome of competition. In some cases, these transcription factors may directly regulate pro- or anti-apoptotic genes, however in others, they appear to act further upstream in the process of competition to define the winner or loser status of a cell. One way in which these factors could affect cell fitness is through the regulation of metabolic pathways, as all these transcription factors have been shown to regulate genes involved in cell metabolism. Indeed, there is accumulating evidence that anabolic differences between neighbouring cells can affect cell fitness in competitive environments. This review will therefore seek to outline what is known about the transcriptional and metabolic regulation of cell competition and to explore the intersection between these two processes.
2. Transcriptional regulation of cell competition
The best-characterised transcriptional regulator of cell competition is Myc, overexpression of which can convert cells into super-competitors. Additionally, a variety of other factors with transcriptional activities, such as p53, NF-κB, TEAD/Scalloped and STAT, have all been shown to either affect the outcome of cell competition or to induce super-competition. Here we will describe what is known about the roles of each one of these factors in competition (summarised in Table 1).
Table 1.
Transcription factors in cell competition. Summary of known transcription factors that play a role in several cell competition contexts as well the activities they produce. Dros. (Drosophila), Mam. (mammals).
| Transcription Factors | Competition context | Model | Activity | Key Studies |
|---|---|---|---|---|
|
p53 |
Irradiated bone marrow | Mam. | Increased p53 in losers | Bondar and Medzhitov [24] |
| Myc overexpression in epithelial cells | Dros. | Increased p53 in winners | de la Cova et al. [28] | |
| Bmpr1a-/- and 4n mESCs | Mam. | Increased p53 in losers | Bowling [26] | |
| ScribkD MDCK cells | Mam. | Increased p53 in losers | Wagstaff [27] | |
| Mdm2+/-, Mdm4+/- embryos | Mam. | Increased p53 in losers | Zhang et al. [25] | |
|
Myc |
Myc overexpression in epithelial cells | Dros. | Increased Myc activity induces super-competition and increases p53 levels | de la Cova [4] Moreno and Basler[3] de la Cova et al. [28] |
| Bmpr1a-/- or Tetraploid mESCs | Mam. | Decreased MYC in losers | Sancho et al. [15] | |
| Scrib- epithelial cells | Dros. | Increased Myc activity rescues loser cell elimination | Chen et al. [9] | |
| Lgl- epithelial cells | Dros. | Decreased Myc levels | Froldi et al. [14] | |
| Myc overexpression in mouse cardiomyocytes and epiblast cells | Mam. | High MYC levels produces super-competition | Clavería et al. [16] Villa del Campo et al. [17,18], Díaz-Díaz et al. [87] |
|
| MYC inhibition in cancer cell lines | Mam. | Decreased MYC induces loser status | Di Giacomo et al. [21] Patel et al. [20] |
|
| Myc overexpression in S2 cells | Dros. | Increased Myc induces winner status | Senoo-Matsuda and Johnston [19] | |
|
NF-κB |
Minute+/-epithelial cells | Dros. | Increased activity in losers | Meyer et al. [54] Germani et al. [55] |
| Myc overexpression in in epithelial cells | Dros. | Increased activity in losers | Meyer et al. [54] Alpar et al. [88] Germani et al. [55] |
|
| Scrib- epithelial cells | Dros. | Increased activity in winners | Katsukawa et al. [56] | |
|
STAT |
Minute+/-; Mahjong+/- epithelial cells | Dros. | STAT promotes winner proliferation | Kolahgar et al. [34] Kucinski et al. [35] |
| Stat92E85C9 epithelial cells and sustained STAT in epithelial cells | Dros. | Decreased STAT induces loser status and STAT activity induces super-competition | Rodrigues et al. [33] | |
|
YAP/Yki TEAD/ Scalloped |
Scrib- epithelial cells | Dros. | Suppressed Yki activity in losers | Chen et al. [9] Yamamoto et al. [39] |
| lgl- epithelial cells | Dros. | Increased Yki activity in winners | Menéndez et al. [38] | |
| Minute+/- epithelial cells | Dros. | Increased Yki activity rescues loser elimination | Tyler et al. [40] | |
| APC-/- epithelial cells | Dros. | Increased Yki activity rescues loser elimination | Suijkerbuijk et al. [42] | |
| Hippo pathway manipulation in epithelial cells | Dros. | Increased Yki activity induces super-competition | Ziosi et al. [43] | |
| Hippo pathway manipulation in embryonic fibroblasts | Mam. | Increased Yki activity induces super-competition, decreased Yki activity induces loser status | Mamada et al. [44] | |
| Hippo pathway manipulation in the epiblast | Mam. | Decreased TEAD activity induces loser status | Hashimoto and Sasaki [45] | |
2.1. Myc
The proto-oncogene MYC is overexpressed in the vast majority of human cancers and thus has been the focus of intense research interest for some time. MYC transcription factors control a wide range of cellular processes, including cell growth, proliferation, ribosomal biogenesis, protein synthesis, glycolysis and mitochondrial biogenesis [1,2]. Moreover, MYC transcriptional activity is modulated in a context-dependent manner by a number of signalling pathways, including by the Hippo pathway, which, as we will discuss later, also regulates cell competition.
Myc was first demonstrated to play a role in competition through its ability to induce super-competition in the Drosophila wing imaginal disc. Here, epithelial cells with four copies of the Myc gene are able to trigger apoptosis in surrounding cells with normal Myc levels. Furthermore, once this elimination has taken place, the Myc-overexpressing cells grow to fill the space left by the dead cells without producing morphological abnormalities [3,4]. This phenotypically silent replacement of loser cells led to the hypothesis that cell competition regulates organ size, an attractive possibility that we have reviewed elsewhere [5].
From this initial role, the importance of Myc in cell competition rapidly expanded through a number of studies that analysed the mechanism of elimination of a range of defective cell types in the Drosophila wing imaginal disc, such as cells carrying lethal giant larvae (lgl−) and scribble (scrib−) gene mutations. Lgl and Scrib are scaffold proteins that bind to the lateral membrane and govern epithelial apical-basal polarity. Mutant flies for these polarity genes demonstrate neoplastic overgrowth that causes large tumourous masses (reviewed in [6]), however, when mutant cells are surrounded by wild-type tissue, they are eliminated by apoptosis [[7], [8], [9], [10], [11], [12], [13]]. Lgl− cells eliminated from wild-type imaginal discs express low Myc levels and Myc overexpression in lgl− or scrib− cells rescues their elimination [9,14]. In contrast to this, when lgl− cells are surrounded by cells carrying the Minute mutation, which makes them haploinsufficient for ribosomal proteins and have lower Myc expression, they are no longer eliminated [14]. Together, these results suggest that relative lower Myc levels is the cause of the elimination of these mutant cell types.
More recently, MYC has also been implicated in cell competition during mouse embryogenesis. In the early embryonic tissue, or epiblast, MYC expression is mosaic and those cells with low MYC levels die by apoptosis [15,16]. Similarly, embryonic stem cells that are eliminated by cell competition, such as those with defective BMP signalling, that are tetraploid or autophagy deficient, all display lower MYC expression than wild-type cells in co-culture [15]. Furthermore, in the epiblast and heart, Myc overexpression turns cells into super-competitors allowing them to replace wild-type cells [[16], [17], [18]]. Therefore, in mouse, just as in Drosophila, relative MYC levels determines the competitive ability of the cell.
Differences in MYC levels have also been suggested to induce cell competition in human cancer cells. The first demonstration that differences in MYC levels can also trigger cell competition between cell lines came in Drosophila, where S2 cells overexpressing Myc outcompete S2 cells that don’t [19]. More recently, in the human breast cancer line MCF7, shRNA inhibition of MYC was shown to lead to out-competition by control cells [20]. Furthermore, analysis of the tumour-stroma interface in a number of human tumour samples found a strong correlation between elevated MYC levels in the tumour and activated caspase-3 expression in the adjacent stroma [21]. This observation was supported by the finding that co-culturing human cancer cell lines with differing levels of MYC expression leads to increased apoptosis in those with lower MYC levels. Together, these results suggest that differences in MYC levels between tumour and stromal cells or within a tumour could aid tumour expansion by inducing super-competition.
From the above studies, it is clear that MYC is an important determinant of relative cell fitness, with winner cells having higher MYC levels than losers. However, in spite of these advances, the precise mechanism by which MYC affects cell fitness is not resolved. Given the importance of MYC in the regulation of cell anabolism, below we will review the different metabolic effects that MYC could have during cell competition.
2.2. P53
P53 is best known as a tumour suppressor that is mutated in about half of human tumours [22]. P53 was originally identified as a mediator of cell cycle arrest or apoptosis in the response to cellular stress. However, in recent years, it has become apparent that P53 can also regulate a plethora of other cellular processes including DNA repair, differentiation, stem cell reprogramming, metabolism and senescence amongst others (reviewed in [23]). The roles that p53 plays are usually complex and like Myc, they are also highly context dependent.
The first description for p53 in cell competition came in 2010, when the competitive behaviour of cells in the hematopoietic system was examined [24]. The authors used ionising radiation (IR) to induce cellular stress in hematopoietic stem and progenitor cells (HSPCs) and then compared their ability to repopulate the bone marrow of irradiated and non-irradiated mice. They observed that irradiated cells showed an increase in p53 expression and that this higher p53 expression caused them to be outcompeted by non-treated hematopoietic cells in the bone marrow. Importantly, the out-competition of the irradiated cells was found to be due to senescence rather than apoptosis [24]. A similar finding was made when mosaic mouse embryos and adult mice were analysed that are haploinsufficient for the Mdm2 and Mdm4 genes, two major negative regulators of p53 activity. Here the authors observed that mutation of these genes throughout the animal led to a moderate increase in p53 levels that had no effect on growth. In contrast to this, mosaic mutation of these genes provided the cells with a competitive disadvantage that was primarily thought to be due to a non-cell autonomous induction of growth arrest [25]. Also in mouse, defective cells eliminated by cell competition during early post-implantation stages show elevated p53 expression. In this context p53 is required for the repression of the mTOR pathway, an important metabolic regulator, in these defective cells specifically in a competitive environment and this repression induces apoptosis [26]. In Madin-Darby Canine Kidney (MDCK) cells, knockdown of scribble (scribKD) in a sub-population of cells leads to their elimination by cell competition [11]. These cells also show elevated p53 expression, which makes them sensitive to tissue crowding. In a competitive environment with wild-type cells, this sensitivity causes mechanical stress and a further elevation of p53 levels that induces their elimination [27].
Differences in p53 levels during cell competition have also been reported in Drosophila. In Myc-induced competition, p53 is required in winner cells to sustain their metabolic and proliferation changes during competition and p53 mutation abolishes their ability to eliminate surrounding wild-type cells [28]. In contrast to this, apoptosis of Minute cells in mosaic imaginal discs was found to be independent of p53 levels as competition was still produced in p53 mutants [29]. Interestingly, Minute cells show elevated expression of the p53 target bZip-domain (Xrp1) during cell competition, which contributes to their reduced translation and cell growth rates and is essential for their competitive elimination [30]. However, this increase in Xrp1 expression is also independent of p53 [31].
In summary, as is evident from the studies described above, the precise role of p53 in competition is highly context dependent, inducing growth arrest in some contexts and regulating apoptosis through the control of the metabolic state or mechanical properties of the cell in others.
2.3. STAT
The Janus kinase/signal transducers and activator of transcription (JAK-STAT) pathway is conserved from Drosophila to humans and is involved in multiple cellular processes including cell division, death, immunity and tumour formation. Activation of STATs occurs downstream of receptor-ligand (usually a cytokine) binding, which triggers JAK-mediated phosphorylation of tyrosine residues. This phosphorylation leads to dimerisation and translocation to the nucleus, where they can bind specific DNA sequences and activate transcription of target genes (reviewed in [32]).
A number of studies in Drosophila have shown that modulating JAK-STAT activity levels can affect competitive cell behaviour. In the wing and eye imaginal discs, wild-type cells induce surrounding cells with deficient JAK-STAT signalling to undergo apoptosis [33]. Conversely, cells with sustained JAK-STAT activation eliminate surrounding wild-type cells through Hid-dependent apoptosis [33]. In the Drosophila posterior midgut, Minute cells, both differentiated and stem cells, are eliminated by apoptosis when surrounded by wild-type cells. This is accompanied by increased stem cell proliferation and symmetric self-renewal of the wild-type cells [34]. During this process, chronic JNK activation in mutant cells induces expression of the JAK-STAT ligand Unpaired-3, which stimulates compensatory proliferation of the wild-type cells via JAK-STAT signalling. In contrast to this, in the imaginal wing disc, mutant cells for Minute and Mahjong (another polarity-associated gene) also display chronic JNK signalling, but this leads to increased JAK-STAT signalling that sustains their proliferation when all the cells in the fly are mutant, as well as promoting the proliferation of surrounding wild-type cells during cell competition [35].
The mechanism by which JAK-STAT signalling mediates cell competition is also still largely unknown. Although JAK-STAT is known to regulate Myc expression, JAK-STAT-induced cell competition appears to be largely independent of Myc, ribosomal biogenesis or other pathways involved in cell competition such as WNT/Wg, BMP/Dpp or Hippo signalling [33]. One intriguing possibility is raised by the fact that most of the JAK-STAT signalling ligands are cytokines (interferons or interleukins) involved in immunity and are regulated by STAT itself with NF-kB signalling cross-talk. Given the recent implication of NF-kB in cell competition that will be discussed below, it is possible that STAT signalling mediates cell competition by cooperating with NF-kB in the regulation of immune-like responses.
2.4. TEAD/Scalloped
TEADs are effectors of the Hippo pathway, a highly conserved kinase cascade involved in the cell-autonomous control of proliferation. Hippo (also known as MST1 and MST2 in humans) phosphorylates and activates large tumour suppressor 1/2 (LATS1/2) kinases, homologues of Warts in Drosophila, which then repress the activity of the transcriptional coactivators YAP and TAZ in mammals or their homologue Yorkie (Yki) in Drosophila. In mammals, the YAP/TAZ complex binds the TEAD family of transcription factors to activate genes involved in promoting proliferation and inhibiting apoptosis. In Drosophila, Yki interacts with the Scalloped transcription factor, which similarly drives the expression of pro-proliferative and pro-survival genes, such as Cyclin E and Myc [36,37]. Thus, repression of the Hippo pathway leads to increased YAP/TAZ or Yki activity and therefore, to increased proliferation and suppressed apoptosis via the regulation of TEAD/Scalloped activity.
Perhaps unsurprisingly, given its role in proliferation and apoptosis, numerous studies have implicated the Hippo pathway in cell competition. Indeed, increased Yki or YAP/TAZ activation is sufficient to rescue the elimination of a range of different loser cell types. For example, in the Drosophila imaginal wing disc, Yki activity is critical for the balance between elimination and overgrowth of polarity-deficient epithelial cells [9,38]. This is illustrated by the observation that Yki activation drives hyperproliferation and the formation of neoplastic masses when the whole tissue is composed of scrib− mutant cells, but if these scrib− cells are surrounded by wild-type cells, Yki activity is suppressed leading to their elimination [9]. Similarly, lgl− mutant cells partially evade elimination by competition if they express an oncogenic constitutively active form of Ras (Ras-V12), which increases Yki activity and therefore the proliferation rate of these double-mutant cells [38].
In the Drosophila adult eye, elimination of scrib− mutant clones by wild-type cells is mediated by the ligand Sas and its receptor PTP10D [39]. Scrib− cells in which PTP10D is knocked-down are not eliminated and demonstrate increased Yki nuclear localisation and expression of target genes. This increased Yki activity is critical for avoiding loser cell status, as in the absence of Sas-PTP10D signalling, JNK signalling switches from driving pro-apoptotic gene expression in loser cells to driving pro-proliferative gene expression through suppression of the Hippo pathway. Hippo pathway signalling has also been implicated in Minute-induced competition in the Drosophila eye. Here, increasing Yki activity in Minute-/+ cells through mutation of negative regulators of the Hippo pathway, including Hippo itself, is sufficient to rescue them from elimination [40]. Similarly, wild-type cells with elevated Yki activity have an enhanced winner phenotype and eliminate Minute-/+ cells more efficiently [41]. Moreover, adenomatous polyposis coli (APC)-null intestinal cells, which behave as super-competitors, also display increased Yki activity, and relative levels of Hippo pathway activity are crucial for determining the outcome of competitive interactions in this context [42]. Thus, the expression of Hippo pathway target genes appears to be a common feature in diverse competition processes.
The importance of Hippo pathway signalling in competition is further highlighted by the observation that elevated expression of Yki/YAP alone, or mutation of other members of the Hippo pathway, is sufficient to convert cells into super-competitors in both Drosophila [43] and mammalian models of competition [44]. Conversely, low Yki/YAP/TAZ activity is sufficient to confer loser status on cells [[44], [45], [46]]. Interestingly, it was recently demonstrated that human glioma cells that express lower levels of YAP are eliminated when surrounded by glioma cells with higher YAP expression [46]. The higher YAP-expressing cells in this system were also found to increase their proliferation and expression of tumourigenic genes specifically in this heterogeneous environment, indicating that these competitive interactions were enhancing tumourigenesis. However, the role of YAP activity in competition appears to be context-dependent as mammalian keratinocytes and MDCK cells expressing constitutively active YAP behave as losers in a competitive environment [47,48].
Myc is a transcriptional target of Yki/YAP/TAZ and thus one explanation for the crucial role of these transcription factors in competition is that they are upstream of Myc-induced competition ([43,49]). Indeed, in the study by Ziosi et al., the cell non-autonomous apoptosis induced by Yki overexpression was abrogated by Myc knock-down. In the pre-implantation mouse embryo, lower TEAD activity also triggers cell elimination by competition [45]. Here Tead1-/- cells show downregulated MYC expression, however reducing MYC activity throughout the embryo using a small molecule inhibitor did not rescue Tead1-/- cell elimination. This suggests that differences in MYC expression with wild-type cells is not the only reason for the elimination of these cells. In summary, although Myc appears to be an important target of TEAD/Scalloped factors during cell competition, it is also likely that these factors have other important targets; an observation which is supported by the fact that Myc-overexpressing cells do not overcome organ size control mechanisms, whereas Yki-overexpressing cells demonstrate unconstrained overgrowth [4,37,[50], [51], [52]].
2.5. NF-kB
The NF-κB transcription factors play a crucial role in the activation of genes involved in cell survival and immunity, such as growth factors and pro-inflammatory cytokines. In both Drosophila and mammals, signalling from Toll-like receptors leads to NF-κB nuclear localisation and transcription of target genes [53]. Intriguingly, a role for Toll receptor signalling and subsequent NF-κB activation in both Minute and Myc-induced models of cell competition has been suggested [54]. In competition induced by Myc overexpression, loser cell elimination was suppressed in a background mutant for either the Drosophila NF-κB homologue, Rel, or for Dredd, the caspase involved in Rel cleavage and activation. Increased Dredd activity in loser cells specifically in competition was found to result in increased Rel-mediated transcription of the pro-apoptotic protein, Hid, which was critical for their elimination. Interestingly, this study also investigated Minute-induced competition and found that the NF-κB homologues, Dif and Dorsal, were required for loser cell elimination in this context through their transcriptional regulation of the pro-apoptotic protein, Reaper. Thus, NF-κB-mediated upregulation of pro-apoptotic genes was required for both forms of cell competition, although each involved distinct NF-κB homologues and pro-apoptotic targets. More recently, another report confirmed the requirement for NF-κB activation in Minute and Myc-induced loser cell elimination in Drosophila and found that inhibition of NF-κB alone was sufficient to confer a growth advantage. However, if the flies were grown under pathogen-free conditions, inhibition of NF-κB activation no longer rescued loser cell elimination in either context [55], presumably because all the cells in the tissue have low NF-κB activity under these conditions. This observation therefore suggests that relative levels of Toll pathway signalling, and thus NF-κB activation, are critical for determining the outcome of competitive interactions.
Intriguingly, Toll signalling has also been implicated in scrib-induced competition. However, in contrast to Minute and Myc-mediated competition, where activation of Toll signalling and NF-κB activity was critical for loser cell elimination, in this context activated Toll signalling blocked competition and loser cell elimination [56]. Here, forced Toll signalling was sufficient to convert loser scrib− cells into super-competitors through activation of Yki. Thus, it appears that NF-κB acts in a context-dependent manner during cell competition, with pro-apoptotic targets in some cell types and pro-proliferation targets in others. Understanding what determines these differential outcomes will be an important avenue of research in the future. It should also be noted that differences exist between mammalian Toll-like receptors, which directly recognise microbial-derived products, and Drosophila Toll receptors, which respond to pathogens indirectly by recognising Spätzle, which is proteolytically processed in response to microbe recognition by distinct circulating receptors [53]. It is therefore unclear whether a similar mechanism could underlie fitness recognition during mammalian cell competition.
3. Metabolic regulation of cell competition
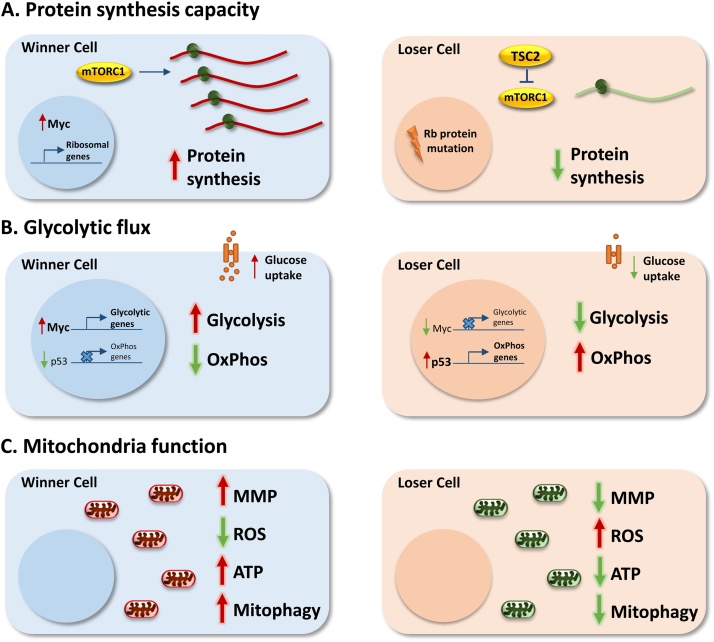
Over the last decade there has been increased evidence for the importance of metabolic changes in cell competition. In many cases this evidence has arisen from the observation that many of the transcription factors implicated in competition (discussed above) play a prominent role in regulating one or more metabolic pathways. Here we will review what is known about these pathways, with special emphasis on three that particularly stand out as likely regulators of competitive cell fitness: the rates of protein synthesis, glycolysis and mitochondrial activity (summarised in Fig. 2).
Fig. 2.
Metabolic determinants of cell fitness in competition.
A. Protein synthesis capacity. Winner cells display increased protein synthesis rates compared to loser cells. This may be due to increased Myc expression or higher mTOR pathway activity in winner cells, or to ribosomal protein mutations in loser cells.
B. Glycolytic Flux. Winner cells display increased glycolytic flux relative to loser cells. This may be driven by transcriptional regulation of metabolic genes by transcription factors such as Myc and p53. Glucose availability in the microenvironment may also affect glycolysis, with winner cells displaying increased uptake of glucose.
C. Mitochondria Function. Loser cells may display impaired mitochondria function, resulting in decreased mitochondrial membrane potential (MMP), ATP production and mitophagy. Reactive oxygen species (ROS) production may also be elevated in loser cells with impaired mitochondrial function.
3.1. Rates of protein synthesis
As indicated above, the first characterised model of cell competition was impaired protein synthesis capacity relative to surrounding cells. In Drosophila, haploinsufficiency for multiple ribosomal protein genes results in a common ‘Minute’ phenotype, characterised by a reduction in the size of surface bristles and a developmental delay associated with reduced protein synthesis and proliferation rate [57]. However, despite these defects, heterozygous flies survive to adulthood and eventually reach a normal body size. In contrast, Minute+/− cell clones are eliminated by apoptosis in the imaginal wing disc in a wild-type background and fail to contribute to the adult wing [[58], [59], [60]]. Ribosomal mutations have also been shown to induce competition in mammals, as mice carrying a heterozygous mutation in the ribosomal protein L24 gene are viable, but heterozygous clones are outcompeted in a wild-type background [61]. These results suggest that impaired protein translation is a conserved mechanism of inducing a lower cell-fitness phenotype.
Further evidence for the importance of protein synthesis rates for cell competition comes from the observation that MYC can promote protein synthesis and ribosome biogenesis through its transcriptional regulation of ribosomal proteins, ribosomal DNA, and initiation factors of translation [62]. In Drosophila, cells overexpressing Myc are no longer able to eliminate surrounding cells with lower Myc levels and are themselves eliminated if they carry the Minute mutation described above, which reduces ribosomal protein levels [3]. Interestingly, MYC has also been shown to regulate protein synthesis during cell competition in the early embryo [16]. Thus, both ribosomal mutations and lower MYC levels would potentially reduce the protein synthesis capacity of a cell. Furthermore, in mouse embryonic stem cells, mutation of Tsc2, a negative regulator of mTORC1 signalling, is sufficient to turn cells into super-competitors [26]. Given that regulation of protein synthesis is one of the most important roles of mTORC1, these results also support the possibility that relative protein synthesis levels could be an important cell-fitness determinant.
3.2. Rate of glycolysis
In addition to a potential role for protein synthesis in cell competition, glycolysis has also emerged as a possible mediator of cell fitness. This is in large part due to Myc, which has a well-characterised role in the promotion of aerobic glycolysis [63]. In Drosophila imaginal discs and S2 cells, Myc overexpression stimulates glucose uptake and glycolysis, and in S2 cells this was shown to be via increased expression of glycolytic genes [28]. Interestingly, this increase in glycolytic flux was accompanied by decreased mitochondrial respiration, strengthening the case that a switch to aerobic glycolysis is occurring in these cells. However, it is worth noting that in Myc-overexpressing S2 and imaginal disc cells, mitochondrial activity is not completely suppressed. Instead, an increase in p53 expression occurs in these cells that promotes oxidative phosphorylation to balance the effects of Myc overexpression. Interestingly, in a competitive environment with wild-type cells, this metabolic flux is accentuated and Myc-overexpressing cells further increase their glycolysis levels in a p53-dependent manner. Importantly, the enhanced metabolism of Myc-overexpressing cells in a competitive environment is required for the expansion of these cells via the elimination of wild-type cells [28], providing compelling evidence for the role of glycolytic flux in cell competition.
A second instance where a shift to aerobic glycolysis affects the outcome of cell competition can be found in the apical extrusion of RasV12-transformed Madin-Darby canine kidney cells (MDCK) cells. During this process, these cells show enhanced glucose uptake, increased lactate dehydrogenase (LDHA) expression and increased lactate secretion, which is accompanied by a loss of mitochondrial activity. This phenotype was not due to a change in mitochondrial mass or number, but rather due to decreased mitochondrial function, as it could be rescued by inhibition of pyruvate dehydrogenase kinase 4 (PDK4), which diverts pyruvate from the Krebs cycle to the production of lactate [64]. Furthermore, this inhibition of PDK4 was sufficient to rescue RasV12 extrusion, highlighting the functional importance of these metabolic changes.
As mentioned above, recent studies have suggested that competitive interactions may be part of the complex interactions occurring between the tumour and stroma or even within the tumour. Tumour cells frequently demonstrate altered metabolism relative to surrounding stromal cells. In particular, higher glycolytic activity is a common feature of many tumours and this can lead to both a depletion of glucose in the microenvironment and to increased lactate release from tumour cells, which acidifies the micro-environment. Additionally, other glycolytic metabolites may also be released, which, along with lactate, are increasingly recognised to have important roles as signalling molecules. The tumour microenvironment is therefore extensively remodelled and there is evidence that this detrimentally affects the proliferation and responsiveness of surrounding stromal and immune cells [65,66].
A well-characterised example of this phenomenon is the effect of lactate on the ability of T and NK cells to mount an anti-tumoural response [65,66]. Lactate accumulation was shown to inhibit T and NK cell activation and infiltration of the tumour in melanomas [67]. One potential mechanism for this could be that lactate export by activated T cells, which themselves have high levels of glycolysis, is inhibited by the presence of high extracellular levels of lactate, thus disturbing their metabolism and function [68]. Additionally, lactate also affects myeloid cell activation, suppressing monocyte activation and dendritic cell differentiation [69,70]. This lactate-induced immune suppression likely has an impact on tumour progression as a correlation can be observed between the concentration of lactate in the tumour and the incidence of metastases and recurrence in cervical cancers [71].
Glucose availability in the tumour micro-environment has also been shown to impact on the responsiveness of infiltrating T cells. In renal carcinoma higher expression of the glucose uptake channel, GLUT-1, was found to correlate with lower CD8+ T cell infiltration of the tumour [72]. Intra-tumoural CD4+ and CD8+ T cells also demonstrate decreased IFNγ production and effector function as a result of glucose restriction in the tumour microenvironment [73,74]. Boosting tumour cell glycolysis by overexpressing MYC, glycolytic enzymes or GLUT-1 increased tumour progression, while suppression of glycolysis by checkpoint blockade therapy restored T cell responses and resulted in tumour regression [73]. Inhibiting glycolysis or targeting glucose uptake channels in tumours could therefore be a potential therapeutic strategy as it would increase the glucose available to circulating immune cells, thus enhancing their function. Additionally, in human glioblastoma samples, the cancer stem cell (CSC) population exhibits an increased efficiency of glucose uptake relative to non-CSCs and this gives them a selective advantage under low glucose conditions [75]. Furthermore, targeting the glucose uptake channel responsible for this advantage reduces the tumourigenic potential and growth of these CSCs. Therefore, nutrient competition both within tumours and between tumour and stromal cells or immune cells can influence tumour progression through mechanisms akin to cell competition.
3.3. Mitochondrial function
Mitochondria are classically seen as the powerhouses of cells, where generation of ATP occurs by oxidative phosphorylation (OXPHOS). Significantly, in addition to supplying energy to cells, mitochondria are also involved in many other cellular processes, such as calcium and reactive oxygen species (ROS) signalling, apoptosis, the immune response and the cell stress response [[76], [77], [78], [79], [80]]. Mitochondrial function was already suggested to have a role in cell-cell communication in Drosophila imaginal discs as mitochondrial defects in Ras-activated epithelial cells lead to oxidative stress that promotes non-cell autonomous growth of surrounding tissue [81,82]. Recently, mitochondria function has also been implicated in cell competition. For example, analysis of the Minute and Mahjong mutations eliminated by competition in the Drosophila wing disc found that both had a common signature of oxidative stress. Mutant cell survival was dependent on the activity of Nrf2, a transcription factor central to the oxidative stress response. The authors observed that, while endogenous Nrf2 expression is required for loser survival in a homogeneous environment, in a competitive context Nrf2 overactivation promotes their elimination [35]. Given that the mitochondria are considered to be the major origin of ROS, these results raise the possibility that defective mitochondrial function is underlying the elimination of Minute and Mahjong mutant cells.
Interestingly, Drosophila Scribble knock-down (scribKD) cells, that are eliminated by cell competition, also display a cell-intrinsic signature of oxidative stress and mitochondrial dysfunction. In the Drosophila wing imaginal discs scribKD cells cause overgrowths and show a loss of mitochondria membrane potential, reduced ATP production, increased ROS and impaired mitophagy (the mechanism of clearing defective mitochondria). Furthermore, the authors found that promoting mitophagy rescues the wing distortion and reduces the death rates of these mutant larvae by half, supporting hypothesis that mitochondria dysfunction plays a role in the dysregulated growth of these cells [83].
Further evidence for the role of mitochondrial activity in determining cell fitness comes from the analysis of the behaviour of immortalized mammalian cell lines. Here, characterisation of stochastically arising sub-clones of human (U2OS), mouse (3T3) and canine (MDCK) cell lines found that the proliferation rates and saturation densities of each cell line predicted their competitive ability, with the faster proliferating cells being invariably the winners. Furthermore, the authors found that the competition between these cells did not require de novo RNA synthesis, but rather was abolished under hypoxia or limiting glutamine concentrations. Both these factors have a direct role in determining OXPHOS output and the observation that dissipating the proton gradient of the electron transport chain with CCCP gave a dose-dependent rescue of loser cell elimination, further supports the possibility that mitochondrial output was a key determinant of cell competition in these cells [84]. Therefore, taken together, all the above studies suggest an important role of mitochondrial performance in the origin of competitive interactions.
4. Conclusion and future perspectives
Understanding the parameters that determine relative cell fitness during cell competition is important as it provides an avenue for the manipulation of this process. Given the plethora of roles that cell competition has been postulated to play, from the optimisation of tissue fitness during ageing [85], to tumour promotion as well as tumour suppression [5], this manipulation has the potential to promote either cell replacement during tissue regeneration or to prevent the expansion of cancer cells. As reviewed above, a number of transcriptional regulators have been shown to profoundly affect the competitive ability of the cell. However, one recurrent theme is that the precise mechanism of action of these transcription factors appears to be highly context dependent and no obvious target for their activity has been identified during cell competition. One interesting possibility that has been put forward is that hypertranscription, or the ability of a cell to globally upregulate their transcriptome during specific transitions, may be important for cell competition [86]. Both MYC and YAP/TAZ can regulate hypertranscription (reviewed in [86]), therefore it is possible that this may provide cells with a competitive advantage over their neighbours. But how cells can measure their relative global transcriptional output is a question that needs resolving.
An alternative possibility that we have explored here is that an important mode by which these transcription factors act during cell competition is through the regulation of one or more metabolic pathways. For example, the mechanism by which Myc confers a competitive cell advantage appears to be through both increased rates of glycolysis as well as via the regulation of protein synthesis. Given the complexity and inter-relatedness of the different cellular metabolic pathways, this raises the intriguing possibility that the metabolic state of the cell may determine its competitive fitness. Following this argument, it is possible that cells may measure their relative fitness levels through the exchange of metabolites. Again, untangling if this is the case and how relative levels of metabolites can be measured between cells is a challenge that will likely fuel the growing cell competition field for years to come.
Declaration of Competing Interest
We have no competing interests.
Acknowledgements
Katerina Lawlor is recipient of an NHLI foundation PhD studentship. Salvador Perez Montero has been a recipient of an EMBO long-term fellowship and a Marie Curie international fellowship. Ana Lima is supported by a British Heart Foundation PhD studentship. Research in the Tristan Rodriguez lab is supported by the MRC project grant (MR/N009371/1) and by being part of a BHF centre for research excellence.
References
- 1.Pelengaris S., Khan M., Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 2.Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno E., Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 4.De La Cova C., Abril M., Bellosta P., Gallant P., Johnston L.A. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 5.Di Gregorio A. Cell Competition and Its Role in the Regulation of Cell Fitness From Development to Cancer. Developmental Cell. 2016;38:621–634. doi: 10.1016/j.devcel.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Humbert P.O. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal N., Kango M., Mishra A., Sinha P. Neoplastic transformation and aberrant cell-cell interactions in genetic mosaics of lethal(2)giant larvae (lgl), a tumor suppressor gene of Drosophila. Dev. Biol. 1995 doi: 10.1006/dbio.1995.0017. [DOI] [PubMed] [Google Scholar]
- 8.Brumby A.M., Richardson H.E. Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;22:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C.-L., Schroeder M.C., Kango-Singh M., Tao C., Halder G. Tumor suppression by cell competition through regulation of the Hippo pathway. Proc. Natl. Acad. Sci. U. S. A. 2012;109:484–489. doi: 10.1073/pnas.1113882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igaki T., Pagliarini R.A., Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr. Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 11.Norman M. Loss of Scribble causes cell competition in mammalian cells. J. Cell. Sci. 2012;125:59–66. doi: 10.1242/jcs.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamori Y. Involvement of Lgl and Mahjong/VprBP in Cell Competition. PLoS Biol. 2010;8:e1000422. doi: 10.1371/journal.pbio.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods D.F., Bryant P.J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 14.Froldi F. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol. 2010;8:33. doi: 10.1186/1741-7007-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho M. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev. Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clavería C., Giovinazzo G., Sierra R., Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 17.Villa del Campo C., Clavería C., Sierra R., Torres M. Cell competition promotes phenotypically silent cardiomyocyte replacement in the mammalian heart. Cell Rep. 2014;8:1741–1751. doi: 10.1016/j.celrep.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Villa del Campo C. Myc overexpression enhances epicardial contribution to the developing heart and promotes extensive expansion of the cardiomyocyte population. Sci. Rep. 2016;6:35366. doi: 10.1038/srep35366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senoo-Matsuda N., Johnston L.A. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18543–18548. doi: 10.1073/pnas.0709021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel M.S., Shah H.S., Shrivastava N. C-myc-Dependent cell competition in human Cancer cells. J. Cell. Biochem. 2017;118:1782–1791. doi: 10.1002/jcb.25846. [DOI] [PubMed] [Google Scholar]
- 21.Di Giacomo S. Human Cancer cells signal their competitive fitness through MYC activity. Sci. Rep. 2017;7:12568. doi: 10.1038/s41598-017-13002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollstein M. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 23.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 24.Bondar T., Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6:309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G. p53 pathway is involved in cell competition during mouse embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 2017;114:498–503. doi: 10.1073/pnas.1617414114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowling S. P53 and mTOR signalling determine fitness selection through cell competition during early mouse embryonic development. Nat. Commun. 2018;9:1763. doi: 10.1038/s41467-018-04167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagstaff L. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 2016;7 doi: 10.1038/ncomms11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De La Cova C. Supercompetitor status of Drosophila myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014;19:470–483. doi: 10.1016/j.cmet.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kale A., Li W., Lee C.H., Baker N.E. Apoptotic mechanisms during competition of ribosomal protein mutant cells: roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ. 2015 doi: 10.1038/cdd.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akdemir F., Christich A., Sogame N., Chapo J., Abrams J.M. p53 directs focused genomic responses in Drosophila. Oncogene. 2007;26:5184–5193. doi: 10.1038/sj.onc.1210328. [DOI] [PubMed] [Google Scholar]
- 31.Lee C.-H. A regulatory response to ribosomal protein mutations controls translation, growth, and cell competition. Dev. Cell. 2018;46:456–469. doi: 10.1016/j.devcel.2018.07.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiu H., Nicholson S.E. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigues A.B. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139:4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolahgar G. Cell competition modifies adult stem cell and tissue population dynamics in a JAK-STAT-Dependent manner. Dev. Cell. 2015;34 doi: 10.1016/j.devcel.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kucinski I., Dinan M., Kolahgar G., Piddini E. Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng Z., Moroishi T., Guan K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J., Wu S., Barrera J., Matthews K., Pan D. The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating yorkie, the drosophila homolog of yap. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Menéndez J., Pérez-Garijo A., Calleja M., Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14651–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto M., Ohsawa S., Kunimasa K., Igaki T. The ligand Sas and its receptor PTP10D drive tumour-suppressive cell competition. Nature. 2017;542 doi: 10.1038/nature21033. [DOI] [PubMed] [Google Scholar]
- 40.Tyler D.M., Li W., Zhuo N., Pellock B., Baker N.E. Genes Affecting Cell Competition in Drosophila. Genetics. 2007;175:643–657. doi: 10.1534/genetics.106.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo P. Nerfin-1 represses transcriptional output of Hippo signaling in cell competition. eLife. 2019;8 doi: 10.7554/eLife.38843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suijkerbuijk S.J.E., Kolahgar G., Kucinski I., Piddini E. Cell Competition Drives the Growth of Intestinal Adenomas in Drosophila. Curr. Biol. 2016;26:428–438. doi: 10.1016/j.cub.2015.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziosi M. dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells. PLoS Genet. 2010;6:e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamada H., Sato T., Ota M., Sasaki H. Cell competition in mouse NIH3T3 embryonic fibroblasts is controlled by the activity of Tead family proteins and Myc. J. Cell Sci. 2015;128 doi: 10.1242/jcs.163675. [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto M., Sasaki H. Epiblast formation by Tead-Yap-dependent expression of pluripotency factors and competitive elimination of unspecified cells. bioRxiv. 2018;449397 doi: 10.1101/449397. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z. Differential YAP expression in glioma cells induces cell competition and promotes tumorigenesis. J. Cell. Sci. 2019 doi: 10.1242/jcs.225714. jcs.225714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiba T. MDCK cells expressing constitutively active Yes-associated protein (YAP) undergo apical extrusion depending on neighboring cell status. Sci. Rep. 2016;6 doi: 10.1038/srep28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishio M. Hippo pathway controls cell adhesion and context-dependent cell competition to influence skin engraftment efficiency. FASEB J. 2019;33(4):5548–5560. doi: 10.1096/fj.201802005R. [DOI] [PubMed] [Google Scholar]
- 49.Neto-Silva R.M., de Beco S., Johnston L.A. Evidence for a Growth-Stabilizing Regulatory Feedback Mechanism between Myc and Yorkie, the Drosophila Homolog of Yap. Dev. Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett F.C., Harvey K.F. Fat Cadherin Modulates Organ Size in Drosophila via the Salvador/Warts/Hippo Signaling Pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 51.Song H. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willecke M. The Fat Cadherin Acts through the Hippo Tumor-Suppressor Pathway to Regulate Tissue Size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 53.Ganesan S., Aggarwal K., Paquette N., Silverman N. NF-κB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer S.N. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science (80-.) 2014;346 doi: 10.1126/science.1258236. 1258236–1258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Germani F., Hain D., Sternlicht D., Moreno E., Basler K. The Toll pathway inhibits tissue growth and regulates cell fitness in an infection-dependent manner. eLife. 2018;7 doi: 10.7554/eLife.39939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katsukawa M., Ohsawa S., Zhang L., Yan Y., Igaki T. Serpin facilitates tumor-suppressive cell competition by blocking toll-mediated yki activation in Drosophila. Curr. Biol. 2018;28:1756–1767. doi: 10.1016/j.cub.2018.04.022. e6. [DOI] [PubMed] [Google Scholar]
- 57.Marygold S.J. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morata G., Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev. Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 59.Simpson P. Parameters of cell competition in the compartments of the wing disc of Drosophila. Dev. Biol. 1979;69:182–193. doi: 10.1016/0012-1606(79)90284-7. [DOI] [PubMed] [Google Scholar]
- 60.Simpson P., Morata G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev. Biol. 1981;85:299–308. doi: 10.1016/0012-1606(81)90261-x. [DOI] [PubMed] [Google Scholar]
- 61.Oliver E.R., Saunders T.L., Tarlé S.A., Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Riggelen J., Yetil A., Felsher D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 63.Dang C.V. MYC on the path to Cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kon S. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat. Cell Biol. 2017 doi: 10.1038/ncb3509. [DOI] [PubMed] [Google Scholar]
- 65.Lyssiotis C.A., Kimmelman A.C. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renner K. Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Front. Immunol. 2017;8:248. doi: 10.3389/fimmu.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brand A. LDHA-associated lactic acid production blunts tumor immunosurveillance by t and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Fischer K. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- 69.Dietl K. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. 2010;184:1200–1209. doi: 10.4049/jimmunol.0902584. [DOI] [PubMed] [Google Scholar]
- 70.Gottfried E. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006;107:2013–2021. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- 71.Walenta S. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–921. [PubMed] [Google Scholar]
- 72.Singer K. Warburg phenotype in renal cell carcinoma: high expression of glucose-transporter 1 (GLUT-1) correlates with low CD8+ T-cell infiltration in the tumor. Int. J. Cancer. 2011;128:2085–2095. doi: 10.1002/ijc.25543. [DOI] [PubMed] [Google Scholar]
- 73.Chang C.-H. Metabolic competition in the tumor microenvironment is a driver of Cancer progression. Cell. 2015 doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ho P.-C. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flavahan W.A. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burke P.J. Mitochondria, bioenergetics and apoptosis in Cancer. Trends in Cancer. 2017;3:857–870. doi: 10.1016/j.trecan.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giorgi C., Marchi S., Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Biol. 2018;19:713–730. doi: 10.1038/s41580-018-0052-8. [DOI] [PubMed] [Google Scholar]
- 78.Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eisner V., Picard M., Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat. Cell Biol. 2018;20:755–765. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohsawa S. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012;490:547–551. doi: 10.1038/nature11452. [DOI] [PubMed] [Google Scholar]
- 82.Nakamura M., Ohsawa S., Igaki T. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nat. Commun. 2014;5:5264. doi: 10.1038/ncomms6264. [DOI] [PubMed] [Google Scholar]
- 83.Yadav A.K., Srikrishna S. scribble (scrib) knockdown induces tumorigenesis by modulating Drp1-Parkin mediated mitochondrial dynamics in the wing imaginal tissues of Drosophila. Mitochondrion. 2019;44:103–110. doi: 10.1016/j.mito.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 84.Penzo-Méndez A.I., Chen Y.-J., Li J., Witze E.S., Stanger B.Z. Spontaneous cell competition in immortalized mammalian cell lines. PLoS One. 2015;10:e0132437. doi: 10.1371/journal.pone.0132437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Merino M.M. Elimination of Unfit Cells Maintains Tissue Health and Prolongs Lifespan. Cell. 2015;160:461–476. doi: 10.1016/j.cell.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Percharde M., Bulut-Karslioglu A., Ramalho-Santos M. Hypertranscription in development, stem cells, and regeneration. Dev. Cell. 2017;40:9–21. doi: 10.1016/j.devcel.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Díaz-Díaz C. Pluripotency Surveillance by Myc-Driven Competitive Elimination of Differentiating Cells. Dev. Cell. 2017;42:585–599. doi: 10.1016/j.devcel.2017.08.011. e4. [DOI] [PubMed] [Google Scholar]
- 88.Alpar L., Bergantiños C., Johnston L.A. Spatially Restricted Regulation of Spñtzle/Toll Signaling during Cell Competition. Dev. Cell. 2018;46:706–719. doi: 10.1016/j.devcel.2018.08.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]