Abstract
Molecular oxygen (O2) sustains intracellular bioenergetics and is consumed by numerous biochemical reactions, making it essential for most species on Earth. Accordingly, decreased O2 concentrations (hypoxia) is a major stressor that generally subverts life of aerobic species and is a prominent feature of pathological states encountered in bacterial infection, inflammation, wounds, cardiovascular defects, and cancer. Therefore, key adaptive mechanisms to cope with hypoxia have evolved in mammals. Systemically, these adaptations include increased ventilation, cardiac output, blood vessel growth, and circulating red blood cell numbers. On a cellular level, ATP consuming reactions are suppressed and metabolism is altered until oxygen homeostasis is restored. A critical question is how mammalian cells sense O2 levels to coordinate diverse biological outputs during hypoxia. The best studied mechanism of response to hypoxia involves hypoxia inducible factors (HIFs), which are stabilized by low oxygen availability and control the expression of a multitude of genes, including those involved in cell survival, angiogenesis, glycolysis, and invasion/metastasis. Importantly, changes in O2 can also be sensed via other stress pathways as well as changes in metabolite levels and the generation of reactive oxygen species (ROS) by mitochondria. Collectively, this leads to cellular adaptations of protein synthesis, energy metabolism, mitochondrial respiration, lipid and carbon metabolism as well as nutrient acquisition. These mechanisms are integral inputs into fine tuning the responses to hypoxic stress.
eTOC
The transcriptional response to hypoxia and the role of hypoxia inducible factors have been extensively studied. Yet, hypoxic cells also adapt to hypoxia by modulating protein synthesis, metabolism and nutrient uptake. Understanding these processes could shed light on pathologies associated with hypoxia, including cardiovascular diseases and cancer, and disease mechanisms such as inflammation and wound repair.
Introduction
Given the central importance of oxygen (O2) in maintaining intracellular ATP levels and serving as an electronic acceptor in a large number of biochemical reactions, it is unsurprising that responses to hypoxia are rapid, important, and highly conserved. Examples of O2 consuming reactions include aerobic respiration, fatty acid desaturation, and those catalysed by a growing number of α-ketoglutarate dioxygenases [G], which are involved in various metabolic reactions, including RNA, DNA, and histone demethylation reactions. Due to vascular insufficiency or overt blood vessel damage and tissue oedema, hypoxia arises in a variety of diseases, including the growth of solid tumours. Once an initially avascular tumour achieves a size extending beyond the natural diffusion limits of O2, hypoxic microdomains develop. For the disease to progress, tumours must acquire blood vessels, either through angiogenesis or vessel co-option. However, tumour blood vessels differ from their normal counterparts in a variety of important phenotypes and perfuse tissue poorly. As such, adaptation to oxygen starvation is a key feature of both primary and metastatic neoplasms (Box 1).
Box 1: Hypoxia and cancer.
Hypoxic regions (partial pressure of oxygen [PO2] < 10 mmHg) arise in tumours through the rapid proliferation of cancer cells in the absence of an efficient vasculature, resulting in the exhaustion of available nutrient and oxygen supplies. As a consequence, hypoxia induces multiple adaptive pathways and genomic changes that enable tumour cells to adapt to poor nutrition and hostile microenvironments for malignant progression140–142. The upregulation of hypoxia-inducible angiogenic factors from hypoxic tumour sites, such as vascular endothelial growth factor (VEGF), triggers tumour mass vascularization to overcome proliferation limitations200. However, the vessels formed during neovascularization are often poorly organised and dysfunctional, either being blunt-ended or having variability in flow velocity or direction. In addition, endothelial cells in normal vessels create a smooth surface permitting laminar flow; however, endothelial cells of tumour-associated vessels have gaps between them, resulting in vascular leakiness, non-laminar flow making blood prone to clotting, and local tissue oedema200–202. Overall, most solid tumours retain hypoxic domains throughout disease progression, selecting for aggressive malignant cells that can withstand the ischaemic stresses of adverse tumour microenvironments.
The biology of hypoxic cancer cells is a product of the interplay between the prevailing oxygen tension, hypoxia-induced signalling (including that of hypoxia-inducible factors; HIFs), interacting genetic defects, and cellular damage by reactive oxygen species (ROS) as discussed in this Review. As such, a solid tumour has dynamic fluctuations in oxygen from mild to severe hypoxia and necrosis, as well areas of acute hypoxia and re-oxygenation. In cancer patients, tumour hypoxia is a therapeutic problem often leading to poor prognosis due to the potential of increased malignancy through clonal selection of hypoxia resistant cancer cells, DNA damage, resistance to chemotherapy and radiation treatment, and an increased likelihood of metastasis203–205. For example, HIFs can enhance the expression of both collagen and extracellular matrix (ECM) remodelling enzymes that promote aberrant collagen containing ECM network formation, leading to tumour cell extravasation, survival in the circulation, and colonization of distant organs206. This makes tumour hypoxia itself an attractive therapeutic target. Over the years, non-toxic hypoxia-activated “prodrugs” have been developed to meet this need207,208. Conceptually, “trigger” units in hypoxic prodrugs are selectively activated in oxygen-starved cells to release toxic “effectors”, capable of killing surrounding tumour cells. These triggers include nitroaromatics, quinones, N-oxides, and transition metals. The N-oxide tirapazamine even entered phase III clinical trials; however, the future translation of tirapazamine and other hypoxic prodrugs remains in doubt209. Other strategies encompass targeting the downstream sequelae of tumour hypoxia, such as angiogenesis and the HIFs themselves.
Cellular hypoxia (0.5%-2% O2) can be transient, due to temporary mismatches between oxygen supply and cellular metabolic demands, or more chronic because of permanent vascular inadequacy, unresolved tissue oedema, and inflammation. Moreover, the degree of O2 deprivation can result in distinct responses, as certain effects on protein folding or O2-consuming biochemical reactions are only observed under severe O2 depletion (anoxic conditions, <0.5% O2). All of these adaptations must be integrated to support essential cellular activities until tissue and organismal responses return cells to appropriate O2 levels.
A major breakthrough in our understanding of cellular responses to changes in O2 levels was the discovery of hypoxia inducible factors (HIFs) and their regulation by the von Hippel–Lindau (VHL) tumour suppressor protein [G] (pVHL) and prolyl hydroxylases (PHD1-3 or EGLN1-3), members of the α-ketoglutarate dioxygenase superfamily. This finding provided a molecular framework for how changes in O2 levels can mount robust transcriptional responses and provides therapeutic targets for cancer, cardiovascular disease, and anaemia. Importantly, it also opened up the burgeoning field of studying α-ketoglutarate dioxygenases that include DNA, RNA, and histone demethylases. This ground-breaking research was justifiably honoured with the 2019 Nobel Prize in Physiology and Medicine awarded jointly to William G. Kaelin Jr, Sir Peter J. Ratcliffe, and Gregg L. Semenza “for their discoveries of how cells sense and adapt to oxygen availability”. Beyond HIFs, responses to decreased O2 levels involve changes in the epigenome, noncoding RNAs, the metabolome, signalling pathways, biochemical reactions, and diverse homeostatic measures to ensure cell survival during hypoxic stress.
In this Review, we discuss current knowledge on how cells respond and adapt to hypoxia. We briefly consider transcriptional responses to hypoxia, mostly dependent on the HIF–PHD–pVHL axis. However, our primary focus is on adaptive mechanisms, involving the impact of hypoxia on protein homeostasis, the functionality of mitochondria (central O2 consuming organelles), metabolism, and nutrient uptake in stressful microenvironments, which include important HIF-independent mechanisms. Of note, we largely focus on cellular responses; critical hypoxic adaptations at the organismal level, including changes in carotid body [G] activity, the cardiopulmonary system, neurological behaviours, erythropoietin [G] production, and other physiological responses are reviewed elsewhere1.
Transcriptional regulation by hypoxia
Hypoxia transcriptionally induces a robust set of genes controlled by HIFs but also a range of other transcription factors, including nuclear factor-κB (NF-κB)2. Nevertheless, the vast majority of O2 sensitive genes are in fact direct HIF targets3. These genes at the cellular and organismal level help adapt to diminishing levels of O2. However, persistent activation of hypoxia-induced genes can result in pathologies, including pulmonary hypertension.
HIF transcription factors and their regulation.
HIF-1 and HIF-2 are major transcription factors involved in the hypoxic response4. HIFs bind to hypoxia response elements (HREs) in the promoter regions of a large number of targets, including those involved in cell survival, angiogenesis, glycolysis, and invasion/metastasis. HIFs are heterodimers consisting of the regulated HIF-α protein subunit which is only expressed during hypoxia, and a constitutively expressed HIF-1β protein subunit. During normoxia, HIF-α subunits are polyubiquitylated by the pVHL–elongin BC-CUL2 [G] complex (referred to as the VHL complex) and targeted for proteasomal degradation3 (Figure 1). The normoxia-dependent interaction between HIF-α subunits and the VHL complex requires hydroxylation of two proline residues within the O2-dependent degradation (ODD) domain of HIF-α3,5. This hydroxylation reaction, which is catalysed by PHDs, is coupled to the oxidative decarboxylation of α-ketoglutarate to succinate and carbon dioxide5 (Figure 1). Importantly, all three PHD enzymes can hydroxylate both HIF-1α and HIF-2α and require O2, iron (Fe2+), and α-ketoglutarate to function6. Mice harbouring individual loss of PHD2 are embryonic lethal compared to loss of PHD1 or PHD3, thereby highlighting distinct functions of PHDs in vivo7. PHD2 is the primary enzyme responsible for HIF-α hydroxylation and subsequent degradation. Hypoxia prevents the hydroxylation of HIF-α subunits and their ubiquitin-mediated proteasomal degradation. As a result, HIF-α subunits dimerize with HIF-1β to form transcriptionally active complexes (Figure 1). The transcriptional activity of HIFs is fine-tuned by another member of the Fe2+ and α-ketoglutarate -dependent dioxygenases family, HIF asparaginyl hydroxylase or FIH-1 (Factor Inhibiting HIF-1)3,5. FIH-1 hydroxylates an asparaginyl residue within the C-terminal transactivating domain of HIF-1α and HIF-2α under normoxia to prevent recruitment of the transcriptional coactivators p300 and CBP, thereby curbing transcriptional output of HIFs8 (Figure 1). It is important to note that FIH-1 has other substrates beyond HIF-α protein subunits and their physiological functions continue to be investigated9,10. By contrast, a recent report by Ratcliffe and colleagues indicates that at least in vitro, PHDs have exquisite specificity towards HIFs11. However, PHDs have been shown to control multiple biological processes independent of HIFs, perhaps by enzyme activity-independent mechanisms12. Moreover, other potential PHD targets might require structures, such as molecular scaffolds, to be efficiently hydroxylated in living cells. It will be important to generate mice harbouring mutations within the PHD hydroxylase C-terminus to render them catalytically inactive to decipher their enzymatic and non-enzymatic roles in vivo.
Figure 1: Transcription regulation induced by hypoxia.
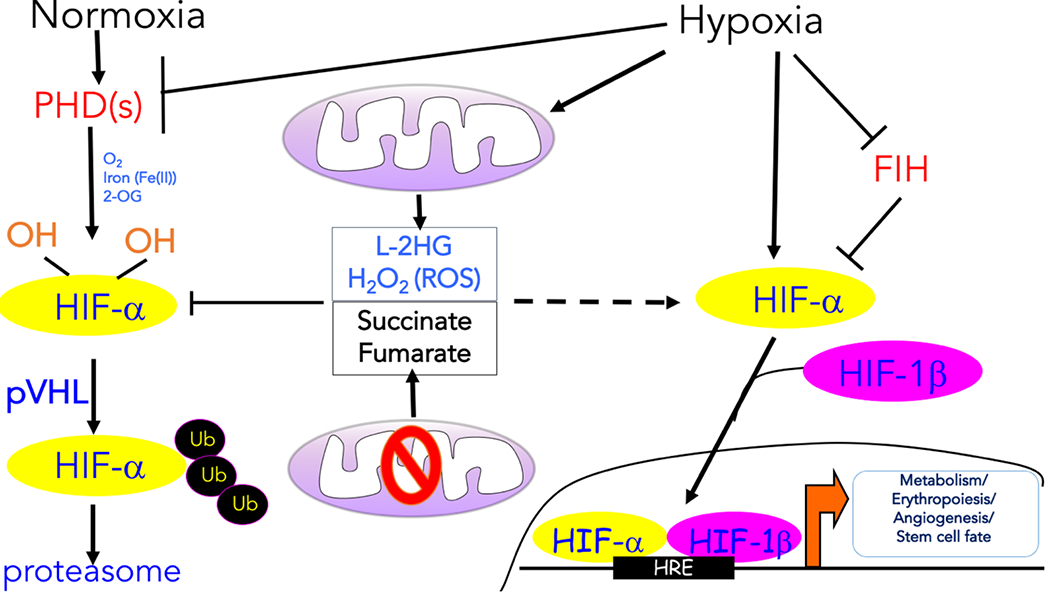
The oxygen-dependent interaction between the hypoxia inducible transcription factor α (HIF-α) subunits and the von Hippel–Lindau (VHL) tumour suppressor protein (pVHL) complex requires hydroxylation of two HIF-α proline residues by a family of α-ketoglutarate -dependent dioxygenases termed prolyl hydroxylases (PHDs), which requires oxygen (O2), iron (Fe2+), and α-ketoglutarate to function. Following hydroxylation, HIF-α subunits are polyubiquitylated by pVHL and targeted for proteasomal degradation. Hypoxia prevents the hydroxylation of the HIF-α protein subunits and their ubiquitin-mediated proteasomal degradation. As a result, the HIF-α protein subunits are allowed to dimerize with the HIF-1β protein subunits to form transcriptionally active complexes that bind to hypoxia response elements (HREs) to coordinate the induction of a large network of genes involved in metabolism, erythropoiesis, angiogenesis, and cell fate. Factor inhibiting HIF-1 (FIH-1) hydroxylates HIF-α subunits under normoxia to prevent recruitment of the transcription coactivators. Various mitochondrial products can also influence the hypoxic response. The production of reactive oxygen species (ROS) by mitochondrial complex III and L-2-hydroxyglutarate (L-2HG) under hypoxia can promote the stabilization of HIF-α protein levels. ROS likely inhibit PHDs by Cys oxidation, while L-2HG competes with α-ketoglutarate. Mutations in tricarboxylic acid (TCA) cycle components result in the accumulation of succinate and fumarate, which also inhibit PHD activity by competing with α-ketoglutarate, thereby causing an accumulation of the HIF-α protein even under normoxia.
Regulation of PHDs.
PHDs are the best characterized O2-sensitive proteins, although various other O2 sensors have been described (Box 2). An intrinsic property of all three PHDs is low affinity for O2 (high Michaelis constant [G] (Km)), making them primed to sense O2 levels and control HIF-α stabilization13. Tissue O2 levels vary, with levels as low as 0.5% O2 in the large intestine and up to 13% O2 in the lungs. However, many major organs have tissue O2 levels at ~3-7%14. Km values for all three PHDs for O2 are similar (230-250 μM) and close to the concentration of dissolved O2 in blood at atmospheric air13. By contrast, FIH-1 has a relatively higher affinity for O2 (Km value ~90 μM)15. Thus, any decrease in O2 levels below atmospheric air (~21% O2) will decrease PHD enzymatic activity5.
Box 2: Cellular oxygen sensing beyond PDH-pVHL-HIF pathway.
As described in the article, the transcriptional response to hypoxia is canonically dependent on the axis comprising prolyl hydroxylases (PHDs), von Hippel–Lindau tumour suppressor protein (pVHL) and hypoxia-inducible factors (HIFs). Although the PHDs are the best characterized O2-sensitive proteins controlled by the family of α-ketoglutarate -dependent dioxygenases (now including ≥70 members), other members of this superfamily could putatively be regulated by variable O2 levels as well as hypoxia-induced changes in ROS and metabolites. Proteins in this superfamily include collagen prolyl 4-hydroxylase I, Jumonji-C domain–containing histone lysine demethylases (JmjC-KDMs), the ten-eleven translocation (TET) family of enzymes involved in DNA demethylation as well as the AlkB homolog 5 (ALKBH5) and Fat Mass and Obesity-Associated (FTO) RNA demethylase enzymes6. Interestingly, these proteins are all inhibited by the accumulation of succinate, fumarate, and L-2- hydroxyglutarate (L-2HG) — metabolites produced by mitochondria upon perturbation of the tricarboxylic acid (TCA) cycle. Furthermore, most of these proteins have a high affinity for O2 and thus, would be active in the hypoxic range. However, there are notable exceptions that demonstrate high Km values for molecular O2 (i.e. low O2 affinities) and are therefore likely to be sensitive to tumour hypoxia, such as the JmjC-KDMs210. Recombinant KDM4B, KDM5A, and KDM6A/UTX exhibit low O2 affinities like the PHD enzymes, while KDM4A, KDM5B, KDM5C, KDM5D, and KDM6A have high O2 affinities211,212. Thus, hypoxia-induced inhibition of KDM4B, KDM5A, or KDM6A/UTX in cell culture results in an increase in a variety of histone methylation marks, including H3K4me3, H3K9me3, H3K27me3, and H3K36me3 (ref. 211), which is independent of the PHD–pVHL-HIF axis. It is important to note that hypoxia induces the transcription of many of the genes encoding JmjC-KDMs, likely to compensate for their decreased activity under low O2 conditions by simply increasing the cellular abundance and output of JmjC-KDM enzymes210.
Recently, the mammalian cysteine oxidase, cysteamine (2-aminoethanethiol) dioxygenase (ADO), was reported to be an O2 sensing enzyme that functions similarly to plant cysteine oxidases (PCOs)213. These enzymes add O2 atoms to N-terminal cysteine thiol groups on target proteins to form sulfinic acid, marking them for ubiquitin-mediated degradation. In plants, this PCO-mediated pathway, the N-degron pathway, acts as an O2-sensing system for hypoxic adaptation214–216. Like PCOs, ADO is not dependent on α-ketoglutarate, but is an iron- and O2-dependent enzyme. Interestingly, ADO can complement the loss of PCO in plants. Under normoxia, ADO promotes proteasomal degradation of RGS4 and RGS5, proteins involved in attenuating G protein signalling, and IL-32, a pro-inflammatory cytokine linked to gastric inflammation. ADO has a low affinity for O2 (apparent Km of O2 > 500 μM) like the PHDs; thus, hypoxia suppresses ADO activity resulting in RGS4, RGS5, and IL-32 protein stabilization213. It will be of interest to elucidate the O2-regulated target proteins of ADO to see whether, beyond O2, metabolites or ROS are able to suppress ADO activity to control physiology or disease. Collectively, the past 25 years of hypoxia research have revealed that multiple O2 sensing mechanisms have evolved to fine tune diverse responses to hypoxia. Beyond the large transcriptional programs regulated by HIFs in most eukaryotes, additional highly conserved O2 sensors must coordinate numerous adaptations required for cell survival under hypoxic stress.
Experimentally in cell culture, HIF-1α protein levels demonstrate a small increase in stability between atmospheric air and 6% O2 followed by an exponential rise as O2 levels approach 0.5% O2 (ref. 16). There are likely multiple inputs into PHDs when O2 levels are below 5% that explain the exponential rise in HIF-1α protein levels, including other sensors of intracellular O2 levels and the production of reactive oxygen species (ROS)17. Manipulating intracellular O2 levels can control PHD activity and thus HIF-α protein stabilization during hypoxia18. Furthermore, the mitochondrial intermembrane space protein coiled-coil helix domain containing protein 4 (CHCHD4; Mia40 in yeast) is necessary for hypoxic induction of HIF-1α protein stabilization by regulating mitochondrial O2 consumption19. ROS are generated during hypoxia by mitochondrial complex III (Figure 1) and their production exponentially increases starting at 5% O2. Decreasing ROS levels genetically or pharmacologically during hypoxia has been shown to diminish HIF-α protein levels20–25. How ROS inactivate PHDs to stabilize the HIF-α protein subunit is not understood. One new hypothesis is that mitochondrial ROS generated during hypoxia promote the oxidation of cysteine residues within PHD2 resulting in oxidative PHD2 homodimerization and inactivation, leading to HIF-α protein stabilization26. Indeed, oxidative modification of cysteine residues is one well-characterized mechanism by which ROS act as signalling molecules27. In response to ROS, redox-sensitive cysteine thiol groups (R-SH) can be oxidized to form disulfide bonds (R-S-S-R) which can mediate structural and functional changes within proteins and thereby regulate their activity. Importantly, PHD2 has several reactive cysteine residues in its C-terminal catalytic domain that may be oxidized by ROS26,28. Interestingly, PHD2 activity requires high intracellular levels of free cysteine, which is regulated by the cysteine dioxygenase (CDO1)28. Free intracellular cysteines may compete with the reactive cysteine residues of PHD2 for ROS-mediated oxidation. Thus, when free intracellular cysteine levels are high, PHD2 cysteine oxidation is prevented; PHD2 is then active and HIF-α protein levels are low. By contrast, limiting the amount of free intracellular cysteine would trigger HIF-α protein accumulation. Currently, the significance of these PHD2 reactive cysteines in the stabilization of HIF-α under physiological hypoxic conditions remains unknown.
Metabolites are a second input that inhibits PHD2 activity (Figure 1). Mutations in tricarboxylic acid (TCA) cycle components succinate dehydrogenase or fumarate hydratase result in the accumulation of the metabolites succinate and fumarate, respectively, and are linked to rare neuroendocrine and renal tumours29. Succinate and fumarate accumulation inhibits PHD2 activity by competing with its substrate α-ketoglutarate, causing an accumulation of HIF-α protein under normoxia (Figure 1)30,31. Independent of these mutations, succinate and fumarate can accumulate and trigger HIF-α stabilization in response to innate immune signals in monocytes to increase the mRNA expression of cytokines, such as IL-1β32. Another competitive antagonist of α-ketoglutarate is L-2- hydroxyglutarate (L-2HG). Hypoxia (0.5% O2) concomitant with acidosis [G] can favour the production of L-2HG and promote the stabilization of the HIF-α protein subunit33,34 (Figure 1). A unifying model to explain HIF activation is the intrinsic decrease in PHD2 activity due to declining O2 levels coupled with added inputs, such as ROS or metabolites, that further diminish PHD2 activity to maximally increase HIF-α protein levels.
Adaptation of protein accumulation
While transcriptional components of hypoxic responses have been relatively well studied, changes in protein synthesis rates under low O2 have been less well characterized. Downregulating translation suppresses energy expensive processes, such as “unnecessary” protein synthesis and prevents the accumulation of stress-induced unfolded and/or misfolded proteins. Hypoxia-induced targeted repression of cap-dependent protein synthesis [G] occurs predominantly at the level of translation initiation, which is normally accomplished by the recruitment of 40S ribosome subunits and initiation tRNAs at the mRNA AUG start codon35,36. Under low O2 levels, this repression is mainly driven by two pathways: downstream of PERK [G] (Protein kinase R (PKR)-like ER kinase) and mTOR [G] (mechanistic target of rapamycin) complex 1 (mTORC1). In addition, there are mechanisms to suppress translation elongation and termination during hypoxia. At the same time, translation of certain transcripts that encode proteins essential for survival in hypoxic environments is increased, thereby allowing adaptation of cells to this stressful condition37.
Inhibition of translation initiation.
Under physiological conditions, eukaryotic initiation factor 2 [G] (eIF2; α, β, and γ components) promotes translation initiation, which is associated with eIF2α being underphosphorylated36. O2 depletion, which can lead to oxidative stress, nutrient deprivation, and signalling disruption, can interfere with protein folding and subsequently result in an accumulation of misfolded as well as unfolded proteins. This has been shown to result in ER stress [G] 38. In order to alleviate ER stress, cells trigger the unfolded protein response (UPR) (Supplementary Box 1) , which serves as a critical cell survival mechanism, involving various adaptations, importantly including reduction of protein load via decreasing protein synthesis (see next section). In this context, one UPR sensor, PERK (Supplementary Box 1), functions as a kinase to phosphorylate eIF2α and inhibit translation initiation. This causes a general suppression of mRNA translational initiation and global protein synthesis to limit the detrimental effects of proteotoxicity39 (Figure 2). The protective role of PERK during hypoxic stress has been shown using mouse embryonic fibroblasts (MEFs) isolated from PERK-null mice, where PERK-deficient MEFs are more sensitive to hypoxic stress than their wildtype counterparts40,41. In addition, initiation of cap-dependent translation requires the binding of eIF4F [G] complex — comprising eIF4A, eIF4G and eIF4E — to the mRNA cap. This is regulated by mTORC1, which phosphorylates eIF4E-binding proteins (4E-BPs). 4E-BPs prevent eIF4E from assimilating into the eIF4F complex and this phosphorylation releases eIF4E allowing eIF4F assembly36. mTORC1 activity is also negatively regulated by hypoxia (Box 3). Accordingly, in hypoxia, eIF4E remains bound to 4E-BPs, resulting in the inhibition of eIF4F assembly and decrease in global translation rates42 (Figure 2). Prolonged hypoxic exposure activates a second, eIF2α– and mTORC1-independent pathway that maintains translation repression, where eIF4F complex breakdown results in eIF4E becoming sequestered in the nucleus by its transporter 4-ET43. This eIF2α regulation shows how varying states of O2 deprivation regulate mRNA translation through distinct mechanisms, each with important contributions to hypoxic gene expression.
Figure 2: Hypoxic adaptations in proteostasis.
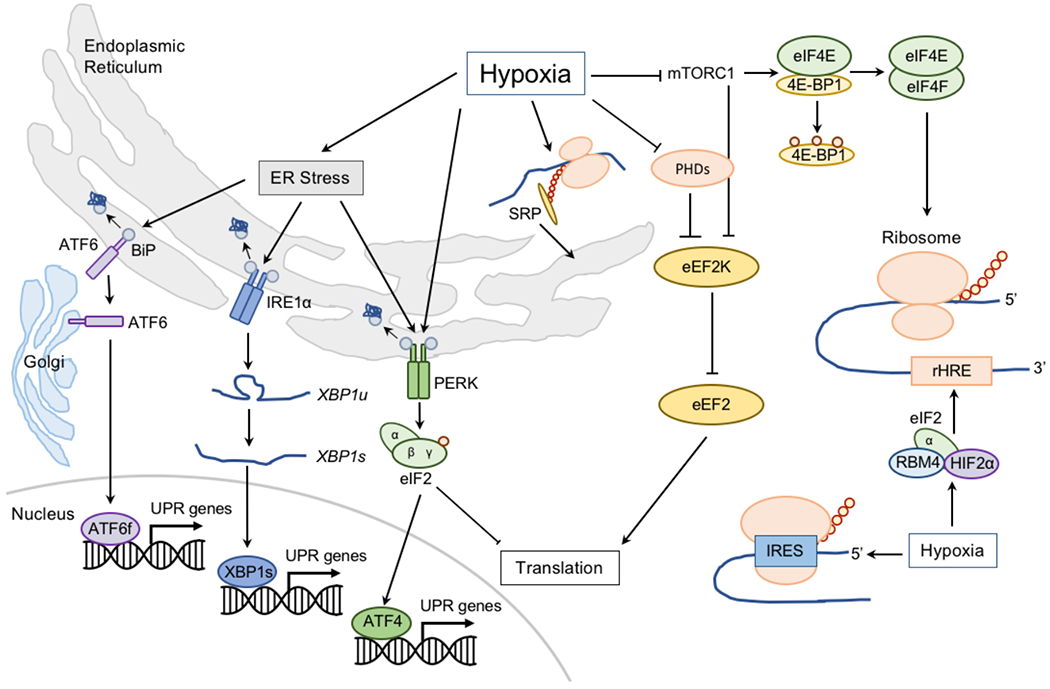
Low oxygen (O2) can induce endoplasmic reticulum (ER) stress and the unfolded protein response (UPR; Supplementary Box 1). Misfolded peptides bind to binding immunoglobulin protein (BiP) and causes it to activate the stress sensors: activating transcription factor 6 (ATF6), inositol-requiring protein 1α (IRE1α), and protein kinase RNA-like ER kinase (PERK) to initiate responses to restore ER homeostasis. ATF6 is transported to the Golgi apparatus, where it is processed to release its active transcriptional form (ATF6f). IRE1α activation and dimerization, triggers its RNase activity, which processes unspliced X box-binding protein 1 (XBP1u) to produce an active transcription factor, spliced XBP1 (XBP1s). Upon activation, PERK phosphorylates the initiation factor eukaryotic translation initiator factor 2α (eIF2α) to attenuate general peptide translation and promote the expression of transcription factor ATF4. Activation of the UPR alleviates the burden of misfolded and/or unfolded proteins, whereas translation inhibition reduces energy expenditure. Hypoxia also negatively regulates translation initiation by controlling the formation of the mRNA cap-binding eIF4F complex, comprising eIF4E, eIF4A and eIF4G. Formation of this complex is promoted by the release of eIF4E from its inhibitors, eIF4E-binding proteins (4E-BPs). This release is regulated by phosphorylation of 4E-BPs by mechanistic target of rapamycin (mTOR). Hypoxia inhibits mTOR activity, thereby interfering with eIF4E release. Hypoxia also promotes nuclear sequestration of eIF4E by its transporter, 4-ET. Elongation of mRNA translation is also regulated during hypoxia by modulating the activity of eukaryotic elongation factor 2 (eEF2) kinase (eEF2K) — an inhibitor of eEF2. eEF2K is negatively regulated by prolyl hydroxylases (PHDs) and mTOR, and inhibition of both during hypoxia increases eEF2K activity, which by phosphorylating eEF2, prevents mRNA elongation. Finally, translation termination is negatively affected by hypoxia due to decreased hydroxylation of eukaryotic release factor 1 (eRF1) by Jumonji domain-containing 4 (JMJD4) — α-ketoglutarate and Fe2+-dependent oxygenase that like PHDs is inhibited in hypoxia. Importantly, some mRNAs need to overcome the translation repression induced by hypoxia, prominently including those encoding mediators of hypoxia, such as hypoxia inducible transcription factor (HIF)-responsive genes. HIF-responsive mRNAs contain hypoxia response element (rHREs). Low O2 stimulates the formation of a complex including HIF-2α, RBM4, and eIF4E2 (eIF4E homolog) that assembles at these rHREs to promote translation initiation. Hypoxia also promotes a formation of hypoxia-specific eIF4F complex (eIF4FH) that binds rHREs. Selective hypoxia-responsive mRNA translation can also occur by direct binding of ribosomes to internal ribosome entry sites (IRES) encoded within the 5′-UTR of certain mRNAs (such as VEGF, eIF4G, and C-MYC). Adaptive protein synthesis during hypoxia is further regulated by the partitioning and recruitment of mRNAs to the ER by signal recognition particles (SRPs) which deliver mRNAs, such as those encoding VEGF, HIF1, and P4HA1 to SRP-binding proteins on the ER membrane.
Box 3: Hypoxic regulation of mTOR.
Mechanistic target of rapamycin (mTOR) controls biomass accumulation and metabolism by modulating key cellular processes. One of the two mTOR complexes, mTOR complex 1 (mTORC1) is inhibited by hypoxia, usually in conjunction with other cell stress stimuli, predominantly through its repressors, tuberous sclerosis protein 1 (TSC1) and TSC2. Acute hypoxic exposure rapidly and reversibly triggers hypophosphorylation of mTORC1 and its effectors eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs), p70 S6 kinase and eukaryotic translation initiation factor 4G (eIF4G), which is independent of AKT (a serine-threonine kinase involved in multiple processes, including metabolism, cell survival, and growth) and AMP-activated protein kinase (AMPK) phosphorylation, ATP levels, and hypoxia-inducible factor 1α (HIF-1α)42,135. In addition, chronic hypoxia exposure results in repression of mTORC1 signalling via two pathways, one being AMPK-dependent and the other through regulated in development and DNA damage response 1 (REDD 1). Upregulation of REDD 1 (refs. 217,218) liberates the TSC2 complex from the chaperone protein 14-3-3, allowing it to associate with TSC1 and repress mTORC1 signalling. Certain mutations in TSC2 confer a growth advantage to cells by repressing hypoxic mTORC1 inhibition and hypoxia-induced cell cycle arrest135. Under conditions of aberrant mTORC1 activation, hypoxia causes dephosphorylation of 4E-BP1/4E-BP2 and increases their association with eIF4E to suppress translation as a means of hypoxia tolerance219. The 3′-untranslated region of REDD 1 possesses microRNA (miRNA) binding sites that further contribute to its post-transcriptional regulation, with miR-7 identified as a repressor of REDD 1 expression. Under hypoxia, miR-7 expression is downregulated, resulting in elevated REDD 1 and consequent inhibition of mTORC1 signalling220.
Hypoxia may also negatively regulate mTORC1 through proteins that interfere with the interaction between mTORC1 and RHEB — a G protein that activates mTORC1 (refs. 221,222). In this case, mitochondrial protein BNIP3, which is induced in hypoxia by HIF-1 activation, directly binds to RHEB to inhibit the mTORC1 pathway223. The sequestering of RHEB away from mTORC1 is also observed when hypoxic cells acidify their microenvironment due to the release of lactate, driving the redistribution of perinuclear lysosomes away from perinuclear RHEB. This suppresses lysosome-bound mTORC1 activity (lysosome is the activation site for mTORC1)65. In addition, hypoxia results in Ataxia Telangiectasia Mutated (ATM)-dependent HIF-1α phosphorylation, which was shown to be required for REDD1 upregulation and downregulation of mTORC1 signalling224. While HIF-1α stability and transcriptional activity are directly regulated by O2 levels, its translation is heavily influenced by mTOR. Inactivating mutations in TSC1/TSC2 or PML — which negatively regulates mTORC1 association with RHEB — and activating mutations in mTOR increase HIF-1α expression to influence the hypoxia-induced transcriptional landscape221,225–227.
Inhibition of translation elongation and termination.
Alongside regulation at the initiation level, elongation of mRNA translation is also regulated during hypoxia. Peptide elongation is mediated by eukaryotic elongation factors (eEFs), and eEF2 has so far been shown to be regulated by variable O2 levels44,45. eEF2 is negatively regulated by eEF2 kinase (eEF2K). eEF2K is normally subject to proteasomal degradation via mTORC1, but mTORC1 inhibition in hypoxia (Box 3) results in eEF2K stabilization, increased eEF2K levels, increased eEF2 phosphorylation, and associated eEF2 inhibition44 (Figure 2) . In addition, PHD2 inhibits eEF2K to promote eEF2 activity46,47. Therefore, in low O2 conditions, when PHD2 activity is impaired, eEF2 becomes phosphorylated and inactivated (Figure 2). Finally, post-translational hydroxylation of eukaryotic release factor 1 (eRF1) by oxygen-sensitiveJMJD4 (Jumonji domain-containing 4) (Box 2) is required for efficient translation termination rates during normoxia48, and during hypoxia, decreased eRF1 hydroxylation leads to inefficient termination (Figure 2).
Adaptive protein synthesis in hypoxia.
Despite global shutdown of translation, it is important to note that not all mRNA translation is halted under hypoxia, which is crucial for cell survival when O2 levels are low. Overcoming translational repression during O2 starvation is needed for de novo synthesis of proteins essential for adaptive responses, such as ATF4 [G] (Activating Transcription Factor 4)49–51. ATF4 directly induces genes involved in protein synthesis, antioxidant response, amino acid transport (AAAT, SLC3A249) and metabolism (ASNS49), as well as autophagy as part of the integrated stress response [G] (ISR)49,52. Concurrently, translation of the HIF family of transcription factors and other proteins necessary for hypoxic responses is maintained to ensure cells have the correct repertoire of stress-responsive factors. Selective hypoxia-responsive mRNA translation can occur by the direct binding of ribosomes to internal ribosome entry sites (IRES) encoded within the 5′-untranslated region (UTR) of mRNAs, which allows specific mRNAs to bypass the requirement for eIF4F formation at the 5′ UTR cap structure53,54 (Figure 2). Genes utilizing IRES-dependent translation in hypoxia include VEGF55, eIF4G56, and C-MYC57. Preferential translation of HIF-α subunits during hypoxia was originally attributed to an IRES as well57,58, but it was later shown that HIF UTRs bind regulatory non-coding RNAs and/or RNA-binding proteins to regulate translation rates by interacting with HIF subunit mRNAs, potentially by creating mRNA loops to facilitate ribosome recycling59,60.
Oxygen levels have also been shown to impact the composition of the eIF4F complex, switching from eIF4A–eIF4E– eIF4G to eIF4A–eIF4E2–eIF4G3 (termed eIF4FH, where eIF4E2 and eIF4G3 are homologues to eIF4E and eIF4G), which was shown to selectively recruit mRNAs regulated by HIFs to the ribosome for translation, irrespective of the overall cellular transcription competency61 (Figure 2). Interestingly, along with its function as a transcription factor, HIF2-α is also a component of a cap-dependent translation initiation complex. Low O2 tension stimulates the formation of a complex that includes HIF2-α, the RNA-binding protein RBM4, and cap-binding eIF4E262,63. This complex assembles at the reverse HREs (rHREs) in RNA, allowing for evasion of hypoxia-induced protein synthesis repression (Figure 2). Furthermore, other physiological stresses associated with hypoxia, such as acidosis due to lactate accumulation, have been shown to affect gene expression changes in a hypoxia-independent manner64,65. This includes the upregulation of genes involved in translation, such as eIF4A2 and ribosomal protein L37 (RPL37). Of note, acidosis alone significantly suppresses peptide synthesis, much like hypoxia64,65.
Adaptive protein synthesis during hypoxia is further regulated by the partitioning and recruitment of mRNAs to the ER, a critical site of peptide synthesis. Signal recognition particles [G] (SRPs) bind to conserved sequences in the UTR and deliver mRNAs to SRP-binding proteins on the ER membrane66 (Figure 2). Examples of mRNAs that specifically localize to the ER in hypoxia for translation include VEGF, HIF1, and P4HA1, which further contribute to hypoxia-specific proteomic adaptations67.
Activation of ER quality control
Exposure to hypoxia, and other cell stresses, can lead to extensive protein modifications, such as protein oxidation, carbonylation and nitrosylation, and, as discussed above, accumulation of unfolded proteins, particularly at the endoplasmic reticulum (ER)68. The control and degradation of misfolded proteins, potentially as a consequence of hypoxia, is crucial to prevent detrimental consequences to cell function. Therefore, in order to alleviate ER stress, the UPR is triggered to serve as a critical cell survival mechanism by reducing protein load via decreasing protein synthesis and augmented protein degradation through ER-associated degradation [G] (ERAD) and autophagy68 through the PERK, IRE1α, and ATF6 signalling pathways69 (Figure 2; Supplementary Box 1).
As described in previous sections, during hypoxia, activated PERK phosphorylates eIF2α to attenuate translation initiation39. This transient halt in protein synthesis allows for energy conservation, decreased ER protein load as well as increased ribosomes available for mRNAs encoding UPR adaptive functions, such as ATF4 (refs. 49,50). Another UPR branch is activated by IRE1α, which following their dimerization and autophosphorylation70, generates a functional spliced XBP1 (XBP1s)71 that as a transcription factor regulates the expression of numerous genes maintaining ER and metabolic homeostasis72–78 (Figure 2). While the XBP1s transactivation domain interacts with RNA polymerase II, XBP1 can also physically interact with many other transcription factors, such as HIF-1α, where it regulates the expression of HIF-1α targets via the recruitment of RNA polymerase II79. Hypoxia induces XBP1 mRNA expression and splicing in a HIF-1α independent manner, resulting in increased levels of activated XBP1 protein, and XBP1 loss severely inhibits tumour growth due to a reduced capacity for tumour cells to survive in hypoxic microenvironments in vitro and in vivo79,80.
In contrast to the other two branches, ATF6, following its activation in the ER, translocates to the Golgi, where it is cleaved by two Golgi-resident proteases81,82. Subsequently, the N-terminal domain (ATF6f), comprising a transcriptional activation domain, basic leucine zipper (bZIP) domain, DNA-binding domain, and nuclear localization signals, translocates to the nucleus where it induces UPR target gene expression to increase ER folding and load capacity, including genes encoding XBP1 and C/EBP homologous protein (CHOP), the latter playing an important role in promoting UPR-induced apoptosis83–87 (Figure 2, Supplementary Box 1). Chronic ER stress mediates a pro-death response — likely via positive feedback loops allowing stabilization of pro-apoptotic transcripts, which are generally unstable88 — indicating that UPR activation plays a role in both adaptive and apoptotic responses under hypoxia40,89,90. Of note, cells with a compromised UPR, such as those with abrogated PERK and eIF2α signalling, are substantially more sensitive to ER-induced cell death compared to their wildtype counterparts, presumably due to proteotoxicity91. Altogether, the activation of UPR plays a major role in the response and adaptation of cells to hypoxia.
Changes to mitochondrial function
Mitochondrial electron transport chain [G] (ETC) in most cell types are the largest consumers of intracellular O2 for generation of ATP, i.e. oxidative phosphorylation. Thus, it can be expected that changes in O2 levels will affect mitochondrial ETC activity. Hypoxia controls ETC function at multiple levels, including the regulation of different mitochondrial ETC complexes and availability of TCA cycle reducing equivalents NADH and FADH2.
Modulation of ETC activity and the TCA cycle under hypoxia.
It is important to note that intracellular O2 levels at 0.3% begin to become rate limiting for ETC activity92. Complex IV (also known as cytochrome c oxidase or COX), the terminal complex within the ETC, donates four electrons to O2 producing two molecules of water. COX has a high affinity for O2 and an apparent Km close to 0.1% O2; thus, the ETC can function at near anoxic levels93 and cells largely maintain their ATP levels during hypoxia. Although hypoxia does not acutely inhibit ETC function, prolonged hypoxia — lasting over hours — can decrease ETC function. Surprisingly, reduction in ETC efficiency is independent of hypoxic limitation of O2 as a substrate for COX activity, but is mediated through the activation of HIF-1-dependent and HIF-1-independent mechanisms94. Of note, hypoxia has been shown to diminish the enzymatic maximal velocity [G] (Vmax) of isolated COX, suggesting an intrinsic O2 dependence of COX during prolonged hypoxia95. However, to ensure that the hypoxia-mediated suppression of COX Vmax does not prevent cells from meeting their metabolic demands, hypoxia induces switching between COX subunits, which is mediated by HIF-1 activation96 (Figure 3). COX is composed of 13 different subunits: 10 regulatory nuclear-encoded subunits and 3 catalytic subunits encoded in the mitochondrial DNA. Hypoxia-mediated HIF-1 activation induces the expression of the nuclear-encoded COX4 isoform 2 (COX4I2) subunit and the mitochondrial protease LON, which targets the alternative COX4 isoform 1 (COX4I1) for proteasomal degradation96. The incorporation of the COX4I2 subunit into COX allows for a more efficient transfer of electrons to O2 during hypoxia.
Figure 3: Impact of hypoxia on mitochondrial function.
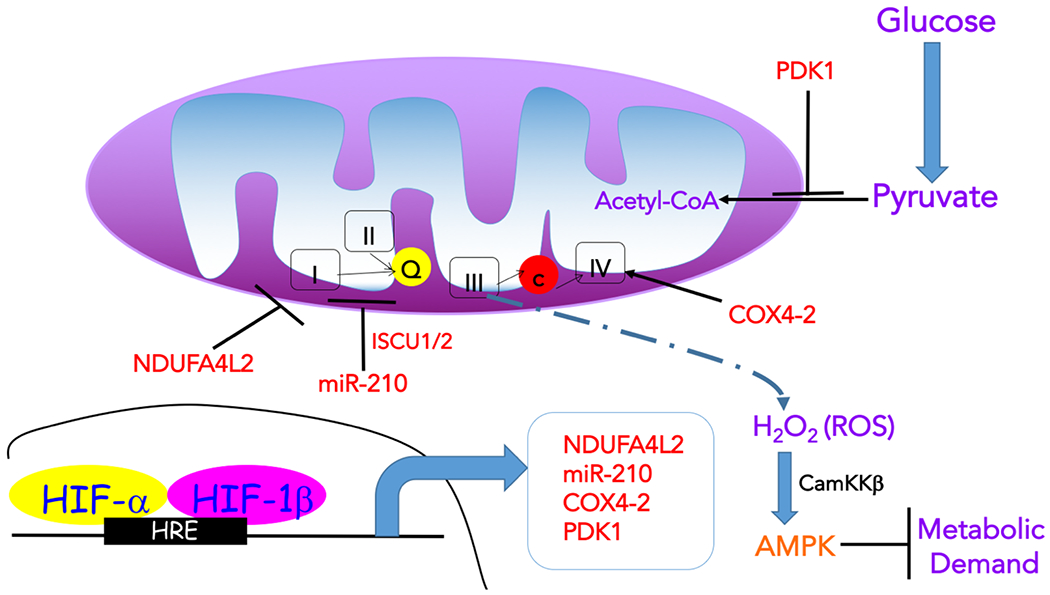
Hypoxia is characterized by decreased flux through the tricarboxylic acid (TCA) cycle in mitochondria, leading to the reduction of metabolites, such as acetyl-CoA and aspartate, required for anabolic processes. Specifically, hypoxia inducible transcription factor 1 (HIF-1) induces the expression of lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase 1 (PDK1), which then negatively regulates the entry of pyruvate to the TCA cycle (by promoting lactate generation and inhibiting its conversion to acetyl-CoA, respectively). Hypoxia also impacts on the activity of the electron transport chain (ETC). Under acute hypoxia, ETC activity is maintained by hypoxia-induced expression of the complex IV (COX) subunit COX4I2, which substitutes the COX4I1 subunit allowing for a more efficient transfer of electrons to oxygen (O2) during hypoxia. Another HIF-1-dependent protein that enhances COX activity through unknown mechanisms is the hypoxia-inducible gene domain family member 1A (HIGD1A). This allows cells to maintain energy homeostasis in the event of short-term stresses. However, under prolonged hypoxia, ETC activity is diminished by inducing NDUFA4L2 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2) in a HIF-1-dependent manner to decrease complex I activity and respiration, as well as several microRNAs (miRNAs), including mir-210, which represses ETC complex assembly. This repression of ETC activity has the potential benefit of decreasing the production of reactive oxygen species (ROS), which are generated by complex III under hypoxia (although mechanisms of hypoxic induction of ROS remain poorly understood) and can be potentially toxic by mediating damage to macromolecules. Nevertheless, hypoxia-induced ROS, and in particular hydrogen peroxide (H2O2), can also have various signalling roles. One of the consequences of mitochondrial ROS increase in hypoxia is the elevation in calcium (Ca2+), which activates Ca2+/calmodulin-dependent protein kinase kinase (CaMKK). CAMKK then activates AMP-activated protein kinase (AMPK) to suppress ATP consuming processes (metabolic demand). Ca2+ is also a signal for firing of carotid body nerves to stimulate ventilation and for vasoconstriction of pulmonary arteries. Moderate amounts of mitochondrial ROS were also shown to have anti-ageing functions. In addition to metabolism, hypoxia modulates mitochondrial dynamics (fission), likely to enhance quality control of damaged mitochondria via mitophagy, which can aid in limiting ROS generation. HRE, hypoxia response element; ISCU1/2, Iron-sulphur cluster assembly scaffold protein 1/2.
Another HIF-1-dependent protein that enhances COX activity through unknown mechanisms is the hypoxia-inducible gene domain family member 1A (HIGD1A)97. By contrast, hypoxia diminishes the activity of the other ETC complexes (I, II, and III). Hypoxia induces the mitochondrial NDUFA4L2 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, 4-like 2) gene in a HIF-1-dependent manner to decrease complex I activity and respiration through unknown mechanisms98. Moreover, hypoxia induces several microRNAs (miRNAs), including mir-210 through HIF-1, which represses ISCU1 and ISCU2, two assembly factors for iron-sulphur (Fe-S) clusters [G], ultimately disrupting the correct assembly of iron-sulphur clusters within ETC complexes I, II, and III99. mir-210 also represses the expression of the complex I subunit NDUFA4, the complex II subunit succinate dehydrogenase subunit D (SDHD), and the COX assembly protein COX10100,101 (Figure 3).
Beyond ETC modulation, hypoxia also controls pyruvate [G] entry into the TCA cycle through HIF-1 induction of lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase 1 (PDK1)102–104. LDHA converts pyruvate to lactate, thus diminishing pyruvate entry into the mitochondrial matrix. Pyruvate dehydrogenase (PDH) converts pyruvate into acetyl-CoA to initiate TCA cycle flux. PDK1 phosphorylates and inactivates the catalytic subunit of PDH, thus preventing conversion of pyruvate to acetyl-CoA. This results in decreased TCA cycle flux which limits the generation of mitochondrial NADH and FADH2. The decrease in the generation of reducing equivalents diminishes electron flux through the ETC (Figure 3). Another important consequence of reduced TCA flux during hypoxia is the diminished generation of aspartate from the TCA cycle metabolite oxaloacetate105. Aspartate is necessary for nucleotide synthesis and cell proliferation. Hypoxia decreases aspartate levels, which was shown to impair cell proliferation in vitro as well as tumour growth in mouse models. Aspartate levels negatively correlate with the markers of hypoxia in primary human tumours. Therefore, aspartate may be a limiting metabolite for tumour growth, and aspartate availability could be targeted for cancer therapy.
A major benefit of hypoxia diminishing respiratory rates is a resultant decrease in production of mitochondrial ROS103. Importantly, ROS at low levels initiate cellular signalling events (see below, the last subsection in this section) but at high levels ROS can initiate cell injury27. Thus, cells exposed to prolonged hypoxia activate HIF-1-dependent transcriptional targets, including HIGD1A, PDK1, COX4I2, and NDUFA4L2, to repress mitochondrial ROS production96,98,103,106. Accordingly, HIF-1 deficient cells that are exposed to hypoxia increase mitochondrial ROS to levels that might induce cell death103.
Changes in mitochondrial morphology in response to hypoxia.
Beyond effects on the TCA cycle and ETC function, hypoxia also controls mitochondrial morphology and quality control. Hypoxia promotes HIF-independent but CHCHD4-mediated perinuclear localization of mitochondria107,108. Mitochondria form tubular networks under normoxia, but undergo fission (fragmentation) under hypoxia109. During hypoxia, the GTPase dynamin-related protein 1 (DRP1) is recruited from the cytosol to the outer mitochondrial membrane by mitochondrial protein fission 1 (FIS1). FIS1, a protein localized to the outer mitochondrial membrane, is required for mitochondrial fission and serves as a receptor for DRP1. Importantly, the scaffolding protein AKAP121 (A Kinase Anchoring Protein 121) can direct cAMP-regulated protein kinase A (PKA)-mediated phosphorylation of DRP1, resulting in disruption of its association with FIS1 (ref. 110). In order to relieve DRP1 inhibition by PKA, hypoxia induces the activity of SIAH2, a E3 ubiquitin ligase that promotes the degradation of AKAP121, allowing for DRP1 to interact with FIS1, resulting in mitochondrial fission110. It is possible that hypoxia promotes mitochondrial fission in order to induce mitophagy, the selective, autophagic elimination of mitochondria that are dysfunctional, to limit ROS production during hypoxia. Indeed, hypoxia can activate mitophagy in certain cell types through Bnip3-like/NIP3-like protein X (BNIP3L/NIX), Bcl-2/Adenovirus E1B 19 kDa-interacting protein 3 (BNIP3), and FUN14 domain-containing protein 1 (FUNDC1)111,112. These proteins localize to the outer mitochondrial membrane and serve as receptors for the mitophagy machinery. Currently, the mechanisms underlying how hypoxia is sensed to trigger mitochondrial fission and mitophagy are not understood.
Hypoxia-induced production of ROS by mitochondria.
For decades, production of superoxide by the mitochondrial ETC and subsequent generation of H2O2 by superoxide dismutases [G]were viewed as a major culprit in causing age-related degeneration of organisms113. One of the pillars to support a mitochondrial free radical theory of ageing was the observation that hypoxia extended the lifespan of mammalian cells in vitro by decreasing ROS114. However, antioxidants (i.e. ROS scavengers) have consistently failed to provide any benefit to normal ageing or age-related diseases in humans or model organisms115. In addition, prolonged hypoxia as well as agents that cause an increase in mitochondrial ROS have now been shown to extend mammalian cell lifespan in vitro, as well as in Caenorhabditis elegans and mice116–118. Indeed, hypoxia increases mitochondrial ROS to induce HIF-dependent induction of human telomerase [G] (hTERT) gene expression to extend the lifespan of mammalian cells119. Moreover, mitochondrial ROS can activate HIF-1 to increase the lifespan of C. elegans120. These data have led to a model where generation of superoxide by mitochondrial ETC can result in H2O2-dependent signalling to trigger adaptation to hypoxia. However, as discussed above, this response has to be finely tuned as high rates of mitochondrial ETC generation of superoxide can eventually result in an overproduction of ROS to incur damage, and cells exposed to prolonged hypoxia will decrease the activity of ETC to limit ROS.
An important physiologic consequence of hypoxia-mediated increases in mitochondrial ROS is the accumulation of Ca2+ observed in cells possessing specialized O2-sensing properties, e.g. carotid body glomus type I cells and human pulmonary smooth muscle cells121. Recent studies demonstrate that these specialized cells express atypical mitochondrial ETC subunits, including NDUFA4L2, COX4I2 and COX8B, which contributes to their sensitivity to hypoxia and allows rapid responses to counteract hypoxic state122,123. In this manner, hypoxia potently induces the firing of carotid body nerves to stimulate ventilation and simultaneously cause vasoconstriction of pulmonary arteries. Conditional loss of NDUFS2 or COX4I2 in carotid body glomus cells diminishes hypoxic increases in mitochondrial ROS production which are necessary to stimulate minute ventilation122,124. Interestingly, Ndufs2-deficient mice show an increased ventilation upon exposure to hypercapnia (5% CO2) demonstrating that a lack of ventilatory response upon impairment of mitochondrial ROS generation is specific to hypoxia124. Genetic deletion of the Rieske-iron sulphur protein (RISP), a subunit of mitochondrial complex III, in pulmonary smooth muscle cells similarly attenuated both hypoxia-induced increases in ROS production and intracellular Ca2+ levels, which are necessary for hypoxia-induced pulmonary artery vasoconstriction125. Moreover, mice with either a smooth muscle-specific deletion of a complex I subunit (NDUFS4loss), a complex III subunit (RISP loss), or a complex IV subunit (COX4I2 loss) lack a hypoxia-induced increase in pulmonary arterial pressure122,126. Thus, mitochondrial ETC-dependent ROS production is required for regulating the function of cells dedicated to O2 sensing for organismal responses.
Interestingly, starting at 3% O2, when O2 is not limiting for ETC function, cells have developed mechanisms to decrease their cellular metabolic demand94. This response does not occur within seconds, but instead takes minutes to hours, involving a combination of transcriptional and post-translational mechanisms. By decreasing demand for ATP by suppressing ATP consuming processes, cells lower their rate of O2 consumption and delay the development of anoxia which, unlike hypoxia, can cause cell death. Work from the 1950’s proposed that cellular ATP utilization is a major determinant of cellular respiratory rate, controlled by cellular ATPases127. Indeed, a plasma membrane-localized sodium-potassium ATPase (Na/K-ATPase) can account for 20–70% of the O2 expenditure of mammalian cells128. The Na/K-ATPase, a heterodimer composed of a catalytic α-subunit and a β-subunit, transports sodium (Na+) and potassium (K+) ions across the plasma membrane to maintain an essential electrochemical gradient [G]. Multiple investigators have reported that hypoxia reversibly rapidly suppresses the activity of the Na/K-ATPase by causing endocytosis of the α-subunit from the plasma membrane. This has been shown to depend on the activation of a key cellular energy sensor, AMP-activated protein kinase (AMPK)129–132. AMPK is a heterotrimeric serine/threonine protein kinase composed of a catalytic α-subunit and two regulatory β- and γ-subunits. Mitochondrial complex III-generated superoxide was shown to potently activate AMPK130–133. AMPK is stimulated either by an increase in AMP-to-ATP ratio or by an increase in Ca2+through the Ca2+/calmodulin-dependent protein kinase kinase (CaMKK)134. Multiple studies have demonstrated, that acute hypoxia (low O2 levels for a few minutes to several hours) does not alter AMP-to-ATP ratio130,135, but rather increases intracellular Ca2+ levels, leading to CaMKK-dependent AMPK activation131,132 (Figure 3). AMPK is typically activated under nutrient limiting conditions to concomitantly suppress ATP consuming processes, such as protein translation, and promote autophagy to provide intracellular nutrients134,135. Thus, hypoxic activation of AMPK is key for reducing cellular metabolic demand under low oxygen conditions.
The mechanisms underlying hypoxic increases in mitochondrial superoxide production are not fully understood. Mitochondrial complex I, II, and III have all been implicated in superoxide generation in multiple cell types; however, complex III is implicated in the majority of these studies20,125,136. In particular, the ubisemiquinone [G] Qo site within mitochondrial complex III is the only known site in the entire ETC capable of release ROS from the mitochondria, producing superoxide into mitochondrial intermembrane spaces as opposed to mitochondrial matrix137. These ROS then enter the cytosol through voltage dependent anion channels (VDAC) located on outer mitochondrial membranes. A recent report proposed an interesting mechanism by which low O2 may increase mitochondrial ROS. It was shown that hypoxia stimulated Na+ entry into the mitochondrial matrix led to reduced mitochondrial inner membrane fluidity via its interaction with lipids138. This resulted in the entrapment and accumulation of ubisemiquinone at the Qo site of mitochondrial complex III to increase superoxide production during hypoxia. Importantly, inhibition of the Na+/Ca2+ exchanger NCLX prevented hypoxia-induced import of Na+ to the mitochondrial matrix, resulting in a decrease in mitochondrial ROS production and HIF-1α protein stabilization. Going forward, the advent of new genetic and pharmacologic tools that diminish the production of mitochondrial superoxide at complex I or III should further demonstrate the in vivo importance of mitochondrial ROS in physiology136,139.
Impact on carbon and lipid metabolism.
Reprogramming intracellular metabolism is a common feature of O2 deprivation140 (Figure 4). These hypoxic mechanisms are of particular relevance to cancer. Disorganized blood vessel formation as well as mismatched rates of cell proliferation and vascular sufficiency lead to stressful microenvironments in solid tumours, where cells are subjected to both O2 and nutrient starvation. Hence, cancer cells take advantage of these metabolic adaptations to fuel survival and/or proliferation to ensure tumour progression in these unfavourable conditions. One of the most prominent adaptations in O2 starved cells is increased glucose uptake and elevated glycolytic flux to promote glucose catabolism. HIF-1 target genes involved in this metabolic reprogramming include those encoding glucose transporters 1 and 3, hexokinase 1 and 2, enolase 1, phosphoglycerate kinase 1, pyruvate kinase M2, and lactate dehydrogenase A141,142. In addition, as mentioned above, PDK1 inhibits the enzymatic activity of pyruvate dehydrogenase, thereby blocking the conversion of pyruvate to acetyl-CoA for entry into the TCA cycle, favouring generation of lactate. Lactate and H+ generated by glycolysis are exported from the cell through the activity of monocarboxylic transporter 4 (MCT4), sodium hydrogen (Na+/H+) exchanger (NHE) isoform 1 (NEH1), and carbonic anhydrase 9 (CAR9). Extracellular lactate can then be taken up by other cancer cells as well as stromal cells, where it is used as a carbon source for the TCA cycle (a mechanism known as anaplerosis)143,144. Strikingly, human non-small-cell lung cancers not only employ lactate as a fuel source, but they incorporate more lactate-derived carbons into TCA cycle intermediates than those from glucose, indicating that lactate can be a preferred anaplerotic substrate143. Furthermore, acidification of the microenvironment by secreted lactate and H+ is significant in that it adversely affects the functionality of infiltrating T cells, resulting in tumour immune evasion140. Similarly, hypoxia induces the uptake of glutamine, a principle anaplerotic substrate for the TCA cycle, by promoting increased expression of glutamine transporters, such as SLC1A5 and SNAT2/SLC38A2 (refs. 145,146). In this way, cells are able to continually produce TCA metabolites, such as citrate, which is subsequently converted into cytosolic acetyl-CoA for anabolic reactions, such as lipid synthesis (lipogenesis). This is critical because of enhanced PDK1 activity and reduced entry of pyruvate-derived acetyl-CoA into the TCA cycle in O2-starved cells; glutamine anaplerosis can therefore maintain lipid homeostasis. Hypoxia, via HIFs, also induces the gene encoding the E3 ubiquitin-protein ligase SIAH1, which triggers ubiquitylation and degradation of the E1 subunit of α-ketoglutarate dehydrogenase, thereby promoting reductive carboxylation of glutamine-derived α-ketoglutarate to citrate, for acetyl-CoA and lipid synthesis146. Finally, HIFs induce the expression of fatty acid synthase (FASN) to stimulate fatty acid synthesis and stearoyl-CoA desaturase (SCD) to drive generation of unsaturated fatty acids142. However, as stated below, SCD activity becomes compromised under hypoxic conditions affecting the ratio of saturated to unsaturated fatty acids, plasma and organelle membrane integrity, and cell function. At the same time, HIF-1 decreases fatty acid β-oxidation and diversion of fatty acids towards ATP production by repressing the expression of acyl-CoA dehydrogenases147.
Figure 4. Overview of sugar and lipid metabolic pathways affected by oxygen availability.
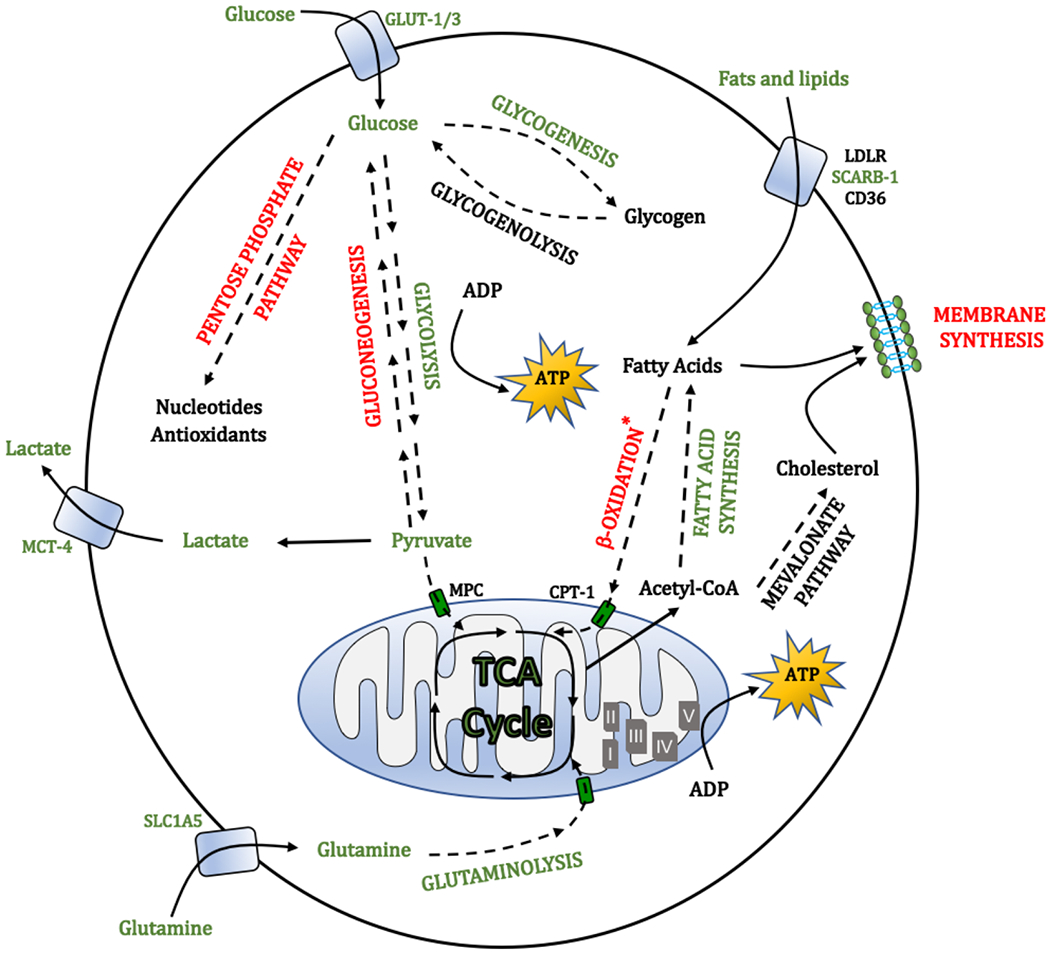
The figure highlights the upregulated (green) and downregulated (red) metabolic pathways under hypoxia. Hypoxic cells increase the uptake and utilization of glucose via glycolysis to obtain energy, while reducing their metabolism in mitochondria (Fig. 3). This leads to the generation of large amounts of lactate, which is secreted and can support metabolism of neighbouring cells. As a protective mechanism, glucose is also diverted towards the serine synthesis pathway to overcome the loss of cellular antioxidant capacity when pentose phosphate pathway activity is decreased. Hypoxia also promotes glycogenesis, which could provide a mechanism of energy storage to survive prolonged stress. Under hypoxic inhibition of the tricarboxylic acid (TCA) cycle, generation of anabolic metabolites, such as acetyl-CoA, which is key for the synthesis of fatty acids, is largely supported by the increased uptake and metabolism of glutamine. Concomitantly, catabolism of fatty acids via β-oxidation is suppressed, while the uptake of lipids from the exterior increased. Lipid desaturation, allowing generation of unsaturated fatty acids is inhibited in hypoxia (owing to the requirement for oxygen by stearoyl-CoA desaturase). To counteract potential lipotoxicity associated with the accumulation of saturated lipids and disruption of membrane structure and integrity, cells increase the uptake of unsaturated lipids from the environment and increase the formation of lipid droplets, which can act as buffers for saturated lipid species. ADP, adenosine diphosphate; ATP, adenosine triphosphate; CD36, cluster of differentiation 36; CPT-1, carnitine palmitoyltransferase 1; ETC, electron transport chain; GLUT-1, glucose transporter 1; LDLR, low density lipoprotein receptor; MCT4, monocarboxylate transporter 4; MPC, mitochondrial pyruvate carrier; SCARB-1, scavenger receptor B1; SLC1A5, solute carrier family 1 (neutral amino acid transporter) member 5.
Lipid availability is important for highly proliferating cells as fatty acids and cholesterol support the production of organelle and plasma membranes and modulation of their fluidity. It is noteworthy that SCD is an O2-consuming enzyme148. Thus, SCD enzymatic activity becomes compromised in low O2 conditions148,149 and to counteract this effect, cancer cells can elevate SCD expression as shown in certain tumours150. Otherwise, build-up of saturated fatty acid precursors is potentially toxic and can impair cellular function, which is associated with disruption of ER membranes and apoptosis148,151. Saturated fatty acid-induced toxicity can be alleviated by supplying exogenous unsaturated lipids, indicating that lipid uptake is an important mechanism for maintaining homeostasis in hypoxic cells148. In addition, recent studies indicate the importance of lipid droplets [G] in alleviating lipotoxicity, including in cancer152. Specifically, the composition of triglycerides is altered to buffer changes in fatty acid saturation, by selectively retaining excess saturated fatty acids in lipid droplets153. That is, unsaturated oleate is preferentially liberated from existing triglycerides to facilitate sequestration of saturated fatty acids and used to counterbalance increased membrane phospholipid saturation153. Transcriptional profiling indicated that the lipid droplet protein perilipin 2 (PLIN2) is elevated in kidney tumours, correlating with increased HIF-2α accumulation, and HIF-2α-dependent PLIN2 expression promotes triglyceride and cholesterol ester storage in lipid droplets, needed for ER membrane integrity and suppression of toxic ER stress responses152. The ER stress resulting from lipotoxicity also engages the IRE1α–XBP1 pathway through multiple molecular mechanisms as shown in a variety of cancers75,148,149,152, suggesting that inhibiting the IRE1α–XBP1 pathway is a useful general strategy for treatment of a variety of tumours experiencing O2 deprivation.
Hypoxic reprogramming of metabolism is also associated with the adaptation to excessive ROS production, which as discussed above, accompanies mitochondrial changes in hypoxia. In this case, hypoxia decreases the expression of glucose-6-phosphate dehydrogenase, thereby decreasing pentose phosphate pathway [G] activity. This inevitably reduces the generation of nucleotides and cell proliferation. However, at the same time, induction of phosphoglycerate dehydrogenase expression by hypoxia allows diversion of glucose towards serine synthesis for a robust antioxidant response, promoting stress resistance154. Glucose can also be diverted towards glycogen synthesis under hypoxia by overexpression of phosphoglucomutase 1 and glycogen synthase 1, which can serve as a preconditioning mechanism that allows the build-up of glucose stores preparing cells for conditions of glucose deprivation141.
Regulation of nutrient use
Due to various intracellular and extracellular stimuli that a cell encounters, including hypoxia and nutrient deprivation, it is critical to maintain cellular protective and adaptive mechanisms to resist stressful changes. Adaptation and survival of cells in such a heterogenic microenvironment requires the coordination of several stress response pathways including regulating nutrient use.
Activation of autophagy.
Macroautophagy (hereafter termed autophagy) is a key process whereby cytosolic components, such as proteins and organelles, are captured in double-membrane vesicles (autophagosomes) and fuse with lysosomes to form autolysosomes. These contents are subsequently catabolized by lysosomal degradative enzymes into products, such as amino acids, carbohydrates and fatty acids that contribute to organelle/protein turnover and nutrient recycling155–157. Under physiological conditions, autophagy is maintained at a low basal rate as part of quality control pathways to remove damaged proteins and organelles158,159. However, it potently responds to external cellular microenvironments and can be influenced by nutrient and O2 availability to promote cell adaptation and survival157,160.
BNIP3 emerged as a HIF-1α target. Accordingly, it is highly elevated in severe hypoxic conditions (~0.1-1% O2) in various cell lines and has pro-survival functions by mediating hypoxia-induced autophagy161–164. Closely-related BNIP3L is also induced by hypoxia162,165, indicating both proteins are necessary for autophagy under these stressful circumstances166. HIF-1α-dependent expression of BNIP3 has also been described as essential in mitophagy as previously mentioned161,167,168.
Severe hypoxic conditions, often accompanied by drastic nutrient depletion, lead to autophagy through HIF-independent mechanisms, such as the UPR (Supplementary Box 1). Upon ER stress, PERK signalling stimulates the translation of ATF4, resulting in the induction of the downstream gene encoding for the transcription factor CHOP, which is responsible for initiation of the apoptotic cascade under prolonged ER stress50,169. Furthermore, AMPK, which is activated during metabolic stress and hypoxia as discussed above, is also a regulator of autophagy170,171. The best studied mechanism by which AMPK regulates autophagy is through the suppression of the mTOR signalling pathway170. AMPK also targets the unc-51-like kinase 1 (ULK1) complex, an initiator kinase during mammalian autophagy. ULK1 is directly phosphorylated by AMPK to maintain autophagic function and mitochondrial homeostasis172–174. Altogether, hypoxia activates autophagy via several mechanisms. This allows the removal of damaged organelles (such as mitochondria through mitophagy, see subsection Changes in mitochondrial morphology in response to hypoxia) and release of nutrients for ongoing cell viability175, that promote cell survival in stressful environments (Figure 5).
Figure 5: Regulation of nutrient acquisition and use under hypoxia.
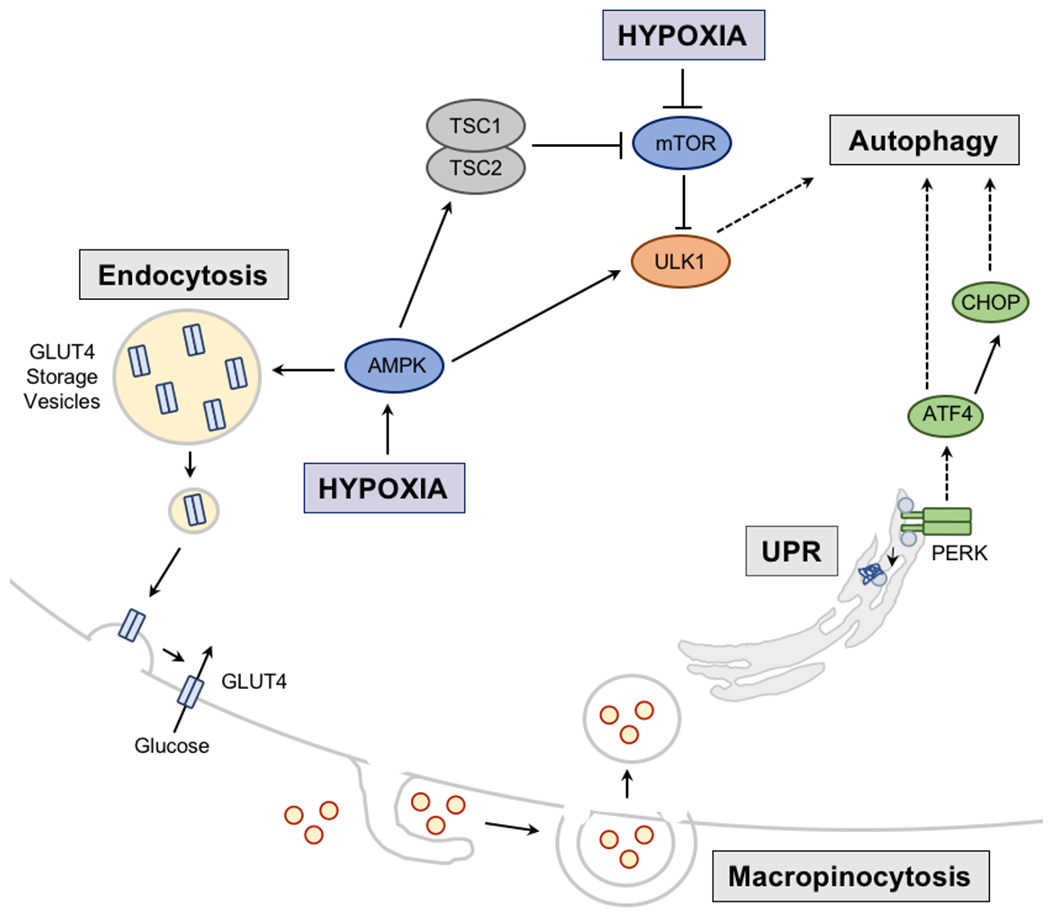
Hypoxia induces nutrient recycling via autophagy. In this case, the unfolded protein response (UPR) resulting from hypoxia, activates protein kinase RNA-like ER kinase (PERK), which induces transcription factor ATF4 and subsequently C/EBP homologous protein (CHOP) to activate autophagic machinery. Hypoxia, via activation of hypoxia inducible factors (HIFs) further regulates autophagy by HIF-mediated regulation of expression of two mitochondrial proteins BNIP3 and BNIP3L that have been linked to autophagy of mitochondria (mitophagy). Autophagy can also be induced during hypoxia via activation of AMP kinase (AMPK) which subsequently activates autophagy directly or indirectly through inhibition of mechanistic target of rapamycin (mTOR), a negative regulator of the Unc-51 like autophagy activating kinase 1 (ULK1). AMPK can also promote the movement of the glucose transporter GLUT4 from its storage vesicles to the cell membrane to promote glucose uptake. Nutrient acquisition is also achieved via macropinocytosis, which is promoted by hypoxia and supports the ingestion of extracellular macromolecules, such as amino acids and fatty acids. TSC1/2, Tuberous sclerosis 1/2.
Regulation of nutrient uptake.
The cellular uptake rate of many nutrients and ions is governed primarily by membrane transporters and receptors that show dynamic localization at both the plasma membrane and defined intracellular membrane compartments. Regulation of the rate of endocytosis and the inverse process, recycling of the endocytosed components back to the plasma membrane, controls the amounts of these cell surface proteins, which in many cases determines nutrient uptake or excretion176. Here we discuss the contribution of endocytic trafficking of glucose transporter GLUT4, which has been well-characterized in hypoxia (Figure 5).
GLUT4 is highly expressed in adipose tissue and skeletal muscle, and is responsible for the majority of glucose uptake to maintain blood glucose levels, thus making it a major regulator of systemic glucose homeostasis. GLUT4 membrane localization is normally tightly regulated by an endosomal pathway, whereby GLUT4 is endocytosed and sequestered to specific intracellular compartments177. Insulin and exercise stimulate GLUT4 redistribution from intracellular compartments to the cell surface to enhance glucose uptake via the AKT pathway178,179. In addition, low energy signalling, such as decreased ATP levels, lead to increased cell surface GLUT4 via activation of AMPK that promotes GLUT4 recycling to the plasma membrane180,181 (Figure 5). Changes in GLUT4 distribution can also be influenced by HIFs and hypoxia. Loss of HIF-1α in myocytes impairs GLUT4 vesicle mobilisation to the plasma membrane due to decreased AMPK activation upon insulin stimulation182. Furthermore, HIF-1α was shown to increase the expression of RAB20 that promotes GLUT4 translocation to the plasma membrane and to negatively regulate TXNIP expression to promote AKT downstream signalling183. In a number of murine studies, long- and short-term chronic intermittent hypoxia triggers AKT and AMPK pathway activation as well as changes in GLUT4 expression in skeletal muscle184–186. Hypoxia can support adaptation of cells to replenish ATP required for cell energetics — such as muscle contraction — by providing ample glucose. By contrast, in adipose tissue, hypoxic response was shown to be linked to obesity and glucose intolerance, as mice with adipose-specific HIF-1β knock-out were protected from age- and diet-induced glucose intolerance, in part due to decreased GLUT4 expression187.
In addition to the recycling of membrane proteins, cells also utilise a form of endocytosis known as pinocytosis (hereafter termed macropinocytosis) to ingest extracellular liquid and nutrients. While macropinocytosis can occur at basal rates, it can also be induced by the GTPase RAS, which has well-studied roles in cell growth, differentiation, and survival, and is a potent oncogene188,189. Notably, in cancer cells harbouring oncogenic RAS mutations, macropinocytosis serves as a mechanism to take up amino acids for cell growth190–192. In addition, hypoxia also promotes the macropinocytosis of unsaturated fatty acids to bypass the inhibition of O2-dependent fatty acid desaturases and prevent lipotoxicity as discussed above148,153 by scavenging serum (Figure 5). Interestingly, oncogenic RAS mutations can mimic the effects of hypoxia on fatty acid scavenging. Overall, given that tumours must sustain growth under nutrient- and O2-deprived conditions, macropinocytosis provides a mechanism for cells to maximize macromolecule uptake from the extracellular space. How RAS mutants and hypoxia regulate macropinocytosis-driven lipid scavenging is currently unknown.
Conclusions and perspective
In summary, acute and chronic hypoxia induce a myriad of responses on a cellular level, and our understanding of how they are coordinated will continue to be investigated. The discovery of HIFs and their regulation by the PHD–pVHL axis has solidified our understanding of how cells autonomously respond to hypoxia at the transcriptional level. However, while the bases of hypoxic gene expression have been described in some detail, new areas of investigation include further dissection of how O2 availability influences O2 sensing, and how ROS and metabolite levels affect multiple distinct but integrated adaptations. We highlight here a few specific areas that in our view are now ripe for investigation. First, it should be addressed which of the >70 α-ketoglutarate-dependent dioxygenases are truly affected by fluctuating oxygen levels encountered in normal and disease states and in which context these sensors are important. In light of links between metabolism and chromatin regulation193 it is warranted to further address how O2-regulated metabolic adaptations affect the epigenome, and whether these changes are functional. Another important question relates to the close integration of O2 and nutrient deprivation and it would be interesting to study how these two stress signals impact on each other and cooperate. As cells in vivo do not reside in isolation, it will be also crucial to understand interactions between multiple cell types in hypoxic microenvironments, which could shed new light on the mechanisms of inflammation, cancer, and other pathologies. Understanding the transcriptional regulation by HIFs has provided therapeutic targets currently under clinical evaluation194,195. Notably, HIF-2α inhibitors are being tested in renal cell carcinoma and pharmacologic inhibition of PHDs is being explored for the treatment of anaemia196,197. However, as conveyed in this article, cellular responses to hypoxia go much beyond the transcriptional networks governed by HIFs. Over the next decade, as more of these mechanisms are elucidated, it is very likely that more clinical opportunities for various diseases will arise. For example, hypoxic exposure increases the survival of mice resembling paediatric diseases linked to mitochondrial dysfunction, such as Leigh syndrome [G], through poorly understood HIF-independent mechanisms198,199. Here, HIF activation is insufficient to rescue disease, and alternative strategies that actually reduce O2 delivery or modulate more specific pathways downstream of hypoxia are needed. Finally, it would be interesting to study mechanisms of response to increased oxygen levels. For example, age-dependent reduction in whole body oxygen consumption can result in tissue hyperoxia198. This raises the question of mechanisms mediating adaptations to this state, particularly in the brain. Addressing these questions will greatly contribute to our understanding of cellular responses to O2 — an integral molecule of aerobic life.
Supplementary Material
Acknowledgements
N.S.C. acknowledges support by R35CA197532, while M.S.C. is supported by P01CA104838 and R35CA220483 from the National Institutes of Health. P.L. is supported by T32AR53461-10 and F32CA217185-02. The authors have no financial interests to disclose. We apologize to those authors whose research could not be directly cited due to space limitations.
Glossary
- Dioxygenases
A group of enzymes that reduce molecular oxygen by incorporating both oxygen atoms into their substrate(s)
- von Hippel–Lindau (VHL) tumour suppressor protein
(pVHL) A protein named for the physicians von-Hippel and Lindau, who characterised patients with highly vascular neoplasia of the kidney, eye, and central nervous system that carried mutations in VHL gene. pVHL is required for the ubiquitylation of hypoxia-inducible factor α and its degradation.
- Carotid body
A cluster of peripheral chemoreceptor cells (glomus type I and glomus type II), which sense oxygen, carbon dioxide and pH levels of blood.
- Erythropoietin
A glycoprotein cytokine secreted by the kidney in response to hypoxia to stimulate erythropoiesis.
- Elongin BC-CUL2
Additional complexes that interact with pVHL. The Elongin BC complex acts as an adaptor connecting Cullin (Cul) proteins.
- The Michaelis constant
(Km) is defined as the substrate concentration at half the maximum reaction velocity.
- Acidosis
The process or condition where there is increased acidity in the blood and other body tissues.
- Cap-dependent protein synthesis
Eukaryotic mRNAs contain a modified guanosine (the cap) at their 5′ ends. Cap-dependent translation requires the binding of an initiation factor, eIF4E, to the cap structure.
- PERK
A sensor of ER stress and stress-induced protein misfolding.
- mTOR
Mechanistic target of rapamycin (mTOR) is a protein kinase that regulates protein synthesis and cell growth in response to growth factors, nutrients, energy levels, and stress.
- eukaryotic initiation factor 2: (eIF2)
A complex comprising α, β, and γ components that integrates a diverse array of stress-related signals to regulate both global and specific mRNA translation.
- ER stress
A condition when the capacity of the endoplasmic reticulum (ER) to fold proteins becomes saturated due to impaired protein glycosylation or disulphide bond formation, or by overexpression of or mutations in proteins entering the secretory pathway
- eIF4F
The cap-binding eukaryotic translation initiation factor 4F (eIF4F) complex consists of three subunits, eIF4A, eIF4E, and eIF4G. eIF4G strongly associates with eIF4E, the protein that directly binds the mRNA cap.
- Activating Transcription Factor 4: (ATF4)
A cAMP-response element binding protein that belongs to the cAMP response element-binding protein (CREB)-2 family of transcription factors.
- Integrated stress response: (ISR)
An adaptive pathway to restore cellular homeostasis by optimizing the cellular response to stress. Its activity is dependent on the cellular context and the type as well as intensity of the stress stimuli.
- Signal recognition particles
Universally conserved ribonucleoproteins that recognizes and targets specific proteins to the endoplasmic reticulum
- ER-associated degradation: (ERAD)
The cellular pathway which targets misfolded proteins of the endoplasmic reticulum for ubiquitination and subsequent degradation by the proteasome.
- Electron transport chain: (ETC)
A series of complexes within the inner mitochondrial membrane that shuttles electrons from NADH and FADH2 to molecular oxygen.
- Enzymatic maximal velocity
(Vmax) The reaction rate when an enzyme is fully saturated by substrate.
- Iron sulphur (Fe-S) clusters
Molecular ensembles of iron and sulfide, often found as components of electron transfer proteins. The ferredoxin proteins are the most common Fe–S clusters in nature.
- Pyruvate
The conjugate base, CH3COCOO−, of pyruvic acid, is a key intermediate in several metabolic pathways throughout the cell.
- Superoxide dismutases
A family of enzymes that catalyze the dismutation of the superoxide (O2−) radical into molecular oxygen (O2) or hydrogen peroxide (H2O2).
- Telomerase
A ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3’ end of a region of repetitive sequences at each end of chromosomes (telomeres).
- Essential electrochemical gradient
A gradient of electrochemical potential, usually for an ion that can move across membranes
- Ubisemiquinone
Ubiquinol is the reduced form of coenzyme Q10 that is oxidized by mitochondrial complex III to the partially reduced form ubisemiquinone and subsequently to the fully oxidized ubiquinone. Ubisemiquionone is a highly unstable free radical that can donate electrons to molecular oxygen to generate superoxide.
- Lipid droplets
Lipid-rich storage organelles that regulate the storage and hydrolysis of neutral lipids. They also serve as a reservoir for cholesterol and acyl-glycerols for membrane formation and maintenance.
- Pentose phosphate pathway
A predominantly anabolic pathway parallel to glycolysis that generates NADPH and precursors for nucleotide synthesis.
- Leigh syndrome
A neurological disorder characterized by progressive loss of mental and motor abilities.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Molecular Cell Biology thanks C. Taylor, M. Ashcroft and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41580-020-0227-y
References
- 1.Pouyssegur J & López-Barneo J Hypoxia in health and disease. Molecular Aspects of Medicine 47–48, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Taylor CT, Doherty G, Fallon PG & Cummins EP Hypoxia-dependent regulation of inflammatory pathways in immune cells. J. Clin. Invest. 126, 3716–3724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keith B, Johnson RS & Simon MC HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 12, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelin WG et al. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402 (2008). [DOI] [PubMed] [Google Scholar]; Reviews by Semenza (2012) and Kaelin et al, (2008) provide an excellent outline of the regulation and importance of HIFs in physiology and diseases.
- 6.Markolovic S, Wilkins SE & Schofield CJ Protein Hydroxylation Catalyzed by 2-Oxoglutarate-dependent Oxygenases. J. Biol. Chem. 290, 20712–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda K et al. Placental but Not Heart Defects Are Associated with Elevated Hypoxia-Inducible Factor Levels in Mice Lacking Prolyl Hydroxylase Domain Protein 2. Mol. Cell. Biol. 26, 8336–8346 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lando D, Peet DJ, Whelan DA, Gorman JJ & Whitelaw ML Asparagine Hydroxylation of the HIF Transactivation Domain: A Hypoxic Switch. Science (80-. ). 295, 858–861 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Zhang N et al. The Asparaginyl Hydroxylase Factor Inhibiting HIF-1α Is an Essential Regulator of Metabolism. Cell Metab. 11, 364–378 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockman ME, Webb JD, Kramer HB, Kessler BM & Ratcliffe PJ Proteomics-based Identification of Novel Factor Inhibiting Hypoxia-inducible Factor (FIH) Substrates Indicates Widespread Asparaginyl Hydroxylation of Ankyrin Repeat Domain-containing Proteins. Mol. Cell. Proteomics 8, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockman ME et al. Lack of activity of recombinant HIF prolyl hydroxylases (PHDs) on reported non-HIF substrates. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee FS Substrates of PHD. Cell Metab. 30, 626–627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI & Myllyharju J Characterization of the Human Prolyl 4-Hydroxylases That Modify the Hypoxia-inducible Factor. J. Biol. Chem. 278, 30772–30780 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Ast T & Mootha VK Oxygen and mammalian cell culture: are we repeating the experiment of Dr. Ox? Nat. Metab 1, 858–860 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Koivunen P, Hirsilä M, Günzler V, Kivirikko KI & Myllyharju J Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J. Biol. Chem. 279, 9899–904 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Jiang BH, Semenza GL, Bauer C & Marti HH Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am. J. Physiol. Physiol 271, C1172–C1180 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Pan Y et al. Multiple Factors Affecting Cellular Redox Status and Energy Metabolism Modulate Hypoxia-Inducible Factor Prolyl Hydroxylase Activity In Vivo and In Vitro. Mol. Cell. Biol. 27, 912–925 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagen T, Taylor CT, Lam F & Moncada S Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science 302, 1975–8 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Yang J et al. Human CHCHD4 mitochondrial proteins regulate cellular oxygen consumption rate and metabolism and provide a critical role in hypoxia signaling and tumor progression. J. Clin. Invest. 122, 600–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandel NS et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 275, 25130–25138 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Brunelle JK et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab 1, 409–414 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Chandel NS et al. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A 95, 11715–11720 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]; A key paper initially linking mitochondria to hypoxic induced gene transcription.
- 23.Mansfield KD et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab 1, 393–399 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzy RD et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab 1, 401–408 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Lin X et al. A chemical genomics screen highlights the essential role of mitochondria in HIF-1 regulation. Proc. Natl. Acad. Sci. 105, 174–179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee G et al. Oxidative Dimerization of PHD2 is Responsible for its Inactivation and Contributes to Metabolic Reprogramming via HIF-1α Activation. Sci. Rep. 6, 18928 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reczek CR & Chandel NS ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33, 8–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs KJ et al. Paracrine Induction of HIF by Glutamate in Breast Cancer: EglN1 Senses Cysteine. Cell 166, 126–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King A, Selak MA & Gottlieb E Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25, 4675–4682 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Selak MA et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7, 77–85 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Pollard PJ et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 14, 2231–2239 (2005). [DOI] [PubMed] [Google Scholar]; Selak et al. (2005) and Pollard et al. (2005) provided initial connection of mitochondrial TCA cycle metabolites to regulation of PHDs.
- 32.Ryan DG et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat. Metab. 1, 16–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Intlekofer AM et al. L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 13, 494–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadtochiy SM et al. Acidic pH Is a Metabolic Switch for 2-Hydroxyglutarate Generation and Signaling. J. Biol. Chem. 291, 20188–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonenberg N & Hinnebusch AG Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson RJ, Hellen CUT & Pestova TV The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Beucken T et al. Translational control is a major contributor to hypoxia induced gene expression. Radiother. Oncol. 99, 379–384 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Feldman DE, Chauhan V & Koong AC The unfolded protein response: A novel component of the hypoxic stress response in tumors. Molecular Cancer Research 3, 597–605 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Brewer JW & Diehl JA PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. 97, 12625–12630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blais JD et al. Perk-Dependent Translational Regulation Promotes Tumor Cell Adaptation and Angiogenesis in Response to Hypoxic Stress. Mol. Cell. Biol. 26, 9517–9532 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koumenis C et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol. Cell. Biol. 22, 7405–16 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arsham AM, Howell JJ & Simon MC A Novel Hypoxia-inducible Factor-independent Hypoxic Response Regulating Mammalian Target of Rapamycin and Its Targets. J. Biol. Chem. 278, 29655–29660 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Koritzinsky M et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 25, 1114–25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connolly E, Braunstein S, Formenti S & Schneider RJ Hypoxia Inhibits Protein Synthesis through a 4E-BP1 and Elongation Factor 2 Kinase Pathway Controlled by mTOR and Uncoupled in Breast Cancer Cells. Mol. Cell. Biol. 26, 3955–3965 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenney JW, Moore CE, Wang X & Proud CG Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv. Biol. Regul. 55, 15–27 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Moore CEJ et al. Elongation Factor 2 Kinase Is Regulated by Proline Hydroxylation and Protects Cells during Hypoxia. Mol. Cell. Biol. 35, 1788–1804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero-Ruiz A et al. Prolyl Hydroxylase-dependent Modulation of Eukaryotic Elongation Factor 2 Activity and Protein Translation under Acute Hypoxia. J. Biol. Chem. 287, 9651–9658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng T et al. Optimal Translational Termination Requires C4 Lysyl Hydroxylation of eRF1. Mol. Cell 53, 645–654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harding HP et al. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Harding HP et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–108 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Han J et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quirós PM et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozak M A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 33, 6593–6602 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King HA, Cobbold LC & Willis AE The role of IRES trans -acting factors in regulating translation initiation. Biochem. Soc. Trans. 38, 1581–1586 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Stein I et al. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 18, 3112–9 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gan W & Rhoads RE Internal Initiation of Translation Directed by the 5′-Untranslated Region of the mRNA for eIF4G, a Factor Involved in the Picornavirus-induced Switch from Cap-dependent to Internal Initiation. J. Biol. Chem. 271, 623–626 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Young RM et al. Hypoxia-mediated Selective mRNA Translation by an Internal Ribosome Entry Site-independent Mechanism. J. Biol. Chem. 283, 16309–16319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lang KJD, Kappel A & Goodall GJ Hypoxia-inducible Factor-1α mRNA Contains an Internal Ribosome Entry Site That Allows Efficient Translation during Normoxia and Hypoxia. Mol. Biol. Cell 13, 1792–1801 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinnebusch AG, Ivanov IP & Sonenberg N Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwerk J & Savan R Translating the Untranslated Region. J. Immunol. 195, 2963–2971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho JJD et al. Systemic Reprogramming of Translation Efficiencies on Oxygen Stimulus. Cell Rep. 14, 1293–1300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uniacke J et al. An oxygen-regulated switch in the protein synthesis machinery. Nature 486, 126–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uniacke J, Kishan Perera J, Lachance G, Francisco CB & Lee S Cancer Cells Exploit eIF4E2-Directed Synthesis of Hypoxia Response Proteins to Drive Tumor Progression. Cancer Res. 74, 1379–1389 (2014). [DOI] [PubMed] [Google Scholar]; Work by Uniacke et al. (2012) and (2014) demonstrated oxygen-mediated changes to protein synthesis via HIF as a translation initiation factor
- 64.Sørensen BS, Busk M, Overgaard J, Horsman MR & Alsner J Simultaneous Hypoxia and Low Extracellular pH Suppress Overall Metabolic Rate and Protein Synthesis In Vitro. PLoS One 10, e0134955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walton ZE et al. Acid Suspends the Circadian Clock in Hypoxia through Inhibition of mTOR. Cell 174, 72–87. e32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hermesh O & Jansen R-P Take the (RN)A-train: Localization of mRNA to the endoplasmic reticulum. Biochim. Biophys. Acta - Mol. Cell Res 1833, 2519–2525 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Staudacher JJ et al. Hypoxia-induced gene expression results from selective mRNA partitioning to the endoplasmic reticulum. Nucleic Acids Res. 43, 3219–3236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hetz C The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Almanza A et al. Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J. 286, 241–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouchecareilh M, Higa A, Fribourg S, Moenner M & Chevet E Peptides derived from the bifunctional kinase/RNase enzyme IRE1α modulate IRE1α activity and protect cells from endoplasmic reticulum stress. FASEB J. 25, 3115–3129 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Liu CY, Schröder M & Kaufman RJ Ligand-independent Dimerization Activates the Stress Response Kinases IRE1 and PERK in the Lumen of the Endoplasmic Reticulum. J. Biol. Chem. 275, 24881–24885 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Calfon M et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Lee K et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.So J-S et al. Silencing of Lipid Metabolism Genes through IRE1α-Mediated mRNA Decay Lowers Plasma Lipids in Mice. Cell Metab 16, 487–499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie H et al. IRE1α RNase–dependent lipid homeostasis promotes survival in Myc-transformed cancers. J. Clin. Invest. 128, 1300–1316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Y et al. Regulation of glucose homeostasis through a XBP-1–FoxO1 interaction. Nat. Med. 17, 356–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J et al. Inflammation Improves Glucose Homeostasis through IKKβ-XBP1s Interaction. Cell 167, 1052–1066. e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y et al. Preventing oxidative stress: a new role for XBP1. Cell Death Differ. 16, 847–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 508, 103–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romero-Ramirez L et al. XBP1 Is Essential for Survival under Hypoxic Conditions and Is Required for Tumor Growth. Cancer Res. 64, 5943–5947 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Haze K, Yoshida H, Yanagi H, Yura T & Mori K Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell 10, 3787–3799 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye J et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–64 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Yamamoto K et al. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6α and XBP1. Dev. Cell 13, 365–376 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Yoshida H, Matsui T, Yamamoto A, Okada T & Mori K XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–91 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Wu J et al. ATF6α Optimizes Long-Term Endoplasmic Reticulum Function to Protect Cells from Chronic Stress. Dev. Cell 13, 351–364 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Shen J, Chen X, Hendershot L & Prywes R ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111 (2002). [DOI] [PubMed] [Google Scholar]
- 87.Fawcett TW, Martindale JL, Guyton KZ, Hai T & Holbrook NJ Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339 ( Pt 1), 135–41 (1999). [PMC free article] [PubMed] [Google Scholar]
- 88.Rutkowski DT et al. Adaptation to ER Stress Is Mediated by Differential Stabilities of Pro-Survival and Pro-Apoptotic mRNAs and Proteins. PLoS Biol. 4, e374 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bi M et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 24, 3470–3481 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rouschop KM et al. PERK/eIF2α signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc. Natl. Acad. Sci. U. S. A. (2013). doi: 10.1073/pnas.1210633110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harding HP, Zhang Y, Bertolotti A, Zeng H & Ron D Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Wilson DF, Rumsey WL, Green TJ & Vanderkooi JM The oxygen dependence of mitochondrial oxidative phosphorylation measured by a new optical method for measuring oxygen concentration. J. Biol. Chem. 263, 2712–8 (1988). [PubMed] [Google Scholar]
- 93.Cooper CE The steady-state kinetics of cytochrome c oxidation by cytochrome oxidase. Biochim. Biophys. Acta - Bioenerg 1017, 187–203 (1990). [DOI] [PubMed] [Google Scholar]
- 94.Wheaton WW & Chandel NS Hypoxia. 2. Hypoxia regulates cellular metabolism. Am. J. Physiol. Physiol 300, C385–C393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chandel NS, Budinger GR & Schumacker PT Molecular oxygen modulates cytochrome c oxidase function. J. Biol. Chem. 271, 18672–7 (1996). [DOI] [PubMed] [Google Scholar]
- 96.Fukuda R et al. HIF-1 Regulates Cytochrome Oxidase Subunits to Optimize Efficiency of Respiration in Hypoxic Cells. Cell 129, 111–122 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Hayashi T et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 112, 1553–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tello D et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting Complex I activity. Cell Metab 14, 768–779 (2011). [DOI] [PubMed] [Google Scholar]
- 99.Chan SY et al. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10, 273–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Z, Li Y, Zhang H, Huang P & Luthra R Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29, 4362–8 (2010). [DOI] [PubMed] [Google Scholar]
- 101.Puisségur M-P et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iyer NV et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1alpha. Genes Dev. 12, 149–162 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim JW, Tchernyshyov I, Semenza GL & Dang CV HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Papandreou I, Cairns RA, Fontana L, Lim AL & Denko NC HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3, 187–197 (2006). [DOI] [PubMed] [Google Scholar]; Work by Kim et al. (2006) and Papandreou et al. provided initial links between HIF-1 target genes to inhibition of mitochondrial metabolism.
- 105.Garcia-Bermudez J et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. (2018). doi: 10.1038/s41556-018-0118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ameri K et al. HIGD1A Regulates Oxygen Consumption, ROS Production, and AMPK Activity during Glucose Deprivation to Modulate Cell Survival and Tumor Growth. Cell Rep. 10, 891–899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomas LW, Staples O, Turmaine M & Ashcroft M CHCHD4 Regulates Intracellular Oxygenation and Perinuclear Distribution of Mitochondria. Front. Oncol. 7, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Al-Mehdi A-B et al. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 5, ra47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fuhrmann DC & Brüne B Mitochondrial composition and function under the control of hypoxia. Redox Biol. 12, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim H et al. Fine-Tuning of Drp1/Fis1 Availability by AKAP121/Siah2 Regulates Mitochondrial Adaptation to Hypoxia. Mol. Cell 44, 532–544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou R, Yazdi AS, Menu P & Tschopp J A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011). [DOI] [PubMed] [Google Scholar]
- 112.Liu L et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 14, 177–85 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Balaban RS, Nemoto S & Finkel T Mitochondria, Oxidants, and Aging. Cell 120, 483–495 (2005). [DOI] [PubMed] [Google Scholar]
- 114.Packer L & Fuehr K Low oxygen concentration extends the lifespan of cultured human diploid cells. Nature 267, 423–5 (1977). [DOI] [PubMed] [Google Scholar]
- 115.Hekimi S, Lapointe J & Wen Y Taking a "good" look at free radicals in the aging process. Trends Cell Biol 21, 569–76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bell EL, Klimova TA, Eisenbart J, Schumacker PT & Chandel NS Mitochondrial Reactive Oxygen Species Trigger Hypoxia-Inducible Factor-Dependent Extension of the Replicative Life Span during Hypoxia. Mol. Cell. Biol. 27, 5737–5745 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schulz TJ et al. Glucose Restriction Extends Caenorhabditis elegans Life Span by Inducing Mitochondrial Respiration and Increasing Oxidative Stress. Cell Metab. 6, 280–293 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Liu X et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 19, 2424–34 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bell EL & Chandel NS Mitochondrial oxygen sensing: regulation of hypoxia-inducible factor by mitochondrial generated reactive oxygen species. Essays Biochem. 43, 17–27 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Lee S-J, Hwang AB & Kenyon C Inhibition of Respiration Extends C. elegans Life Span via Reactive Oxygen Species that Increase HIF-1 Activity. Curr. Biol. 20, 2131–2136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weir EK, López-Barneo J, Buckler KJ & Archer SL Acute oxygen-sensing mechanisms. N. Engl. J. Med. 353, 2042–55 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sommer N et al. Mitochondrial Complex IV Subunit 4 Isoform 2 Is Essential for Acute Pulmonary Oxygen Sensing. Circ. Res. 121, 424–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moreno-Domínguez A et al. Acute O2 sensing through HIF2α-dependent expression of atypical cytochrome oxidase subunits in arterial chemoreceptors. Sci. Signal. (2019). doi: 10.1126/scisignal.aay9452 [DOI] [PubMed] [Google Scholar]
- 124.Fernández-Agüera MC et al. Oxygen Sensing by Arterial Chemoreceptors Depends on Mitochondrial Complex I Signaling. Cell Metab. 22, 825–837 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Waypa GB et al. Superoxide generated at mitochondrial complex III triggers acute responses to hypoxia in the pulmonary circulation. Am. J. Respir. Crit. Care Med. 187, 424–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; Genetic evidence that the mitochondrial respiratory chain is necessary for organismal acute responses to hypoxia.
- 126.Schleifer G et al. Impaired hypoxic pulmonary vasoconstriction in a mouse model of Leigh syndrome. Am. J. Physiol. Cell. Mol. Physiol 316, L391–L399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.CHANCE B & WILLIAMS GR The respiratory chain and oxidative phosphorylation. Adv. Enzymol. Relat. Subj. Biochem. 17, 65–134 (1956). [DOI] [PubMed] [Google Scholar]
- 128.Milligan LP & McBride BW Energy costs of ion pumping by animal tissues. J. Nutr. 115, 1374–82 (1985). [DOI] [PubMed] [Google Scholar]
- 129.Helenius IT, Dada LA & Sznajder JI Role of ubiquitination in Na,K-ATPase regulation during lung injury. Proc. Am. Thorac. Soc. 7, 65–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Emerling BM et al. Hypoxic activation of AMPK is dependent on mitochondrial ROS but independent of an increase in AMP/ATP ratio. Free Radic. Biol. Med. 46, 1386–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mungai PT et al. Hypoxia triggers AMPK activation through reactive oxygen species-mediated activation of calcium release-activated calcium channels. Mol. Cell. Biol. 31, 3531–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gusarova GA et al. Hypoxia leads to Na,K-ATPase downregulation via Ca(2+) release-activated Ca(2+) channels and AMPK activation. Mol. Cell. Biol. 31, 3546–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laderoute KR et al. 5’-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol. Cell. Biol. 26, 5336–47 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garcia D & Shaw RJ AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol. Cell 66, 789–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu L et al. Hypoxia-Induced Energy Stress Regulates mRNA Translation and Cell Growth. Mol. Cell 21, 521–531 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orr AL et al. Suppressors of superoxide production from mitochondrial complex III. Nat. Chem. Biol. 11, 834–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muller FL, Liu Y & Van Remmen H Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279, 49064–49073 (2004). [DOI] [PubMed] [Google Scholar]
- 138.Hernansanz-Agustín P et al. Mitochondrial Na+ import controls oxidative phosphorylation and hypoxic redox signalling. bioRxiv 385690 (2018). doi: 10.1101/385690 [DOI] [Google Scholar]
- 139.Szibor M et al. Broad AOX expression in a genetically tractable mouse model does not disturb normal physiology. Dis. Model. Mech. 10, 163–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Samanta D & Semenza GL Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochimica et Biophysica Acta - Reviews on Cancer 1870, 15–22 (2018). [DOI] [PubMed] [Google Scholar]; Timely review article summarizing how HIFs regualate immune cells within the tumour microenvironment.
- 141.Nakazawa MS, Keith B & Simon MC Oxygen availability and metabolic adaptations. Nat. Rev. Cancer 16, 663–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xie H & Simon MC Oxygen availability and metabolic reprogramming in cancer. J. Biol. Chem. 292, 16825–16832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faubert B et al. Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jang C et al. Metabolite Exchange between Mammalian Organs Quantified in Pigs. Cell Metab. 30, 594–606. e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morotti M et al. Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates endocrine resistance in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 116, 12452–12461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sun RC & Denko NC Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab 19, 285–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; Important paper indicating why hypoxia promotes reductive carboxylation of α-ketoglutarate via isocitrate dehydrogenase by degrading an α-ketoglutarate dehydrogenase complex subunit, and altered use of glutamine carbons for lipid synthesis.
- 147.Huang D et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 8, 1930–1942 (2014). [DOI] [PubMed] [Google Scholar]
- 148.Kamphorst JJ et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. 110, 8882–8887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Young RM et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 27, 1115–1131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; Kamphorst et al. (2013) and Young et al. (2013) demonstrated that tumour relevant oxygen levels can inhibit enzymatic activity of the prominent fatty acyl desaturase, Stearoyl CoA Desaturase 1 (SCD1).
- 150.Peck B & Schulze A Lipid desaturation - the next step in targeting lipogenesis in cancer? FEBS J. 283, 2767–2778 (2016). [DOI] [PubMed] [Google Scholar]
- 151.Shimabukuro M, Zhou Y-T, Levi M & Unger RH Fatty acid-induced cell apoptosis: A link between obesity and diabetes. Proc. Natl. Acad. Sci. 95, 2498–2502 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qiu B et al. HIF2 -Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 5, 652–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ackerman D et al. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep. 24, 2596–2605. e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Samanta D & Semenza GL Serine synthesis helps hypoxic cancer stem cells regulate redox. Cancer Research 76, 6458–6462 (2016). [DOI] [PubMed] [Google Scholar]
- 155.Levine B & Klionsky DJ Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Developmental Cell (2004). doi: 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 156.Feng Y, He D, Yao Z & Klionsky DJ The machinery of macroautophagy. Cell Res. 24, 24–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kimmelman AC & White E Autophagy and Tumor Metabolism. Cell Metab. 25, 1037–1043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mazure NM & Pouysségur J Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 22, 177–180 (2010). [DOI] [PubMed] [Google Scholar]
- 159.Choi AMK, Ryter SW & Levine B Autophagy in Human Health and Disease. N. Engl. J. Med. 368, 651–662 (2013). [DOI] [PubMed] [Google Scholar]
- 160.Poillet-Perez L & White E Role of tumor and host autophagy in cancer metabolism. Genes Dev. 33, 610–619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang H et al. Mitochondrial Autophagy Is an HIF-1-dependent Adaptive Metabolic Response to Hypoxia. J. Biol. Chem. 283, 10892–10903 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162.Bellot G et al. Hypoxia-Induced Autophagy Is Mediated through Hypoxia-Inducible Factor Induction of BNIP3 and BNIP3L via Their BH3 Domains. Mol. Cell. Biol. 29, 2570–2581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pouysségur J, Dayan F & Mazure NM Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441, 437–443 (2006). [DOI] [PubMed] [Google Scholar]
- 164.Azad MB et al. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 4, 195–204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fei P et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell 6, 597–609 (2004). [DOI] [PubMed] [Google Scholar]
- 166.Mazure NM & Pouysségur J Atypical BH3-domains of BNIP3 and BNIP3L lead to autophagy in hypoxia. Autophagy 5, 868–869 (2009). [DOI] [PubMed] [Google Scholar]
- 167.Band M, Joel A, Hernandez A & Avivi A Hypoxia-induced BNIP3 expression and mitophagy: in vivo comparison of the rat and the hypoxia-tolerant mole rat, Spalax ehrenbergi. FASEB J 23, 2327–2335 (2009). [DOI] [PubMed] [Google Scholar]
- 168.Chourasia AH & Macleod KF Tumor suppressor functions of BNIP3 and mitophagy. Autophagy 11, 1937–1938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rozpedek W et al. The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress. Curr. Mol. Med. 16, 533–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mihaylova MM & Shaw RJ The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biology (2011). doi: 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hardie DG AMPK and autophagy get connected. EMBO J. 30, 634–635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang C et al. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat. Commun 9, 3492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Russell RC et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Egan DF et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science (80-. ) 331, 456–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Frezza C et al. Metabolic Profiling of Hypoxic Cells Revealed a Catabolic Signature Required for Cell Survival. PLoS One 6, e24411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Antonescu CN, McGraw TE & Klip A Reciprocal regulation of endocytosis and metabolism. Cold Spring Harb. Perspect. Biol. 6, a016964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang S & Czech MP The GLUT4 Glucose Transporter. Cell Metab. 5, 237–252 (2007). [DOI] [PubMed] [Google Scholar]
- 178.Herman MA & Kahn BB Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J. Clin. Invest. 116, 1767–75 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Foley K, Boguslavsky S & Klip A Endocytosis, Recycling, and Regulated Exocytosis of Glucose Transporter 4. Biochemistry 50, 3048–3061 (2011). [DOI] [PubMed] [Google Scholar]
- 180.Fazakerley DJ et al. Kinetic Evidence for Unique Regulation of GLUT4 Trafficking by Insulin and AMP-activated Protein Kinase Activators in L6 Myotubes. J. Biol. Chem. 285, 1653–1660 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mu J, Brozinick JT, Valladares O, Bucan M & Birnbaum MJ A Role for AMP-Activated Protein Kinase in Contraction- and Hypoxia-Regulated Glucose Transport in Skeletal Muscle. Mol. Cell 7, 1085–1094 (2001). [DOI] [PubMed] [Google Scholar]
- 182.Sakagami H et al. Loss of HIF-1α impairs GLUT4 translocation and glucose uptake by the skeletal muscle cells. Am. J. Physiol. Metab 306, E1065–E1076 (2014). [DOI] [PubMed] [Google Scholar]
- 183.Görgens SW et al. Hypoxia in Combination With Muscle Contraction Improves Insulin Action and Glucose Metabolism in Human Skeletal Muscle via the HIF-1α Pathway. Diabetes 66, 2800–2807 (2017).28811274 [Google Scholar]
- 184.Li G, Wang J, Ye J, Zhang Y & Zhang Y PPARα Protein Expression Was Increased by Four Weeks of Intermittent Hypoxic Training via AMPKα2-Dependent Manner in Mouse Skeletal Muscle. PLoS One 10, e0122593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Siques P et al. Long-Term Chronic Intermittent Hypobaric Hypoxia Induces Glucose Transporter (GLUT4) Translocation Through AMP-Activated Protein Kinase (AMPK) in the Soleus Muscle in Lean Rats. Front. Physiol 9, 799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Y et al. Effects of four weeks intermittent hypoxia intervention on glucose homeostasis, insulin sensitivity, GLUT4 translocation, insulin receptor phosphorylation, and Akt activity in skeletal muscle of obese mice with type 2 diabetes. PLoS One 13, e0203551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee KY, Gesta S, Boucher J, Wang XL & Kahn CR The Differential Role of Hif1β/Arnt and the Hypoxic Response in Adipose Function, Fibrosis, and Inflammation. Cell Metab. 14, 491–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bloomfield G & Kay RR Uses and abuses of macropinocytosis. J. Cell Sci. 129, 2697–705 (2016). [DOI] [PubMed] [Google Scholar]
- 189.Recouvreux MV & Commisso C Macropinocytosis: A Metabolic Adaptation to Nutrient Stress in Cancer. Front. Endocrinol. (Lausanne) 8, 261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Commisso C et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Palm W et al. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 162, 259–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kamphorst JJ et al. Human Pancreatic Cancer Tumors Are Nutrient Poor and Tumor Cells Actively Scavenge Extracellular Protein. Cancer Res. 75, 544–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li X, Egervari G, Wang Y, Berger SL & Lu Z Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nature Reviews Molecular Cell Biology 19, 563–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen W et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cho H et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Courtney KD et al. HIF-2 Complex Dissociation, Target Inhibition, and Acquired Resistance with PT2385, a First-in-Class HIF-2 Inhibitor, in Patients with Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. (2019). doi: 10.1158/1078-0432.ccr-19-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Maxwell PH & Eckardt KU HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat. Rev. Nephrol. 12, 157–168 (2016). [DOI] [PubMed] [Google Scholar]
- 198.Jain IH et al. Leigh Syndrome Mouse Model Can Be Rescued by Interventions that Normalize Brain Hyperoxia, but Not HIF Activation. Cell Metab 30, 824–832. e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ast T et al. Hypoxia Rescues Frataxin Loss by Restoring Iron Sulfur Cluster Biogenesis. Cell 177, 1507–1521. e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Krock BL, Skuli N & Simon MC Hypoxia-Induced Angiogenesis: Good and Evil. Genes and Cancer 2, 1117–1133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wong BW, Marsch E, Treps L, Baes M & Carmeliet P Endothelial cell metabolism in health and disease: impact of hypoxia. EMBO J. 36, 2187–2203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Eales KL, Hollinshead KER & Tennant DA Hypoxia and metabolic adaptation of cancer cells. Oncogenesis 5, e190–e190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Höckel M et al. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother. Oncol. 26, 45–50 (1993). [DOI] [PubMed] [Google Scholar]
- 204.Brizel DM et al. Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res. 56, 941–943 (1996). [PubMed] [Google Scholar]
- 205.Vaupel P, Kelleher DK & Höckel M Oxygenation status of malignant tumors: Pathogenesis of hypoxia and significance for tumor therapy. Semin. Oncol 28, 29–35 (2001). [DOI] [PubMed] [Google Scholar]
- 206.Eisinger-Mathason TSK et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 3, 1190–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sun JD et al. Selective tumor hypoxia targeting by hypoxia-activated prodrug TH-302 inhibits tumor growth in preclinical models of cancer. Clin. Cancer Res. 18, 758–70 (2012). [DOI] [PubMed] [Google Scholar]
- 208.Denny WA The role of hypoxia-activated prodrugs in cancer therapy. Lancet Oncology 1, 25–29 (2000). [DOI] [PubMed] [Google Scholar]
- 209.Baran N & Konopleva M Molecular Pathways: Hypoxia-Activated Prodrugs in Cancer Therapy. Clin. Cancer Res. 23, 2382–2390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shmakova A, Batie M, Druker J & Rocha S Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochem. J. 462, 385–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chakraborty AA et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science (80-. ) 363, 1217–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Batie M et al. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science (80-. ) 363, 1222–1226 (2019). [DOI] [PubMed] [Google Scholar]
- 213.Masson N et al. Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science (80-. ). 365, 65–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Licausi F et al. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 (2011). [DOI] [PubMed] [Google Scholar]
- 215.Gibbs DJ et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Weits DA et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brugarolas J et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sofer A, Lei K, Johannessen CM & Ellisen LW Regulation of mTOR and Cell Growth in Response to Energy Stress by REDD1. Mol. Cell. Biol. 25, 5834–5845 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ding M et al. The mTOR Targets 4E-BP1/2 Restrain Tumor Growth and Promote Hypoxia Tolerance in PTEN-driven Prostate Cancer. Mol. Cancer Res. 16, 682–695 (2018). [DOI] [PubMed] [Google Scholar]
- 220.Seong M, Lee J & Kang H Hypoxia-induced regulation of mTOR signaling by miR-7 targeting REDD1. J. Cell. Biochem. 120, 4523–4532 (2019). [DOI] [PubMed] [Google Scholar]
- 221.Bernardi R et al. PML inhibits HIF-1α translation and neoangiogenesis through repression of mTOR. Nature 442, 779–785 (2006). [DOI] [PubMed] [Google Scholar]
- 222.Sancak Y et al. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science (80-. ) 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li Y et al. Bnip3 Mediates the Hypoxia-induced Inhibition on Mammalian Target of Rapamycin by Interacting with Rheb. J. Biol. Chem. 282, 35803–35813 (2007). [DOI] [PubMed] [Google Scholar]
- 224.Cam H, Easton JB, High A & Houghton PJ mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol. Cell 40, 509–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Brugarolas JB, Vazquez F, Reddy A, Sellers WR & Kaelin WG TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 4, 147–58 (2003). [DOI] [PubMed] [Google Scholar]
- 226.Hudson CC et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 22, 7004–14 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Thomas GV et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat. Med. 12, 122–127 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


