Abstract
Human immunodeficiency virus type 1 (HIV-1) infection remains a severe public health problem worldwide. In this study, we investigated the distribution of HIV-1 subtypes and the prevalence of drug resistance mutations (DRMs) among patients with HIV-1 infection in Henan Province, China. HIV-1 strains in blood samples taken from inpatients and outpatients visiting the Sixth People’s Hospital of Zhengzhou from August 2017 to July 2019 with a viral load (VL) greater than 1000 copies/ml were subjected to subtype and DRMs analysis. Out of a total of 769 samples, subtype and DRM data were obtained from 657 (85.43%) samples. Phylogenetic analysis based on partial pol gene sequences indicated that the most commonly found genotype was subtype B (45.51%, 299/657), followed by CRF01_AE (28.61%, 188/657), CRF07_BC (15.68%, 103/657), CRF08_BC (0.76%, 5/657), C (0.61%, 4/657), A (0.30%, 2/657), and others (8.52%, 56/657). Circulating recombinant forms (CRFs) were most commonly found in patients who were naïve to antiretroviral treatment (ART) (68.67%, 160/233). The percentage of patients with one or more major drug-resistance mutations was 50.99% (335/657), and it was 6.44% (15/233) in ART-naive patients that were primarily infected with subtype B (17.74%). Resistance mutations were most common at codons 65, 103, 106, 184, and 190 of the reverse transcriptase gene and codon 46 of the protease gene. Our study provides detailed information about the distribution of HIV-1 subtypes and the incidence of drug resistance mutations of different subtypes in ART-experienced and naïve patients. This can guide policymakers in making decisions about treatment strategies against HIV-1.
Introduction
Human immunodeficiency virus type 1 (HIV-1) is one of the most important communicable infectious diseases worldwide. It presents a great threat to public health in China and abroad [1, 2]. Since the first domestic HIV-1 outbreak was detected among injecting drug users (IDUs) in the city of Ruili in Yunnan Province in 1989 [3], China has made great efforts toward controlling and preventing this disease. Following the country’s social and economic development and the implementation of policies regarding blood donation, the primary transmission routes of HIV-1 in China have shifted from blood donation and injection drug use (IDU) to hetero- or homosexual activity [2]. There are about 1.3 million people infected with HIV-1 as of December 31, 2018, in China, and the number of newly diagnosed HIV-1 cases has increased rapidly in recent years [4, 5]. A major feature of HIV-1 is its high level of molecular polymorphism, which is due to its high replication rate and the lack of proofreading activity in its reverse transcriptase. At present, more than 10 subtypes of HIV, which can be divided into groups M, N, and O, have been reported worldwide [6]. The distribution of different subtypes has been found to exhibit geographic-specific patterns. HIV-1 subtype B is widespread globally, especially in Europe and the United States, but it only accounts for approximately 10% of HIV infections in the other parts of the world [7]. In China, an increased frequency of recombinant strains has been observed nationwide [8].
Antiretroviral therapy (ART) has significantly slowed the progression of immunodeficiency in patients infected with HIV-1 [9]. However, the emergence of drug resistance mutations (DRMs) due to natural and drug-induced selection poses as a great challenge for successful ART [10]. Previous studies have demonstrated that genotypic resistance is frequently associated with the failure of ART [11–13]. HIV-1 drug resistance can also be transmitted to another person. The prevalence of transmitted drug resistance (TDR) in North America and western Europe ranges from 4% to 20% [14–16]. Drug resistance testing is widely used in clinical practice to guide rational drug use. The HIV-1 pol gene is generally used to identify mutations associated with resistance to reverse transcriptase (RT) or protease (PR) inhibitors and for HIV-1 subtyping [17]. Genetic typing of HIV-1 strains is important because different subtypes exhibit different patterns of drug resistance and treatment responses [18].
In this study, blood samples were collected from HIV-positive inpatients and outpatients visiting the Sixth People’s Hospital of Zhengzhou from August 2017 to July 2019 in Henan Province, China, and the distribution of HIV-1 subtypes and the prevalence of DRMs were analyzed.
Materials and methods
The study population included HIV-positive inpatients and outpatients visiting the Sixth People’s Hospital of Zhengzhou from August 2017 to July 2019 in Henan Province, China. The criteria for inclusion were that the HIV-1 viral load in plasma sample was greater than 1000 copies/ml [19]. Data about the patient's general condition, transmission route, HIV-1 RNA level, CD4+ T-cell count, and history of ART were collected. Blood samples were collected into EDTA-containing tubes, and plasma samples were prepared by centrifugation. The HIV-1 RNA viral load (VL) in the plasma samples was determined using a Human Immunodeficiency Virus (HIV-1) RNA Quantitative Diagnostic Kit (Northeast Pharmaceutical Group, Shenyang, China) according to the manufacturer’s protocol [20–22]. Briefly, viral RNA was extracted from plasma samples using magnetic beads, and real-time fluorescence quantitative RT-PCR detection of HIV-1 was performed by a TaqMan probe method. An internal standard was used to detect the presence of PCR inhibitors in the samples to avoid false-negative results.
Whole blood was collected either at the same time as sample collection for VL measurement or afterward, and plasma was separated by centrifugation as described above. HIV-1 RNA was isolated from 140 μl of plasma using a magnetic-bead-based Virus RNA Extraction Kit (Zhijiang, Shanghai, China) according to the manufacturer’s protocol. An HIV-1 pol gene sequence including the protease gene and part of the reverse transcriptase gene was reverse transcribed, amplified, and sequenced as described previously, with modifications [23, 24]. Briefly, the HIV-1 pol gene was reverse transcribed using the specific primer R5073 and a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, MA, USA). The target sequence (approximately 1,300 bp) was amplified by nested PCR using LA Taq™ Version 2.0 (Takara, Shiga, Japan) with two primer sets (PRO-F1 and RT-R1 for first-round PCR and PRO-F2 and RT-R2 for second-round PCR). Each amplified fragment was sequenced with two primer sets (CF1, CF3, CR2, and CR4 as the primary sequencing primer set; CF2, CF4, CR1, and CR3 as the backup sequencing primer set) (Table 1). For analysis of drug resistant mutations and antiretroviral susceptibility, sequences were submitted to the Stanford HIV-1 drug resistance database (https://hivdb.stanford.edu/), which undergoes frequent updates.
Table 1.
Primers used in this study
| Name | Sequence | HXB2 location |
|---|---|---|
| RT primer | ||
| R5073 | 5′-CCACACAATCATCACCTGCC-3′ | 5073-5092 |
| Amplification primer sets | ||
| PRO-F1 | 5′-TTGGAAATGTGGAAAGGAAGGAC-3′ | 2028-2050 |
| RT-R1 | 5′-CTGTATTTCTGCTATTAAGTCTTTTGATGGG-3′ | 3509-3539 |
| PRO-F2 | 5′-CAGAGCCAACAGCCCCACCA-3′ | 2147-2166 |
| RT-R2 | 5′-CTGCCAGTTCTAGCTCTGCTTC-3′ | 3441-3462 |
| Sequencing primer sets | ||
| CF1 | 5′-CACTCTTTGGCAACGACCC-3′ | 2260-2278 |
| CF3 | 5′-AATTGGGCCTGAAAATCC-3′ | 2694-2711 |
| CR2 | 5′-GGATTTTCAGGCCCAATT-3′ | 2600-2618 |
| CR4 | 5′-GGAATATTGCTGGTGATCC-3′ | 3014-3033 |
| CF2 | 5′-GTTGACTCAGATTGGTTG-3' | 2519-2537 |
| CF4 | 5′-TGGAGAAAATTAGTAGATTT-3' | 2760-2780 |
| CR1 | 5′-ATAAAACCTCCAATTCC-3′ | 2396-2413 |
| CR3 | 5′-CTGTCCTTTTTCTTTAT-3′ | 2736-2753 |
RT, Reverse transcription
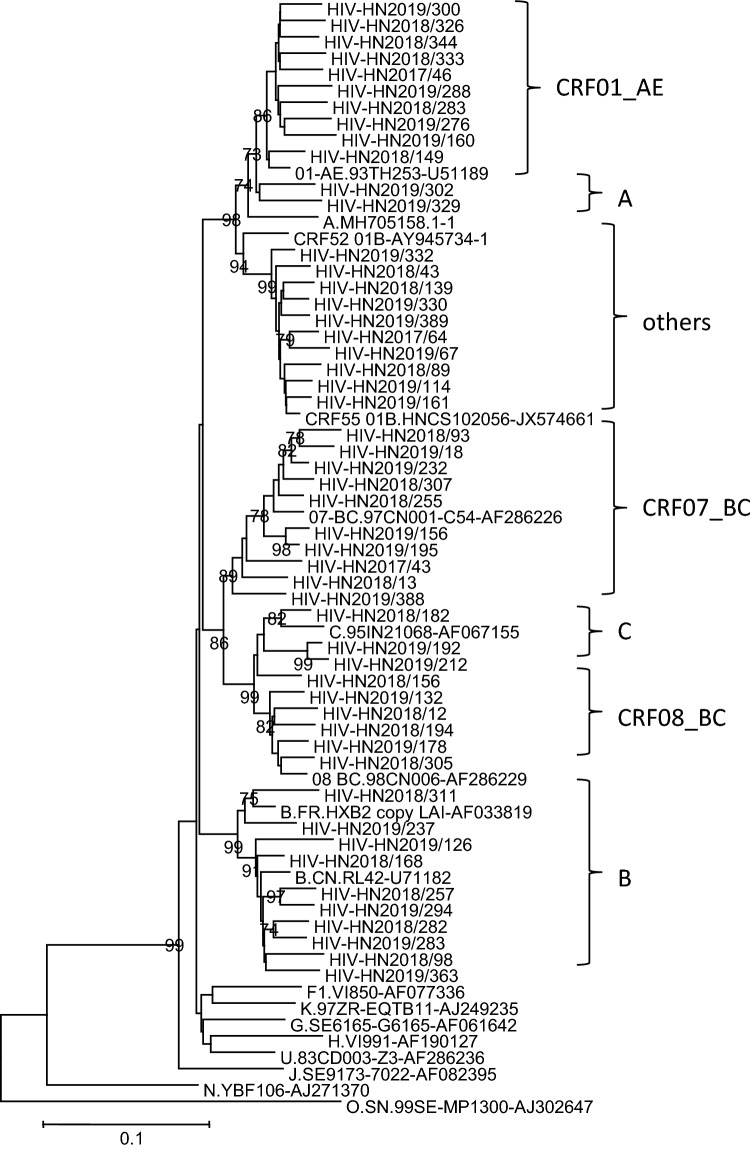
The pol region has been identified as a reliable region for HIV-1 subtyping [25, 26]. For subtyping analysis, the HIV-1 genotype was determined using the REGA HIV-1 Subtyping Tool (Version 3.0; https://dbpartners.stanford.edu:8080/RegaSubtyping/ stanford-hiv/typingtool/) and confirmed by phylogenetic analysis. Newly determined pol gene sequences were aligned with subtype reference sequences downloaded from the Los Alamos HIV Sequence Database (https://www.hiv.lanl.gov/), using CLUSTAL W. For phylogenetic analysis, 10 representative sequences or all sequences from each subtype were selected, and a phylogenetic tree was constructed using Molecular Evolutionary Genetic Analysis (MEGA) software (version 6) based on the neighbor-joining method and the Tamura-Nei model with 1000 bootstrap replicates. The Kimura two-parameter model was used with a transition-transversion ratio of 2.0 and the scale bar represented a genetic distance of 1% [27, 28]. Bootstrap values higher than 70% are shown at the corresponding nodes. The clustering of specific subtypes is indicated.
Statistical analysis was carried out using the SPSS statistics program (version 20) for Windows (SPSS, Inc., USA). Differences in rates of drug resistance among groups were analyzed by chi-square test, and a p-value less than 0.05 was considered statistically significant.
The sequences determined in this study were deposited in the GenBank database under the accession numbers MT042116 - MT042772.
Results
A total of 769 HIV-1 patients were tested using the protocol given above, and the reverse transcriptase and protease regions were successfully amplified for 657 patients. Demographic characteristics of these patients (n = 657) are shown in Table 2. The median age of all patients was 38 years (ranging from 5 to 81). Of these patients, 80.8% were male, with a median age of 37 years (ranging from 5 to 81), and 19.2% were females, with a median age of 45 years (ranging from 8 to 75). In addition, 99.2% of the patients were of Han ethnicity, and 96.8% came from Henan Province. Among these patients, men having sex with men (MSM) and heterosexual activity (HSX) were the two major transmission routes, accounting for 21.6% and 18.0% of the infections in the study population, respectively, and they increased to 38.2% and 31.7%, respectively, after patients with unspecified information were removed. Almost all HIV-1 patients under the age of 15 had been infected by mother-to-child transmission. It was also observed that the rate of infection by plasmapheresis and blood transfusion increased with age and peaked in the age range of 46–60 years (Fig. 1). Medication history showed that almost all ART-experienced patients had been treated with tenofovir (TDF) + lamivudine (3TC) + efavirenz (EFV)/nevirapine (NVP), as those drugs were provided for free, and the administration of those drugs was supervised by local rural physicians (Table 3).
Table 2.
Baseline demographic and viral characteristics
| Characteristic | No. of patients [N = 657] |
|---|---|
| Sex | |
| Male - no. (%) | 531 (80.82) |
| Female - no. (%) | 126 (19.18) |
| Age at diagnosis (years) | |
| Male (median/range) | 37/(5-81) |
| Female (median/range) | 45/(8-75) |
| Race/ethnicity | |
| Han ethnicity - no. (%) | 652 (99.24) |
| Non-Han ethnicity - no. (%) | 5 (0.76) |
| Geographic location of subjects | |
| Henan - no. (%) | 636 (96.80) |
| Other - no. (%) | 21 (3.20) |
| Risk factor for HIV | |
| MSM - no. (%) | 142 (21.61) |
| HSX - no. (%) | 118 (17.96) |
| MTCT - no. (%) | 33 (5.02) |
| PL - no. (%) | 39 (5.94) |
| BT - no. (%) | 36 (5.48) |
| IDU - no. (%) | 4 (0.61) |
| OTH - no. (%) | 285 (43.38) |
| CD4+ T cell count (cells/µl) | |
| CD4+, median (min/max) | 52 (1/1196) |
| HIV Baseline viral load (copies/ml plasma) | |
| VL, median (min/max) | 49077 (1151/4280000) |
MSM, men who have sex with men; HSX, heterosexual orientation; IDU, injection drug use; PL, plasmapheresis; BT, blood transfusion; MTCT, mother-to-child transmition; OTH, others, including patients whose risk factors were unknown or patients who did not provide information
Fig. 1.
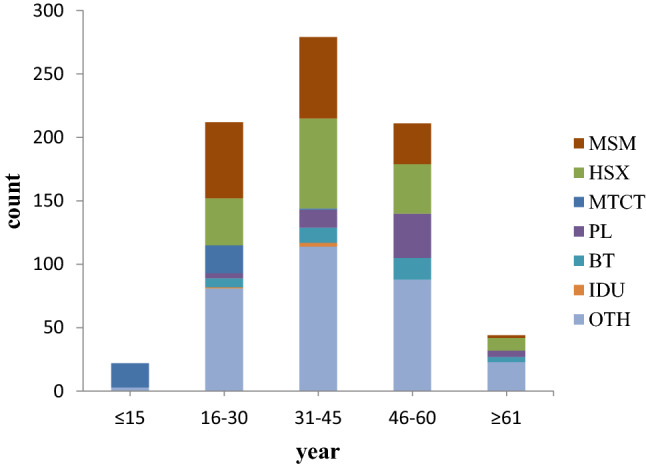
The characteristics of HIV transmission in the population of Henan. This graph shows the proportion of different transmission routes in each age group. MSM, men who have sex with men; HSX, heterosexual orientation; IDU, injection drug use; PL, plasmapheresis; BT, blood transfusion; MTCT, mother-to-child transmission; OTH, others, including patients whose risk factors were unknown or were unwilling to provide information
Table 3.
Drug resistance analysis of drugs and mutations
| Drug category | Drugs | DR (no.) | DRMs* |
|---|---|---|---|
| NRTIs | ABC | 225 | K65R, L74VI, Y115F, Q151M |
| AZT | 59 | K70R, T215FY, Q151M | |
| 3TC | 281 | K65R, M184VI | |
| TDF | 169 | K65R | |
| NNRTIs | EFV | 325 | K103NS, L100I, K101P, V106AM, Y181CIV, Y188LCH, G190ASEQ |
| NVP | 327 | K103NS, L100I, K101EP, V106AM, Y181CIV, Y188LCH, G190ASEQ | |
| PIs | LPV/r | 11 | I47A, L76V, V82A, I84V |
NRTIs, nucleoside reverse transcriptase inhibitors; NNRTIs, non-nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors; ABC, abacavir; AZT, zidovudine; 3TC, lamivudine; TDF, tenofovir; EFV, efavirenz; NVP, nevirapine; LPV/r, lopinavir/r
*Only mutations associated with medium or high drug resistance are listed
Phylogenetic analysis of the pol gene showed that the strains isolated in Henan Province could be divided into six known subtypes and one mixed and unspecified subtype group (others) (Fig. 2). The distribution of each subtype was as follows: B, 45.5% (299/657); CRF01_AE, 28.6% (188/657); CRF07_BC, 15.7% (103/657); CRF08_BC, 0.761% (5/657); C, 0.609% (4/657); A, 0.304% (2/657); and other, 8.52% (56/657). Recombinants accounted for 53.6% (352/657) of all subtypes, with CRF01_AE and CRF07_BC being the major recombinant strains (Table 4). Based on clinical information, 233 cases were in ART-naïve patients, accounting for 35.5% (233/657) of the study population. In these ART-naïve patients, the most prevalent subtypes were CRF01_AE (33.5%, 78/233), CRF07_BC (30.0%, 70/233), and type B (26.6%, 62/233).
Fig. 2.
Phylogenetic tree based on pol sequences, constructed using Molecular Evolutionary Genetic Analysis (MEGA) software (version 6) based on the neighbor-joining method and the Tamura-Nei model with 1000 bootstrap replicates. Bootstrap values higher than 70% were shown at the corresponding nodes. The clustering of specific subtypes is indicated
Table 4.
Distribution and prevalence of DR among HIV-1 subtypes
| Subtype | Patients n (%) |
DR n (%) |
ART-naive n (%) |
ART-naive DR n (%) |
|---|---|---|---|---|
| B | 299 (45.51) | 202 (67.56) | 62 (26.61) | 11 (17.74) |
| CRF01_AE | 188 (28.61) | 88 (46.81) | 78 (33.48) | 3 (4.29) |
| CRF07_BC | 103 (15.68) | 23 (22.33) | 70 (30.04) | 1 (1.43) |
| CRF08_BC | 5 (0.76) | 3 (60.00) | 1 (0.43) | 0 (0.00) |
| C | 4 (0.61) | 0 (0.00) | 2 (0.86) | 0 (0.00) |
| A | 2 (0.30) | 1 (50.00) | 1 (0.43) | 0 (0.00) |
| Others | 56 (8.52) | 18 (32.14) | 19 (8.15) | 0 (0.00) |
| Total | 657 (100) | 335 (50.99) | 233 (100) | 15 (6.44) |
CRF, circulating recombinant forms; DR, drug resistance; “Others” include recombinants including CRF55_01B, A1/B, B/C, CRF07_BC/A, and CRF07_BC/D, A1/C/G,, A1/C/B, C/A1, CRF01_AE/A1
The percentage of patients with one or more major DRMs was 51.0% (335/657). Information about DRMs corresponding to specific drugs is listed in Table 3. Resistance mutations at codons 65, 103, 106, 184, and 190 of the reverse transcriptase gene and codon 46 of the protease gene were the most prevalent. M184VI, a mutation that leads to high-level resistance against emtricitabine (FTC)/3TC and low-level resistance against abacavir (ABC), was the most prevalent, resulting a resistance rate of 37.6% (247/657) (Table 5). The drug resistance rate in ART-naïve patients was 6.44% (15/233), and the major drug-resistance mutations were V106AMI, M230L, and K219QE.
Table 5.
Prevalence of drug-class-specific major mutations in treatment-experienced and naïve patients
| PIs (n = 8) | NRTIs (n = 295) | NNRTIs (n = 314) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mutation | Prevalence n (%) | Mutation | Prevalence n (%) | Mutation | Prevalence n (%) | |||
| Pa | P-naïveb | Pa | P-naïveb | Pa | P-naïveb | |||
| V32I | 2 (0.30) | 0 (0) | M41L | 44 (6.70) | 0 (0) | L100I | 17 (2.59) | 0 (0) |
| M46I | 8 (1.22) | 0 (0) | K65R | 120 (18.26) | 1 (0.83) | K101EP | 56 (8.52) | 0 (0) |
| I47A | 2 (0.30) | 0 (0) | D67N | 47 (7.15) | 0 (0) | K103NS | 119 (18.11) | 3 (2.52) |
| I54V | 5 (0.76) | 0 (0) | T69D | 7 (1.07) | 0 (0) | V106AMI | 107 (16.29) | 9 (8.41) |
| L76V | 1 (0.15) | 0 (0) | K70ER | 43 (6.54) | 0 (0) | Y181CIV | 98 (14.92) | 1 (1.02) |
| V82A | 4 (0.76) | 0 (0) | L74VI | 39 (5.94) | 0 (0) | Y188LCH | 32 (4.87) | 0 (0) |
| I84V | 2 (0.30) | 0 (0) | Y115F | 49 (7.46) | 0 (0) | G190ASEQ | 113 (17.20) | 0 (0) |
| M184VI | 247 (37.60) | 1 (0.40) | M230L | 15 (2.28) | 1 (6.67) | |||
| L210W | 26 (3.96) | 0 (0) | ||||||
| T215FY | 48 (7.31) | 0 (0) | ||||||
| K219QE | 31 (4.72) | 2 (6.45) | ||||||
| Q151M | 0 (0) | 0 (0) | ||||||
DRMs, drug resistance mutations; NNRTIs, non-nucleoside reverse transcriptase inhibitors; NRTIs, nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors
aPopulation prevalence of drug resistance mutations
bPrevalence of drug resistance mutations in treatment-naïve HIV patients
Major and minor DRMs affecting protease inhibitors (PIs) were observed in 1.22% (8/657) and 2.89% (19/657) of cases, respectively. The major mutation sites were M46I, I54V, and V82A, with V82M as a rare nonpolymorphic variant leading to potential low-level atazanavir (ATV/r) resistance and low-level lopinavir (LPV/r) resistance. For nucleoside reverse transcriptase inhibitors (NRTIs), major and minor DRMs were found in 44.9% (295/657) and 10.1% (66/657) of the cases, respectively. The major mutations affecting NRTIs were K65R and M184VI. K70GQNT is similar to K70E in that it can lead to low-level ABC/TDF resistance and potential low-level FTC/3TC resistance [29]. T215ANCD, a revertant of T215YF that arises in the absence of NRTIs, leads to low-level zidovudine (AZT)/ stavudine (D4T) resistance [30]. K219NR, an accessory thymidine analog mutation (TAM), leads to potential low-level AZT/D4T resistance [31, 32]. For non-nucleoside reverse transcriptase inhibitors (NNRTIs), major and minor resistance-associated mutation rates were found in 47.8% (314/657) and 33.5% (220/657) of the cases, respectively. The major mutations affecting NNRTIs were K103NS, V106AMI, and G190ASEQ (Tables 5 and 6).
Table 6.
Prevalence of drug-class-specific minor mutations in treatment-experienced and naïve patients
| PIs (n = 19) | NRTIs (n = 66) | NNRTIs (n = 220) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mutation | Prevalence n (%) | Mutation | Prevalence n (%) | Mutation | Prevalence n (%) | |||
| Pa | P-naïveb | Pa | P-naïveb | Pa | P-naïveb | |||
| L10F | 7 (1.07) | 2 (28.57) | E44AED | 23 (3.50) | 0 (0) | A98G | 11 (1.67) | 0 (0) |
| K20T | 1 (0.15) | 0 (0) | A62V | 37 (5.63) | 0 (0) | K101H | 7 (1.07) | 0 (0) |
| L23I | 1 (0.15) | 0 (0) | K65EN | 2 (0.30) | 1 (50.00) | V108I | 26 (3.96) | 1 (3.85) |
| L24I | 2 (0.30) | 0 (0) |
K70T NQG |
24 (3.65) | 0 (0) | E138A/GKQ | 20 (3.04) | 1 (5.00) |
| L33F | 6 (0.91) | 0 (0) |
K70 Deletion |
1 (0.15) | 0 (0) | V179DE/F | 131 (19.94) | 23 (17.56) |
| K43T | 1 (0.15) | 0(0) | V75M/I | 11 (1.67) | 0 (0) | Y188F/D | 2 (0.30) | 0 (0) |
| F53L | 1 (0.15) | 0 (0) | F77L | 1 (0.15) | 0 (0) | G190C | 3 (0.46) | 0 (0) |
| Q58E | 7 (1.07) | 4 (57.14) | Y115F | 2 (0.30) | 0 (0) | H221Y | 42 (6.39) | 1 (2.38) |
| T74P | 1 (0.15) | 0 (0) | T215ACDN | 6 (0.91) | 0 (0) | P225H | 20 (3.04) | 0 (0) |
| V82M | 1 (0.15) | 0 (0) | K219NR | 20 (3.04) | 0 (0) | F227L | 34 (5.18) | 0 (0) |
| L89V | 1 (0.15) | 0 (0) | M230I | 1 (0.15) | 0 (0) | |||
| L234I | 1 (0.15) | 0 (0) | ||||||
| K238T | 9 (1.37) | 1 (11.11) | ||||||
| P236L | 1 (0.15) | 1 (100) | ||||||
DRMs, drug resistance mutations; NNRTIs, non-nucleoside reverse transcriptase inhibitors; NRTIs, nucleoside reverse transcriptase inhibitors; PIs, protease inhibitors
aPopulation prevalence of DRMs
bPrevalence of drug resistance mutations in treatment-naïve HIV patients
When analyzed by group, the rates of drug resistance were found to be significantly higher in treatment-experienced patients for NRTIs, NNRTIs, and NRTIs plus NNRTIs (P < 0.05), but not for PIs (P > 0.05). When analyzed by subtype, the rate of drug resistance in subtype B was found to be significantly higher than that in subtypes CRF01_AE and CRF07_BC (P < 0.05). When analysis was restricted to treatment-experienced patients, the rate of drug resistance in subtype B was significantly higher than that in subtype CRF07_BC (P < 0.05), but not higher than in subtype CRF01_AE (p > 0.05). The rate of drug resistance in subtypes CRF01_AE and CRF07_BC was significantly different in treatment-experienced patients (P < 0.05).
Discussion
HIV/AIDS patients in Henan Province were some of the first in China to benefit from the national free ART program that was initiated in 2003 [2]. Unfortunately, this has inevitably led to drug resistance after long-term treatment [33]. Drug resistance not only leads to failure of treatment but also poses a risk of transmission of drug-resistant strains, which can occur by different routes [14]. Previous studies have demonstrated transmission of viruses with resistance to drugs from one or more classes (multidrug resistance) [34–37]. No large-scale HIV-1 drug resistance tests have been conducted in Henan Province, and the demand from patients and clinicians is urgent. Under such circumstances, we adopted an in-house method to conduct HIV-1 drug resistance tests. Alongside drug resistance profiles, HIV-1 subtypes were also analyzed in this study due to their importance in pathogenesis, vaccine development, transmission, disease progression, and treatment response [18, 38].
Genotyping and drug resistance analysis could not be performed on some of the samples (14.5%, 112/769) because the targeted sequences had not been successfully amplified, possibly due to a poor match between primers that were used and the HIV-1 variants present in those samples. Genotyping analysis indicated that subtype B was the dominant subtype among all subjects, which is consistent with a previous report [39]. The dominant subtypes shifted to CRF01_AE and CRF07_BC when only treatment-naïve patients were considered. This phenomenon may be explained by possible immigrant-mediated transmission, as the CRF01_AE and CRF07_BC strains are dominant in other provinces (e.g., Guangdong, Yunnan, Zhejiang, Liaoning, and Gansu), and CRF01_AE was not found in Henan Province before 2006 [40].
Drug resistance analysis showed that the percentage of patients with one or more major drug-resistance mutations was 51.0%, which is higher than those (40.2% and 45.4%) reported by others [41, 42]. This difference may be due to the dominance of subtype B, the large proportion of older patients, the longer duration of medication regimens, and the greater number of patients with advanced HIV/AIDS disease. Most ART-experienced HIV-1 patients in our study were infected through illicit blood and plasma donation in the mid-1990s and have since then been on medication. Genotype B has been the dominant subtype since the epidemic in Henan Province, and natural evolution and selection of drug-resistant mutants due to poor compliance and limited drug diversity in the early days of treatment may have led to severe drug resistance within this genotype. NNRTI resistance mutations were dominant in our study, while NRTIs dominated in Yunnan and Guangxi [39]. This difference may have been due to differences in subtype distribution, because HIV DRMs are subtype-specific [43, 44]. One previous study showed that some NNRTI resistance mutations could significantly increase the replication capacity of subtype B strains [45]. This may be the reason why NNRTI resistance mutations were more common than NRTI resistance mutations in Henan Province. The proportion of drug resistance mutants varied significantly among the different subtypes derived from treatment-naïve patients, with the largest proportion in subtype B (17.7%, 11/62), followed by CRF01_AE (4.29%, 3/70), and CRF07_BC (1.43%, 1/70). This is consistent with the history of the HIV epidemic in Henan Province and may be due to the transmission of drug resistance [46].
In summary, our study illustrates the current state of the HIV/AIDS epidemic in Henan Province. Subtype B predominates, but with a gradually decreasing percentage, suggesting that the previously most common route of HIV-1 infection, illicit blood donation, has been brought under control to a certain extent and that more attention should be paid to emerging groups, such as those infected by MSM and HSX. Nationwide, free drug resistance testing is financially supported by the government and only covers patients who have exhibited clinical signs of drug resistance. The rate of drug resistance in treatment-naïve patients has reached a moderate level, and drug resistance testing should be considered mandatory before initiating ART in such patients.
Author contributions
YH and QZ conceived and designed the study. ZY, SW, JL, and JP performed the experiments. LX, YS, XD, XY, CL, and SM collected clinical samples. ZY and JL analyzed the data. ZY prepared the manuscript. YH revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study was approved by the Institutional Ethics Committee of The Sixth People’s Hospital of Zhengzhou, China.
Informed consent
Signed informed consent was obtained from each patient before the collection of blood samples.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qingxia Zhao, Email: zhqx666@163.com.
Yuqi Huo, Email: 1246105971@qq.com.
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS). World Health Organization. https://www.unaids.org/en/resources/documents/2019/aids-by-the-numbers
- 2.Zhang F, Haberer JE, Wang Y, Zhao Y, Ma Y, Zhao D, et al. The Chinese free antiretroviral treatment program: challenges and responses. AIDS (London, England) 2007;21(8):S143–S148. doi: 10.1097/01.aids.0000304710.10036.2b. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Li Z, Zhang K, et al. HIV was first discovered among injection drug users in China. Chin J Epidemiol. 1990;11(3):184–185. [Google Scholar]
- 4.Li G, Jiang Y, Zhang L (2019) HIV upsurge in China's students. American Association for the Advancement of Science [DOI] [PubMed]
- 5.Tang Q, Lu HZ. HIV/AIDS responses in China should focus on the impact of global integration. Biosci Trends. 2018;12(5):507–509. doi: 10.5582/bst.2018.01269. [DOI] [PubMed] [Google Scholar]
- 6.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–57. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 7.Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11(1):45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 8.Yuan R, Cheng H, Chen LS, Zhang X, Wang B. Prevalence of different HIV-1 subtypes in sexual transmission in China: a systematic review and meta-analysis. Epidemiol Infect. 2016;144(10):2144–2153. doi: 10.1017/S0950268816000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 10.Chimukangara B, Samuel R, Naidoo K, de Oliveira T. Primary HIV-1 drug resistant minority variants. AIDS Rev. 2017;19(2):89–96. [PubMed] [Google Scholar]
- 11.Bartlett JA, Buda JJ, von Scheele B, Mauskopf JA, Davis EA, Elston R, et al. Minimizing resistance consequences after virologic failure on initial combination therapy: a systematic overview. J Acquir Immune Defic Syndr (1999) 2006;41(3):323–331. doi: 10.1097/01.qai.0000197070.69859.f3. [DOI] [PubMed] [Google Scholar]
- 12.Phillips AN, Dunn D, Sabin C, Pozniak A, Matthias R, Geretti AM, et al. Long term probability of detection of HIV-1 drug resistance after starting antiretroviral therapy in routine clinical practice. AIDS (London, England) 2005;19(5):487–494. doi: 10.1097/01.aids.0000162337.58557.3d. [DOI] [PubMed] [Google Scholar]
- 13.von Wyl V, Yerly S, Böni J, Bürgisser P, Klimkait T, Battegay M, et al. Emergence of HIV-1 drug resistance in previously untreated patients initiating combination antiretroviral treatment: a comparison of different regimen types. Arch Intern Med. 2007;167(16):1782–1790. doi: 10.1001/archinte.167.16.1782. [DOI] [PubMed] [Google Scholar]
- 14.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 15.Pillay D. Current patterns in the epidemiology of primary HIV drug resistance in North America and Europe. Antivir Ther. 2004;9(5):695–702. doi: 10.1177/135965350400900514. [DOI] [PubMed] [Google Scholar]
- 16.Machnowska P, Meixenberger K, Schmidt D, Jessen H, Hillenbrand H, Gunsenheimer-Bartmeyer B, et al. Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS ONE. 2019;14(1):e0209605. doi: 10.1371/journal.pone.0209605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasquier C, Millot N, Njouom R, Sandres K, Cazabat M, Puel J, et al. HIV-1 subtyping using phylogenetic analysis of pol gene sequences. J Virol Methods. 2001;94(1–2):45–54. doi: 10.1016/S0166-0934(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 18.Lessells RJ, Katzenstein DK, de Oliveira T. Are subtype differences important in HIV drug resistance? Curr Opin Virol. 2012;2(5):636–643. doi: 10.1016/j.coviro.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fund Global, World Health Organization (2017) HIV drug resistance report 2017: World Health Organization
- 20.Lewin SR, Vesanen M, Kostrikis L, Hurley A, Markowitz M. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol. 1999;73(7):6099–6103. doi: 10.1128/JVI.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim BC, Ju MK, Dan-Chin-Yu A, Sommer P. Quantitative detection of HIV-1 particles using HIV-1 neutralizing antibody-conjugated beads. Anal Chem. 2009;81(6):2388–2393. doi: 10.1021/ac802267u. [DOI] [PubMed] [Google Scholar]
- 22.Saha BK, Tian B, Bucy RP. Quantitation of HIV-1 by real-time PCR with a unique fluorogenic probe. J Virol Methods. 2001;93(1–2):33–42. doi: 10.1016/S0166-0934(00)00288-3. [DOI] [PubMed] [Google Scholar]
- 23.Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet (London, England) 1999;353(9171):2195–2199. doi: 10.1016/S0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 24.Baxter JD, Mayers DL, Wentworth DN, Neaton JD, Hoover ML, Winters MA, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS (London, England). 2000;14(9):F83–93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- 25.Jahanbakhsh F, Hattori J, Matsuda M, Ibe S, Monavari SH, Memarnejadian A, et al. Prevalence of transmitted HIV drug resistance in Iran between 2010 and 2011. PLoS ONE. 2013;8(4):e61864. doi: 10.1371/journal.pone.0061864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch RM, Shen T, Gnanakaran S, Derdeyn CA. Appreciating HIV type 1 diversity: subtype differences in Env. AIDS Res Hum Retrovir. 2009;25(3):237–248. doi: 10.1089/aid.2008.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro-Nallar E, Pérez-Losada M, Burton GF, Crandall KA. The evolution of HIV: inferences using phylogenetics. Mol Phylogenet Evol. 2012;62(2):777–792. doi: 10.1016/j.ympev.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megens S, De Wit S, Bernatchez J, Dekeersmaeker N, Vinken L, Covens K, et al. Characterization of amino acids Arg, Ser and Thr at position 70 within HIV-1 reverse transcriptase. Acta Clin Belg. 2014;69(5):348–357. doi: 10.1179/2295333714Y.0000000038. [DOI] [PubMed] [Google Scholar]
- 30.Mitsuya Y, Varghese V, Wang C, Liu TF, Holmes SP, Jayakumar P, et al. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol. 2008;82(21):10747–10755. doi: 10.1128/JVI.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melikian GL, Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, et al. Standardized comparison of the relative impacts of HIV-1 reverse transcriptase (RT) mutations on nucleoside RT inhibitor susceptibility. Antimicrob Agents Chemother. 2012;56(5):2305–2313. doi: 10.1128/AAC.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkhout B, Back NK, de Ronde A, Jurriaans S, Bakker M, Parkin NT, et al. Identification of alternative amino acid substitutions in drug-resistant variants of the HIV-1 reverse transcriptase. AIDS (London, England) 2006;20(11):1515–1520. doi: 10.1097/01.aids.0000237367.56864.75. [DOI] [PubMed] [Google Scholar]
- 33.Szubert AJ, Prendergast AJ, Spyer MJ, Musiime V, Musoke P, Bwakura-Dangarembizi M, et al. Virological response and resistance among HIV-infected children receiving long-term antiretroviral therapy without virological monitoring in Uganda and Zimbabwe: observational analyses within the randomised ARROW trial. PLoS Med. 2017;14(11):e1002432. doi: 10.1371/journal.pmed.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecht FM, Grant RM, Petropoulos CJ, Dillon B, Chesney MA, Tian H, et al. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339(5):307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 35.Imrie A, Beveridge A, Genn W, Vizzard J, Cooper DA. Transmission of human immunodeficiency virus type 1 resistant to nevirapine and zidovudine. J Infect Dis. 1997;175(6):1502–1506. doi: 10.1086/516487. [DOI] [PubMed] [Google Scholar]
- 36.Yerly S, Kaiser L, Race E, Bru J, Clavel F, Perrin L. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet. 1999;354(9180):729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]
- 37.Brodine SK, Shaffer RA, Starkey MJ, Tasker SA, Gilcrest JL, Louder MK, et al. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann Intern Med. 1999;131(7):502–506. doi: 10.7326/0003-4819-131-7-199910050-00004. [DOI] [PubMed] [Google Scholar]
- 38.Luft LM, Gill MJ, Church DL. HIV-1 viral diversity and its implications for viral load testing: review of current platforms. Int J Infect Dis. 2011;15(10):e661–e670. doi: 10.1016/j.ijid.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Sun G, Liang S, Li J, Li T, Wang Z, et al. Different distribution of HIV-1 subtype and drug resistance were found among treatment naïve individuals in Henan, Guangxi, and Yunnan province of China. PLoS ONE. 2013;8(10):e75777. doi: 10.1371/journal.pone.0075777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He X, Xing H, Ruan Y, Hong K, Cheng C, Hu Y, et al. A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS ONE. 2012;7(10):e47289. doi: 10.1371/journal.pone.0047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao Y, Tian D, Zheng YY, Xi HL, Liu D, Yu M, et al. Characteristics of HIV-1 natural drug resistance-associated mutations in former paid blood donors in Henan Province, China. PLoS ONE. 2014;9(2):e89291. doi: 10.1371/journal.pone.0089291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JY, Li HP, Li L, Li H, Wang Z, Yang K, et al. Prevalence and evolution of drug resistance HIV-1 variants in Henan, China. Cell Res. 2005;15(11–12):843–849. doi: 10.1038/sj.cr.7290356. [DOI] [PubMed] [Google Scholar]
- 43.Wainberg MA, Brenner BG. Role of HIV subtype diversity in the development of resistance to antiviral drugs. Viruses. 2010;2(11):2493–2508. doi: 10.3390/v2112493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wainberg MA, Brenner BG. The impact of HIV genetic polymorphisms and subtype differences on the occurrence of resistance to antiretroviral drugs. Mol Biol Int. 2012;2012(undefined):256982. doi: 10.1155/2012/256982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Li L, Li HP, Zhuang DM, Liu SY, Liu YJ, et al. Competitive capacity of HIV-1 strains carrying M184I or Y181I drug-resistant mutations. Chin Med J. 2009;122(9):1081–1086. [PubMed] [Google Scholar]
- 46.Liao L, Xing H, Shang H, Li J, Zhong P, Kang L, et al. The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J Acquir Immune Defic Syndr (1999) 2010;53(1):S10–S14. doi: 10.1097/QAI.0b013e3181c7d363. [DOI] [PMC free article] [PubMed] [Google Scholar]