Abstract
The inhibitory activity of (±)-citalopram on human (h) α3β4, α4β2, and α7 nicotinic acetylcholine receptors (AChRs) was determined by Ca2+ influx assays, whereas its effect on rat α9α10 and mouse habenular α3β4* AChRs by electrophysiological recordings. The Ca2+ influx results clearly establish that (±)-citalopram inhibits (IC50's in μM) hα3β4 AChRs (5.1 ± 1.3) with higher potency than that for hα7 (18.8 ± 1.1) and hα4β2 (19.1 ± 4.2) AChRs. This is in agreement with the [3H]imipramine competition binding results indicating that (±)-citalopram binds to imipramine sites at desensitized hα3β4 with >2-fold higher affinity than that for hα4β2. The electrophysiological, molecular docking, and in silico mutation results indicate that (±)-citalopram competitively inhibits rα9α10 AChRs (7.5 ± 0.9) in a voltage-independent manner by interacting mainly with orthosteric sites, whereas it inhibits a homogeneous population of α3β4* AChRs at MHb (VI) neurons (7.6 ± 1.0) in a voltage-dependent manner by interacting mainly with a luminal site located in the middle of the ion channel, overlapping the imipramine site, which suggests an ion channel blocking mechanism. In conclusion, (±)-citalopram inhibits α3β4 and α9α10 AChRs with higher potency compared to other AChRs but by different mechanisms. (±)-Citalopram also inhibits habenular α3β4*AChRs, supporting the notion that these receptors are important endogenous targets related to their anti-addictive activities.
Keywords: Nicotinic acetylcholine receptor, (±)-Citalopram, Selective serotonin reuptake inhibitor, Medial habenula, Brain slices
1. Introduction
(±)-Citalopram is a selective serotonin reuptake inhibitor (SSRI) used for the treatment of depressive disorders (Varia and Rauscher, 2002), and off-label for alcohol withdrawal (Angelone et al., 1998) and hot flashes as well as eating, anxiety, premenstrual dysphoria, and post-traumatic stress disorders. In several European countries, (±)-citalopram is also approved for panic and obsessive-compulsive disorders (Stein et al., 2001).
In addition to being able to selectively inhibit serotonin transporters, SSRIs behave as noncompetitive antagonists (NCAs) of several nicotinic acetylcholine receptors (AChRs) (reviewed in (García-Colunga et al., 2016). The majority work on this area have concerned SSRIs such as fluoxetine, paroxetine, and sertraline (Andreasen et al., 2011a; Arias et al., 2010a; Fryer and Lukas, 1999; Garcia-Colunga et al., 1997), whereas there is less data on (±)-citalopram, especially on its interaction with α9α10 AChRs. The most compelling evidence that (±)-citalopram modulates AChR activity is based on animal studies indicating that non-selective (e.g., nicotine) (Popik et al., 2003) and α7-selective (e.g., PNU-282987) agonists (Andreasen et al., 2012, 2011b) enhance the activity of this antidepressant. These results are in agreement with the hypercholinergic hypothesis of depression, where an excessive cholinergic tone over the noradrenergic system may develop in depressive states, and consequently antidepressant-induced AChR inhibition could be part of their mechanisms of action [reviewed in (Arias et al., 2014; García-Colunga et al., 2016; Mineur and Picciotto, 2010)]. The observation of a higher rate of smoking in depressed patients compared to the general population also supports this hypothesis [reviewed in (Mineur and Picciotto, 2010)].
A first attempt to establish whether there is a relationship between (±)-citalopram's clinical effects and its AChR selectivity is to determine the activity of this antidepressant at different AChR subtypes. For example, inhibition of α3β4-containing (α3β4*) AChRs expressed in the habenulo-interpeduncular cholinergic pathway might be related to the beneficial activity of (±)-citalopram to alleviate depression (Varia and Rauscher, 2002) and/or alcohol withdrawal (Angelone et al., 1998). Habenular α3β4* AChRs are considered important targets for several anti-addictive compounds (Glick et al., 2002; Maisonneuve and Glick, 2003; McCallum et al., 2012). Compounds with relatively higher selectivity for this receptor subtype such as 18-methoxycoronaridine (Arias et al., 2017) have anti-addictive properties and decrease alcohol intake in rodents (Rezvani et al., 2016), whereas bupropion and mecamylamine have antidepressant activity (Arias et al., 2018a, 2014). Nevertheless, no direct evidence of (±)-citalopram-induced habenular α3β4* AChRs inhibition has been demonstrated so far. In this regard, we sought to determine the selectivity of (±)-citalopram for different AChR subtypes, and its activity on MHb by electrophysiology recordings of ventral inferior (VI) MHb neurons that strongly express α3β4* AChRs (Quick et al., 1999; Shih et al., 2014). To further determine its mechanisms of action, the interaction of (±)-citalopram with imipramine sites at α3β4 and α4β2 AChRs is compared by radioligand competition binding experiments, whereas the functional and structural interactions with α3β4 AChRs is respectively resolved by voltage-dependence and molecular docking studies.
A correlation between ligand-induced α9α10 AChR inhibition and anti-pain and anti-inflammatory activity has been observed (McIntosh et al., 2009; Romero et al., 2017). Since these AChRs are not expressed in the brain (Elgoyhen et al., 2001, 1994; Morley et al., 2018) but in outer hair cells (Elgoyhen and Katz, 2012; Goutman et al., 2015) and different immune cells (Peng et al., 2004), it is possible that the observed anti-inflammatory activity is mediated by inhibition of α9α10 AChRs expressed in lymphocytes. Since a direct correlation between depression and inflammation has also been shown (Christmas et al., 2011), it is plausible that the antidepressant (Varia and Rauscher, 2002) and anti-inflammatory (Sacre et al., 2010) effects of (±)-citalopram might be mediated, at least partially, by its inhibitory activity on α9α10 AChRs. However, the functional interaction of (±)-citalopram with α9α10 AChRs has not previously determined. Since this is a prerequisite to decipher this relationship, the activity of (±)-citalopram was studied on α9α10 AChRs expressed in Xenopus oocytes by voltage-clamp recordings and its mechanism of action determined by voltage-dependence, ligand competition, molecular docking, and in silico mutation experiments.
A better understanding of the functional interaction and selectivity of SSRIs for different AChR subtypes, especially α3β4 and α9α10 AChRs, is crucial to develop novel analogs for safer pharmacotherapies.
2. Materials and methods
2.1. Material
[3H]Imipramine hydrochloride (47.5 Ci/mmol) was obtained from PerkinElmer Life Sciences Products, Inc. (Boston, MA) and stored at −20°C. (±)-Citalopram hydrobromide was purchased from MedChemExpress USA (New Jersey, USA). (±)-Epibatidine hydrochloride and QX-314 were obtained from Tocris Bioscience (Ellisville, MO, USA). Fluo-4 was obtained from Molecular Probes (Eugene, Oregon, USA). Euthasol (sodium pentobarbital, 100 mg/kg; sodium phenytoin, 12.82 mg/kg) was obtained from LeVet Pharma (Oudewater, Netherlands). Polyethylenimine, acetylcholine (ACh), probenecid, atropine, imipramine hydrochloride, and bovine serum albumin (BSA), were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Salts were of analytical grade.
2.2. Ca2+ influx measurements in cells expressing hα3β4, hα4β2 or hα7 AChRs
Ca2+ influx measurements were performed on HEK293-hα3β4, HEK293-hα4β2, and GH3-hα7 cells as previously described (Arias et al., 2018a, 2018b, 2017, 2016, 2010a, 2010b, 2010c). Briefly, 5 x 104 cells per well were seeded 72 h prior to the experiment on black 96-well plates (Costar, New York, USA) and incubated at 37°C in a humidified atmosphere (5% CO2/95% air). Under these conditions, the majority of expressed hα4β2 and hα3β4 AChRs have the (α4/3)3(β2/4)2 stoichiometry (see Arias et al., 2016, and references therein). 16–24 h before the experiment, the medium was changed to 1% BSA in HEPES-buffered salt solution (HBSS) (130 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 0.8 mM MgSO4, 0.9 mM NaH2PO4, 25 mM glucose, 20 mM Hepes, pH 7.4). On the day of the experiment, the medium was removed by flicking the plates and replaced with 100 μL HBSS/1% BSA containing 2 μM Fluo-4 and 2.5 mM probenecid. The cells were then incubated at 37°C in a humidified atmosphere (5% CO2/95% air) for 1 h.
To determine the antagonistic activity of (±)-citalopram (Fig. 1), plates were flicked to remove excess of Fluo-4, washed twice with HBSS/1% BSA, refilled with 100 μL of HBSS containing different concentrations of (±)-citalopram, and incubated for 5 min. Plates were finally placed in the cell plate stage of the fluorescent imaging plate reader (Molecular Devices, Sunnyvale, CA, USA), and 0.1 μM (±)-epibatidine was added from the agonist plate to the cell plate using the 96-tip pipettor simultaneously to fluorescence recordings for 3 min. A baseline consisting of 5 measurements of 0.4 s each was recorded. The laser excitation and emission wavelengths are 488 and 510 nm, at 1 W, and a CCD camera opening of 0.4 s.
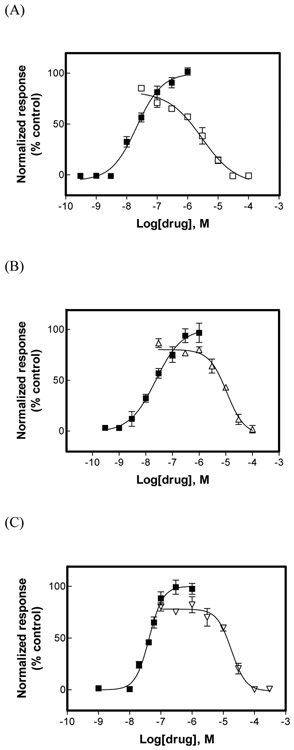
Fig. 1.
Effect of (±)-citalopram on (±)-epibatidine-induced Ca2+ influx in HEK293-hα3β4 (A), HEK293-hα4β2 (B), and GH3-hα7 (C) cells. Increased concentrations of (±)-epibatidine (■) activated each hα3β4 (A), hα4β2 (B), and hα7 (C) AChR. Subsequently, cells were pre-treated (5 min) with several concentrations of (±)-citalopram (□), followed by addition of 0.1 μM (+)-epibatidine. Response was normalized to the maximal (±)-epibatidine response which was set as 100%. The plots are representative of 4-5 determinations, where the error bars are the S.D. The calculated IC50 and nH values are summarized in Table 1.
2.3. Voltage clamp recordings on oocytes expressing rα9α10 AChRs
Rat α9 and α10 subunits were expressed in Xenopus oocytes as previously described (Ballestero et al., 2005). Electrophysiological recordings were performed at −70 mV using two-electrode voltage-clamp (Arias et al., 2018b; Ballestero et al., 2005). Oocytes were pre-incubated for 2 min with (±)-citalopram before adding acetylcholine (ACh) and (±)-citalopram. The average peak amplitude of three control ACh responses just before the exposure to (±)-citalopram was used to normalize the amplitude of each test response in the presence of the drug.
To further determine the mechanism of inhibition of (±)-citalopram, two approaches were used: (1) the EC50 values for ACh were obtained in the absence and presence of 8.0 μM (±)-citalopram (close to its experimental IC50 value; see Table 1), and (2) current-voltage (I-V) relationships were obtained by applying 2-s voltage ramps from −120 to +50 mV, 10-s after the peak response to 10 μM ACh, in the presence and absence of 10 μM (±)-citalopram, from a holding potential (Vhold) of −70 mV (Arias et al., 2018b). Leakage correction was performed by subtraction of the I-V curve obtained before the application of ACh.
Table 1.
Inhibitory potency (IC50) of (±)-citalopram at different AChR subtypes.
| AChR subtype | Method | IC50, μM | nHf |
|---|---|---|---|
| hα3β4 a | (±)-Epibatidine-induced Ca2+ influx in HEK293-hα3β4 cells | 5.1 ± 1.3 | 1.00 ± 0.10 |
| hα4β2 b | (±)-Epibatidine-induced Ca2+ influx in HEK293-hα4β2 cells | 19.0 ± 4.2 | 1.24 ± 0.19 |
| hα7 c | (±)-Epibatidine-induced Ca2+ influx in GH3-hα7 cells | 18.8 ± 1.1 | 1.44 ± 0.22 |
| rα9α10 d | Voltage-clamp recordings of ACh-activated rα9α10 AChRs expressed in Xenopus oocytes | 7.5 ±0.9 | 1.35 ± 0.10 |
| Habenular mα3β4* e | Patch-clamp recordings of ACh-activated HMb (VI) neurons | 7.6 ± 1.0 | 0.90 ± 0.10 |
Values were obtained from Figures 1Aa, 1Bb, 1Cc, 2Bd, and 3Be, respectively.
Hill coefficient.
2.4. Patch-clamp recordings on brain slices
An animal study protocol pertaining to this study (#IS00003604) was reviewed and approved by the Northwestern University Institutional Animal Care and Use Committee. Procedures also followed the guidelines for the care and use of animals provided by the National Institutes of Health (NIH) Office of Laboratory Animal Welfare. Mice were housed at 22 °C on a 12-h light/dark cycle with food and water ad libitum. Mice were weaned on postnatal day 21 and housed with same-sex littermates. Experiments were conducted on C57BL/6J mice obtained from Jackson Laboratories. All studies were restricted to male mice, age 8–24 weeks.
Brain slices were prepared as previously described (Arias et al., 2017; Shih et al., 2014). Mice were anesthetized with Euthasol (sodium pentobarbital, 100 mg/kg; sodium phenytoin, 12.82 mg/kg) before trans-cardiac perfusion with oxygenated (95% O2/5% CO2), 4 °C N-methyl-d-glucamine (NMDG)-based recovery solution that contains (in mM): 93 NMDG, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 10 MgSO4·7H2O, and 0.5 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4). Brains were immediately dissected after the perfusion and held in oxygenated, 4 °C recovery solution for one minute before cutting a brain block containing the MHb and sectioning the brain with a vibratome (VT1200S; Leica). Coronal slices (250 μm) were sectioned through the medial habenula and transferred to oxygenated, 33 °C recovery solution for 12 min. Slices were then kept in holding solution (containing in mM: 92 NaCl, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 2 MgSO4·7H2O, and 2 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4) for 60 min or more before recordings.
Brain slices were transferred to a recording chamber being continuously superfused at a rate of 1.5–2.0 mL/min with oxygenated 32 °C recording solution. The recording solution contained (in mM): 124 NaCl, 2.5 KCl, 1.2 NaH2PO4, 24 NaHCO3, 12.5 glucose, 2 MgSO4·7H2O, and 2 CaCl2·2H2O; 300–310 mOsm; pH 7.3–7.4). Patch pipettes were pulled from borosilicate glass capillary tubes (1B150F-4; World Precision Instruments, USA) using a programmable microelectrode puller (P-97; Sutter Instrument, USA). Tip resistance ranged from 4.5 to 8.0 MΩ when filled with internal solution. The following internal solution was used for the concentration-response experiments (in mM): 135 potassium gluconate, 5 EGTA, 0.5 CaCl2, 2 MgCl2, 10 HEPES, 2 MgATP, and 0.1 GTP; pH adjusted to 7.25 with Tris base. This internal solution also contained QX-314 (2 mM) for improved voltage control. The following internal solution was used for the voltage dependence experiments (in mM): 117 CsCH3SO3, 20 HEPES, 0.4 EGTA, 2.8 NaCl, 5 TEA-Cl, 2.5 MgATP, 0.1 spermine, and 0.25 MgGTP. The osmolarity of internal solutions were adjusted to 290 mOsm with sucrose.
Neurons within brain slices were visualized with infrared or visible differential interference contrast (DIC) optics. Neurons in the ventral inferior (VI) aspect of the MHb were targeted for recordings, as previously described (Arias et al., 2017; Shih et al., 2014). Electrophysiology experiments were conducted using a Scientifica SliceScope or Nikon FN-1 upright microscope. A computer running pCLAMP 10 software was used to acquire whole-cell recordings along with an Axopatch 200B amplifier and an A/D converter (Digidata 1440A). pClamp software and acquisition hardware were from Molecular Devices. Data were sampled at 10 kHz and low-pass filtered at 1 kHz. Immediately prior to gigaseal formation, the junction potential between the patch pipette and the superfusion medium was nulled. Series resistance was uncompensated.
To record physiological events following local application of drugs, a drug-filled pipette was moved to within 20–40 μm of the recorded neuron using a second micromanipulator. The drug (dissolved in recording solution) was dispensed onto the recorded neuron by using a Picopump (World Precision Instruments) at an ejection pressure of 12 psi for 250 ms. The ejection volume varied depending on the goal of the experiment. Atropine (1 μM) was present in the superfusion medium when using ACh application to prevent activation of muscarinic AChRs. To determine the voltage-dependence of (±)-citalopram-induced inhibition, voltage ramps (200 ms) were applied from the holding potential of −60 mV to a final value of 50 mV before returning to −60 mV. For such experiments, ACh was puff-applied and the ramp was executed during steady-state AChR currents. AChR-mediated currents were measured at both −60 mV and +50 mV in the same neuron before and after superfusion of (±)-citalopram (60 μM).
The voltage dependence of an inhibiting agent is related with the electrical distance of its binding site, measured from the external side of the membrane channel. Thus, the fraction of the electrical field sensed at the citalopram's binding site within the receptor’s ion channel (i.e., δ), was subsequently calculated using the one-site blocking model (López-Valdés and García-Colunga, 2001):
| (1) |
where, [Citalopram] is the concentration of the ligand, IC50(0) is the ligand concentration to produce 50% inhibition at 0 mV, Vm is the applied membrane potential, z is the valence of the blocking molecule, F is the Faraday constant, R is the gas constant, and T is the absolute temperature.
2.5. [3H]Imipramine competition binding experiments using either hα3β4 or hα4β2 AChR-containing membranes
To determine whether (±)-citalopram binds to the imipramine sites at hα3β4 and hα4β2 AChRs, [3H]imipramine competition binding experiments were performed using either hα3β4 or hα4β2 AChR-containing membranes prepared from the respective HEK293-hα3β4 and HEK293-hα4β2 cells, as previously described (Arias et al., 2010a, 2010b, 2010c). In this regard, hα3β4 or hα4β2 AChR-containing membranes (1.5 mg/mL) were suspended in binding saline buffer (50 mM Tris–HCl, 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, pH 7.4) and pre-incubated with 15.2 nM [3H]imipramine in the presence of 0.1 μM (±)-epibatidine (receptors are mainly in the desensitized state) for 30 min at room temperature (RT). The total volume was divided into aliquots, and increasing concentrations of the ligand under study were added to each tube and incubated for 2 h at RT. The nonspecific binding was determined in the presence of 100 μM imipramine.
AChR-bound [3H]imipramine was then separated from free ligand by a filtration assay using a 48-sample harvester system with GF/B Whatman filters (Brandel Inc., Gaithersburg, MD, USA), previously soaked with 0.5% polyethylenimine for 30 min. The membrane-containing filters were transferred to scintillation vials with 3 mL of Bio-Safe II (Research Product International Corp, Mount Prospect, IL, USA), and the radioactivity was determined using a Beckman 6500 scintillation counter (Beckman Coulter, Inc., Fullerton, CA, USA).
2.6. Analysis methods
The concentration–response results from heterologous cells and MHb neurons, as well as from the radioligand competition binding experiments were curve-fitted by nonlinear least squares analysis using the Prism software (GraphPad Software, San Diego, CA), and the respective EC50, IC50, and nH values calculated. The obtained IC50 values for (±)-citalopram were transformed into inhibition constants (Ki) using the Cheng–Prusoff relationship (Cheng and Prusoff, 1973):
| (2) |
where [[3H]imipramine] is the initial concentration of [3H]imipramine, and Kdimipramine is the dissociation constant for [3H]imipramine at the hα3β4 [0.41 μM (Arias et al., 2010c)] and hα4β2 [0.83 μM (Arias et al., 2010b)] AChRs.
The binding affinity and voltage-dependence differences were determined by Student's t-test analysis.
2.7. Molecular Docking and Molecular Dynamics Simulations
Since the functional experiments were performed with hα3β4 AChRs in the (α3)3(β4)2 stoichiometry (Arias et al., 2016), and (α9)2(α10)3 is the most probable form (Boffi et al., 2017; Plazas et al., 2005), these two AChR stoichiometries were first built using the X-ray structure (PDB ID: 5KXI) of the human α4β2 AChR at 3.9 Å resolution (Morales-Perez et al., 2016) as the homologous template using the MODELLER program (Šali et al., 1995) as implemented in the Accelrys Discovery Studio 2.5 software.
(+)-Citalopram in the protonated state (i.e., protonated at physiological pH) was modeled using VEGA ZZ and subsequently docked at each AChR model using AutoDock Vina (Trott and Olson, 2010). Protocols for minimization, partial charge calculations and docking were carried out as previously described (Arias et al., 2018a, 2018b, 2016).
To determine the stability of each pose within its predicted docking site, 20-ns molecular dynamics (MD) simulations were performed as previously described (Arias et al., 2018a, 2018b, 2016), using NAMD (Phillips et al., 2005) and CHARMM force field, and VEGA ZZ (Pedretti et al., 2004) as interface. Poses with RMSD variance (VAR) <1 during the last third of the MD were used.
2.8. Calculation of the theoretical binding energies
Theoretical binding energies (TBE), measured from the individual poses at the end of the MD, were calculated using molecular mechanics as in (Arias et al., 2018a, 2018b). The TBE values are estimations used only for comparative purposes among receptors and its respective sites, and do not intend to represent absolute binding energies. More negative TBE values indicate higher theoretical binding affinities (TBA).
2.9. In silico mutations
To structurally explain the different binding behavior of escitalopram between (α3)3(β4)2 and (α9)2(α10)3 models, in silico mutations were performed on those amino acid positions involved in orthosteric and luminal binding, respectively, using homologous residues from subunits α3, α9, or α10. Mutations were implemented using the Build Mutants module implemented in the Accelrys Discovery Studio 2.5 software which also use the MODELLER algorithms (Šali et al., 1995) for this purpose.
3. Results
3.1. AChR selectivity for (±)-citalopram
The activation potency of (±)-epibatidine on each human AChR was first determined by assessing the fluorescence change in the respective AChR-expressing cells after (±)-epibatidine stimulation. The respective EC50 values for (±)-epibatidine are in the same range as previous determinations (Arias et al., 2018a, 2018b, 2017, 2016, 2010a, 2010b, 2010c).
The inhibitory activity of (±)-citalopram was subsequently assessed by pre-incubating (±)-citalopram with the respective hα3β4- (Fig. 1A), hα4β2- (Fig. 1B), and hα7-expressing cells (Fig. 1C), for 5 min before (±)-epibatidine stimulation (0.1 μM). Interestingly, (±)-citalopram inhibited (±)-epibatidine-induced hα3β4 AChR activity (IC50 = 5.1 ± 1.3 μM) with higher potency compared to that for the hα4β2 (19.1 ± 1.3 μM) and hα7 (18.8 ± 1.3 μM) AChRs (Table 1). The results showing that the nH values for (±)-citalopram are near unity (Table 1) indicate that the inhibitory process is mediated by a non-cooperative mechanism. A non-cooperative mechanism, in turn, suggests that there is potentially only one binding site or several sites with similar affinity.
3.2. (±)-Citalopram inhibits rα9α10 AChRs in a concentration-dependent and voltage-independent manner
Voltage-clamp experiments showed that (±)-citalopram inhibits ACh (10 μM)-evoked rα9α10 AChR activity in a concentration-dependent manner (Figs. 2A,B), giving an IC50 value of 7.5 ± 0.9 μM (Table 1). The observed nH value close to unity (Table 1) indicated that the inhibitory process is mediated by a non-cooperative mechanism. To further study the inhibitory mechanism elicited by (±)-citalopram, two approaches were used. First, the activity elicited by increasing concentrations of ACh was determined in the absence and presence of 8.0 μM (±)-citalopram (Fig. 2B). The competition curves showed that (±)-citalopram produced a parallel rightward shift of ACh-evoked responses. A significant (p = 0.0001) increase of the ACh EC50 value (7.5-fold) was observed in the presence of (±)-citalopram, with no changes in agonist maximal responses and nH values (Table 2), supporting a competitive mechanism of inhibition. Secondly, the inhibitory activity of 10 μM (±)-citalopram was determined at different membrane potentials, as shown in the representative I/V curve (Fig. 2C). The ACh responses were inhibited by (±)-citalopram at both negative (−90 mV) and positive (+40 mV) potentials with similar percentage (46.0 ± 4.7% and 43.3 ± 4.9%, respectively; Student’s t-test; p = 0.1) (Fig. 2D), indicating that the observed inhibition is mainly voltage-independent and consistent with a competitive mode of action.
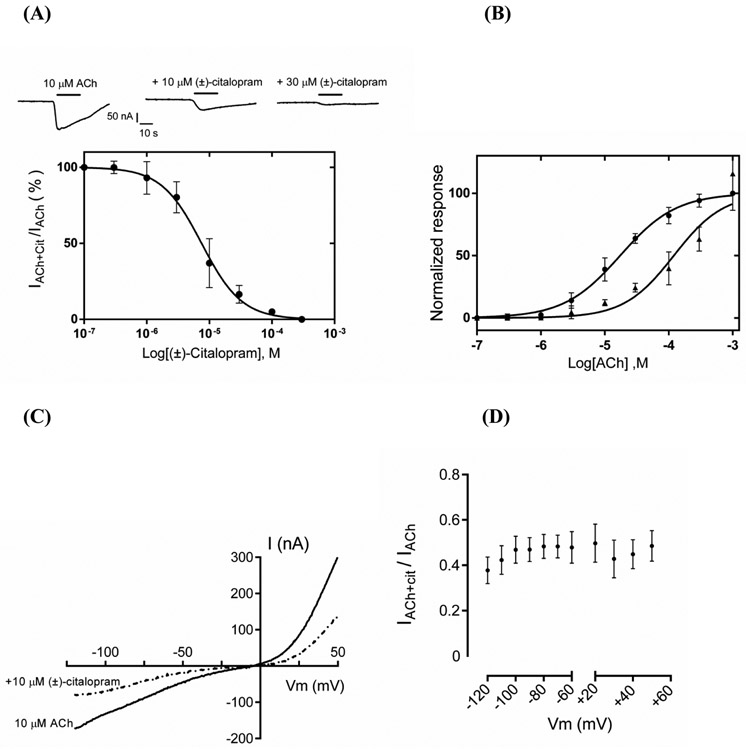
Fig. 2.
Effect of (±)-citalopram on acetylcholine (ACh)-evoked activity at rα9α10 AChRs expressed in Xenopus oocytes. (A) Responses of rα9α10 AChRs elicited by 10 μM ACh are diminished by increasing concentrations of (±)-citalopram. The inhibition curve was obtined by the co-application of 10 μM ACh and increasing concentrations of (±)-citalopram (r2 = 0.96; n = 7). Responses (mean ± SEM) were normalized to that elicited by 10 μM ACh (its EC50 value) which was set as 100%. The calculated IC50 and nH values are summarized in Table 1. (B) Concentration-response curves for ACh in the absence (●) and presence (▲) of 8 μM (±)-citalopram (n = 6). The EC50 values for ACh in the absence and presence of (±)-citalopram are summarized in Table 2. A statistical difference was obtained (p = 0.0001). (C) A representative current-voltage response (n = 7) obtained by applying 2-s voltage ramps from −120 to +50 mV, 10-s after the peak response to 10 μM ACh from a holding potential (Vhold) of −70 mV, in the presence and absence of 10 μM (+)-citalopram. (D) The comparison of (±)-citalopram-induced inhibition at different membrane potentials showed no statistical difference (Student’s t-test; p = 0.1), indicating a voltage-independent mechanism (n = 7).
Table 2:
Potency of ACh (EC50) in the absence and presence of (±)-citalopram at the α9α10 AChR.
| ACh | EC50 (μM) | nH |
|---|---|---|
| No (±)-citalopram | 17.8 ± 1.8 | 0.98 ± 0.2 |
| 8.0 μM (±)-citalopram | 134.6 ± 16.5 | 1.11 ± 0.6 |
Values obtained from Figure 2B.
3.3. (±)-Citalopram inhibits ACh-evoked currents from MHb (VI) neurons in a concentration- and voltage-dependent manner
Patch-clamp recordings on MHb (VI) neurons showed that 100 μM ACh puffs activated endogenous AChRs (see control traces in Fig. 3A). MHb (VI) neurons were identified primarily by their close proximity (<50-70 μm) to the 3rd ventricle within the ventral aspect of the MHb. They were secondarily distinguished from other nearby brain areas (e.g., thalamus and lateral habenula) via the presence of slow (1-8 Hz) tonic firing (Shih et al., 2014).
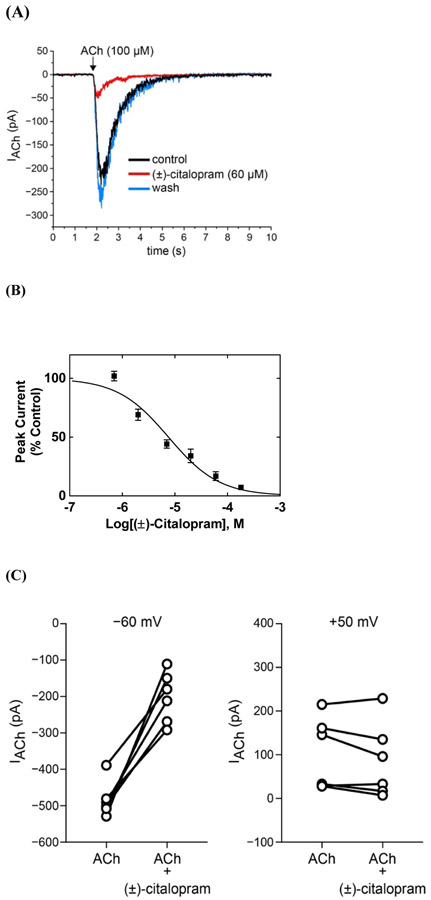
Fig. 3.
Inhibitory potency of (±)-citalopram on ACh-evoked currents from MHb (VI) neurons. (A) ACh puffer (100 μM)-evoked currents from MHb (VI) neurons are decreased by 60 μM (±)-citalopram. The puffer was performed for 250 ms at a pressure of 12 psi. After washing, the peak amplitude completely recovered, indicating a reversible inhibition. (B) Concentration-response relationship for the inhibitory activity of (±)-citalopram on ACh-evoked currents from MHb (VI) neurons. Response was normalized to the maximal ACh response which was set as 100%. The plot (r2 = 0.90) is representative of 5-8 determinations (mean ± SEM). The calculated IC50 and nH values are summarized in Table 1. (C) Voltage-dependence of (±)-citalopram-induced inhibition of ACh-evoked currents from MHb (VI) neurons. Steady-state ACh puffer (100 μM)-evoked currents from MHb (VI) neurons were recorded at −60 mV and +50 mV in the same cell, in the absence and presence of 60 μM (±)-citalopram. Data plots show ACh-activated currents before and after (±)-citalopram superfusion at the indicated membrane potential. Paired Student's t-test analyses indicated that (±)-citalopram reduced ACh-evoked response amplitudes by 42.4 ± 6.1% at −60 mV (p = 0.0006) and 74.6 ± 13.6% at +50 mV (p = 0.1494).
The observed inward currents elicited by ACh were reduced by superfusion of 60 μM (±)-citalopram (Fig. 3A), and drug washout resulted in complete recovery of the original response amplitude. Full recovery after washout confirms that the recording remained stable during drug applications. A concentration-dependent inhibition was determined for (±)-citalopram (Fig. 3B) by using a wide range of concentrations (i.e., 0.07-180 μM), giving an inhibitory potency of 7.6 ± 1.0 μM (Table 1). The observed nH value close to unity (Table 1) suggested a non-cooperative mechanism.
To further examine the antagonistic mechanism of (±)-citalopram on native MHb AChRs, the inhibitory activity of this drug was compared at a holding potential of −60 mV and +50 mV in the same cell. (±)-Citalopram (60 μM) inhibited ACh-evoked responses by 42 ± 6 % at negative potential (−60 mV; paired Student's t-test: p = 0.0002), whereas its effect at positive potential (+50 mV) was not statistically significant (84 ± 12 %; p = 0.2536) compared with the values for ACh alone (Fig. 3C). Nevertheless, a significant difference was observed between these two extreme potentials (p = 0.0088). These results suggest that (±)-citalopram preferentially blocks habenular α3β4* AChRs in a voltage-dependent fashion.
Considering that (±)-citalopram's activity is voltage-dependent, the electrical distance (δ) of its binding site along the ion channel, was subsequently calculated using Eq. (1). The determined IC50(0) (92 μM) and δ (0.40) values suggested that citalopram isomers interact with a binding site located within the pore, close to the middle region of the ion channel.
3.4. Binding affinity of (±)-citalopram for the [3H]imipramine sites at either hα3β4 or hα4β2 AChRs
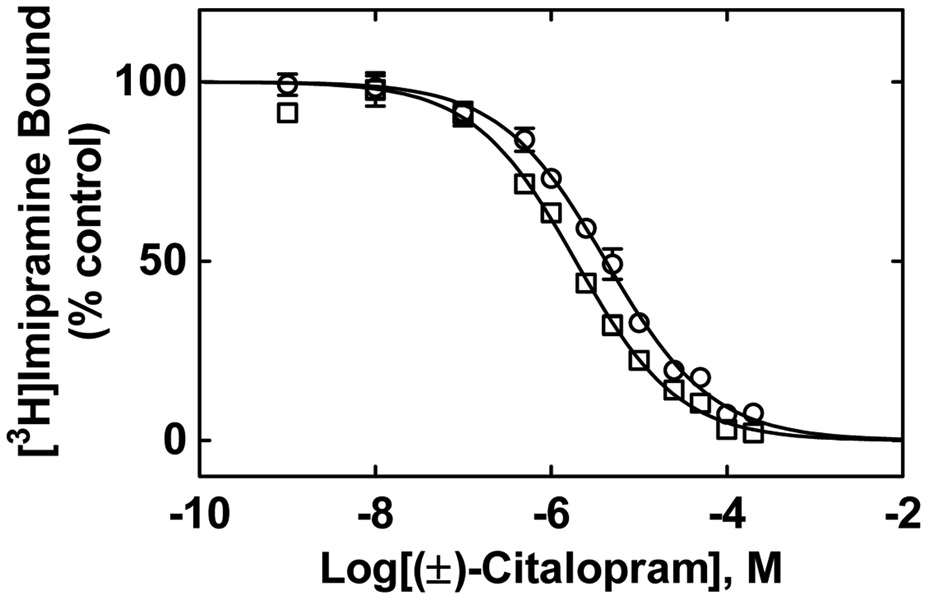
To determine whether (±)-citalopram binds to the [3H]imipramine at either hα3β4 or hα.4β2 AChRs, the effect of this antidepressant on [3H]imipramine binding was determined on desensitized AChRs [i.e., in the presence of (±)-epibatidine] (Fig. 4). The results indicated that the binding affinity of (±)-citalopram for hα3β4 AChRs (Ki = 1.8 ± 0.1 μM) was >2-fold higher than that for hα4β2 AChRs (4.1 ± 0.3 μM) (paired Student's t-test, p = 0.0009) (Table 3). Since the binding affinity for resting hα4β2 AChRs [i.e., in the presence of 0.1 κ-bungarotoxin; (Arias et al., 2010a, 2010b, 2010c)] (5.9 ± 0.4 μM) was in the same range as that in the desensitized state, no additional experiments were performed on resting hα3β4 AChRs.
Figure 4.
(±)-Citalopram-induced inhibition of [3H]imipramine binding to either hα3β4 (□) or hα4β2 (○) AChRs in the desensitized state. Each AChR-containing membrane (1.5 mg/mL) was pre-incubated (30 min) with 15.2 nM [3H]imipramine in the presence of 0.1 μM (±)-epibatidine (receptors are mainly in the desensitized state), and then equilibrated with increasing concentrations of (±)-citalopram. Nonspecific binding was determined at 100 μM imipramine. The plots are combinations of 2-4 experiments, each performed in triplicate, where the error bars are the S.D. The IC50 and nH values were obtained by nonlinear least-squares fit of the plots (r2 = 0.95 for both). The Ki values, calculated using Eq. (1), were summarized in Table 2.
Table 3.
Binding affinity of (±)-citalopram for the [3H]imipramine sites at the respective hα3β4 and hα4β2 AChRs in the desensitized state.
The observed nH values (i.e., close to unity) (Table 3) indicated that (±)-citalopram inhibits [3H]imipramine binding to either hα3β4 or hα4β2 AChR by non-cooperative mechanisms. This suggest, in turn, that there is potentially only one binding site or several sites with similar binding affinity on each AChR subtype.
3.5. Molecular Docking and Molecular Dynamics Simulations of S-(+)-citalopram (escitalopram) to the h(α3)3(β4)2 and h(α9)2(α10)3 AChRs
In the h(α3)3(β4)2 AChR model, two luminal sites for escitalopram were found by molecular docking (Fig. 5A), and their stability confirmed by molecular dynamics (Table 4; Fig. 5D). The docking procedure did not find any conformer at the orthosteric sites, but two non-orthosteric sites (not shown for simplicity), which presumably do not directly influence nicotine binding.
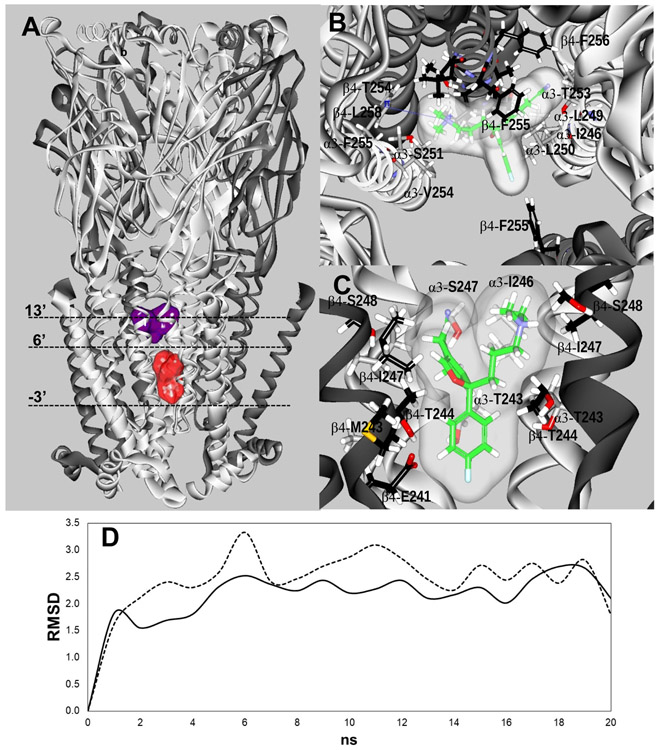
Figure 5.
(A) Docking sites for S-(+)-citalopram (escitalopram) at the h(α3)3(β4)2 AChR model. Escitalopram docked to two luminal sites (surface model), a high-affinity site located closer to the extracellular ion channel’s mouth (blue) and a low-affinity site located closer to the cytoplasmic side (red). α3 (white) and β4 (dark grey) subunits are represented as solid ribbons. Dotted lines indicate the positions of Gly (position −3’), Ser (position 6’), and Val (position 13’) rings along the ion channel. (B) In the high-affinity site, escitalopram (as sticks and its transparent surface model, colored by atoms with carbons in green) interacted with M2 residues located between positions 5' and 16', forming a strong H-bond with β4-T254 (position 12') (dotted black line), and a cation-π interaction with α3-F255 (position 14’) (solid blue line). (C) In the low-affinity site, escitalopram interacted with M2 residues located between positions −3' and 6'. A complete list of residues is summarized in Table 4. The interacting residues (as sticks) are labeled by their subunit, residue one letter code, and amino acid sequence number, and colored by atoms, including carbons (grey for the α3 subunit, and black for the β4 subunit), nitrogens (blue), oxygens (red), and hydrogen (white). (D) Molecular dynamics simulations (20 ns) of escitalopram interacting with the high- (—) and low-affinity (-----) sites, respectively, at the h(α3)3(β4)2 model.
Table 4.
Residues involved in the docking of S-(+)-citalopram (escitalopram) to luminal sites at the h(α3)3(β4)2 AChR.
| Luminal Site |
TBE (Kcal/mol) |
RMSD Mean / Var |
α3 | β4 | Homologous residues a |
M2 Position (nng) |
|
|---|---|---|---|---|---|---|---|
| α9 | α10 | ||||||
| High-affinity | −14.31 | 2.30 / 0.02 | I246* | V248 | V248 | 5′ | |
| S248* | T249 | T249 | 6′ (Ser) | ||||
| L249 | L251 | L251 | 8′ | ||||
| L250 | L251 | L252 | L252 | 9′ (Leu) | |||
| S251 | A252 | A253 | A253 | 10′ | |||
| T253 | T254 (H-bond donor) | T255 | T255 | 12′ | |||
| V254 | F255 | V256 | V256 | 13′ (Val) | |||
| F255(Cation-π) | F256 | F257 | F257 | 14′ | |||
| L258 | L259 | L259 | 16′ | ||||
| Low-affinity | −4.27 | 2.55 / 0.07 | G239 | G240 | G241 | G241 | −3′ (Gly) |
| E240 | E241 | E242 | E242 | −2′ | |||
| V242 | M243 | V244 | V244 | 1′ | |||
| T243 | T244 | S245 | S245 | 2′ (Thr) | |||
| L245 | L246 | L246 | 3′ | ||||
| 1246* | 1247 | V248 | V248 | 5′ | |||
| S247 | S248* | T249 | T249 | 6′ (Ser) | |||
Residues from the high- and low-affinity sites belong to different α3 and β4 indicating no overlapping between both sites.
The homologous residues at α9 and α10 subunits are also included for comparative purposes.
The high-affinity binding site (TBE = −14.31 Kcal/mol; Table 4) is located between positions 5’ and 16’, in the middle of the ion channel but toward the extracellular ion channel’s mouth (Figs. 5A-E). The M2 residues at each position are: α3-I246 (5’), β4-S248 (6’; Ser ring), α3-L249 (8’), α3-L250 and β4-L251 (9’; Leu ring), α3-S251 and β4-A252 (10’), α3-T253 and β4-T254 (12’); α3-V254 and β4-F255 (13’; Val ring); α3-F255 and β4-F256 (14’), and β4-L258 (16’). Favoring its higher affinity is the presence of a strong H-bond with the β4-T254 side chain oxygen. In addition, a cation-π interaction is established between the N+ atom of escitalopram and the aromatic moiety of α3-F255. The low-affinity binding site is located closer to the cytoplasmic side, between positions - 3’ and 6’ (Table 4). The M2 residues at each position are: α3-G239 and β4-G240 (−3’; Gly ring), α3-E240 and β4–E241 (−2’), α3-V242 and β4-M243 (1’), α3-T243 and β4-T244 (2’; Thr ring), β4-L245 (3’), α3-I246 and β4-I247 (5’), and α3-S247 and β4-S248 (6’). No H-bond nor cation-π interactions were detected at this site. Residues α3-I246 (5’) and β4-S248 (6’) from this site belong to different subunit copies respect to those belonging to the high-affinity site at the pentameric h(α3)3(β4)2 receptor.
In the h(α9)2(α10)3 model, escitalopram interacted with three orthosteric binding sites (Fig. 6A) located in the interface between (+)α10 (principal component) and (−)α9 or another (−)α10 (complementary component) (Boffi et al., 2017). Their stabilities were confirmed by molecular dynamics (Fig. 6D; Table 5). The conformers at sites 1 and 2 found at the (+)α10/(−)α9 interface showed TBA values than that for site 3. (Table 5). Several additional docking sites were found but none of them positioned in such a way to sterically block the ion channel (i.e., at or near the middle of the channel). Moreover, no conformers were found at the non-orthosteric binding sites [i.e., (−)α10/(+)α9].
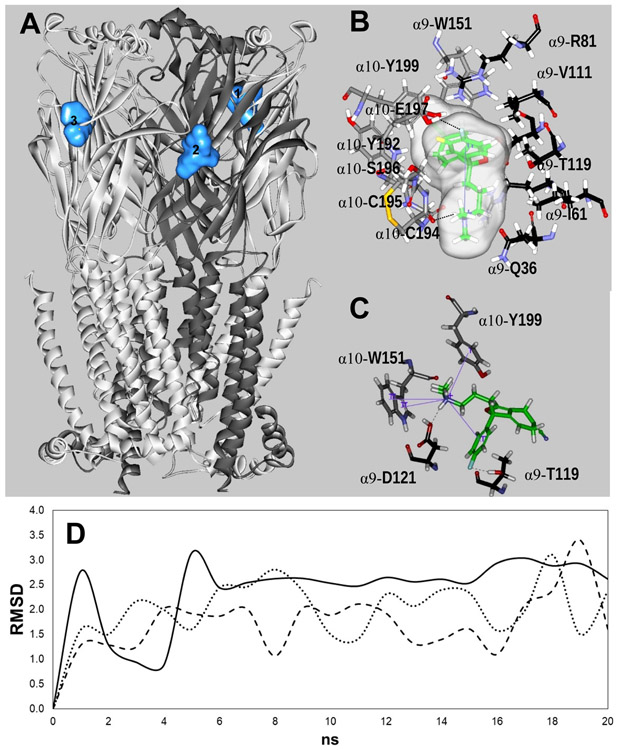
Figure 6.
Docking sites for S-(+)-citalopram (escitalopram) at the h(α9)2(α10)3 AChR model. (A) Escitalopram interacted with three possible orthosteric sites located at the interface between the (+)α10 (principal component) and (−)α9 [or another (−)α10] subunit (complementary component) (light blue surface models). α10 (white) and α9 (dark grey) subunits are represented as solid ribbons. (B) In site 1, escitalopram (as sticks and its transparent surface model, colored by atoms with carbons in green) formed a strong H-bond with two oxygens, one on the α10-C194 main chain and another on the α10-E197 side chain (dotted black line). Interestingly, an intramolecular cation-π interaction is formed in escitalopram, between N+ and its fluorophenyl ring (solid blue line). (C) In site 2, escitalopram (as sticks colored by atoms with carbons in green) formed a network of H-bond and cation-π interactions. Other details are included in Figure 5. The complete list of residues interacting at each site is summarized in Table 5. (D) Molecular dynamics simulations (20 ns) of escitalopram interacting with sites 1 (-----), 2 (—), and 3 (…··), respectively, at the h(α9)2(α10)3 model.
Table 5.
Residues involved in the docking of S-(+)-citalopram (escitalopram) to the agonist binding sites at the h(α9)2(α10)3 AChR.
| Orthosteric Site | 1 | 2 | 3 | ||
|---|---|---|---|---|---|
| TBE (Kcal / mol) | −15.22 | −14.85 | −12.13 | ||
| RMSD (Mean / Var) | 1.75 / 0.14 | 2.58 / 0.019 | 2.34 / 0.15 | Homologous residues* | |
| Secondary Structure | Subunit and Residues | ||||
| (−)α9 | (−)α9 | (−)α10 | (−)β4 | ||
| β1 Sheet | T34 | ||||
| Q36 | Q36 | E36 | Q38 | ||
| β2 Sheet | W57(−) | W57(−) | |||
| R59 (−) | R59(−) | R59(−) | K61 | ||
| N60 | |||||
| I61 | I61 | E61 | E63 | ||
| β3 Sheet | S79 | ||||
| R81 | R81 | R81 | R83 | ||
| β5 Sheet | V111(−) | V111(−) | V111(−) | I113 | |
| R113(−) | |||||
| β6 Sheet | T119(−) | T119(−) | R119(−) | L121 | |
| W120 | W120 | ||||
| D121(−) | D121(−) | D121(−) | L123 | ||
| (+)α10 | (+)α10 | (+)α10 | (+)α9 | (+)α3 | |
| β4-β5 Loop | Y95(+) | Y95(+) | Y95(+) | Y95 | Y93 |
| β7 Sheet | S150 | S150 | S150 | S150 | S148 |
| β7-β8 Loop | W151(+) | W151(+) | W151(+) | W151 | W149 |
| T152 | T152 | ||||
| G154 | G154 | G154 | N154 | D152 | |
| β9-β10 loop | Y192 | Y192 | Y192 | Y192 | Y190 |
| G193 | G193 | G193 | G193 | N191 | |
| C194(+) | C194(+) | C194(+) | C194 | C192 | |
| C195(+) | C195(+) | C195(+) | C195 | C193 | |
| S196 | S196 | S196 | S196 | E194 | |
| E197 | E197 | E197 | E197 | E195 | |
| β10 Sheet | Y199(+) | Y199(+) | Y199(+) | Y199 | Y197 |
Bold: residues forming H-bonds; Italics: residues forming cation-π interactions.
Principal (+) or complementary (−) canonical component.
The homologous residues at α3 and β4 subunits are also included for comparative purposes.
The three sites have residues coming from the same receptor domains. The complementary component [(−)α9] is formed by six common residue positions from the β1, β2, β3, β5 and β6 sheets. Four of them coincide with those for (−)-nicotine binding (i.e., canonical positions) (Brejc et al., 2001), including R59 (β2 sheet), V111 (β5 sheet), α9-T119-α10-R119 and D121 (β6 sheet) (Table 5), and two novel positions at α9-Q36-α10-E36 and V111. Sites 1 and 2 have two common positions, W57 (a canonical site), and W120. Other residues involved are (−)α10-T34 (site 3), N60, S79, and the canonical R113 (site 2). Eleven common residues, coming only from the (+)α10 subunit, form the principal component. Five of them agree with canonical (−)-nicotine binding positions (Brejc et al., 2001), including Y95 (β4-β5 loop), W151 (β7-β8 loop), C194-C195 (β9-β10 loop), and Y199 (β10 sheet) (Table 5). Other common residues include S150, G154, Y192, G193, S196, and E197. In addition, T152 is shared by sites 2 and 3.
At site 1, α10-C194 establishes a H-bond between its main chain O and a H of one of the methyl groups attached to the ammonium moiety of escitalopram (Fig. 6B), whereas another H-bond is formed between the E197 side chain O and a H of the phenyl group. At this site, escitalopram has a conformation that enables it to establish an intramolecular cation-π interaction between N+ and the fluorophenyl ring. At site 2, two H-bonds are formed, one between escitalopram’s fluorine and the H of the α9-T119 side chain, and another between escitalopram’s ammonium H and the hydroxyl O of the α9-D121 side chain. In addition, there is a network of cation-π interactions: the intramolecular interaction also seen at site 1 for escitalopram, and the intermolecular interaction between the N+ of escitalopram and both rings of α10-W151 and the phenyl ring of α10-Y199 (Fig. 7). Finally, site 3 is the only one composed by two adjacent α10 subunits. At this site, escitalopram establishes a cation-π interaction with R119 and a H-bond between the O of the α10-G193 main chain and the ammonium H of escitalopram.
Figure 7.
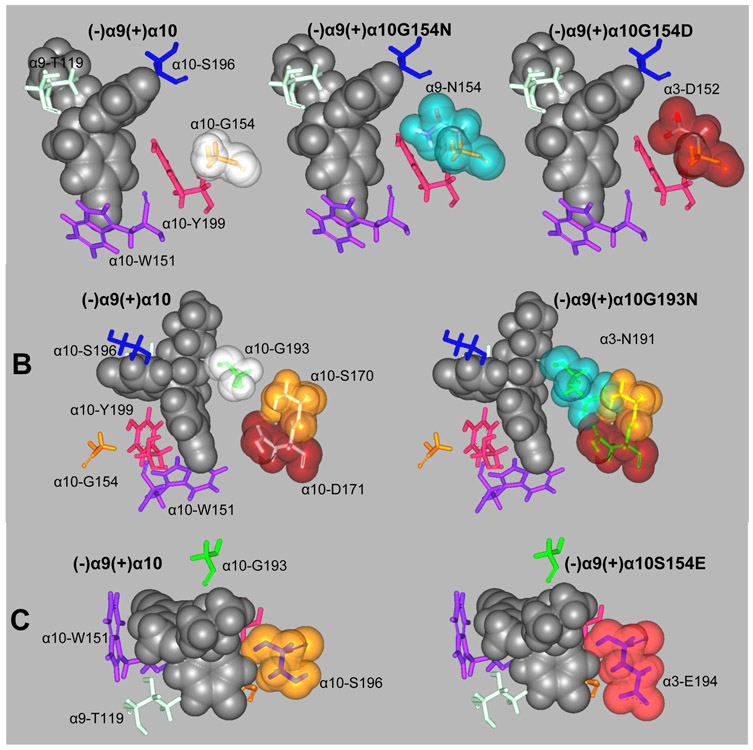
In silico mutations of (+)α10 residues involved in escitalopram binding to orthosteric sites to their respective homologous residues at α9 and α3. (A) α10-G154 mutations to its α9 (α10G154N) and α3 (α10G154D) homologous residues. (B) α10-G193 mutation to its α3 homologous residue (α10G193N). (C) α10-S196 mutation to its α3 homologous residue (α10S196E). Escitalopram is represented by its van der Waal solid surface in black. Mutated residues are represented as sticks surrounded by their van der Waal transparent surfaces. Other important binding site residues are represented as sticks.
3.6. Structural differences between escitalopram docked to the h(α3)3(β4)2 and h(α9)2(α10)3 AChRs
Considering that both h(α3)3(β4)2 and h(α9)2(α10)3 AChR models are based on the same template, differences in docking results must be due to, and should be explained by alterations in the amino acid sequences as follow.
3.6.1. Luminal sites
Two luminal sites for escitalopram were characterized at the h(α3)3(β4)2 but none was observed at the h(α9)2(α10)3 (Fig. 5). To find out the structural reasons of this difference at the amino acid level, a comparison was made between homologous M2 residues involved in escitalopram binding at both receptors (Table 4). Both receptors have the same amino acids at positions 8’, 9’, 12’, 14’, and 16’, where the high-affinity site for escitalopram is located. However, I246 (position 5’) is only found at α3, whereas V248, the homologous residue at α9 and α10, is also hydrophobic but slightly smaller. At ring 6’, β4-S248 is substituted by the slightly larger but also polar residue T249 in α9 and α10. Although these differences may have some influence on escitalopram binding, there is a more important modification at position 10’ between the polar α3-S251 and its homologous residue, the hydrophobic α9-A253 (both β4 and α10 have Ala at this position). The key difference between both receptors is centered on ring 13’, where β4 has a Phe residue (F255), while all other subunits have Val. Figure 5B shows that F255 protrudes to the middle of the ion channel from one side making a direct contact with escitalopram docked at the opposite side, supporting a total blockage of the lumen. This interaction is structurally not possible at the h(α9)2(α10)3 AChR, since the homologous residue, V254, is smaller and does not contact escitalopram.
In the low-affinity site, escitalopram is stacked at the cytoplasmic and narrowest end of the h(α3)3(β4)2 channel. There are three M2 sequence differences between both receptors: at position 1’, β4 has a Met residue (M243), whilst the other subunits have Val as the homologous residue. However, since the larger side chain of M243 is not facing the lumen, it has no influence on ligand binding (Fig. 5C). There are two important differences between both receptors. One is at ring 2’, where a larger Thr residue is present in h(α3)3(β4)2 (Fig. 5C) compared to Ser at h(α9)2(α10)3 (Table 4). The other is at ring 5’, where h(α3)3(β4)2 has a larger Iie residue compared to a Val present at h(α9)2(α10)3.
To determine the importance of these structural differences, receptor mutants were constructed, and escitalopram docked as described previously. The h(α3)3(β4)2 mutations included β4F255V (to test the high-affinity luminal site), and α3T243S, α3I246V, β4T244S, β4I244V (to test the low-affinity luminal site). The h(α9)2(α10)3 mutations included α9V256F (high-affinity site), and α9α10S245T and α9α10V248I (low-affinity site). The h(α3)3(β4)2 mutations abolished the docking of escitalopram at both luminal sites. When h(α9)2(α10)3 carried only the α9V256F mutation, escitalopram was able to dock to a locus similar to the high-affinity, and when the remaining α9α10S245T and α9α10V248I mutations were added, escitalopram could also dock to a locus similar to the low-affinity site.
3.6.2. Orthosteric Binding Sites
Our in silico studies indicated that escitalopram docked at the orthosteric binding sites of the h(α9)2(α10)3 (Fig. 6), but not h(α3)3(β4)2, AChRs. In addition, escitalopram docked to non-orthosteric binding sites at the latter receptor but not to the former. Escitalopram does not bind to “non-orthosteric” sites in the h(α9)2(α10)3 AChR because the (+)α9 side is not capable of behaving as does (+)α10, even considering that it has the characteristic adjacent double Cys residues at the binding site.
In order to explain why escitalopram does not bind to h(α3)3(β4)2 orthosteric sites, we compared the amino acid sequence between the (+) sides of α3 and α10, the (−) sides of α9 or α10 [since the latter is also able to behave as a (−) side in site 3] and β4, or both types of differences (Table 5). For the (+) side, a comparison with (+)α9 residues is also shown.
Essential positions, useful for comparison analysis, are those that are common to the three sites at the h(α9)2(α10)3. There are seven such positions at the (−) side, none of which can explain the absence of binding at h(α3)3(β4)2. More specifically: (1) Same homologous residues [e.g., α9-Q36 and β4-Q38 (1st position), α10-E61 and β4-E63 (3rd position), and Arg in all subunits (4th position)]; (2) Structurally similar homologous residues that maintain the same basic functions [e.g., charged α9- and α10-R59 vs β4-K61 (2nd position), hydrophobic α9- and α10-V111 vs β4-I113 (5th position); (3) Different homologous residues where neither charge nor steric hindrance play an important role [e.g., polar α9-T119 is not involved in H-bonding, and charged α10-R119 is similarly bulky as the hydrophobic residue β4-L121 at site 1 (6th position)]; (4) Although there is a difference between α9-/α10-D121 and β4-L123 (7th position), Asp is involved in H-bonding only at site 2, so polarity is no essential and both type of residues are similar in size.
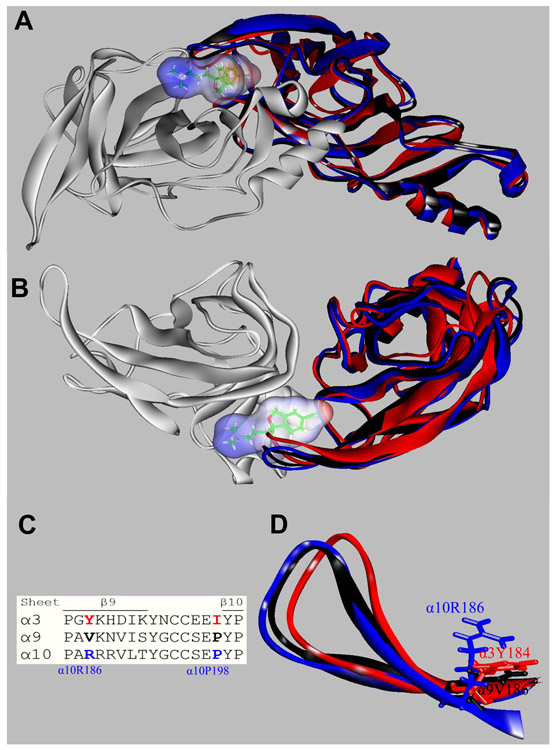
At the (+) side there are twelve positions, eleven of which have the same residue at the α9 and α10 subunits (Table 5). There is only one position where both α9 and α3 subunits differ from α10. The homologous residues of α10-G154 are α9-N154 and α3-D152, respectively. α3 also differs from α10 at α10-G193, where the former has an Asn191 residue, and at α10-S196, where α3 has a Glul94 residue (Table 5). To test if these residues are the basis of escitalopram binding at the orthosteric sites, in silico mutants were constructed and molecular docking performed as previously explained. The mutations α3D152G, α3N191G, and α3E194S did not enable escitalopram binding to h(α.3)3(β4)2, Likewise, the mutation α9N194G did not enable escitalopram binding to (+)α9. Conversely, the mutations α10G154D, α10G193N, or α10S196E, which we expected should be important because they could promote an overlapping with escitalopram (Fig. 7), did not abolish escitalopram binding to h(α9)2(α10)3. Therefore, other structural reasons must be responsible for the preference of escitalopram binding to the orthosteric sites at h(α9)2(α10)3 but not at h(α3)3(β4)2 receptors. Consequently, we visually inspected the superposed α3, α9, and α10 orthosteric binding sites with escitalopram docked at (+)α10 (Figs. 8A,B). We found that the β9-β10 loop, which contains the Cys pair typical of α subunits, was widely open in α10 allowing escitalopram to fit into the binding site. This loop is closer to the receptor center in α9, and it is even closer in α3, and causes escitalopram to overlap the main chain atoms of these subunits when is at the (+)α10 docking position. During model construction, loops are optimized according to the surrounding residues. Figure 8C shows a sequence comparison of the surrounding residues. We found at least two possible residues (in blue) that could force the β9-β10 loop in α10 to set apart from the receptor. From the β9 sheet (Fig. 8D), α10R186, which points to the middle of the β9-β10 loop and is bulkier than α9V186 and α3Y184, and from the β10 sheet, α9α10Ρ198, which alters the backbone conformation with respect to α3I196.
Figure 8.
Orthosteric binding sites at the superposed (+)α3 (red), (+)α9 (black), (+)α10 (blue), and (−)α9 (white) subunits. Escitalopram is shown as sticks surrounded by its molecular surface. The β9-β10 loop at the α3 and α9 subunits are closer to the receptor center than that at α10 (A: Extracellular view; B: Cytoplasmic view), and consequently there is no room for escitalopram to fit in the agonist binding site in α3 and α9. The α3- and α9-β9-β10 loops overlap the ligand when is docked as in the (α9)2(α10)3 receptor. (C) Amino acid sequence comparison between α3, α9, and α10 subunits at the level of the β9-β10 loop. Blue: amino acids identified as the cause of the different β9-β10 loop conformations (see text). (D) A detailed side chain view at α10R186, α9V186, and α3Y184 positions, showing the differences of side chains occupied volume that would force the α10-β9-β10 loop to set apart from the receptor.
The binding of escitalopram to non-orthosteric sites at h(α3)3(β4)2 is fairly different to that described above for the orthosteric sites. However, none of these differences seem to be essential to explain the absence of this type of binding at the h(α9)2(α10)3 AChR. At the (+)β4 side, however, R151 forms a cation-π interaction that might be indispensable for escitalopram binding, that is lacking at α9 and α10 where G149 is the homologous residue.
4. Discussion
This work demonstrates the selectivity of (±)-citalopram for different AChR subtypes, the different mechanisms of inhibition between α9α10 AChRs and native α3β4* AChRs expressed in MHb (VI) neurons, and whether this antidepressant shares the same binding site(s) as that for imipramine.
The present Ca2+ influx results indicate the following AChR selectivity for (±)-citalopram (IC50s in μM): hα3β4 (5.1 ± 1.3) > hα7 (18.8 ± 1.1) ~ hα4β2 (19.1 ± 4.2). Interestingly, the same preference for hα3β4 AChRs was observed for other SSRIs (Arias et al., 2010a) as well as for structurally different antidepressants such as tricyclic antidepressants (Arias et al., 2018b, 2010b, 2010c) and bupropion (Arias et al., 2018a, 2014) using the same assay (i.e., Ca2+ influx). Based on previous studies of several SSRIs at hα3β4 AChRs (Arias et al., 2010a), the following rank order of inhibitory potencies was obtained: fluoxetine (2.0 ± 0.4) ~ paroxetine (2.6 ± 0.3) > (±)-citalopram (5.1 ± 1.3).
Our competition binding results indicated that (±)-citalopram inhibits [3H]imipramine binding to desensitized hα3β4 AChRs with >2-fold higher affinity than that for desensitized hα4β2 AChRs. By comparing with other SSRIs (Arias et al., 2010a), the following rank order of affinities (Ki's in μM) for the hα3β4 AChR was obtained: (±)-citalopram (1.8 ± 0.1) > fluoxetine (4.8 ± 0.5) > paroxetine (6.9 ± 0.6), indicating that although (±)-citalopram binds with relatively higher affinity to its allosteric hα3β4 AChR sites, its cellular response is less efficient compared to that for other used SSRIs.
The results from a variety of methods coincide with a luminal location for citalopram's binding site(s) at habenular α3β4* AChRs. First, the patch-clamp results demonstrated that (±)-citalopram inhibits ACh-evoked currents in MHb (VI) neurons with relatively high potency (7.6 ± 1.0 μM) in a voltage-dependent manner, by interacting with a binding site located close to the middle portion of the ion channel [i.e., electrical distance (δ) = 0.40]. Second, the molecular docking and in silico results using the h(α3)3(β4)2 model showed two luminal sites for escitalopram, compatible with an ion channel blockade mechanism. Interestingly, the high-affinity luminal site for escitalopram where β4-F255 is crucial was situated between positions 5’ and 16’, in agreement with the calculated electrical distance and at the same level of the imipramine locus within the α3β4 ion channel found in previous studies (Arias et al., 2010a). Since our radioligand binding results showed direct competition between [3H]imipramine and (±)-citalopram, we can conclude that citalopram isomers overlap the binding site for imipramine. Given that the high-affinity site for citalopram is located in the middle of the ion channel compared to the low-affinity site, which is closer to the cytoplasmic side of the ion channel, a potential scenario can be considered where once the high-affinity site is occupied by citalopram, the probability of the low-affinity site to be occupied is considerably diminished. This mutually exclusive mechanism is compatible with the observed nH value close to unity, suggesting a non-cooperative mechanism.
The majority of the current response at MHb (VI) neurons has been ascribed to α3β4* AChRs (Quick et al., 1999; Shih et al., 2014), strongly suggesting that the observed inhibition is mediated by this receptor subtype. Interestingly, this value is similar to that obtained by Ca2+ influx experiments where (±)-citalopram inhibits HEK293-hα3β4 cells expressing only the hα3β4 AChR subtype. Although a direct comparison of the calculated potencies between heterologous cells and MHb (VI) neurons cannot be done due to intrinsic differences in the used methods [e.g., see (Arias et al., 2018b, 2017)], it is possible to suggest that (±)-citalopram inhibits a homogenous population of α3β4* AChRs in MHb (VI) neurons. These results contrast with that obtained with (+)-catharanthine and (±)-18-methoxycoronaridine which apparently inhibit a heterogenous population of α3β4* AChRs (Arias et al., 2017). Since the mouse brain concentration of R-(−)- and S-(+)-citalopram (i.e., escitalopram) after acute treatment with 10 mg/kg (±)-citalopram was 0.5 and 1.1 μM, respectively (Karlsson et al., 2013), it is plausible that part of its clinical activity is mediated by inhibition of habenular α3β4* AChRs.
The voltage-clamp results also demonstrated that (±)-citalopram inhibits ACh-evoked rα9α10 currents (7.5 ± 0.9 μM) in a voltage-independent and competitive manner. Previous studies showed that imipramine (Arias et al., 2018b) and the serotoninergic antagonist ICS-205,930 (Rothlin et al., 2003) inhibited α9α10 AChRs by a competitive mechanism. These results add to a wide variety of compounds that block α9α10 AChRs with different potencies (Rothlin et al., 2000, 1999; Verbitsky et al., 2000), and support the notion that structurally and functionally different antidepressants inhibit α9α10 AChRs by a competitive mechanism, opposite to the noncompetitive mechanism observed at other AChR subtypes. The molecular docking studies also supported the experimental results indicating a competitive mechanism of inhibition for (±)-citalopram. In particular, escitalopram formed three stable interactions with orthosteric, but not luminal, sites at the h(α9)2(α10)3 AChR, which is compatible with the observed nH value greater than unity, suggesting a cooperative mechanism between multiple sites. In silico mutations also showed that the β9-β10 loop (i.e., “loop C”, which carries the characteristic double Cys in the α subunits) is fundamental for orthosteric binding to h(α9)2(α10)3, whereas alterations in the loop conformation, especially on α10R186 and α10P198 homologous positions, are responsible for the lack of orthosteric binding to h(α3)3(β4)2.
The results indicating that (±)-citalopram has higher selectivity for α3β4 AChRs and inhibits habenular α3β4* AChRs could be related to its clinical effects. This possibility is based on the observed relationship that compounds with relatively higher selectivity for α3β4 AChRs such as bupropion and mecamylamine present antidepressant activity (Arias et al., 2018a, 2014). The observation that the same compounds act in a synergistic manner with 18-methoxycoronaridine (Glick et al., 2002) and all of them alleviate alcohol and nicotine withdrawal effects (Arias et al., 2014; Chi and De Wit, 2003), might be related with the beneficial effects elicited by (±)-citalopram during alcohol withdrawal (Angelone et al., 1998).
Our findings clearly demonstrate that (±)-citalopram presents receptor selectivity, preferably inhibiting α3β4 and α9α10 AChRs but by different mechanisms. The results showing that (±)-citalopram inhibits MHb (VI) neurons with potency similar to that found at hα3β4 AChRs support that concept that this antidepressant interacts with a homogeneous population of native α3β4* AChRs.
HIGHLIGHTS.
(±)-Citalopram inhibits hα3β4 with higher potency than that for hα7 and hα4β2 AChRs
(±)-Citalopram inhibits α9α10 AChRs in a voltage-independent manner
(±)-Citalopram inhibits habenular α3β4* AChRs in a voltage-dependent manner
Radioligand binding and molecular docking (MD) support a luminal location at α3β4
MD differentiates competitive (α9α10) vs noncompetitive (α3β4) inhibitory mechanisms
Acknowledgements
This work was supported by grants from NIH (DA040626) (to R.M.D.), National Agency for Scientific and Technologic Promotion, Argentina (to A.B.E.), and Dirección General de Asuntos del Personal Académico, UNAM, Mexico (PASPA grant) (to J.G-C).
Abbreviations:
- AChR
nicotinic acetylcholine receptor
- SSRI
selective serotonin reuptake inhibitor
- ACh
acetylcholine
- NCA
noncompetitive antagonist
- RT
room temperature
- MHb
medial habenula
- VI
ventral inferior
- Ki
inhibition constant
- Kd
dissociation constant
- IC50
ligand concentration that produces 50% inhibition of binding (or of agonist activation)
- EC50
agonist concentration that produces 50% receptor activation
- nH
Hill coefficient
- r2
goodness-of-fit for the linear regression
- DMEM
Dulbecco's Modified Eagle Medium
- BSA
bovine serum albumin
- HBSS
HEPES-buffered salt solution
- (±)-citalopram
1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-5-isobenzofurancarbonitrile
References
- Andreasen JT, Henningsen K, Bate S, Christiansen S, Wiborg O, 2011a. Nicotine reverses anhedonic-like response and cognitive impairment in the rat chronic mild stress model of depression: Comparison with sertraline. J. Psychopharmacol. 10.1177/0269881110391831 [DOI] [PubMed] [Google Scholar]
- Andreasen JT, Nielsen EO, Christensen JK, Olsen GM, Peters D, Mirza NR, Redrobe JP, 2011b. Subtype-selective nicotinic acetylcholine receptor agonists enhance the responsiveness to citalopram and reboxetine in the mouse forced swim test. J. Psychopharmacol 10.1177/0269881110364271 [DOI] [PubMed] [Google Scholar]
- Andreasen JT, Redrobe JP, Nielsen EO, 2012. Combined α7 nicotinic acetylcholine receptor agonism and partial serotonin transporter inhibition produce antidepressant-like effects in the mouse forced swim and tail suspension tests: A comparison of SSR180711 and PNU-282987. Pharmacol. Biochem. Behav 10.1016/j.pbb.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Angelone SM, Bellini L, Di Bella D, Catalano M, 1998. Effects of fluvoxamine and citalopram in maintaining abstinence in a sample of Italian detoxified alcoholics. Alcohol Alcohol, 10.1093/oxfordjournals.alcalc.a008371 [DOI] [PubMed] [Google Scholar]
- Arias HR, Biała G, Słomka MK-, Targowska-Duda KM, Biala G, Kruk-Slomka M, 2014. Interaction of nicotinic receptors with bupropion: Structural, functional, and pre-clinical perspectives. Recept. Clin. Investig 1, 30–45. 10.14800/rci.65; [DOI] [Google Scholar]
- Arias HR, Feuerbach D, Ortells MO, 2018a. Bupropion and (±)-SADU-3-72 inhibit human α3β4 nicotinic acetylcholine receptors by luminal and non-luminal interactions. Neurotransmitter e1631 10.14800/nt.1631 [DOI] [Google Scholar]
- Arias HR, Feuerbach D, Ortells MO, 2016. Bupropion and its photoreactive analog (±)-SADU-3-72 interact with luminal and non-luminal sites at human α4β2 nicotinic acetylcholine receptors. Neurochem. Int 100, 67–77. 10.1016/j.neuint.2016.08.013 [DOI] [PubMed] [Google Scholar]
- Arias HR, Feuerbach D, Targowska-Duda KM, Russell M, Jozwiak K, 2010a. Interaction of selective serotonin reuptake inhibitors with neuronal nicotinic acetylcholine receptors. Biochemistry 49, 5734–5742. 10.1021/bi100536t [DOI] [PubMed] [Google Scholar]
- Arias HR, Jin X, Feuerbach D, Drenan RM, 2017. Selectivity of coronaridine congeners at nicotinic acetylcholine receptors and inhibitory activity on mouse medial habenula. Int. J. Biochem. Cell Biol 92, 202–209. 10.1016/j.biocel.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias HR, Rosenberg A, Targowska-Duda KM, Feuerbach D, Jozwiak K, Moaddel R, Wainer IW, 2010b. Tricyclic antidepressants and mecamylamine bind to different sites in the human α4β2 nicotinic receptor ion channel. Int. J. Biochem. Cell Biol 42, 1007–1018. 10.1016/j.biocel.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias HR, Targowska-Duda KM, Feuerbach D, Sullivan CJ, Maciejewski R, Jozwiak K, 2010c. Different interaction between tricyclic antidepressants and mecamylamine with the human α3β4 nicotinic acetylcholine receptor ion channel. Neurochem. Int 56, 642–649. 10.1016/j.neuint.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Arias HR, Vázquez-Gómez E, Hernández-Abrego A, Gallino S, Feuerbach D, Ortells MO, Elgoyhen AB, García-Colunga J, 2018b. Tricyclic antidepressants inhibit hippocampal α7* and α9α10 nicotinic acetylcholine receptors by different mechanisms. Int. J. Biochem. Cell Biol 100 10.1016/j.biocel.2018.04.017 [DOI] [PubMed] [Google Scholar]
- Ballestero JA, Plazas PV, Kracun S, Gomez-Casati ME, Taranda J, Rothlin CV, Katz E, Millar NS, Elgoyhen AB, 2005. Effects of quinine, quinidine, and chloroquine on α9α10 nicotinic cholinergic receptors. Mol. Pharmacol 68, 822–829. 10.1124/mol.105.014431.fusion [DOI] [PubMed] [Google Scholar]
- Boffi JC, Marcovich T, Gill-Thind JK, Corradi J, Collins T, Lipovsek MM, Moglie M, Plazas PV, Craig PO, Millar NS, Bouzat C, Elgoyhen AB, 2017. Differential Contribution of Subunit Interfaces to α 9 α 10 Nicotinic Acetylcholine Receptor Function . Mol. Pharmacol, 10.1124/mol.116.107482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K, Van Dijk WJ, Klaassen RV, Schuurmans M, Van Der Oost J, Smit AB, Sixma TK, 2001. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature, 10.1038/35077011 [DOI] [PubMed] [Google Scholar]
- Cheng Y-C, Prusoff WH, 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (150) of an enzymatic reaction. Biochem. Pharmacol 22, 3099–3108. 10.1016/0006-2952(73)90196-2 [DOI] [PubMed] [Google Scholar]
- Chi H, De Wit H, 2003. Mecamylamine attenuates the subjective stimulant-like effects of alcohol in social drinkers. Alcohol. Clin. Exp. Res 10.1097/01.ALC.0000065435.12068.24 [DOI] [PubMed] [Google Scholar]
- Christmas DM, Potokar JP, Davies SJC, 2011. A biological pathway linking inflammation and depression: Activation of indoleamine 2,3-dioxygenase. Neuropsychiatr. Dis. Treat [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen a B., Vetter DE, Katz E, Rothlin CV, Heinemann SF, Boulter J, 2001. α10: a Determinant of Nicotinic Cholinergic Receptor Function in Mammalian Vestibular and Cochlear Mechanosensory Hair Cells. Proc. Natl. Acad. Sci. U. S. A 98, 3501–3506. 10.1073/pnas.051622798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S, 1994. α9: An acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell 79, 705–715. 10.1016/0092-8674(94)90555-X [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Katz E, 2012. The efferent medial olivocochlear-hair cell synapse. J. Physiol. Paris 106, 47–56. 10.1016/jjphysparis.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Lukas RJ, 1999. Antidepressants noncompetitively inhibit nicotinic acetylcholine receptor function. J. Neurochem 10.1046/j.1471-4159.1999.0721117.x [DOI] [PubMed] [Google Scholar]
- Garcia-Colunga J, Awad JN, Miledi R, 1997. Blockage of muscle and neuronal nicotinic acetylcholine receptors by fluoxetine (Prozac) (antidepressantsXenopus oocytescholinergic-serotonergic interactionacetylcholine receptor modulation), Pharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Colunga J, Targowska-Duda KM, Arias HR, 2016. Functional and structural interactions between selective serotonin reuptake inhibitors and nicotinic acetylcholine receptors. Neurotransmitter 3, 1293 10.14800/nt.1293 [DOI] [Google Scholar]
- Glick SD, Maisonneuve IM, Kitchen BA, 2002. Modulation of nicotine self-administration in rats by combination therapy with agents blocking α3β4 nicotinic receptors. Eur. J. Pharmacol 10.1016/S0014-2999(02)01944-1 [DOI] [PubMed] [Google Scholar]
- Goutman JD, Elgoyhen AB, Gómez-Casati ME, 2015. Cochlear hair cells: The sound-sensing machines. FEBS Lett. 589, 3354–3361. 10.1016/j.febslet.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve IM, Glick SD, 2003. Anti-addictive actions of an iboga alkaloid congener: A novel mechanism for a novel treatment. Pharmacol. Biochem. Behav 10.1016/S0091-3057(03)00119-9 [DOI] [PubMed] [Google Scholar]
- McCallum SE, Cowe MA, Lewis SW, Glick SD, 2012. α3β4 nicotinic acetylcholine receptors in the medial habenula modulate the mesolimbic dopaminergic response to acute nicotine in vivo. Neuropharmacology. 10.1016/j.neuropharm.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JM, Absalom N, Chebib M, Elgoyhen AB, Vincler M, 2009. α9 nicotinic acetylcholine receptors and the treatment of pain. Biochem. Pharmacol 10.1016/j.bcp.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineur YS, Picciotto MR, 2010. Nicotine receptors and depression: revisiting and revising the cholinergic hypothesis. Trends Pharmacol. Sci 31, 580–586. 10.1016/j.tips.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Perez CL, Noviello CM, Hibbs RE, 2016. X-ray structure of the human α4β2 nicotinic receptor. Nature 538 10.1038/naturel9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley BJ, Whiteaker P, Elgoyhen AB, 2018. Commentary: Nicotinic Acetylcholine Receptor α9 and α10 Subunits Are Expressed in the Brain of Mice. Front. Cell. Neurosci 10.3389/fncel.2018.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedretti A, Villa L, Vistoli G, 2004. VEGA - An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J. Comput. Aided. Mol. Des 18, 167–173. 10.1023/B:JCAM.0000035186.90683.f2 [DOI] [PubMed] [Google Scholar]
- Peng H, Ferris RL, Matthews T, Hiel H, Lopez-Albaitero A, Lustig LR, 2004. Characterization of the human nicotinic acetylcholine receptor subunit alpha (α) 9 (CHRNA9) and alpha (α) 10 (CHRNA10) in lymphocytes. Life Sci. 10.1016/j.lfs.2004.05.031 [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K, 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas PV, Katz E, Gomez-Casati ME, Bouzat C, Elgoyhen AB, 2005. Stoichiometry of the α9α10 nicotinic cholinergic receptor. J. Neurosci 10.1523/JNEUROSCI.3805-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popik P, Kozela E, Krawczyk M, 2003. Nicotine and nicotinic receptor antagonists potentiate the antidepressant-like effects of imipramine and citalopram. Br. J. Pharmacol 10.1038/sj.bjp.0705359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick MW, Ceballos RM, Kasten M, McIntosh JM, Lester RAJ, 1999. α3β4 subunit-containing nicotinic receptors dominate function in rat medial habenula neurons. Neuropharmacology, 10.1016/S0028-3908(99)00024-6 [DOI] [PubMed] [Google Scholar]
- Rezvani AH, Cauley MC, Slade S, Wells C, Glick S, Rose JE, Levin ED, 2016. Acute oral 18-methoxycoronaridine (18-MC) decreases both alcohol intake and IV nicotine self-administration in rats. Pharmacol. Biochem. Behav 10.1016/j.pbb.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero HK, Christensen SB, Di Cesare Mannelli L, Gajewiak J, Ramachandra R, Elmslie KS, Vetter DE, Ghelardini C, Iadonato SP, Mercado JL, Olivera BM, McIntosh JM, 2017. Inhibition of α9α10 nicotinic acetylcholine receptors prevents chemotherapy-induced neuropathic pain. Proc. Natl. Acad. Sci 114, E1825–E1832. 10.1073/pnas.1621433114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin CV, Katz E, Verbitsky M, Vetter DE, Heinemann SF, Elgoyhen AB, 2000. Block of the α9 nicotinic receptor by ototoxic aminoglycosides. Neuropharmacology. 10.1016/S0028-3908(00)00056-3 [DOI] [PubMed] [Google Scholar]
- Rothlin CV, Lioudyno MI, Silbering AF, Plazas PV, Casati ME, Katz E, Guth PS, Elgoyhen AB, 2003. Direct Interaction of Serotonin Type 3 Receptor Ligands with Recombinant and Native alpha 9alpha 10-Containing Nicotinic Cholinergic Receptors. Mol. Pharmacol 10.1124/mol.63.5.1067 [DOI] [PubMed] [Google Scholar]
- Rothlin CV, Katz E, Verbitsky M, Elgoyhen AB, 1999. The alpha9 nicotinic acetylcholine receptor shares pharmacological properties with type A gamma-aminobutyric acid, glycine, and type 3 serotonin receptors. Mol. Pharmacol [DOI] [PubMed] [Google Scholar]
- Sacre S, Medghalchi M, Gregory B, Brennan F, Williams R, 2010. Fluoxetine and citalopram exhibit potent antiinflammatory activity in human and murine models of rheumatoid arthritis and inhibit toll-like receptors. Arthritis Rheum, 10.1002/art.27304 [DOI] [PubMed] [Google Scholar]
- Šali A, Potterton L, Yuan F, van Vlijmen H, Karplus M, 1995. Evaluation of comparative protein modeling by MODELLER. Proteins Struct. Funct. Bioinforma 10.1002/prot.340230306 [DOI] [PubMed] [Google Scholar]
- Shih P-Y, Engle SE, Oh G, Deshpande P, Puskar NL, Lester HA, Drenan RM, 2014. Differential Expression and Function of Nicotinic Acetylcholine Receptors in Subdivisions of Medial Habenula. J. Neurosci 10.1523/JNEUROSCI.0476-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DJ, Montgomery SA, Kasper S, Tanghoj P, 2001. Predictors of response to pharmacotherapy with citalopram in obsessive-compulsive disorder. Int. Clin. Psychopharmacol 10.1097/00004850-200111000-00007 [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ, 2010. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem 31, 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varia L, Rauscher F, 2002. Treatment of generalized anxiety disorder with citalopram. Int. Clin. Psychopharmacol [DOI] [PubMed] [Google Scholar]
- Verbitsky M, Rothlin CV, Katz E, Belén Elgoyhen A, 2000. Mixed nicotinic-muscarinic properties of the α9 nicotinic cholinergic receptor. Neuropharmacology. 10.1016/S0028-3908(00)00124-6 [DOI] [PubMed] [Google Scholar]