Abstract
Circular RNAs (circRNAs) have recently been identified as a new member of endogenous noncoding RNAs. CircRNAs exhibit high stability and can thus can be used as valuable biomarkers for monitoring the occurrence and development of hepatocellular carcinoma (HCC). The present study sought to explore the diagnostic significance of plasma circRNAs in hepatitis B virus (HBV)-related HCC. Plasma circRNAs from 10 patients with hepatitis B (HBV)-related HCC and 5 patients with HBV-related liver cirrhosis were investigated by microarray to screen differentially expressed circRNAs, 157 upregulated and 161 downregulated circRNAs were found. Twenty-four circRNAs were further investigated via quantitative reverse-transcriptase–polymerase chain reaction assay in a training cohort (n = 48), hsa_circ_0027089 exhibited the highest significance and further distinguished 64 HCC patients from 40 cirrhosis patients and 72 healthy participants in a validation cohort. These results indicate that plasma hsa_circ_0027089 can serve as a new marker for the diagnosis of HBV-related HCC.
Plasma hsa_circ_0027089 can serve as a new marker for the diagnosis of hepatitis B virus–related HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world, ranking fifth in global tumors, and its mortality rate is the second highest among malignant tumors (1). The outcome remains poor due to a high postoperative recurrence rate (2–4).
At present, the diagnosis of HCC mainly depends on imaging examination and serum marker screening. The main serum markers are alpha-fetoprotein (AFP), abnormal prothrombin, and phosphatidyl proteoglycan-3 and the like (5). AFP has been widely used for serodiagnosis and screening of HCC given its clinical performance (6). However, the sensitivity of AFP in the diagnosis of HCC is only 40–65%, and the specificity is between 76 and 96% (7). Therefore, it is imperative to explore better serum diagnostic markers.
Over the last two decades, several new cutting-edge technologies, such as next-generation sequencing (8,9) and microarray technologies (10), have emerged, leading the search for biomarkers into a new era of ‘omics' (11). Using these technologies, the analysis of tens of thousands of molecular targets has become affordable and operable. Currently, numerous circulating markers and tissue markers have been identified (10,12–14). For instance, we recently identified a plasma microRNA panel, which has considerable clinical value in diagnosing early-stage HCC (10).
Circular RNA is a new member of endogenous noncoding RNAs, which are characterized by covalently closed loop structures (15,16). Recent studies have found that a large number of circular RNAs play important roles in lung (17), breast (18), gastric cancer (19) and HCC (20–23). Circular RNAs are insensitive to ribonuclease and are more stable than other linear RNA molecules due to the special circular closed structure (15,16,24). Therefore, circular RNAs (circRNAs) can act as potential biomarkers for the diagnosis of HCC.
To our knowledge, no research has investigated the role of plasma circRNAs in early diagnosis of HCC. This study sought to explore the diagnostic significance of plasma circRNAs in hepatitis B virus (HBV)-related HCC.
Methods
Study design and patients
The purpose of this study was to identify plasma circRNAs as valuable biomarkers for monitoring and diagnosis of HBV-related HCC. The strategy was to screen differentially expressed circRNAs through microarray in a discovery phase, then narrow the candidates down in a training phase, and the selected circRNA was finally evaluated in a validation phase.
In total, 239 participants admitted to Zhongshan hospital between January 2015 and December 2015 were enrolled in the study. The participants were allocated to three phases (Figure 1). The investigational protocol was approved by local institutional review boards, and informed consent was obtained from all participants. Basic information and blood samples were collected. The inclusion and exclusion criteria were as follows:
Figure 1.
Study design.
General inclusion criteria: (i) age ≥ 18 years and ≤ 80 years, (ii) the capability to give informed consent, (iii) not severely ill in the intensive care unit. Healthy group: (i) had the medical checkup at Zhongshan hospital, (ii) healthy condition without malignancy. HBV-related liver cirrhosis: (i) with HBV infection, (ii) liver biopsy diagnosed by two experienced pathologists or diagnosis must be supported by two image reports (ultrasound B, computed tomography or magnetic resonance imaging). HBV-related HCC patients: (i) with HBV infection, (ii) diagnosed by two experienced pathologists, (iii) no pre-operative chemotherapy, radiotherapy, transarterial chemoembolization or ablation.
The characteristics of the participants (Table 1) were well balanced among three cohorts (discovery, training and validation). There was no significant discrepancy in the distribution of age, sex or tumor characteristics.
Table 1.
Characteristics of study subjects in the training and validation cohort
| Variables | n (%) | n (%) | P |
|---|---|---|---|
| HCC | Training (N = 24) | Validation (N = 64) | |
| Age, years | 0.93 | ||
| ≤50 | 11 (45.8) | 30 (46.9) | |
| >50 | 13 (54.2) | 34 (53.1) | |
| Gender | 0.88 | ||
| Male | 22 (91.7) | 58 (90.6) | |
| Female | 2 (8.3) | 6 (9.4) | |
| Serum AFP, ng/mL | 0.45 | ||
| ≤20 | 10 (41.7) | 20 (31.3) | |
| >20 | 14 (58.3) | 44 (68.7) | |
| Tumor diameter (cm) | 0.43 | ||
| ≤3 | 9 (37.5) | 17 (26.6) | |
| >3 | 15 (62.5) | 47 (73.4) | |
| Tumor number | 0.59 | ||
| Single | 19 (79.2) | 46 (71.9) | |
| Multiple | 5 (20.8) | 18 (28.1) | |
| Edmonson grade | 0.075 | ||
| I/II | 20 (83.3) | 40 (62.5) | |
| III/IV | 4 (16.7) | 24 (37.5) | |
| BCLC stage | 0.057 | ||
| 0/A | 10 (62.5) | 13 (20.3) | |
| B/C | 14 (37.5) | 51 (79.7) | |
| Cirrhosis | Training (N = 24) | Validation (N = 40) | |
|---|---|---|---|
| Age, years | 0.95 | ||
| ≤50 | 11 (45.8) | 18 (45) | |
| >50 | 13 (54.2) | 22 (55) | |
| Gender | 0.89 | ||
| Male | 16 (66.7) | 26 (65) | |
| Female | 8 (33.3) | 14 (35) | |
| Serum AFP, ng/mL | 0.72 | ||
| ≤20 | 20 83.3) | 35 (87.5) | |
| >20 | 4 (16.7) | 5 (12.5) |
BCLC, Barcelona clinic liver cancer.
Discovery phase
Plasma circRNAs from 10 patients with HBV-related HCC and 5 patients with HBV-related liver cirrhosis (Supplementary Table S1) were investigated through microarray to screen differentially expressed circRNAs. Arraystar Human circRNA Arrays V2.0 (8x15K, Arraystar), which consists of 13 617 known circRNAs was used in this study.
Plasma preparation and RNA isolation
Plasma preparation and RNA isolation was performed as previously described (10). The purity and concentration of the RNA samples were determined by NanoDrop ND-1000 (NanoDrop Technologies, Waltham, MA). RNA integrity was assessed by electrophoresis on a denaturing agarose gel. Total RNAs from each sample were digested with Rnase R (Epicentre) to remove linear RNAs and enrich circular RNAs.
Sample labeling and array hybridization
Sample labeling and array hybridization were performed according to the manufacturer's protocol (Arraystar). Briefly, the enriched circular RNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were purified using the RNeasy Mini Kit (Qiagen) and then hybridized onto the Arraystar Human circRNA Arrays V2.0 (8x15K, Arraystar). The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven. Then the hybridized arrays were washed, fixed and scanned using the Agilent Scanner G2505C.
Data analysis
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the R software limma package. Differentially expressed circRNAs between two samples with statistical significance were identified through fold-change filtering (>2). Hierarchical clustering was performed to demonstrate distinguishable circRNA expression pattern among samples.
Training phase
By screening the microarray data based on a false discovery rate < 0.05, fold-change > 4, and P-value < 0.05, 24 candidate circRNAs were chosen for further analysis by quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) in 24 patients with HBV-related HCC and 24 patients with HBV-related liver cirrhosis (training cohort). The low-abundance candidates in plasma were excluded, and the candidate with the highest fold-change and significant difference (hsa_circ_0027089) was further evaluated in a validation cohort.
Validation phase
The diagnostic performance of the selected circRNA (hsa_circ_0027089) was then evaluated in 64 HCC patients, 40 cirrhosis and 72 healthy participants.
Quantitative reverse transcriptase–polymerase chain reaction
qRT–PCR was performed as described previously (24). Hsa-miR-1228 was used as the internal control (10). Primers were listed in Supplementary File 1 (available at Carcinogenesis online).
Statistical analysis
Statistical analysis was performed with SPSS 23.0 software (SPSS, Chicago, IL). The Mann–Whitney unpaired test was used for the comparison between HCC and control (cirrhosis, hepatic cyst and hepatic hemangioma) (10). The cutoff point of the surrogate circRNA was identified using MedCalc. A receiver-operating characteristic (ROC) curve was constructed according to the predicted probability of the surrogate circRNA, and the area under the ROC curve (AUC) was used to evaluate the diagnostic ability of the circRNA. A two-tailed P-value < 0.05 was used to indicate a significant result.
Results
Screening of circRNAs in discovery cohort
To investigate the expression of circRNA in the plasma of patients with HCC, we collected plasma from 10 patients with HBV-related HCC and 5 patients with HBV-related cirrhosis (as a negative control) for microarray detection.
After homogenization analysis of the raw data of the chip, the results showed that the 15 samples were homogeneous (Supplementary Figure S1A, available at Carcinogenesis online). Using fold-change > 2 and P < 0.05 as the screening criteria, we found a total of 157 upregulated circRNAs and 161 downregulated circRNAs (Supplementary File 2, available at Carcinogenesis online). Further heatmap analysis revealed that we were able to clearly distinguish between HCC and cirrhosis samples using 86 circRNAs (Figure 2A).
Figure 2.
(A) Heatmap analysis revealed that circRNAs can easily distinguish between HCC and cirrhosis samples, arrow indicated the location of hsa_circ_0027089. (B) qRT–PCR results of the candidate circRNAs in training cohort. *P < 0.05; **P < 0.01; n.s., not significant.
Further testing and verification of candidate circRNAs in the training cohort
By screening the microarray results based on false discovery rate < 0.05, fold-change > 4 and P < 0.05, we selected 24 differentially expressed circRNAs (20 upregulated and 4 downregulated circRNAs) for further investigation.
We measured the relative expressions of the 24 circRNAs via qRT–PCR in 24 patients with HCC and 24 patients with cirrhosis. The results showed that 16 candidates exhibited low abundance in plasma and were therefore not suitable for clinical detection. Two candidates (hsa_circ_0005198 and hsa_circ_000882) exhibited no significant differences between HCC and cirrhosis. Six circRNAs (hsa_circ_0077930, hsa_circ_0027089, hsa_circ_0001818, hsa_circ_0011883, hsa_circ_0001070, hsa_circ_0026337) exhibited high abundance in plasma and were differentially expressed (Figure 2B; Supplementary File 3, available at Carcinogenesis online). We performed Sanger sequencing verification on the six candidate circRNAs. The results showed that the product sequence is singular and identical to the circRNA reverse splicing sequence in the circbase database (Supplementary Figure S1B, available at Carcinogenesis online). PCR results showed that hsa_circ_0027089 had the highest fold-change; therefore, we further investigated the clinical significance of hsa_circ_0027089.
Validation of hsa_circ_0027089 for HCC versus control
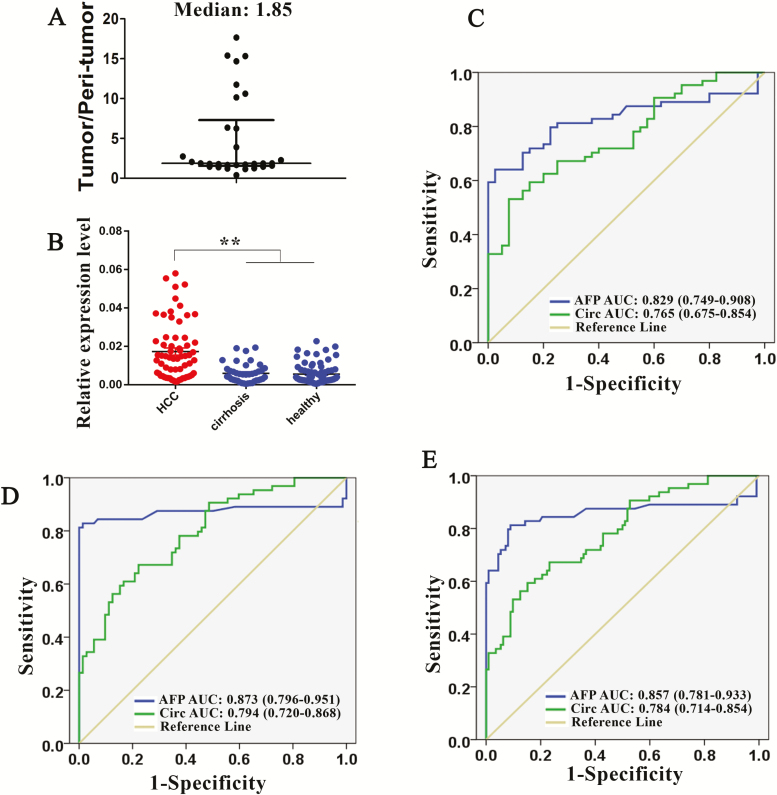
We measured the relative expression of hsa_circ_0027089 in tumor and peri-tumor tissues of 30 HCC patients. The results showed that HCC tissues exhibit higher hsa_circ_0027089 level than peripheral normal tissues, with a median fold-change of 1.85 (Figure 3A).
Figure 3.
(A) Ratio of tumor/peri-tumor level of hsa_circ_0027089 measured by qRT–PCR, the median ratio is 1.85. (B) qRT–PCR results of hsa_circ_0027089 in validation cohort. (C–E) Receiver operating characteristic curve analysis (ROC) for AFP and hsa_circ_0027089 in the validation cohort. Area under the curve (AUC) estimation in (C) HCC versus cirrhosis; (D) HCC versus Healthy; and (E) HCC versus non-HCC (cirrhosis + healthy). **P < .01.
We then measured hsa_circ_0027089 level in a validation cohort of 64 HCC patients, 40 cirrhosis patients and 72 healthy participants using qRT–PCR (Supplementary File 4, available at Carcinogenesis online). Hsa_circ_0027089 was significantly upregulated in the plasma of patients with HCC compared with cirrhosis patients and healthy participants (P < 0.01) (Figure 3B).
We performed the ROC analysis for hsa_circ_0027089 and AFP in HCC versus cirrhosis (AUC: hsa_circ_0027089 versus AFP, 0.765 versus 0.829) (Figure 3C), HCC versus healthy (AUC: hsa_circ_0027089 versus AFP, 0.794 versus 0.873) (Figure 3D) and HCC versus non-HCC (cirrhosis + healthy) (AUC: hsa_circ_0027089 versus AFP, 0.784 versus 0.857) (Figure 3E). The results showed that the AUC of hsa_circ_0027089 is comparable to that of AFP.
Using MedCalc to analyze the PCR data (ddCt value) of the 48 patients (training cohort), we identified the cutoff point of hsa_circ_0027089 as 0.011 (ddCt value), i.e., serum level of hsa_circ_0027089 in HCC patients tends to be higher than 0.011. Using 0.011 as the cutoff point of hsa_circ_0027089, and 20 ng/ml as the cutoff point of AFP, we further compared their diagnostic value in the validation cohort. The diagnostic accuracy of AFP was slightly better than hsa_circ_0027089 in HCC versus cirrhosis (AFP [sensitivity: 68.75%, specificity: 87.5%] versus hsa_circ_0027089 [sensitivity: 57.81%, specificity: 85%]), HCC versus healthy (AFP [sensitivity: 68.75%, specificity: 100%] versus hsa_circ_0027089 [sensitivity: 57.81%, specificity: 84.7%]) and HCC versus non-HCC (AFP [sensitivity: 68.75%, specificity: 95.5%] versus hsa_circ_0027089 [sensitivity: 57.81%, specificity: 84.8%]).
We then combined hsa_circ_0027089 and AFP in diagnosing HCC, the combination exhibited better sensitivity, but poorer specificity (HCC versus cirrhosis [sensitivity: 79.69%, specificity: 75%], HCC versus healthy [sensitivity: 79.69%, specificity: 86.11%], and HCC versus non-HCC [sensitivity: 79.69%, specificity: 82.14%]).
Discussion
In this study, we explored the diagnostic significance of plasma circRNAs in HBV-related HCC through a series of experiments. First, we performed circRNA microarray on 10 HCC patients and 5 cirrhosis patients to screen differentially expressed circRNAs. Twenty-four circRNAs were further verified via qRT–PCR in a training cohort (n = 48). The selected circRNA (hsa_circ_0027089) was then evaluated in a validation cohort (n = 176) and further distinguished 64 HCC patients from 40 cirrhosis patients and 72 healthy participants.
In China, the majority of HCC cases develop with a history of hepatitis B virus (HBV)-related hepatitis in which there is continuous inflammation and regeneration of hepatocytes (25). The coexistence of inflammation and cirrhosis makes the early diagnosis of HCC more difficult partly due to the overwhelming number of circRNAs released by inflammation and hepatocyte regeneration. Therefore, we compared HBV-related HCC with HBV-related cirrhosis to screen HCC associated circRNAs in this study, which could exclude aberrant circRNAs from the cirrhosis groups.
CircRNAs have currently attracted increasing attention due to its critical function in HCC carcinogenesis and progression (20–23). Han et al. (20) revealed that circMTO1 suppresses HCC progression by acting as the sponge of oncogenic miR-9 to promote p21 expression. Zhang et al. (26) showed that exosome circRNAs secreted from adipocytes promote tumor growth and reduce DNA damage by suppressing miR-34a and activating the USP7/Cyclin A2 signaling pathway. Recent studies have found abnormal expression of circRNAs in the plasma of HCC patients (26,27), given its special structural characteristics and stability, plasma circRNAs exhibit the potential to serve as a novel biomarker in HCC diagnosis.
By analyzing raw data of the chip, we found a total of 157 upregulated circRNAs and 161 downregulated circRNAs. We have mapped the distribution of these differentially expressed circRNAs on chromosomes (Supplementary Figure S2A, available at Carcinogenesis online). Each chromosome contains differentially expressed circRNAs, and 4 circRNAs derived from mitochondrial DNA are also differentially expressed in the plasma of HCC patients. Through genetic analysis, we found that differentially expressed circRNAs are mainly concentrated in exons, partly from introns and minimally from other locations in the genome (Supplementary Figure S2B, available at Carcinogenesis online). Heatmap analysis showed that we were able to distinguish between HCC patients and cirrhosis patients with 86 circRNAs, which implies the diagnostic value of plasma circRNAs in HCC.
Using a validation cohort of 176 participants, we confirmed that hsa_circ_0027089 can discriminate HCC from cirrhosis and healthy participants. Through genomic alignment, we found that hsa_circ_0027089 is located in the genomic region chr12:57059987-57064148 (GRCh37), which is formed by reverse splicing of exons 5 and 6 of the PTGES3 gene (Supplementary Figure S2C, available at Carcinogenesis online). Using Arraystar's homemade miRNA target prediction software, we found that hsa_circ_0027089 had binding sites for several microRNAs, such as hsa-miR-15b-3p, hsa-miR-141-5p, hsa-miR-197-3p, hsa-miR-205-3p and hsa-miR-670-3p (Supplementary Figure S2D, available at Carcinogenesis online). Previous studies have shown that these microRNAs are closely related to the development of malignant tumors. Li et al. (28) revealed that hsa-miR-15b-3p can regulate HCC cell growth and metastasis through targeting OIP5; Ji et al. (29) found that high expression of miR-15b predicts poor prognosis for HCC patients. Jiang et al. (30) reported that hsa-miR-205-3p is abnormally expressed in tissues and serum of patients with non-small cell lung cancer. High-throughput sequencing studies have found that hsa-miR-141-5p and hsa-miR-205-3p are dysregulated in prostate cancer tissues (31,32). Those studies indicate that hsa_circ_0027089 may participate in various biological processes of tumor by regulating these microRNAs or as a microRNA carrier transferring between tissue cells.
Currently, AFP is widely used for diagnosis and surveillance of HCC (6). We therefore compared the diagnostic significance of hsa_circ_0027089 with AFP. ROC analysis showed that the diagnostic accuracy of AFP was slightly better than hsa_circ_0027089. And the combination of hsa_circ_0027089 and AFP possessed better ability to identify HCC patients. The results imply that plasma circRNA is a promising marker for diagnosis of HBV-related HCC.
There are several limitations in our research. First, the number of HCC patients recruited is relatively small, the result might be overinterpreted. Large-scale studies are needed to confirm these findings in the future. Second, there is no consensus on endogenous control for the circulating circRNAs in HCC. Therefore, we used miR-1228, which was demonstrated to be stable in our former study of circulating biomarkers (10), as an internal control. Further studies are needed to identify stable circulating circRNAS as internal control.
In summary, we identified a plasma circRNA (hsa_circ_0027089) that can discriminate HBV-related HCC from HBV-related cirrhosis and healthy participants, and its diagnostic performance is comparable to that of AFP. Plasma hsa_circ_0027089 can serve as a new marker for the diagnosis of HBV-related HCC.
Funding
This work was supported by National Natural Science Funds of China (81402376, 81572296, 81830102, 81572823, 81772578, 81502028, 31601114), Shanghai Hospital Development Center (SHDC12015104), National Key Research and Development Program (2016YFC0902400).
Author contributions
K.Z., H.Z. and Y.P. contributed equally to this work.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- AFP
alpha-fetoprotein
- AUC
area under the ROC curve
- circRNAs
circular RNAs
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- ROC
receiver-operating characteristic
References
- 1. Siegel R.L., et al. (2019) Cancer statistics, 2019. CA. Cancer J. Clin., 69, 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Bruix J., et al. (2014) Hepatocellular carcinoma: clinical frontiers and perspectives. Gut, 63, 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruix J., et al. (2016) Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology, 150, 835–853. [DOI] [PubMed] [Google Scholar]
- 4. Bruix J., et al. ; RESORCE Investigators. (2017) Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet, 389, 56–66. [DOI] [PubMed] [Google Scholar]
- 5. Rich N., et al. (2014) Hepatocellular carcinoma tumour markers: current role and expectations. Best Pract. Res. Clin. Gastroenterol., 28, 843–853. [DOI] [PubMed] [Google Scholar]
- 6. Zhang B.H., et al. (2004) Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol., 130, 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu X.W., et al. (2013) Expression and role of icam-1 in the occurrence and development of hepatocellular carcinoma. Asian Pac. J. Cancer Prev., 14, 1579–1583. [DOI] [PubMed] [Google Scholar]
- 8. Cho W., et al. (2012) Emerging personalized oncology: sequencing and systems strategies. Future Oncol., 8, 637–641. [DOI] [PubMed] [Google Scholar]
- 9. Meyerson M., et al. (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet., 11, 685–696. [DOI] [PubMed] [Google Scholar]
- 10. Zhou J., et al. (2011) Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol., 29, 4781–4788. [DOI] [PubMed] [Google Scholar]
- 11. Marquardt J.U., et al. (2012) Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J. Hepatol., 56, 267–275. [DOI] [PubMed] [Google Scholar]
- 12. Zhu K., et al. (2013) MiR-146a enhances angiogenic activity of endothelial cells in hepatocellular carcinoma by promoting PDGFRA expression. Carcinogenesis, 34, 2071–2079. [DOI] [PubMed] [Google Scholar]
- 13. Zhu K., et al. (2014) MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci. Rep., 4, 5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu K., et al. (2013) Biomarkers for hepatocellular carcinoma: progression in early diagnosis, prognosis, and personalized therapy. Biomark. Res., 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeck W.R., et al. (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Memczak S., et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495, 333–338. [DOI] [PubMed] [Google Scholar]
- 17. Kumar M.S., et al. (2014) HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature, 505, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Liu Z., et al. (2019) Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis., 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li P., et al. (2017) Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer, 116, 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han D., et al. (2017) Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology, 66, 1151–1164. [DOI] [PubMed] [Google Scholar]
- 21. Halgand B., et al. (2018) Hepatitis B virus pregenomic RNA in hepatocellular carcinoma: a nosological and prognostic determinant. Hepatology, 67, 86–96. [DOI] [PubMed] [Google Scholar]
- 22. Shi L., et al. (2017) Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis., 8, e3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang M., et al. (2018) CircRNAs as biomarkers of cancer: a meta-analysis. BMC Cancer, 18, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen T.B., et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. [DOI] [PubMed] [Google Scholar]
- 25. Villanueva A., et al. (2013) Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat. Rev. Gastroenterol. Hepatol., 10, 34–42. [DOI] [PubMed] [Google Scholar]
- 26. Zhang H., et al. (2019) Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene, 38, 2844–2859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Zhang X., et al. (2018) circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis., 9, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., et al. (2017) OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget, 8, 18129–18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ji W.B., et al. (2016) High expression of miR-15b predicts poor prognosis for hepatocellular carcinoma after curative hepatectomy. Oncol. Rep., 36, 1901–1908. [DOI] [PubMed] [Google Scholar]
- 30. Jiang M. et al. (2013) Relative expressions of miR-205-5p, miR-205-3p, and miR-21 in tissues and serum of non-small cell lung cancer patients. Mol. Cell. Biochem., 383, 67–75. [DOI] [PubMed] [Google Scholar]
- 31. Daniel R., et al. (2017) A Panel of microRNAs as diagnostic biomarkers for the identification of prostate cancer. Int. J. Mol. Sci., 18, 1281–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paziewska A., et al. (2018) Candidate diagnostic miRNAs that can detect cancer in prostate biopsy. Prostate, 78, 178–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.