Abstract
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases. In most cases, the development of the disease is sporadic and is not associated with any currently known mutations associated with PD. It is believed that changes associated with the epigenetic regulation of gene expression may play an important role in the pathogenesis of this disease. The study of individuals with an almost identical genetic background, such as monozygotic twins, is one of the best approaches to the analysis of such changes. A whole-transcriptome analysis of dermal fibroblasts obtained from three pairs of monozygotic twins discordant for PD was carried out in this work. Twenty-nine differentially expressed genes were identified in the three pairs of twins. These genes were included in seven processes within two clusters, according to the results of an enrichment analysis. The cluster with the greatest statistical significance included processes associated with the regulation of the differentiation of fat cells, the action potential, and the regulation of glutamatergic synaptic transmission. The most significant genes, which occupied a central position in this cluster, were PTGS2, SCN9A, and GRIK2. These genes can be considered as potential candidate genes for PD.
Electronic supplementary material
The online version of this article (10.1007/s12031-019-01452-3) contains supplementary material, which is available to authorized users.
Keywords: Parkinson’s disease, Whole-transcriptome analysis, Twins, Fibroblasts, Gene expression
Introduction
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases. It is caused by the disruption of the functioning of various neurotransmitter systems with a distinct predominance of dopaminergic (DAergic) system deficiency (Gasser 2009; Lesage and Brice 2009). A long presymptomatic period of development is a characteristic feature of PD pathogenesis. Motor symptoms are manifested only after the loss of 50–60% of DAergic neurons in the substantia nigra and a 70–80% decrease in dopamine levels in the striatum (Cookson et al. 2008; Kalia and Lang 2015).
It has been convincingly demonstrated in recent years that genetic factors play an important role in the development of PD (Kalinderi et al. 2016; Lesage and Brice 2012; Soto-Ortolaza and Ross 2016). In most cases, however, the development of the disease is sporadic and is not associated with any currently known mutations associated with PD (Singleton et al. 2013). In this regard, the picture of the PD etiopathogenesis is not completely understood. Moreover, it is clear that, in addition to changes in the primary structure of DNA, changes associated with the epigenetic regulation of gene expression may also play an important role in the pathogenesis of PD. Changes in the epigenetic pattern can be associated with the effect of environmental factors, the assessment of the action of which can be carried out via the study of individuals with a genetic background that is almost identical, including monozygotic twins discordant for PD.
Obviously, the main changes are observed in the cells of the central nervous system during the development of PD. However, it is known that the all identified genes of the monogenic forms of this disease are not specific to nerve cells and are expressed in various cell types. Therefore, changes that occur during the development of this pathology in neurons can be observed in other types of cells. Skin fibroblasts are among the cells that are available for research. Currently, only one transcriptome study of skin biopsies obtained from 12 patients with PD has been reported. In that study, processes that are characteristic of PD were identified, including mitochondrial dysfunction and impaired protein metabolism. The degeneration observed in the central nervous system was concluded to be systemic and also manifests itself in peripheral tissues (Planken et al. 2017). Based on these findings, it can be concluded that homeostatic imbalance and stress can lead to increased susceptibility to external and internal factors in patients, which may be reflected in the functioning of peripheral tissues.
The results of this work suggest that fibroblasts can be considered as a cellular model to study PD pathogenesis, search for biomarkers, and select therapies for PD. Here, an analysis of whole-transcriptome changes in fibroblast cultures obtained from three pairs of twins discordant for PD was carried out to confirm this hypothesis.
Materials and Methods
Twins Discordant for PD
Three pairs of phenotypically and genetically monozygotic twins discordant for PD were enrolled in the study. These individuals (of Russian ethnic origin and residing in European Russia) were diagnosed at the State Public Health Institution Primorsk Regional Clinical Hospital No. 1 and did not have any family history of PD. Participants were studied according to the International Parkinson and Movement Disorder Society-sponsored Unified Parkinson’s Disease Rating Scale (Fahn et al. 1987) and Hoehn–Yahr scores (Goetz et al. 2004). The diagnosis of PD was based on the UK Parkinson’s Disease Society Brain Bank criteria (Hughes et al. 1992). Patients were at stages 2–4 of the Hoehn–Yahr scale and had a mixed form of PD. The disease duration was at least 7 years. The healthy siblings did not have any signs of PD at the time of collection of the biological material. The twins lived in the same area, and their work was not associated with dangerous factors, such as pesticides or heavy metals. There was no information about the presence of head injuries in their history. Sex, age, and stage of the disease are presented in Table 1.
Table 1.
Characteristic of twin pairs
| Pair no. | PD status | Sex | Year of birth | Disease onset date | PD stage (Hoehn–Yahr scale) |
|---|---|---|---|---|---|
| Pair no. 1 | Healthy | Female | 1956 | — | — |
| PD | Female | 1956 | 2002 | 2 | |
| Pair no. 2 | Healthy | Female | 1947 | — | — |
| PD | Female | 1947 | 2007 | 4 | |
| Pair no. 3 | Healthy | Female | 1949 | — | — |
| PD | Female | 1949 | 2011 | 3 |
Skin Biopsy
A skin biopsy was performed to obtain fibroblasts in compliance with complete anonymity. The skin explant was taken from the upper third of the forearm. The size of the explant was 5–8 mm. After the collection of the biomaterial, the wound was aseptically treated and closed with an antibacterial patch. Recommendations for the care of the wound surface were given.
Obtaining the Primary Culture of Skin Fibroblasts
After biopsy, skin explants were placed in Dulbecco’s modified Eagle’s medium (DMEM) (Paneco, Russia), containing 20% fetal bovine serum (FBS) (HyClone, USA), 1 mM glutamine (ICN Biomedicals, USA), and penicillin–streptomycin (150 U/ml; 150 μg/ml) (Paneco, Russia). Tubes with explants were placed on ice and delivered to the laboratory within 24 h. Each explant was then divided into ten parts in a separate Petri dish (35 mm) pretreated with 0.1% gelatin. A sterile cover glass was pressed on top of the explants.
DMEM culture medium (4–5 ml) (Paneco, Russia) containing 20% FBS (HyClone, USA), 1 mM glutamine (ICN Biomedicals, USA), and penicillin–streptomycin (100 U/ml; 100 μg/ml) (Paneco, Russia) was poured in the Petri dish. The explant was observed under a microscope for infection by bacterial flora without changing the medium for 5 days. Cells began to “leave” the explant actively within 1 week. At this time, the medium was replaced with fresh DMEM (Paneco, Russia) containing 20% FBS (HyClone, USA), 1 mM glutamine (ICN Biomedicals, USA), and penicillin–streptomycin (50 U/ml; 50 μg/ml) (Paneco, Russia). Cells were scattered at a rate of 1:3 in fibroblast culture medium (DMEM (Paneco, Russia) containing 10% FBS (HyClone, USA)), 1 mM glutamine (ICN Biomedicals, USA), and penicillin–streptomycin (50 U/ml; 50 μg/ml) (Paneco, Russia) after reaching the monolayer stage. The medium was changed once every 2 days, with further cultivation. Fibroblasts were expanded for 2–3 weeks and subjected to cryopreservation in an amount of at least ten ampoules from each donor.
RNA Isolation and Sequencing
Total RNA from skin fibroblasts was isolated using TRIzol (Invitrogen, USA) according to the manufacturer’s recommendations. The quality and quantity of the isolated total RNA were assessed using a Bioanalyzer instrument and an RNA 6000 Nano Kit (Agilent, USA).
For RNA sequencing, the poly(A) fraction was extracted from total RNA. Sequencing libraries were prepared from the poly(A) fraction using the NEBNext® mRNA Library Prep Reagent Set (NEB, USA). Sequencing was conducted using HiSeq1500 (Illumina, USA), generating at least 15 million 50-base-long reads per library.
RNA-seq Data Analysis
Raw FASTQ files were processed by trimming, to remove ambiguous nucleotides and bases with poor quality. Quality filtering and removal of adapters were performed using AdapterRemoval V2 software (Schubert et al. 2016).
Trimmed FASTQ files were aligned on the transcriptome that was obtained from the GRCH38 genome and GRCH38.92 gene annotation using the rsem-prepare-reference command (Li and Dewey 2011) with the star option enabled, to also generate STAR indices (Dobin et al. 2013).
Alignment was performed by STAR and RSEM. The pseudocounts obtained were normalized using the TMM algorithm realized in the R package “edgeR,” command “calcNormFactors” (Robinson et al. 2010), and CPM normalization and realized in the “limma” R package (Ritchie et al. 2015) and “voom” command. Normalized reads were processed using the “voom” (mean/dispersion ratio estimation, to apply weights for observations), “lmfit” (to create a linear model to describe observations), and eBayes (to establish parameters for the linear model) commands from the limma package.
Differentially expressed genes (DEGs) were obtained between each pair of twins separately. DEGs were selected based on a fold change (FC) threshold (FC > 1.5) and an FDR-corrected P of a moderated t test <0.05, as assessed using limma. DEGs that were significantly differentially expressed between all three pairs of twins in the same direction were selected exclusively for further analysis.
GO Enrichment Analysis
A Gene Ontology term enrichment (The Gene Ontology 2019) was carried out using the apps ClueGO v. 2.5.3 (Bindea et al. 2009) and CluePedia v. 1.5.3 (Bindea et al. 2013) for Cytoscape v. 3.6.1.
Significantly enriched terms were selected based on a Bonferroni-corrected hypergeometric test P < 0.05. Term groups were selected by ClueGO based on the number of common genes/terms (>50%). Term clusters were selected based on common genes.
Gene-interaction networks were built using Pathway Studio® v. 12.1.0.9 (Elsevier, Netherlands) (Hettne et al. 2016). A network of selected metabolic processes and DEGs was built using CluePedia v. 1.5.3.
Expression Analysis of Individual Candidate Genes
Total RNA from skin fibroblasts was used for gene expression analysis (see “RNA Isolation and Sequencing” section).
Gene expression analysis was performed using reverse transcription and qPCR with TaqMan probes. The reverse transcription reaction was performed on a T3 Thermocycler amplifier (T3 Thermoblock, Biometra, Germany) using the RevertAid™ H Minus Reverse Transcriptase kit (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendations. A mixture of random hexamer primer (Thermo Fisher Scientific, USA) and Oligo (dT)18 primer (“Thermo Fisher Scientific,” USA) was used in a ratio of 3: 2, respectively.
The sequences of the primers and probes for expression analysis of the candidate genes were designed using Beacon Designer 7.0 software (Premier Biosoft, USA) and the nucleotide sequences of the PTGS2, SCN9A, and GRIK2 genes and the housekeeping genes SARS and PSMD6. The sequences of gene-specific primers and probes are presented in Table 2.
Table 2.
Nucleotide sequences of gene-specific primers and probes
| Gene | Nucleotide sequence |
|---|---|
|
PTGS2 NM__000963.1* |
Probe: 5’-VIC- GGTGAAACTCTGGCTAGACAGCGTAAACT-BHQ2-3' Forward primer: 5'-GCCAGCTTTCACCAACGG-3' Reverse primer: 5'-TGACTGTGGGAGGATACATCTCT-3' |
|
SCN9A ENST00000409672.5** |
Probe: 5’-VIC- GTCTACCCCCAATCAGTCACCACTCAGC-BHQ2-3' Forward primer: 5'-GGCATAGGCGAGCACATGAAA-3' Reverse primer: 5'-TCGGCAAATTCAGTCTCAGATCCTA-3' |
|
GRIK2 ENST00000369138.5 |
Probe: 5’-VIC- GGTGCTTTCAGTTTTTGTGGCAGTGGG-BHQ2-3' Forward primer: 5'-TGGTGGCATCTTCATTGTTCTG-3' Reverse primer: 5'-TAGGGAGGAATACATAGTCTTGAGGTA-3' |
|
SARS |
Probe: 5’-FAM-TCGCCACTCGCTGTCTGCCTTCACCA-BHQ1-3' Forward primer: 5'-CCCAGCCCTCATCCGAGAG-3' Reverse primer: 5'-TGTTCAAGTTGTCTGCCCGAAATC-3' |
|
PSMD6 |
Probe: 5’-VIC- AGGCGGTTTCTCCTGTCCCAGTCTCCTC -BHQ2-3' Forward primer: 5'- AACACAGAAAAGGCCAAAAGCTTAAT -3' Reverse primer: 5'- AATAGCCACACAATAAAGACCCTGAT -3' |
*Numbers in the database GenBank (accession numbers).
**Accession numbers in the Ensembl database.
FAM and VIC, fluorescent dyes; BHQ1 and BHQ2, fluorescence quenchers.
For real-time PCR, cDNA obtained in the reverse transcription reaction was used as a template. Before being added to the reaction mix, cDNA was diluted in an aqueous solution of tRNA from Escherichia coli (100 ng/μl) (Suslov and Steindler 2005) to concentration of 0.02 ng/μl. Real-time PCR was performed using the StepOnePlus™ System (Applied Biosystems, USA). The composition of 30 μl of the reaction mixture consisted of 5 μl of cDNA (0.02 ng/μl), 3 μl of PCR buffer (×10) (Synthol, Russia), 3 μl 25 mM MgCl2, 10 pM primers (Evrogen, Russia), 2.5 pM probe (DNA Synthesis, Russia), 200 μM of each dNTP, and 1 enzyme unit Taq DNA polymerase (Synthol, Russia). Thermal cycling was performed as follows: 50 °C, 60 s; 95 °C, 600 s; further 40 cycles of 95 °C, 20 s; and 61 °C, 50 s. Each sample was analyzed three times to correct for differences in sample quality and the efficiency of the reverse transcription reaction.
Statistical and Bioinformatics Analyses
Nucleotide sequences of gene-specific primers and probes were designed using the Biosoft International Beacon designer 7.0 program (Palo Alto, USA) and the corresponding sequences from NCBI database. Checking the specificity of primers and probes was performed using the resource Primer3 and BLAST (Wheeler et al. 2003) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), as well as the base IDT OligoAnalyzer 3.1 (http://eu.idtdna.com/calc/analyzer).
The relative levels of the transcripts in the test groups were calculated as R = 2−ΔΔCt (Livak and Schmittgen 2001). Statistical data processing was performed using the “Statistica for Windows 8.0” software package (StatSoft, Inc. (2007)), STATISTICA (version 8.0. www.statsoft.com), and MS Excel 2013 software (Microsoft). Data were analyzed using the nonparametric Mann–Whitney U test. Gene-interaction networks were made using the Pathway Studio® 12.0 (“Elsevier”) program (Hettne et al. 2016).
Results
A whole-transcriptome analysis of fibroblasts obtained from three pairs of monozygotic twins discordant for PD was performed.
Bioinformatics analysis of RNA-seq data was carried out separately for each pair of twins. Genes that were differentially expressed between all three pairs of twins in the same direction were selected exclusively for further analysis. This analysis identified 29 DEGs (Appendix A).
A Gene Ontology term enrichment was conducted for these genes. We considered “metabolic process” terms with a P value on a hypergeometric test <0.05 (Bonferroni corrected) and all types of GO-term-to-gene connections. Significant terms are presented in Table 3.
Table 3.
Functional clusters selected according to the results of the bioinformatics analysis
| Group | GO ID | GO term | P val | Gene |
|---|---|---|---|---|
| Group 1 | GO:0045600 | Positive regulation of fat cell differentiation | 4.43E-05 | INHBA |
| PTGS2 | ||||
| RARRES2 | ||||
| SFRP2 | ||||
| Group 2 | GO:0019228 | Neuronal action potential | 3.64E-04 | COMP |
| GRIK2 | ||||
| SCN9A | ||||
| GO:0051966 | Regulation of synaptic transmission, glutamatergic | 1.22E-03 | GRIK2 | |
| PTGS2 | ||||
| SCN9A | ||||
| Group 3 | GO:0048016 | Inositol phosphate-mediated signaling | 1.22E-03 | AKAP6 |
| LMCD1 | ||||
| NFATC2 | ||||
| GO:0051155 | Positive regulation of striated muscle cell differentiation | 1.33E-03 | AKAP6 | |
| EFNB2 | ||||
| NFATC2 | ||||
| GO:0097720 | Calcineurin-mediated signaling | 6.93E-04 | AKAP6 | |
| LMCD1 | ||||
| NFATC2 | ||||
| GO:0033173 | Calcineurin–NFAT signaling cascade | 8.48E-04 | AKAP6 | |
| LMCD1 | ||||
| NFATC2 |
Table 3 shows that seven metabolic process terms were identified, two of which were directly related to the functioning of the nervous system (GO:0019228, “neuronal action potential”; GO:0051966, “regulation of synaptic transmission, glutamatergic”).
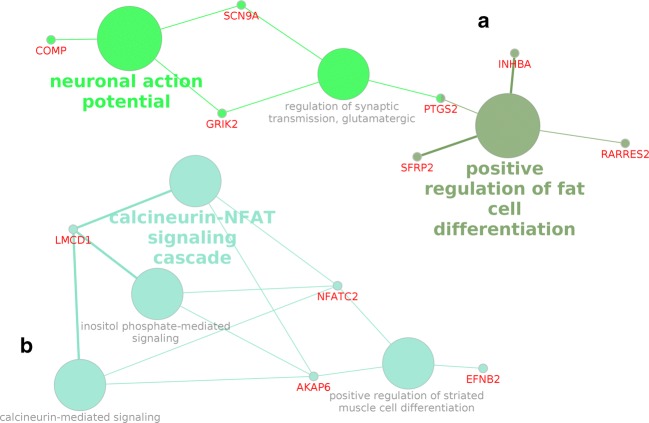
A network of the selected metabolic processes and DEGs was built during the enrichment analysis (Fig. 1).
Fig. 1.
Network of selected metabolic processes and DEGs (obtained using CluePedia v. 1.5.3). Cluster A consists of the processes included in groups 1 and 2. Cluster B consists of the processes included in group 3
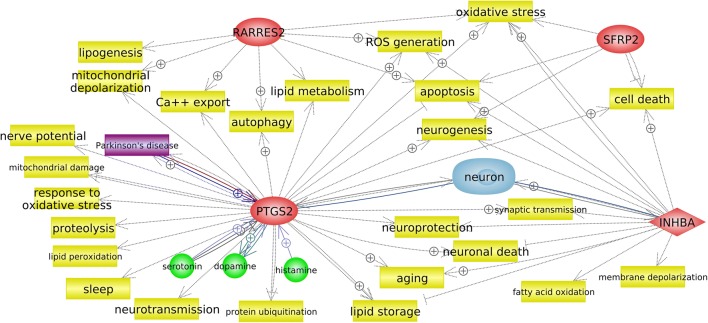
Figure 1 shows that the identified processes were divided into two large clusters that were not connected with each other. Cluster A included the process “positive regulation of fat cell differentiation,” which exhibited the greatest statistical significance. It should be noted that the genes involved in this process are associated not only with the differentiation of fat cells but also with other processes, including neuronal ones (Fig. 2).
Fig. 2.
Gene network of positive regulation of fat cell differentiation (obtained using Pathway Studio v. 12.1.0.9). Processes are highlighted in yellow, genes are in red, disease (PD) is in purple, and cell (neuron) is in blue. The colored arrows indicate the relationship between the gene and the object: regulatory pathways are in gray, genetic changes are in burgundy, arrows marked with + indicate positive links
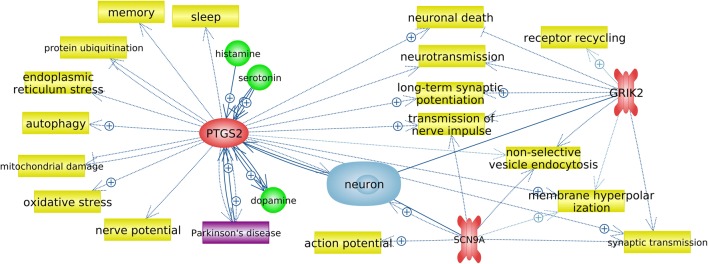
The metabolic process “positive regulation of fat cell differentiation” shared a common gene (PTGS2) with the metabolic process “regulation of synaptic transmission, glutamatergic,” and the SCN9A and GRIK2 genes were included in the processes “regulation of synaptic transmission, glutamatergic” and “neuronal action potential” (Table 3, Fig. 1). A network of functional interactions was built for the most significant genes, which occupied a central place in cluster A (Fig. 3).
Fig. 3.
Gene network of regulation of synaptic transmission, glutamatergic (obtained using Pathway Studio v. 12.1.0.9). Processes are highlighted in yellow, genes are in red, disease (PD) is in purple, and cell (neuron) is in blue; arrows marked with + indicate positive links
Also, for genes PTGS2, SCN9A, and GRIK2, an analysis of the change in mRNA level of these genes was performed using real-time PCR. The results of this analysis are presented in Table 4.
Table 4.
Relative mRNA levels of the genes studied in dermal fibroblasts, derived from three pairs of monozygotic twins, discordant for Parkinson’s disease
| Gene | Pair no. 1 | Pair no. 2 | Pair no. 3 | |||
|---|---|---|---|---|---|---|
| RNA seq1,2 | Real-time PCR1 | RNA seq | Real-time PCR | RNA seq | Real-time PCR | |
| PTGS2 | 0.563 |
0.274 (0.15–0.42)5 |
0.24 |
0.48 (0.26–0.71) |
0.34 |
0.64 (0.27–0.95) |
| GRIK2 | 5.84 |
2.55 (1.24–3.81) |
2.89 |
2.89 (1.46–3.31) |
4.08 |
2.03 (0.99–3.27) |
| SCN9A | 0.19 |
0.41 (0.20–0.64) |
0.19 |
0.39 (0.17–0.62) |
0.46 |
0.46 (0.21–0.65) |
1The data presented in the table have the P value means < 0.05. 2FDR-corrected P of a moderated t test < 0.05.
3Fold change > 1.5; 4median. 525–75 quartiles. The levels of the mRNA of the genes studied in the control group were taken as 1.
As it can be seen from Table 4, statistically significant changes in expression were revealed for all analyzed genes. Moreover, the data obtained in the study of changes in mRNA levels of these genes to the full extent reflect the results of whole-transcriptome analysis.
Discussion
To date, only two articles have been published on the analysis of monozygotic twins who are discordant for PD (Kaut et al. 2017; Woodard et al. 2014). Woodard et al. analyzed one pair of twins who were carriers of the N370S mutation in the GBA gene. The authors analyzed only the efficiency of the differentiation of neuronal cells in DAergic neurons and evaluated the activity of the GBA enzyme and the level of α-synuclein in cultured cells (Woodard et al. 2014). Kaut et al. analyzed the changes in the methylation profile of the peripheral blood of twins discordant for PD (Kaut et al. 2017).
Thus, presently, there are no studies of the changes in the transcriptome profile of monozygotic twins who are discordant for the sporadic form of PD. In this regard, a whole-transcriptome analysis of three pairs of twins who were discordant for this disease and whose genomes had no mutations associated with PD was conducted.
Processes directly related to the functioning of the nervous system (GO:0019228, “neuronal action potential”; GO:0051966, “regulation of synaptic transmission, glutamatergic”) were identified as a result of bioinformatics processing of RNA-seq data. These processes, together with the process “positive regulation of fat cell differentiation,” which had the highest statistical significance, formed a cluster of processes, cluster A. An enrichment analysis revealed that the process of differentiation of fat cells was located separately from the other two processes in the cluster (Table 3). However, Fig. 1 shows that these processes are related through the PTGS2 gene. In general, genes from the process “positive regulation of fat cell differentiation” are involved in other processes (Fig. 2). Therefore, it is necessary to refer to various bioinformatics resources to obtain more complete information when conducting this type of research.
In cluster B, processes related to the functioning of calcineurin-mediated signaling (GO: 0097720) and calcineurin–NFAT signaling cascade (GO: 0033173) are noteworthy. Calcineurin is known to be Ca2+/calmodulin-dependent serine/threonine-specific protein phosphatase, widely represented in neurons (Klee et al. 1979). NFATC2, one of the nuclear factors of activated T-cell (NFAT) family of transcription factors, is a target of calcineurin (Crabtree and Olson 2002). These proteins move to the nucleus and participate in the regulation of gene expression in most cases through other transcription factors, when NFAT proteins are dephosphorylated by calcineurin (Macian 2005). It is currently known that these proteins are involved in the regulation of neuronal survival, synaptic plasticity, and axon growth in addition to participating in the functioning of the immune system (Graef et al. 2003; Nguyen and Di Giovanni 2008; Schwartz et al. 2009). Besides, it was shown in transgenic mice that A53T mutant α-synuclein activates calcineurin and subsequent transfer of NFAT proteins to the nucleus in dopaminergic neurons of the midbrain. According to the authors, their data indicate the active involvement of calcineurin- and NFAT-mediated signaling pathways in the α-synuclein-mediated degeneration of dopaminergic neurons in PD (Luo et al. 2014). Based on this hypothesis, we can conclude that the processes identified by us can be directly related to the development of PD.
The process “regulation of synaptic transmission, glutamatergic” occupied a central place in cluster A (Fig. 1). A network of gene functional interactions was built for PTGS2, GRIK2, and SCN9A, which were included in this process (Fig. 3). The network obtained revealed that all three genes are associated with the functioning of neurons and are involved in processes that, when functionally changed, may be associated with the pathogenesis of PD.
COX-2, which is encoded by the PTGS2 gene, is expressed in response to various stimuli; in particular, it is induced in response to NMDA-associated synaptic activity (Yamagata et al. 1993). COX-2 is involved in the synthesis of several prostaglandins, each of which affects the survival and death of neurons in various ways. It was shown that the activation of COX-2 and subsequent synthesis of prostaglandins occur in a wide range of acute and chronic neurodegenerative pathologies and are associated with neuronal death (Wu et al. 2007). The dependence of COX-2 expression on the natural excitatory synaptic activity is confirmed by the presence of immunoreactivity of this protein in distal dendrites and dendritic spines, which are involved in synaptic signaling, and its exceptional localization in excitatory glutamatergic neurons (Kaufmann et al. 1997). It has been shown that the expression of this gene may be associated with neurodegenerative diseases such as Alzheimer’s disease, amyotrophic lateral sclerosis, and PD (Minghetti 2004). An increase in COX-2 expression has been found in postmortem samples of DAergic neurons in the substantia nigra of patients with PD and in a mouse model that was generated using MPTP. As suggested by the authors, the involvement of increased COX-2 expression in neurodegeneration in PD can be confirmed by the fact that MPTP-induced neurodegeneration decreases in mice in which this gene has been knocked out (Teismann et al. 2003). Conversely, a decrease in COX-2 expression leads to a decrease in the production of prostaglandins, which are necessary for the normal functioning of the brain (Shewchuk 2014). COX-2 inhibition has been shown to lead to neuronal death via autophagy (Niranjan et al. 2018). Our expression analysis showed an 1.8–4.2-fold decrease (median = (from 0.27 to 0.64), Table 4) in the level of the PTGS2 transcript. This may indicate that this gene is involved in the neurodegenerative process in PD and that its downregulation may lead to neuronal death through autophagy.
The identified GRIK2 gene encodes the GLUK2 receptor, which belongs to the kainate group of glutamate receptors. These receptors are involved in synaptic transmission and play a modulatory role in maintaining the balance of excitation and inhibition (Contractor et al. 2000; Swanson and Sakai 2009). The modulatory effect of kainate receptors occurs through several mechanisms that affect the presynaptic (Kullmann 2001; Lauri et al. 2001) and postsynaptic terminals (Castillo et al. 1997), the rhythmic activity of the neural network (Fisahn et al. 2004), the functioning of the astroglial network, and the neuron–glia interaction (Verkhratsky and Kirchhoff 2007). It has been shown that parkin interacts and ubiquitinates GLUK2 and regulates the levels of this subunit. The expression of mutant parkin correlates with GLUK2 accumulation and excitotoxicity. The accumulation of GLUK2 was detected in postmortem brain samples of patients with PD, as well as in mice with a Q311X mutation in the Prkn gene (Maraschi et al. 2014). We also found a 2.9–5.8-fold increase in the expression of GRIK2 (median = (from 2.03 to 2.89), Table 4). This may affect the development of excitotoxicity and the subsequent degeneration of neurons in PD, which points to a possible role for GRIK2 in the pathogenesis of PD.
SCN9A encodes the α-subunit of the ninth potential-dependent sodium channel (Nav1.7 channel). This channel plays an important role in nociception and in the generation and distribution of action potential (Hoffmann et al. 2018), among several other processes (Fig. 3). Nav1.7 channels are expressed in olfactory neurons (Cregg et al. 2010). In the absence of functional Nav1.7 channels, olfactory neurons continue to produce action potentials after being excited by odors but cannot initiate synaptic signaling (Weiss et al. 2011). Therefore, anosmia is a common co-phenotype in patients with variants of loss of SCN9A function (Cox et al. 2006; Weiss et al. 2011). It should be noted that one of the nonmotor signs of the early stages of PD is the disruption of smelling (Hawkes et al. 1997; Muller et al. 2002). Our expression analysis revealed a 2.2–5.4-fold decrease in the expression of SCN9A (median = (from 0.39 to 0.46), Table 4).
Thus, it can be concluded that the PTGS2, GRIK2, and SCN9A genes may be considered as candidate genes for PD; however, their role in the pathogenesis of this disease needs further verification.
Conclusions
The basis of this work was the analysis of an extremely rare phenomenon – monozygotic twins discordant for PD. An RNA-seq analysis of dermal fibroblasts obtained from three pairs of such twins was carried out. Twenty-nine DEGs in all three pairs of twins were identified in the analysis. These genes were included in seven processes within two clusters, according to the results of an enrichment analysis. The cluster with the greatest statistical significance included processes associated with the regulation of the differentiation of fat cells, the action potential, and the regulation of glutamatergic synaptic transmission. The most significant genes, which occupied a central position in this cluster, were PTGS2, SCN9A, and GRIK2. These genes can be considered as potential candidate genes for PD.
Electronic supplementary material
(XLSX 14 kb)
Acknowledgments
The work was carried out using equipment of the Center of Cellular and Gene Technology of the Institute of Molecular Genetics of the Russian Academy of Sciences.
Abbreviations
- PD
Parkinson’s disease
- DAergic
Dopaminergic
- FBS
Fetal bovine serum
- DEGs
Differentially expressed genes
Funding Information
This work was supported by the Russian Foundation for Basic Research (project no. 18-315-20009).
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no competing interests.
Statement of Informed Consent
Written informed consent for the use of biological material was obtained from all study participants.
The study was conducted in accordance with the World Medical Assembly Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects. This work was approved by the Ethics Committee of the Institute of Molecular Genetics of Russian Academy of Sciences.
Footnotes
Highlights
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases.
A highly rare phenomenon is described: i.e., monozygotic twins discordant for PD.
Processes directly related with the functioning of the nervous system were revealed.
PTGS2, SCN9A, and GRIK2 can be considered as potential candidate genes for PD.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anelya Kh. Alieva, Email: anelja.a@gmail.com
Margarita M. Rudenok, Email: margaritamrudenok@gmail.com
Ekaterina V. Novosadova, Email: novek-img@mail.ru
Ivan N. Vlasov, Email: invlasov@mail.ru
Elena L. Arsenyeva, Email: arslena@mail.ru
Anna V. Rosinskaya, Email: rosinskaya@bk.ru
Igor A. Grivennikov, Email: igorag@img.ras.ru
Petr A. Slominsky, Email: slomin@img.ras.ru
Maria I. Shadrina, Email: shadrina@img.ras.ru
References
- Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–186. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- Contractor A, Swanson GT, Sailer A, O'Gorman S, Heinemann SF. Identification of the kainate receptor subunits underlying modulation of excitatory synaptic transmission in the CA3 region of the hippocampus. J Neurosci. 2000;20:8269–8278. doi: 10.1523/JNEUROSCI.20-22-08269.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR, Hardy J, Lewis PA. Genetic neuropathology of Parkinson’s disease. Int J Clin Exp Pathol. 2008;1:217–231. [PMC free article] [PubMed] [Google Scholar]
- Cox JJ, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Cregg R, Momin A, Rugiero F, Wood JN, Zhao J. Pain channelopathies. J Physiol. 2010;588:1897–1904. doi: 10.1113/jphysiol.2010.187807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn BS, Elton R, M.o.t.U.D. Committee . Unified Parkinson’s disease rating scale. Recent developments in Parkinson’s disease. Florham Park: Macmillan Health Care Information; 1987. [Google Scholar]
- Fisahn A, Contractor A, Traub RD, Buhl EH, Heinemann SF, McBain CJ. Distinct roles for the kainate receptor subunits GluR5 and GluR6 in kainate-induced hippocampal gamma oscillations. J Neurosci. 2004;24:9658–9668. doi: 10.1523/JNEUROSCI.2973-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T. Molecular pathogenesis of Parkinson disease: insights from genetic studies. Expert Rev Mol Med. 2009;11:e22. doi: 10.1017/S1462399409001148S1462399409001148. [DOI] [PubMed] [Google Scholar]
- Goetz CG, et al. Movement Disorder Society Task Force report on the Hoehn and Yahr staging scale: status and recommendations. Mov Disord. 2004;19:1020–1028. doi: 10.1002/mds.20213. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Hawkes CH, Shephard BC, Daniel SE. Olfactory dysfunction in Parkinson’s disease. J Neuronal Neurosurg Psychiat. 1997;62:436–446. doi: 10.1136/jnnp.62.5.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettne KM, et al. The implicitome: a resource for rationalizing gene-disease associations. PLoS One. 2016;11:e0149621. doi: 10.1371/journal.pone.0149621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, et al. NaV1.7 and pain: contribution of peripheral nerves. Pain. 2018;159:496–506. doi: 10.1097/j.pain.0000000000001119. [DOI] [PubMed] [Google Scholar]
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Lang AE. Parkinson's disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- Kalinderi K, Bostantjopoulou S, Fidani L. The genetic background of Parkinson's disease: current progress and future prospects. Acta Neurol Scand. 2016;134:314–326. doi: 10.1111/ane.12563. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Andreasson KI, Isakson PC, Worley PF. Cyclooxygenases and the central nervous system. Prostaglandins. 1997;54:601–624. doi: 10.1016/S0090-6980(97)00128-7. [DOI] [PubMed] [Google Scholar]
- Kaut O, et al. Epigenome-wide DNA methylation analysis in siblings and monozygotic twins discordant for sporadic Parkinson's disease revealed different epigenetic patterns in peripheral blood mononuclear cells. Neurogenetics. 2017;18:7–22. doi: 10.1007/s10048-016-0497-x. [DOI] [PubMed] [Google Scholar]
- Klee CB, Crouch TH, Krinks MH. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc Natl Acad Sci U S A. 1979;76:6270–6273. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann DM. Presynaptic kainate receptors in the hippocampus: slowly emerging from obscurity. Neuron. 2001;32:561–564. doi: 10.1016/S0896-6273(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Lauri SE, Delany C, J Clarke VR, Bortolotto ZA, Ornstein PL, Isaac J TR, Collingridge GL. Synaptic activation of a presynaptic kainate receptor facilitates AMPA receptor-mediated synaptic transmission at hippocampal mossy fibre synapses. Neuropharmacology. 2001;41:907–915. doi: 10.1016/S0028-3908(01)00152-6. [DOI] [PubMed] [Google Scholar]
- Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- Lesage S, Brice A. Role of mendelian genes in "sporadic" Parkinson's disease. Parkinsonism Relat Disord. 2012;18(Suppl 1):S66–S70. doi: 10.1016/S1353-8020(11)70022-0. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo J, et al. A calcineurin- and NFAT-dependent pathway is involved in alpha-synuclein-induced degeneration of midbrain dopaminergic neurons. Hum Mol Genet. 2014;23:6567–6574. doi: 10.1093/hmg/ddu377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Maraschi A, et al. Parkin regulates kainate receptors by interacting with the GluK2 subunit. Nat Commun. 2014;5:5182. doi: 10.1038/ncomms6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minghetti L. Cyclooxygenase-2 (COX-2) in inflammatory and degenerative brain diseases. J Neuropathol Exp Neurol. 2004;63:901–910. doi: 10.1093/jnen/63.9.901. [DOI] [PubMed] [Google Scholar]
- Muller A, Mungersdorf M, Reichmann H, Strehle G, Hummel T. Olfactory function in Parkinsonian syndromes. J Clin Neurosci. 2002;9:521–524. doi: 10.1054/jocn.2001.1071. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Di Giovanni S. NFAT signaling in neural development and axon growth. Int J Dev Neurosci. 2008;26:141–145. doi: 10.1016/j.ijdevneu.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjan R, Mishra KP, Thakur AK. Inhibition of cyclooxygenase-2 (COX-2) initiates autophagy and potentiates MPTP-induced autophagic cell death of human neuroblastoma cells, SH-SY5Y: an inside in the pathology of Parkinson's disease. Mol Neurobiol. 2018;55:8038–8050. doi: 10.1007/s12035-018-0950-y. [DOI] [PubMed] [Google Scholar]
- Planken A, Kurvits L, Reimann E, Kadastik-Eerme L, Kingo K, Koks S, Taba P. Looking beyond the brain to improve the pathogenic understanding of Parkinson's disease: implications of whole transcriptome profiling of Patients' skin. BMC Neurol. 2017;17:6. doi: 10.1186/s12883-016-0784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Neural activity regulates synaptic properties and dendritic structure in vivo through calcineurin/NFAT signaling. Neuron. 2009;62:655–669. doi: 10.1016/j.neuron.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Shewchuk BM. Prostaglandins and n-3 polyunsaturated fatty acids in the regulation of the hypothalamic-pituitary axis. Prostaglandins Leukot Essent Fat Acids. 2014;91:277–287. doi: 10.1016/j.plefa.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer MJ, Bonifati V. The genetics of Parkinson's disease: progress and therapeutic implications. Mov Disord. 2013;28:14–23. doi: 10.1002/mds.25249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Ortolaza AI, Ross OA. Genetic susceptibility variants in parkinsonism. Parkinsonism Relat Disord. 2016;22(Suppl 1):S7–S11. doi: 10.1016/j.parkreldis.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33:e181. doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Sakai R. Ligands for ionotropic glutamate receptors. Prog Mol Subcell Biol. 2009;46:123–157. doi: 10.1007/978-3-540-87895-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teismann P, et al. Cyclooxygenase-2 is instrumental in Parkinson's disease neurodegeneration. Proc Natl Acad Sci U S A. 2003;100:5473–5478. doi: 10.1073/pnas.0837397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology C The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A, Kirchhoff F. Glutamate-mediated neuronal-glial transmission. J Anat. 2007;210:651–660. doi: 10.1111/j.1469-7580.2007.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, et al. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature. 2011;472:186–190. doi: 10.1038/nature09975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. L. Database resources of the National Center for Biotechnology. Nucleic Acids Research. 2003;31(1):28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard CM, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson's disease. Cell Rep. 2014;9:1173–1182. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wang Q, Liang X, Andreasson K. Divergent effects of prostaglandin receptor signaling on neuronal survival. Neurosci Lett. 2007;421:253–258. doi: 10.1016/j.neulet.2007.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 14 kb)