Abstract
With its identification as a proto-oncogene in chronic lymphocytic leukaemia and central role in regulating NF-κB signalling, it is perhaps not surprising that there have been an increasing number of studies in recent years investigating the role of BCL-3 (B-Cell Chronic Lymphocytic Leukaemia/Lymphoma-3) in a wide range of human cancers. Importantly, this work has begun to shed light on our mechanistic understanding of the function of BCL-3 in tumour promotion and progression. Here, we summarize the current understanding of BCL-3 function in relation to the characteristics or traits associated with tumourigenesis, termed ‘Hallmarks of Cancer’. With the focus on colorectal cancer, a major cause of cancer related mortality in the UK, we describe the evidence that potentially explains why increased BCL-3 expression is associated with poor prognosis in colorectal cancer. As well as promoting tumour cell proliferation, survival, invasion and metastasis, a key emerging function of this proto-oncogene is the regulation of the tumour response to inflammation. We suggest that BCL-3 represents an exciting new route for targeting the Hallmarks of Cancer; in particular by limiting the impact of the enabling hallmarks of tumour promoting inflammation and cell plasticity. As BCL-3 has been reported to promote the stem-like potential of cancer cells, we suggest that targeting BCL-3 could increase the tumour response to conventional treatment, reduce the chance of relapse and hence improve the prognosis for cancer patients.
In recent years there has been an increasing number of studies investigating the role of the atypical IκB protein BCL-3 in a wide range of human cancers; here we review its function in relation to the ‘Hallmarks of Cancer’.
Introduction
Globally, cancer is one of the most common causes of mortality, with colorectal cancer the third most common malignancy and fourth highest cause of cancer death (1). Tumourigenesis is driven through acquisition of somatic mutations, epigenetic modifications (such as alterations in alternative splicing) and regulation by non-coding RNA molecules that function to regulate the expression of tumour suppressor and oncogenes. These changes lead to phenotypic alterations that mark the transformation of normal cells to cancer cells, known as the ‘Hallmarks of Cancer’, as originally defined by Hanahan and Weinberg (2). These Hallmarks are evading apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, limitless replicative potential, sustained angiogenesis and tissue invasion and metastasis. An updated version of the hallmarks describes a further two ‘emerging hallmarks’; deregulating cellular energetics and avoiding immune destruction, along with two ‘enabling characteristics’; genome instability and tumour promoting inflammation (3).
NF-κB is a critical inflammatory signalling pathway
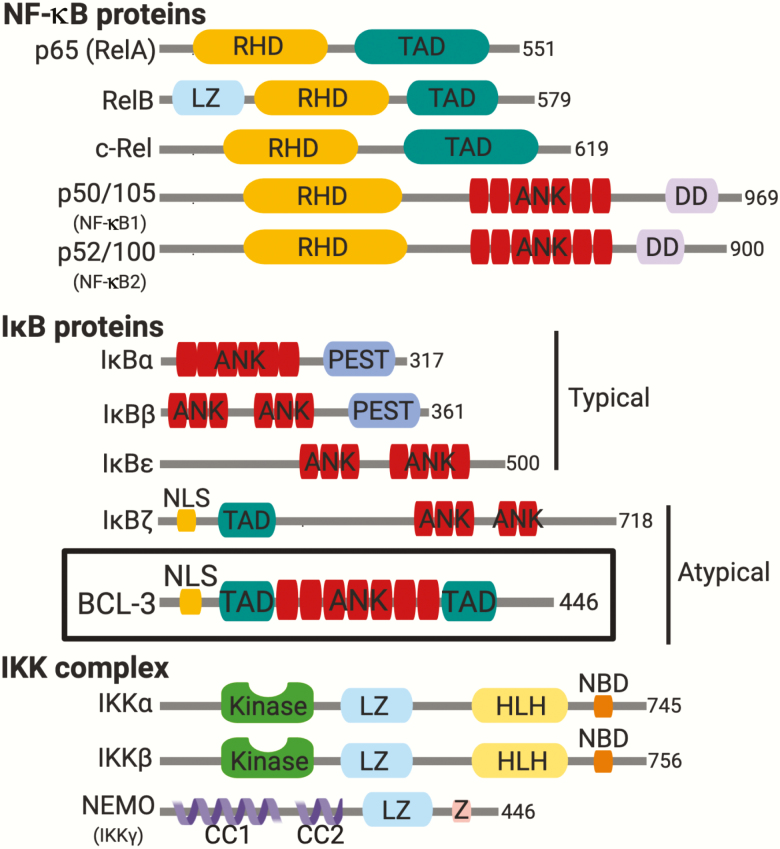
The NF-κB family of transcription factors is quickly activated following numerous stress stimuli including infection (4), ionizing radiation (5), chemical/physical stress and pro-inflammatory signals—including those related to cancer (6, 7). NF-κB signalling relies on three principal protein families: NF-κB proteins, inhibitor of kappa B (IκB) proteins and the IKK complex (Figure 1). NF-κB proteins are a group of structurally related subunits: RelA (p65), RelB, c-Rel, NF-κB1 (p105/p50) and NF-κB2 (p100/p52) (8, 9). All NF-κB subunits contain an N-terminal Rel homology domain, which mediates DNA binding, dimerization of subunits and inhibitory protein binding (10). P65, RelB and c-Rel possess a transactivation domain which enables transcriptional activation following stimulus. Dimerization of NF-κB subunits into homo- or heterodimers follows activation through either the canonical or non-canonical pathways, although this is perhaps an oversimplification as up to 15 different NF-κB family complexes can be formed (11, 12). The canonical and non-canonical signalling pathways have been the subject of many elegant reviews and are not covered here.
Figure 1.
Principal components of NF-κB signalling. NF-κB signalling relies on three principal proteins: NF-κB proteins, IκB proteins and the IKK complex. The five members of the NF-κB family: p65 (RelA), RelB, c-Rel, p50/105 (NF-κB1) and p50/105 (NF-κB2) all share a N-terminal Rel homology domain that is responsible for DNA binding and dimerization. RelA, p65 and RelB also have a C-terminal transcriptional activation domain (TADs) which mediates interactions with co-factors but is not present in p50/100 or p52/105. The IκB proteins: IκBα, IκBβ, IκBε, BCL-3 and IκBζ are characterized by the presence of multiple ankyrin repeat domains (ANK = red). These domains assemble into elongated cylinders and associate with the DNA-binding domains of NF-κB dimers. Classically, IκB proteins sequester NF-κB dimers in the cytoplasm rendering them transcriptionally inactive. Atypical IκB proteins including BCL-3, the focus of this review, contain additional nuclear localization signal and transactivation domains. Unlike typical IκB proteins, these atypical nuclear IκB proteins can both inhibit and activate NF-κB target gene expression. The IKK complex contains two kinase subunits IKKα and Inhibitor of Nuclear Factor Kappa B Kinase Subunit Beta and a regulatory subunit NEMO (NF-κB essential modifier) or IKKγ. The total number of amino acids in each protein is indicated on the right-hand side. Leucine-zipper-like motif (LZ), death domain (DD), domain rich in proline (P), glutamate (E), serine (S) and threonine (T) (PEST), Kinase domain (Kinase), helix–loop–helix domain (HLH), NEMO-binding domain (NBD), coiled-coil domain (CC) and zinc-finger domain (Z). Figure created with BioRender.com.
The term ‘atypical NF-κB pathway’ involves NF-κB dimers and their associated activation that do not fall under the umbrella of the canonical or non-canonical pathways: included in this atypical calssification are the p50/p52 homodimers and their associated co-regulatory protein, B-Cell Lymphoma-3 (BCL-3), which will be the focus of this review. This pathway is a critical component of the broader NF-κB response and plays a role in fine-tuning the canonical and non-canonical responses to an inflammatory stimulus. However, it is important to point out that BCL-3 may elicit functional effects through all branches of NF-κB signalling due to its various nuclear and cytoplasmic roles, as explored throughout this review.
BCL-3
The BCL-3 gene was first found in chronic lymphocytic leukaemia after sequencing of recurring t(14;19)(q32.3;q13.1) translocations which result in transcriptional upregulation of BCL3 (13). This revealed a protein with seven ankyrin repeat domains, a proline-rich N terminal domain and a serine- and proline-rich C-terminal domain with a total molecular weight of around 47 kDa (14). Studies in normal blood cells showed increased expression of BCL-3 following mitogenic stimulation and also linked the structure of BCL-3 to those of known cell cycle regulators, prompting the authors to label it as a candidate proto-oncogene (14). Shortly after its discovery, a flurry of papers from multiple groups linked BCL-3 to the NF-κB signalling pathway (15–18) and BCL-3 was subsequently classified as an atypical member of the IκB family due to its contrasting roles in NF-κB-mediated transcription.
BCL-3 as a regulator of an atypical NF-κB signalling pathway
Functionally, BCL-3 differs from the other IκB proteins (IκB-α, IκB-β and IκB-ε) which bind to NF-κB proteins in the cytoplasm, inhibiting their nuclear translocation and subsequent transcriptional activity. In both non-tumour and tumour cells, BCL-3 binds to processed p50 and p52 homodimers to activate or repress a subset of NF-κB regulated genes (Figure 2). P50 and p52 are thought to form strong homodimers with high affinity compared with other homodimer species (19). These homodimers bind to the majority of κB sites at gene promoter regions and binding is known to occur at multiple sites within the same promoter (20). In contrast to other NF-κB subunits, p50 and p52 lack transactivation domains and subsequently require co-factors to induce activation of transcription (21). First thought to inhibit nuclear translocation of p50 homodimers (22), binding of BCL-3 to p50 homodimers was later shown to unmask the nuclear localization signal of p50 allowing translocation of the complex into the nucleus (23, 24).
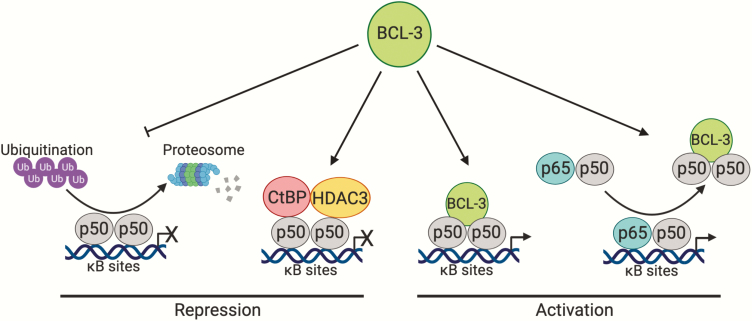
Figure 2.
BCL-3 regulation of the atypical NF-κB signalling pathway. The atypical pathway is defined as the activation of the NF-κB p50 and p52 homodimers. BCL-3 acts as a regulator of the atypical NF-κB pathway by binding to processed p50 and p52 homodimers to repress or activate a subset of NF-κB regulated genes. In terms of transcriptional regulation, BCL-3 stabilizes repressive p50 homodimers bound to κB sites by inhibiting ubiquitination and subsequent proteolytic degradation of p50 homodimers. Additionally, BCL-3 can also recruit co-repressors CtBP and HDAC3 to repress transcription of NF-κB target genes. Conversely, BCL-3 directly activates transcription of NF-κB target genes by associating with p50 and p52 homodimers, providing these homodimers with two transactivation domains that they otherwise lack. BCL-3 can also remove repressive p50 homodimers situated at κB sites on DNA, allowing NF-κB heterodimers (p65/p50) associated with canonical signalling to activate transcription at these sites. Whilst BCL-3 is known to directly interact with p52 homodimers, the mechanism by which BCL-3 regulates p52 homodimer activation remains to be elucidated however it is speculated to follow a similar mechanism to p50 homodimers. Figure created with BioRender.com.
Transactivator or transrepressor
There have been many studies investigating the role of BCL-3 in the regulation of NF-κB target genes. Interestingly, BCL-3 has been demonstrated to both activate and repress transcription of NF-κB targets in a context-dependent manner with respect to the promoter, cell type or stimuli analysed. In terms of transcriptional repression, early work from Kerr et al. described inhibition of NF-κB reporter activity in the presence of BCL-3 in Drosophila S2 cells and Jurkat T-cells (18). In support of this, using cells from Bcl3−/− mice it was demonstrated that Bcl-3 could enhance repression of NF-κB target genes. This was shown to be through inhibition of ubiquitination and subsequent proteolytic degradation of p50 homodimers bound to κB sites at promoters of certain genes; thus, extending the half-life of repressive p50 homodimers (25, 26). In addition, BCL-3 can recruit co-repressors CtBP and HDAC3 to repress transcription of NF-κB target genes in transformed keratinocytes (27). Therefore, BCL-3 can repress transcription by stabilizing repressive p50 homodimers or via recruitment of co-repressors.
Conversely, BCL-3 may activate transcription using a number of different mechanisms. Franzoso et al. demonstrated an interaction between BCL-3 and p50 and observed an increase in NF-κB reporter activity through BCL-3-mediated reversal of p50 homodimer-induced repression in transfected NTERA2 cells (17). A further mechanism of transcriptional activation by BCL-3 is the direct removal of repressive p50 homodimers already situated at κB sites on DNA, allowing NF-κB subunits associated with canonical signalling to bind and activate transcription at these sites (17, 28). As an alternative to reversing repression at κB sites, BCL-3 may also directly transactivate genes in multiple cell types when associated with p50 or p52 homodimers; owing to BCL-3 possessing both N- and C-terminal transactivation domains (14, 29, 30).
Interaction of BCL-3 with co-regulatory proteins
BCL-3 can recruit other co-regulators to influence gene expression, as reported by Dechend et al. who discovered interactions between BCL-3 and Pirin, Tip60, Jab1 and Bard1. Of these interactors, Tip60 and Jab1 were found to enhance transcription of the P-selectin promoter in Drosophila SL2 cells when co-transfected with Bcl-3 and p50 (31). This adds a further dimension to the regulation of transcription by BCL-3, with some co-regulators resulting in repression of target genes and others in transactivation. Further protein–protein interactions have been documented such as with Hsp70, HDAC1 and 3, CtBP1 and CtBP2 (27, 32) and more recently β-catenin in colorectal cancer cells (33).
Acetylation is known to play an important role in NF-κB family subunits (34), although it is unknown if the bridging role that BCL-3 plays with proteins such as Tip60 and HDAC1/3 facilitates acetylation of p50 or p52 homodimers. Evidence exists to show that p50 can be acetylated and that this acetylation results in enhanced binding to κB sites in T cells (35), it is interesting to speculate that Tip60 and HDAC1/3 interactions could be influential in determining homodimer transactivation activity in the nucleus.
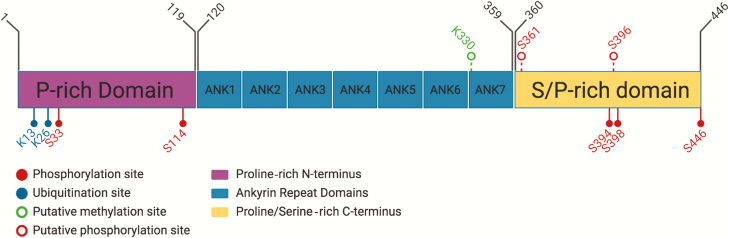
Although early work on BCL-3 focussed on its transcriptional regulation in relation to its interaction with NF-κB homodimeric subunits, more recent work has centred on how post-translational modifications affect these interactions and downstream transcriptional regulation (Figure 3 summarizes the key modifications of BCL-3). BCL-3 is extensively phosphorylated at its C-terminus, which regulates p52 (36) and p50 (24, 37) homodimer binding. In NIH3T3 cells, Viatour et al. demonstrated phosphorylation of BCL-3 at residues S394 and S398 by glycogen synthase kinase 3 promoted proteasomal degradation of BCL-3 (38). A recent mass spectrometry study in HEK293T cells has revealed 27 phosphorylation sites on BCL-3 (39). Subsequent characterization revealed phosphorylation at S33 by Akt shifts polyubiquitination from K48 to K63, resulting in resistance to degradation, translocation to the nucleus and activation of p52 homodimers (39). S446 phosphorylation by IKK1/2 and S114 phosphorylation by Erk2 both stabilized the BCL3:p52 complex on DNA to enhance transcriptional activation (39). Therefore, whether BCL-3 acts as a co-activator or co-repressor may be determined by the combination of post-translational modifications—brought about by the particular cell type or stimuli—that alter the function and/or localization of BCL-3.
Figure 3.
Gene map of BCL-3 highlighting important sites of post-translational modification. BCL-3 is 446 amino acids in length with a proline-rich N-terminus, an ankyrin repeat region consisting of seven ankyrin repeats and a proline- and serine-rich C-terminus. In the C-terminus, serine residues S361 and S396 are GSK3β targets and result in protein degradation. S33, S114 and S446 are phosphorylated by AKT, ERK2 and IKK1/2, respectively, leading to stabilization of BCL-3. Lysine residues K13 and K26 undergo ubiquitination with either stabilizing K48 or degradative K63 ubiquitin chains. Other putative PTM sites in BCL-3 are lysine residue K330 that is thought to be monomethylated in resting cells and two potential sites for ATM phosphorylation, S361 and S396, both of which are followed by the typical glycine residue found in ATM target sites. Figure created with BioRender.com.
In summary, BCL-3 has multiple functions ranging from activation of p50/p52 subunits in the cytoplasm, their nuclear import and modulation of homodimer transactivation through facilitating binding of different transcriptional co-regulators. It is also suggested that BCL-3 has contrasting functions depending on the pathological model used; in particular there is a significant reversal of function in inflammation models (40) which could be explained by the availability of co-factors and/or post-translational modification of chromatin surrounding NF-κB dimers (41).
BCL-3 and cancer
Given its identification as a proto-oncogene in chronic lymphocytic leukaemia and central role in regulating NF-κB signalling, it is perhaps not surprising that there have been an increasing number of studies in recent years investigating the role of BCL-3 in a variety of human solid cancers (42). There are multiple post-translational modifications that affect the function or localization of BCL-3, as discussed above, and may promote the oncogenic potential of BCL-3.
The clinical utility of BCL-3 as a marker of prognosis is highlighted by a growing number of clinical studies: increased protein and mRNA expression has been associated with adverse clinicopathological characteristics in hepatocellular carcinoma patients (43). Increased BCL-3 expression was also correlated with adverse prognostic features and reduced survival rate in cervical cancer (44). Niu et al. suggest that BCL3 is a promising molecular biomarker of paediatric acute myeloid leukaemia with unfavourable prognosis (45), and the use of BCL-3 expression levels has also been proposed to predict response to alkylating agents in glioma (46). Interestingly, BCL-3 is part of a four gene signature suggested for use in determining the prognosis of patients with clear-cell renal-cell carcinoma (47). These and other studies have begun to shed light on our mechanistic understanding of the function of BCL-3 in tumour promotion and progression, including the role BCL-3 plays in a number of the Hallmarks of Cancer (3), as discussed below.
Sustaining proliferative signalling
Cancer cells can disregard the homeostatic cues that govern normal cells ability to grow and, importantly, stop growing. Cancers acquire the ability for self-sustaining proliferation through a number of different means, including dysregulation of cell cycle, increased growth factor production, stimulation of cells within the microenvironment (resulting in increased pro-proliferative paracrine signalling) and the deregulation of growth factor receptors leading to hypersensitivity to growth factors (3, 48). The effect of BCL-3 on tumour growth/proliferation has been observed in different tumour types. In colorectal cancer cells, BCL-3 induces the post-translational stabilization of c-MYC (mediated by ERK1/2) increasing tumour xenograft size (49), while BCL-3 overexpression was shown to induce cell cycle progression, mediated by Cyclin D1 in hepatocellular carcinoma (43, 50), malignant melanoma (51) and breast cancer (52). The converse was also true, as repression of Cyclin D1 by p53 was shown to be dependent on BCL-3 suppression in H1229 lung cancer and U2-01 sarcoma cell lines (53). Furthermore, proliferation of skin cancer cells was abrogated when nuclear translocation of BCL-3 was blocked by its upstream regulator, CYLD (54).
Activating invasion and metastasis
Invasion into surrounding tissues is a defining feature of malignant cells followed by metastasis to distant organ sites, which is often the fatal event of solid malignancy (3). In breast cancer models it has been demonstrated that BCL-3 drives metastasis of tumour cells (55, 56): In HER2-positive breast tumour cells, BCL-3 knockout (KO) resulted in an 80% reduction of metastatic burden in mice following tail-vein injection of tumour cells (55). While in the MMTV-PyMT mouse model of mammary adenocarcinoma, Bcl-3 suppression using Dox-inducible shRNA resulted in reduction of lung metastases, through targeting Smad3 stability in the transforming growth factor-β signalling pathway (56). Additionally, a clinical study examined immunohistochemistry from paired normal and tumour tissue in hepatocellular carcinoma and discovered BCL-3 expression resulted in advanced Tumour, Node, Metastasis stage; this was shown to have contributed to the poorer prognosis observed in these patients (43).
Evasion of apoptosis
BCL-3 regulates cellular apoptosis in a variety of models. For example, data have shown BCL-3 regulates apoptosis in colorectal and cervical tumour cell lines following UV-radiation (57) and protects breast carcinoma cells from undergoing apoptosis following UV-radiation (58). BCL-3 is thought to inhibit proteins such as Smac/Diabolo and p53 through upregulation of HDM2 (59–62). However, even in p53-null backgrounds BCL-3 suppression was able to initiate apoptosis through targeting the expression of DNA-PKcs (59). BCL-3 is a potent survival factor in colorectal cancer (63, 64), and activates the pro-survival AKT/PKB pathway (64). In hepatocellular carcinomas, BCL-3 is frequently overexpressed in tumour tissue compared with normal tissue, in conjunction with p50 and p52 NF-κB subunits (65). However, recently published data using a hepatocyte-specific BCL-3 overexpression mouse model revealed that BCL-3 expression promoted hepatocyte death following an inflammatory insult. As a result, these mice developed fewer hepatocellular carcinomas (66). This study also highlighted that BCL-3 overexpression reduced the influx of certain populations of immune cells into the liver (CD8+ T cells, B cells and leucocytes), protecting against induced inflammation. It may be that hepatocellular carcinomas are particularly sensitive to alterations in canonical NF-κB signalling and that repression of canonical NF-κB occurs when BCL-3 is overexpressed in combination with atypical NF-κB homodimers, leading to abrogation of apoptosis in this context (26). Interestingly, a recent study by Zou et al. described BCL-3-mediated increase in checkpoint marker PD-L1 expression, enhancing proliferation of ovarian cancer cells (67). PD-L1 is known to inhibit tumour infiltrating lymphocytes (68); therefore, it is tempting to speculate that BCL-3 could play a similar role in protecting against tumour cell apoptosis via regulation of tumour infiltrating lymphocytes in both hepatocellular carcinoma and ovarian cancer.
The role of BCL-3 in inflammation and immunity
NF-κB signalling is recognized to have contrasting function depending on the biological context (69), consistent with the role of BCL-3 in cancer compared with its role in non-malignant, inflammatory models. Interestingly, the presence of BCL-3 appears to attenuate inflammation in non-malignant models. BCL-3 KO mice have defects in lymphoid organs, including aberrant development of Peyer’s patches, which would affect intestinal immune surveillance and response to certain pathogens (70).
Effects of BCL-3 KO have been studied in a variety of different inflammatory models, including pancreatitis, colitis, dermatitis and rheumatoid arthritis. In a mouse model of acute pancreatitis, where pancreatic inflammation is stimulated using cerulin or sodium taurocholate, results showed that Bcl-3−/− mice had increased levels of oedema and necrosis in their pancreata (71). The observation that Bcl-3 KO increases the severity of inflammation has been corroborated in an inflammatory bowel disease model (Crohn’s disease and ulcerative colitis) (40). In a contact hypersensitivity model of atopic dermatitis, Bcl-3−/− mice had worsened inflammation following topical oxalazone treatment, which appeared to be mediated through increased cytokine production (72). Earlier evidence for this mechanism came from Carmody et al. via examining the expression of pro-inflammatory cytokines in murine immune cells (macrophages and dendritic cells) devoid of BCL-3 (26). BCL-3 acted to repress canonical NF-κB transcription of target cytokines (such as TNF-α, CXCL1, CXCL2, IL-1β and IL-10), leading to significantly increased expression following inflammatory stimuli in the Bcl-3 KO cells, corroborating previous data (73). This study was particularly interesting as it demonstrated that not all cytokines responded in the same way to Bcl-3 KO. For cytokines that showed an early spike in transcription following lipopolysaccharide, Bcl-3 KO had a significant impact on their production, while cytokines such as IL-6 showed a slower increase in transcription that was unchanged in Bcl-3−/− cells compared with Bcl-3 wild-type controls. These data suggest that Bcl-3 KO affects different aspects (early and late) of the NF-κB-driven response to inflammation. Interestingly, Inhibitor of Nuclear Factor Kappa B Kinase Subunit Beta KO in intestinal epithelial cells worsens the histological severity of colonic inflammation and led to the animals losing greater amounts of weight (74). If Bcl-3 KO leads to increased binding of canonical NF-κB dimers (26), then it is unclear why the phenotype observed with Bcl-3 KO is similar to the phenotype observed when canonical NF-κB has been inactivated by Inhibitor of Nuclear Factor Kappa B Kinase Subunit Beta deletion. It may be that long-term KO and transient knockdown have different effects on the canonical (transiently activated) and non-canonical pathway (sustained activation). Additionally, the various models and cell types used in these studies may account for some of the context-dependent function of Bcl-3.
As a further illustration of the potential context-dependent function of BCL-3, another recent study showed that Bcl-3 overexpression, specifically in CD4+ T cells (including T regulatory cells), results in a pancolitis of the large bowel of mice (75). This cell-type specific effect was corroborated by work in CD4+ cells using a rheumatoid arthritis model, which showed overexpression of Bcl-3 in these cells was implicated in the pathogenesis of rheumatoid arthritis (76). Interestingly, Bcl-3−/− T cells failed to induce colitis when transferred into Rag1−/− mice (77), suggesting an inability of Bcl-3−/− T cells to respond to the microbiota-derived and antigen-specific signals that drive colitis in this model.
Following induction of inflammation there is a concomitant induction of BCL-3 in a variety of tissue types. Induction of BCL-3 in response to canonical NF-κB signalling was first observed by Brasier et al. in a hepatocellular carcinoma background (78). More recent work has shown the importance of the alternative NF-κB pathway in regulation of BCL-3 transcription in colorectal cancer (63). This is likely to represent a feedback loop, in place to regulate pathways such as NF-κB and modulate their function following a stimulus. Evidence for this comes from a number of sources; in respiratory syncytial virus infection of airway epithelial cells, BCL-3 is initially upregulated leading to inhibition of NF-κB and STAT transcription by acting as a bridging factor to HDAC1, which is transcriptionally repressive (32). Furthermore, BCL-3 represses STAT3 regulating proteins (79); repression of STAP2 by BCL-3 and p50 homodimers in conjunction with CtBP on the STAP2 promoter can diminish STAP2-dependent BRK kinase signalling that phosphorylates and activates STAT3 (80). BCL-3 is upregulated in tissue from patients with inflammatory bowel disease (75), although further data from inflammatory bowel disease patients have shown that CpG sites in the BCL3 gene are commonly methylated and therefore repressed in B cells isolated from diseased tissue (81). Bcl-3−/− granulocytes, isolated following pulmonary transplant in mice, had higher levels of apoptosis as measured by annexin V (82). This links to other data showing Bcl-3 KO cells in mice colons displayed increased caspase-3 cleavage following dextran-sodium sulphate-induced colitis (40), suggesting these cells were undergoing more apoptosis. Despite these data showing the role of BCL-3 as both a pro-inflammatory and an anti-inflammatory mediator in non-cancer tissue, it does still appear that BCL-3 has similar control over regulation of apoptosis in non-malignant tissue compared with the role of BCL-3 in cancers.
In summary, BCL-3 has alternative roles in inflammation depending on the cell type it is expressed in and whether all tissues have lost or gained expression. It is suggested that KO of Bcl-3 reduces the ability of cells to upregulate survival programmes following an inflammatory insult, leading to concomitant rise in apoptosis. This results in a worse grade of histological inflammation and tissue damage, thereby propagating inflammation. This indicates that it may be the pro-survival role of BCL-3 following stress or insult which is critical to its function.
BCL-3 in colorectal cancer
Importantly, BCL-3 has been shown to play multiple roles in the promotion of colorectal cancer. It was discovered by Puvvada et al. that BCL-3 nuclear expression in primary tumours was negatively associated with survival. This was calculated through analysis of staining and microarray data gathered from tumour specimens of 23 patients that had undergone colorectal cancer resection (83). Strong nuclear BCL-3 staining was also reported in 33% of 270 tumour samples, and was suggested to be an important diagnostic determinant (84). These findings were recently corroborated by Legge et al. who (using publically available datasets) also reported that BCL-3 expression is associated with poor prognosis in colorectal cancer (33). Various mechanisms of BCL-3-mediated tumour promotion have been observed in colorectal cancer. One of these is suggested to be through regulation of Cyclin D1 in SW480 cells, after it was observed that expression of BCL-3 and Cyclin D1 was reduced following treatment with COX-2 inhibitor NS398 (85). Work from our group has shown that BCL-3 promotes colorectal tumour cell survival and growth in vivo, through activation of the AKT pathway via phosphoinositide 3-kinase and mammalian target of rapamycin (mTOR) signalling (64). Moreover, BCL-3 also stabilizes c-MYC—one of the key oncogenes in early stage colorectal tumourigenesis following adenomatous polyposis coli inactivation—via extracellular signal-regulated kinase signalling (49).
Conversely, a recent paper has suggested Bcl-3 may play a protective role in colitis-associated colorectal cancers. This observation was made in the mouse model of colitis associated colorectal cancers, where tumours are induced through the addition of azoxymethane/dextran-sodium sulphate. Tang et al. demonstrated that mice with conditional deletion of Bcl-3 in the intestine developed significantly more polyps than wild-type controls, though interestingly polyps from both animals were the same size and staining of proliferation markers were comparable, indicating Bcl-3 loss was promoting tumour initiating capacity. This was suggested to be mediated through TNF-α, as tumour burden was not increased in Bcl-3/TNF-α double KO mice (86). Interestingly, earlier work by O’Carroll et al. revealed increased proliferation of intestinal epithelium in Bcl-3−/− mice compared with controls, showing that this preserved intestinal tissue architecture, protecting against colitis (40). It is tempting to speculate that the increased proliferation in Bcl-3−/− intestines following dextran-sodium sulphate administration may contribute to tumour development observed in the Tang et al. study. It is important to remember that in the O’Carroll study, germline Bcl-3 KO will affect the immune cell compartment which will play a key role in colitis associated colorectal cancers, as discussed in the previous section.
BCL-3 and colorectal cancer stem cells
Cancers are thought to arise from stem (or stem-like) cells that gain a competitive advantage over their neighbours. With time this clonal expansion of mutant cells will eventually give rise to a tumour (87). The cancer stem cell hypothesis states there are cells within tumours capable of self-renewal, in addition to producing other heterogeneous, differentiated cell types that constitute the tumour mass (88). In the colon, deregulation of Wnt signalling is recognized as the key initiating event in colorectal cancer; activation of Wnt/β-catenin signalling, most commonly through inactivating mutations in adenomatous polyposis coli, leads to the formation of benign polyps, the first stage in the adenoma to carcinoma sequence (89). Important studies in stem cell biology have begun to identify the mechanisms underpinning the expansion of the adenomatous polyposis coli mutant stem cells that contribute to the earliest stages of colorectal tumour development (90), which may represent novel targets for therapeutic intervention.
Previous data have highlighted the importance of the interplay between NF-κB signalling and the Wnt/β-catenin pathway in colorectal carcinogenesis; in particular that non-stem cells engineered to exhibit high levels of Wnt and NF-κB signalling can de-differentiate, initiating tumours in mice (91). Recently we have shown that BCL-3 is an important co-activator of β-catenin/T-cell factor-mediated transcriptional activity in colorectal cancer cells, increasing expression of Wnt-regulated intestinal stem cell genes. BCL-3 was demonstrated to bind to β-catenin; RNAi-mediated BCL-3 suppression reduced β-catenin/T-cell factor-dependent transcription and the expression of intestinal stem cell genes and Wnt targets LGR5 and ASCL2, both widely accepted colorectal cancer stem cell markers (92–96). Furthermore, we showed that BCL-3 promotes the stem cell phenotype in colorectal cancer cells by increasing colorectal spheroid and tumoursphere formation in 3D culture conditions, indicating BCL-3 may promote tumour initiation in vivo and may be an effective target for reducing cancer stem cell plasticity (33). Results from our study complement findings by Chen et al. who suggest BCL-3 promotes stem-like activity and contributes to the maintenance of naive pluripotency in mouse embryonic stem cells (97). Due to its role in regulating both β-catenin and NF-κB signalling, it is interesting to speculate that BCL-3 may enhance the de-differentiation of non-stem cells (91), therefore aiding reconstitution of the tumour when the cancer stem cells have been deleted (94).
Summary
In conclusion, BCL-3 may represent an exciting new route for targeting the Hallmarks of Cancer; in particular by limiting the impact of the enabling hallmarks of tumour promoting inflammation and cell plasticity. Excitingly, targeting stem cell plasticity offers the possibility of overcoming some of the limitations of directly targeting cancer stem cell highlighted in recent studies (94). We suggest that targeting BCL-3 function (through suppressing the stem-like potential of cancer cells) would increase the tumour response to conventional treatment, reduce the chance of relapse and hence improve the prognosis for colorectal cancer patients. Given the emerging role of BCL-3 in a number of different cancers, we are hopeful that the new class of BCL-3 inhibitors currently under development will prove effective in a wide range of human cancers.
Funding
This work was supported by PhD studentships from Bowel & Cancer Research PhD studentship (D.N.L., P.T.), an MRC clinical research training fellowship (MR/N001494/1 to A.C.C.), a Wellcome Trust Four Year PhD Programme in Dynamic Molecular Cell Biology (203988/Z/16Z to C.P.), an MRC Research grant (MR/R017247/1 to A.C.W., T.J.C.) and by the John James Bristol Foundation.
Conflict of Interest Statement: The authors have declared no conflicts of interest.
Glossary
Abbreviations
- BCL-3
B-Cell Lymphoma-3
- IκB
inhibitor of kappa B
References
- 1. Arnold M., et al. (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut, 66, 683–691. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., et al. (2000) The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 4. Ghosh S., et al. (1998) NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- 5. Brach M.A., et al. (1991) Ionizing radiation induces expression and binding activity of the nuclear factor kappa B. J. Clin. Invest., 88, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoesel B., et al. (2013) The complexity of NF-κB signaling in inflammation and cancer. Mol. Cancer, 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colotta F., et al. (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis, 30, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 8. Schmid R.M., et al. (1991) Cloning of an NF-kappa B subunit which stimulates HIV transcription in synergy with p65. Nature, 352, 733–736. [DOI] [PubMed] [Google Scholar]
- 9. Bours V., et al. (1992) A novel mitogen-inducible gene product related to p50/p105-NF-kappa B participates in transactivation through a kappa B site. Mol. Cell. Biol., 12, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q., et al. (2017) 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell, 168, 37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huxford T., et al. (2009) A structural guide to proteins of the NF-kappaB signaling module. Cold Spring Harb. Perspect. Biol., 1, a000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perkins N.D. (1997) Achieving transcriptional specificity with NF-kappa B. Int. J. Biochem. Cell Biol., 29, 1433–1448. [DOI] [PubMed] [Google Scholar]
- 13. McKeithan T.W., et al. (1990) Identification of a transcriptional unit adjacent to the breakpoint in the 14;19 translocation of chronic lymphocytic leukemia. Genes Chromosomes Cancer, 1, 247–255. [DOI] [PubMed] [Google Scholar]
- 14. Ohno H., et al. (1990) The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell, 60, 991–997. [DOI] [PubMed] [Google Scholar]
- 15. Hatada E.N., et al. (1992) The ankyrin repeat domains of the NF-kappa B precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-kappa B DNA binding. Proc. Natl. Acad. Sci. USA, 89, 2489–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wulczyn F.G., et al. (1992) Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-kappa B. Nature, 358, 597–599. [DOI] [PubMed] [Google Scholar]
- 17. Franzoso G., et al. (1992) The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-kappa B-mediated inhibition. Nature, 359, 339–342. [DOI] [PubMed] [Google Scholar]
- 18. Kerr L.D., et al. (1992) The proto-oncogene bcl-3 encodes an I kappa B protein. Genes Dev., 6, 2352–2363. [DOI] [PubMed] [Google Scholar]
- 19. Hoffmann A., et al. (2006) Transcriptional regulation via the NF-kappaB signaling module. Oncogene, 25, 6706–6716. [DOI] [PubMed] [Google Scholar]
- 20. Wessells J., et al. (2004) BCL-3 and NF-kappa B p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J. Biol. Chem., 279, 49995–50003. [DOI] [PubMed] [Google Scholar]
- 21. Southern S.L., et al. (2012) BAG-1 interacts with the p50-p50 homodimeric NF-κB complex: implications for colorectal carcinogenesis. Oncogene, 31, 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naumann M., et al. (1993) The NF-kappa B precursor p105 and the proto-oncogene product Bcl-3 are I kappa B molecules and control nuclear translocation of NF-kappa B. EMBO J., 12, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q., et al. (1994) BCL3 encodes a nuclear protein which can alter the subcellular location of NF-kappa B proteins. Mol. Cell. Biol., 14, 3915–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nolan G.P., et al. (1993) The bcl-3 proto-oncogene encodes a nuclear I kappa B-like molecule that preferentially interacts with NF-kappa B p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol., 13, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collins P.E., et al. (2014) Inhibition of transcription by B cell Leukemia 3 (Bcl-3) protein requires interaction with nuclear factor κB (NF-κB) p50. J. Biol. Chem., 289, 7059–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carmody R.J., et al. (2007) Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science, 317, 675–678. [DOI] [PubMed] [Google Scholar]
- 27. Keutgens A., et al. (2010) The repressing function of the oncoprotein BCL-3 requires CtBP, while its polyubiquitination and degradation involve the E3 ligase TBLR1. Mol. Cell. Biol., 30, 4006–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franzoso G., et al. (1993) The oncoprotein Bcl-3 can facilitate NF-kappa B-mediated transactivation by removing inhibiting p50 homodimers from select kappa B sites. EMBO J., 12, 3893–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bours V., et al. (1993) The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell, 72, 729–739. [DOI] [PubMed] [Google Scholar]
- 30. Fujita T., et al. (1993) The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev., 7, 1354–1363. [DOI] [PubMed] [Google Scholar]
- 31. Dechend R., et al. (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene, 18, 3316–3323. [DOI] [PubMed] [Google Scholar]
- 32. Jamaluddin M., et al. (2005) Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-kappaB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J. Virol., 79, 15302–15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Legge D.N., et al. (2019) BCL-3 promotes a cancer stem cell phenotype by enhancing beta-catenin signalling in colorectal tumour cells. Dis. Models Mech. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiernan R., et al. (2003) Post-activation turn-off of NF-kB dependent transcription is regulated by acetylation of p65. J. Biol. Chem., 278, 2758–2766. [DOI] [PubMed] [Google Scholar]
- 35. Furia B., et al. (2002) Enhancement of nuclear factor-kB acetylation by coactivator p300 and HIV-1 TAT proteins. J. Biol. Chem. 277, 4973–4980. [DOI] [PubMed] [Google Scholar]
- 36. Bundy D.L., et al. (1997) Diverse effects of BCL3 phosphorylation on its modulation of NF-kappaB p52 homodimer binding to DNA. J. Biol. Chem., 272, 33132–33139. [DOI] [PubMed] [Google Scholar]
- 37. Caamaño J.H., et al. (1996) Constitutive expression of Bc1-3 in thymocytes increases the DNA binding of NF-kappaB1 (p50) homodimers in vivo. Mol. Cell. Biol., 16, 1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viatour P., et al. (2004) GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol. Cell, 16, 35–45. [DOI] [PubMed] [Google Scholar]
- 39. Wang V.Y., et al. (2017) Bcl3 phosphorylation by Akt, Erk2, and IKK is required for its transcriptional activity. Mol. Cell, 67, 484–497.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O’Carroll C., et al. (2013) Bcl-3 deficiency protects against dextran-sodium sulphate-induced colitis in the mouse. Clin. Exp. Immunol., 173, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim J.H., et al. (2005) Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature, 434, 921–926. [DOI] [PubMed] [Google Scholar]
- 42. Maldonado V., et al. (2011) Role of Bcl-3 in solid tumors. Mol. Cancer, 10, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tu K., et al. (2016) BCL-3 promotes the tumor growth of hepatocellular carcinoma by regulating cell proliferation and the cell cycle through cyclin D1. Oncol. Rep. [DOI] [PubMed] [Google Scholar]
- 44. Zhao H., et al. (2016) BCL3 exerts an oncogenic function by regulating STAT3 in human cervical cancer. Onco. Targets Ther., 9, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Niu Y., et al. (2019) BCL3 expression is a potential prognostic and predictive biomarker in acute myeloid leukemia of FAB subtype M2. Pathol. Oncol. Res., 25, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu L., et al. (2018) BCL3 expression promotes resistance to alkylating chemotherapy in gliomas. Sci. Transl. Med., 10, eaar2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai J., et al. (2016) A four-gene signature predicts survival in clear-cell renal-cell carcinoma. Oncotarget, 7, 82712–82726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Asghar U., et al. (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov., 14, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Z., et al. (2013) The IκB family member Bcl-3 stabilizes c-Myc in colorectal cancer. J. Mol. Cell Biol., 5, 280–282. [DOI] [PubMed] [Google Scholar]
- 50. Park S.G., et al. (2006) Up-regulation of cyclin D1 by HBx is mediated by NF-kappaB2/BCL3 complex through kappaB site of cyclin D1 promoter. J. Biol. Chem., 281, 31770–31777. [DOI] [PubMed] [Google Scholar]
- 51. Massoumi R., et al. (2009) Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med., 206, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Westerheide S.D., et al. (2001) The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol. Cell. Biol., 21, 8428–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rocha S., et al. (2003) p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol. Cell. Biol., 23, 4713–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Massoumi R., et al. (2006) Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell, 125, 665–677. [DOI] [PubMed] [Google Scholar]
- 55. Wakefield A., et al. (2013) Bcl3 selectively promotes metastasis of ERBB2-driven mammary tumors. Cancer Res., 73, 745–755. [DOI] [PubMed] [Google Scholar]
- 56. Chen X., et al. (2016) Bcl-3 regulates TGFβ signaling by stabilizing Smad3 during breast cancer pulmonary metastasis. Cell Death Dis., 7, e2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ren X., et al. (2013) Rhomboid domain containing 1 inhibits cell apoptosis by upregulating AP-1 activity and its downstream target Bcl-3. FEBS Lett., 587, 1793–1798. [DOI] [PubMed] [Google Scholar]
- 58. Choi H.J., et al. (2010) Bcl3-dependent stabilization of CtBP1 is crucial for the inhibition of apoptosis and tumor progression in breast cancer. Biochem. Biophys. Res. Commun., 400, 396–402. [DOI] [PubMed] [Google Scholar]
- 59. García I., et al. (2013) Bcl-3 regulates UVB-induced apoptosis. Hum. Cell, 26, 47–55. [DOI] [PubMed] [Google Scholar]
- 60. Bauer A., et al. (2006) The NF-kappaB regulator Bcl-3 and the BH3-only proteins Bim and Puma control the death of activated T cells. Proc. Natl. Acad. Sci. USA, 103, 10979–10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hosono N., et al. (2010) Glutathione S-transferase M1 inhibits dexamethasone-induced apoptosis in association with the suppression of Bim through dual mechanisms in a lymphoblastic leukemia cell line. Cancer Sci., 101, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kashatus D., et al. (2006) Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev., 20, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tao Y., et al. (2018) Alternative NF-kappaB signaling promotes colorectal tumorigenesis through transcriptionally upregulating Bcl-3. Oncogene. [DOI] [PubMed] [Google Scholar]
- 64. Urban B.C., et al. (2016) BCL-3 expression promotes colorectal tumorigenesis through activation of AKT signalling. Gut, 65, 1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O’Neil B.H., et al. (2007) Expression of nuclear factor-kappaB family proteins in hepatocellular carcinomas. Oncology, 72, 97–104. [DOI] [PubMed] [Google Scholar]
- 66. Gehrke N., et al. (2017) Hepatic B cell leukemia-3 suppresses chemically-induced hepatocarcinogenesis in mice through altered MAPK and NF-κB activation. Oncotarget, 8, 56095–56109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zou Y., et al. (2018) The proto-oncogene Bcl3 induces immune checkpoint PD-L1 expression, mediating proliferation of ovarian cancer cells. J. Biol. Chem., 293, 15483–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kythreotou A., et al. (2018) PD-L1. J. Clin. Pathol., 71, 189–194. [DOI] [PubMed] [Google Scholar]
- 69. Perkins N.D., et al. (2006) Good cop, bad cop: the different faces of NF-kappaB. Cell Death Differ., 13, 759–772. [DOI] [PubMed] [Google Scholar]
- 70. Paxian S., et al. (2002) Abnormal organogenesis of Peyer’s patches in mice deficient for NF-kappaB1, NF-kappaB2, and Bcl-3. Gastroenterology, 122, 1853–1868. [DOI] [PubMed] [Google Scholar]
- 71. Song L., et al. (2016) BCL3 reduces the sterile inflammatory response in pancreatic and biliary tissues. Gastroenterology, 150, 499–512.e20. [DOI] [PubMed] [Google Scholar]
- 72. Tassi I., et al. (2015) The NF-κB regulator Bcl-3 modulates inflammation during contact hypersensitivity reactions in radioresistant cells. Eur. J. Immunol., 45, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kuwata H., et al. (2003) IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-alpha production in macrophages. Blood, 102, 4123–4129. [DOI] [PubMed] [Google Scholar]
- 74. Greten F.R., et al. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell, 118, 285–296. [DOI] [PubMed] [Google Scholar]
- 75. Reissig S., et al. (2017) Elevated levels of Bcl-3 inhibits Treg development and function resulting in spontaneous colitis. Nat. Commun. 8, 15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meguro K., et al. (2015) Role of Bcl-3 in the development of follicular helper T cells and in the pathogenesis of rheumatoid arthritis. Arthritis Rheumatol., 67, 2651–2660. [DOI] [PubMed] [Google Scholar]
- 77. Tang W., et al. (2014) The oncoprotein and transcriptional regulator Bcl-3 governs plasticity and pathogenicity of autoimmune T cells. Immunity, 41, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Brasier A.R., et al. (2001) NF-kappa B-inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-kappa B1 residence. J. Biol. Chem., 276, 32080–32093. [DOI] [PubMed] [Google Scholar]
- 79. Keutgens A., et al. (2010) BCL-3 degradation involves its polyubiquitination through a FBW7-independent pathway and its binding to the proteasome subunit PSMB1. J. Biol. Chem., 285, 25831–25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ikeda O., et al. (2010) Interactions of STAP-2 with Brk and STAT3 participate in cell growth of human breast cancer cells. J. Biol. Chem., 285, 38093–38103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lin Z., et al. (2012) Identification of disease-associated DNA methylation in B cells from Crohn’s disease and ulcerative colitis patients. Dig. Dis. Sci., 57, 3145–3153. [DOI] [PubMed] [Google Scholar]
- 82. Kreisel D., et al. (2011) Bcl3 prevents acute inflammatory lung injury in mice by restraining emergency granulopoiesis. J. Clin. Invest., 121, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Puvvada S.D., et al. (2010) NF-kB and Bcl-3 activation are prognostic in metastatic colorectal cancer. Oncology, 78, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Saamarthy K., et al. (2015) Early diagnostic value of Bcl-3 localization in colorectal cancer. BMC Cancer, 15, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang S.M., et al. (2010) Effect of COX-2 inhibitor on the expression of BCL-3 and cyclin D1 in human colon cancer cell line SW480. Zhonghua Wei Chang Wai Ke Za Zhi, 13, 612–615. [PubMed] [Google Scholar]
- 86. Tang W., et al. (2016) The B-cell tumor promoter Bcl-3 suppresses inflammation-associated colon tumorigenesis in epithelial cells. Oncogene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Philpott A., et al. (2014) Lineage selection and plasticity in the intestinal crypt. Curr. Opin. Cell Biol., 31, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Clarke M.F., et al. (2006) Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res., 66, 9339–9344. [DOI] [PubMed] [Google Scholar]
- 89. Vogelstein B., et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koo B.K., et al. (2014) Stem cells marked by the R-spondin receptor LGR5. Gastroenterology, 147, 289–302. [DOI] [PubMed] [Google Scholar]
- 91. Schwitalla S., et al. (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell, 152, 25–38. [DOI] [PubMed] [Google Scholar]
- 92. Barker N., et al. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature, 457, 608–611. [DOI] [PubMed] [Google Scholar]
- 93. Hirsch D., et al. (2014) LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis, 35, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shimokawa M., et al. (2017) Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. [DOI] [PubMed] [Google Scholar]
- 95. Ziskin J.L., et al. (2013) In situ validation of an intestinal stem cell signature in colorectal cancer. Gut, 62, 1012–1023. [DOI] [PubMed] [Google Scholar]
- 96. de Sousa e Melo F., et al. (2017) A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature, 543, 676–680. [DOI] [PubMed] [Google Scholar]
- 97. Chen CY., et al. (2015) Bcl3 bridges LIF-STAT3 to Oct4 signaling in the maintenance of naive pluripotency. Stem cells (Dayton, Ohio). [DOI] [PubMed] [Google Scholar]