Abstract
Background
A sudden rise of Candida krusei candidemia cases was noticed in our hospital within 1 year with maximum cases from paediatric unit. The present study reports the results of epidemiological investigation of possible outbreak of candidemia by C. krusei in paediatric unit at our tertiary care centre.
Methods
Clinical characteristics and risk factors associated with C. krusei candidemia were evaluated. Yeast identification and antifungal susceptibility testing was performed according to standard protocol. To find the potential source of C. krusei in hospital environment and hand colonization, swabs were collected from different fomites (n = 40) and hand washings from 24 health care workers (HCW), respectively. Infection control and prevention practices were intensified following the recognition of outbreak. Genetic typing was done by fluorescent amplified fragment length polymorphism (FAFLP) technique. Case–control comparison was performed with C. tropicalis and C. pelliculosa cases.
Results
Candida krusei fungaemia significantly affected paediatric group (82/186, 44%) as compared to adults (14/130, 10.8%; p < 0.001). Among paediatric group, maximum isolation was reported from neonatal unit of paediatric emergency (NUPE). C. krusei was isolated from hands of one HCW and washbasin in NUPE. FAFLP revealed clonality between blood and environmental isolates indicating cross-transmission of C. krusei. Gastrointestinal disease (p = 0.018), previous antibiotics (p = 0.021) especially to carbapenems (p = 0.039), was significant among C. krusei candidemia cases compared to C. pelliculosa cases.
Conclusion
We report the largest outbreak of C. krusei candidemia in paediatric unit within 1 year with isolation of related strains from environment and hands of HCW. Routine screening of hand hygiene practices revealed non-compliance to standard practices leading to the increase in C. krusei candidemia cases.
Keywords: Candida krusei, Candidemia, Outbreak, Paediatrics, Molecular typing
Introduction
Candidemia, the fourth most common cause of nosocomial sepsis accounts for high attributable mortality (25–60%) despite the availability of advanced diagnostic and management strategies [1]. The developing countries report a 4–15 times higher rate of candidemia than developed countries although systematic surveillance data are lacking [2]. Paediatric intensive care units (ICUs) exhibit extremely high incidence of candidemia in developing nations when compared to developed countries (42.7 vs. 0.043–0.47 per 1000 admissions) [1]. Among Candida species causing candidemia, non-albicans Candida (NAC) species are the leading agents in developing countries with a noticeable decrease of C. albicans in developed nations too [3]. C. glabrata is the major species prevalent in the USA and Europe, while C. tropicalis and C. parapsilosis constitute maximum cases in developing countries [4–7]. There has been a notable emergence of rare yeasts causing fungaemia worldwide. Such scenario has been observed in India as well. Few outbreaks caused by Pichia anomala, Kodamaea ohmeri and C. auris have been reported from India [8–10]. Although relatively uncommon, C. krusei is implicated in causing candidemia outbreaks worldwide with a very few case numbers. Earlier, two C. krusei candidemia outbreaks have been reported from India in neonatal ICU (NICU) with seven and six cases [11, 12]. The rise of C. krusei is of concern due to its intrinsic resistance to fluconazole. Few studies suggest that C. krusei infection is acquired in the hospital settings although mechanisms are not delineated [13]. We noticed a sudden and sustained surge of candidemia cases by C. krusei (n = 82) at paediatrics unit of our centre during a 1-year period which is the highest number recorded in any C. krusei outbreak worldwide. The present study reports the results of the epidemiological investigation of the prolonged outbreak of candidemia by C. krusei at our tertiary care centre.
Methods
Outbreak Setting
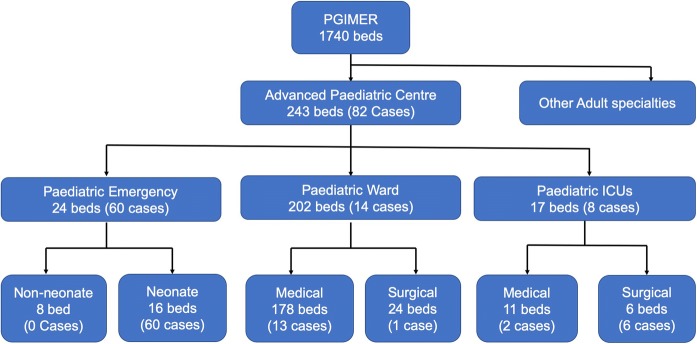
Postgraduate Institute of Medical Education and Research, Chandigarh, is a 1740 bedded public sector tertiary care hospital in North India. Advanced paediatric centre (APC), a separate building comprising 243 beds, faces a huge rush of patients with around 15–16 thousand admissions every year. The detailed set-up of APC along with the number of C. krusei candidemia cases identified in different units is provided in Fig. 1. The neonatal unit of paediatric emergency (NUPE) is a high-dependency unit consisting of 16 beds in which neonates born in other hospitals and visiting our hospital are admitted.
Fig. 1.
Flowchart showing different wards with the number of beds and C. krusei cases in each ward and ICUs
Diagnosis of Candidemia
Candidemia was defined as the isolation of Candida species from at least one blood culture in the presence of clinical features of sepsis. Aseptically drawn peripheral venous blood (1–2 mL paediatrics) was inoculated into BACTEC blood culture bottles (BD BACTEC™ 9240, New Jersey, USA) and incubated according to manufacturer’s instructions. The samples which beeped a positive signal were subjected to Gram staining and inoculated onto blood agar, Sabouraud dextrose agar and incubated at 37 °C for 24 h. Candida species were identified by phenotypic methods (corn meal agar, sugar fermentation and assimilation and germ tube test) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Billerica, MA, USA). Few isolates were subjected to molecular identification for confirmation by Sanger sequencing targeting internal transcribed spacer (ITS) region of ribosomal DNA [14].
Case Definition
A case patient was defined as any patient admitted in paediatric ward during 1 January 2014 to 31 December 2014, with clinical features of sepsis and isolation of C. krusei from blood culture. Blood culture positive from same patient within 30 days after first positive culture was considered as a single case. A neonate was defined as an infant ≤ 28 days of age. Medical records of these paediatric patients were examined to obtain detailed clinical history.
Case–Control Analysis
A retrospective case–control study was performed in the paediatric ward to determine the clinical characteristics and risk factors for C. krusei candidemia. Patients with C. tropicalis candidemia (n = 36) and C. pelliculosa candidemia (n = 27) admitted in paediatric unit in same period were included as controls as these were the next common species isolated after C. krusei during that year.
Environmental Surveillance
To find the potential source of C. krusei, environmental surveillance was carried out as follows: (a) samples from different surfaces in paediatric wards (n = 40) such as bed railing, washbasins, taps, medicine trolley and ventilator surfaces were collected using swabs, (b) whereas hand washings of health care workers (HCW) (n = 24) posted in NUPE were collected using standard bag broth technique. All the samples were processed according to the method described by Strausbaugh et al. [15].
Infection Control and Preventive Intervention
Following the recognition of the rising numbers of C. krusei candidemia cases, a dedicated infection control nurse (ICN) was posted in NUPE to oversee the infection control and prevention (ICP) practices. The other ICP practices including surface and equipment decontamination protocols were reviewed and strengthened. Infection control nurses, trained in observation and recording of hand hygiene (HH) activities, conducted a prospective audit in the paediatric unit. The nurse spent 20 min during a busy period each day observing and recording compliance of all health care staff and patient attendants in the unit. HH opportunities included before and after patient care, before an invasive procedure, when hands were visibly soiled with blood or body fluids of patients, and before and after removing gloves. To minimize the Hawthorne effect, the participants did not know that they were being observed for HH compliance. Both washing hands with soap and water and rubbing hands with alcohol products counted as HH performed. The total number of opportunities and actual HH performed was recorded by the observer. Compliance was expressed as percentage (observed HH/total opportunities for HH).
Antifungal Susceptibility Testing (AFST) of Isolates
The minimum inhibitory concentrations (MICs) were determined for all the C. krusei isolates for amphotericin B (Sigma-Aldrich, India), fluconazole (Sigma-Aldrich, India), voriconazole (Pfizer Central Research, Tadworth, UK), itraconazole (Sigma-Aldrich, India), posaconazole (Merck Sharp and Dohme, Gurgaon, India), caspofungin (Merck Sharp and Dohme, India), micafungin (Astellas Pharma, USA) and anidulafungin (Pfizer Central Research, Tadworth, UK) using the microbroth dilution method as per the M27-A3 protocol of Clinical and Laboratory Standards Institute (CLSI) [16].
Molecular Typing
Fluorescent amplified fragment length polymorphism (FAFLP) technique was carried out to evaluate the clonality of the clinical and environmental isolates from various sources [9, 17]. Clonality is defined as the isolates having its origin from the single source/cell and has low degree of genetic exchange due to asexual reproduction [18]. C. albicans and C. tropicalis were used as an out-group, and C. krusei ATCC 6258 was used as control. Additionally, representative isolates of C. krusei isolated from 2014–2018 were also subjected to molecular typing to determine whether the outbreak strains are still circulating in the hospital. DNA extraction from the isolates and its quality check was performed as described earlier. DNA was digested using a pair of restriction enzymes HpyCH4IV and MseI (New England Biolabs, Ipswich, MA, USA). DNA ligase enzyme was used to ligate respective adapters to the fragments generated. Pre-selective amplification was done using per-selective primers of HpyCH4IV (5′-GTAGACTGCGTACCCGT-3′) and MseI (5′-GATGAGTCCTGACTAA-3′). For selective amplification, HpyCH4IV primer with one selective residue (5′-GTAGACTGCGTACCCGTC-3′) and MseI primer (5′-GATGAGTCCTGACTAACA-3′) with two selective residues were used and the primers of HpyCH4IV were labelled with 6-FAM. Capillary electrophoresis of the amplified products along with LIZ 500 (size standard marker) was performed in an ABI automated DNA Sequencer 3130 (Applied Bioscience, Foster City, CA, USA). Typing data were then imported to BIONUMERICS v 7.6 software (Applied Maths, Ghent, Belgium). The fingerprint curves were converted into bands, and normalized bands for each lane were assigned using the band position of the reference dye (LIZ500). The similarity coefficient was estimated by Pearson correlation with negative similarities clipped to zero. Cluster analysis was performed by the unweighted pair group method with arithmetic mean (UPGMA) [17]. Isolates showing > 96% similarity were considered to exhibit clonal origin.
Statistical Analysis
The data were analysed using SPSS 22.0 (SPSS Inc, Chicago, IL). Descriptive analysis is presented as frequencies; medians and range; or means and standard deviation (SD), as appropriate. Pearson’s Chi-square test and Fisher’s exact test were used to compare categorical variables, while the Student’s t-test, ANOVA, Mann–Whitney or Kruskal–Wallis tests were employed for continuous data. Two-tailed p values < 0.05 were taken as significant. Normalcy of the three groups was determined by Kolmogorov–Smirnov test. A p value < 0.05 was considered statistically significant.
Results
A total of 316 cases of candidemia were diagnosed during a period of 1 year between 1 January and 31 December 2014. The majority (58.8%, 186/316) cases were below 18 years of age, while adults constituted 41.2% (130/316) of cases. Among the paediatric group, 74.7% (139/186) were neonates. Overall, C. krusei (30.3%, 96/316) was the commonest isolate followed by C. tropicalis (27.8%, 88/316), C. albicans (14.8%, 47/316) and C. pelliculosa (9.8%, 31/316). Among the paediatric population (n = 186), C. krusei (44%, 82/186) formed the majority followed by C. tropicalis (19.4%, 36/186), C. pelliculosa (14.5%, 27/186), C. albicans (8.6%, 16/186), C. parapsilosis (4.3%, 8/186), C. guilliermondii (2.2%, 4/186), K. ohmeri (1.6%, 3/186), C. orthopsilosis (1%, 2/186), T. asahii (1%, 2/186), Cyberlindnera jadinii (1%, 2/186), C. auris (0.5%, 1/186), L. elongisporus (0.5%, 1/186), C. fabianii (0.5%, 1/186) and C. glabrata (0.5%, 1/186).
Candida krusei fungaemia significantly affected paediatric group (44%, 82/186) as compared to adults (10.8%, 14/130; p < 0.001). The median age of the paediatric patients was 13 days (IQR = 6–28 days) with the majority being male (70.7%, 58/82). Among 82 C. krusei cases, 65 (79.3%, 65/82) were neonates and 72.3% (47/65) of them were males. The median age of neonates was 9 days (IQR 6–28 days). Only 13.8% (9/65) were premature. In-hospital mortality data available for 74 patients showed mortality at 22.94% (17/74).
The month-wise distribution of paediatric candidemia cases by C. krusei, C. tropicalis and C. pelliculosa during the study period is depicted in Fig. 2. The majority of the cases clustered in the months of September and October 2014. Maximum cases were reported from NUPE (73.1%, 60/82) followed by paediatric medicine wards (15.9%, 13/82), paediatric surgery ICU (7.3%, 6/82), paediatric medicine ICU (2.4%, 2/82) and paediatric surgery ward (1.2%, 1/82) (Fig. 3).
Fig. 2.
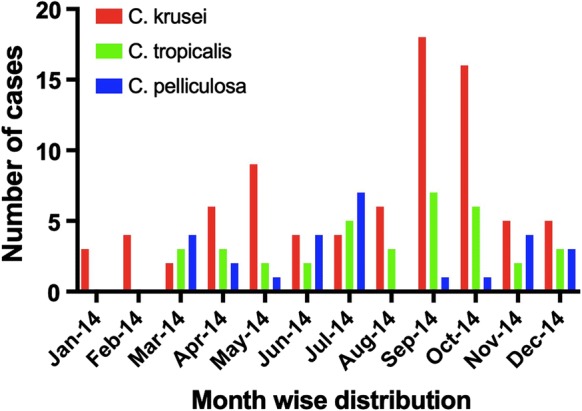
Month-wise distribution of paediatric candidemia cases by C. krusei, C. tropicalis and C. pelliculosa
Fig. 3.
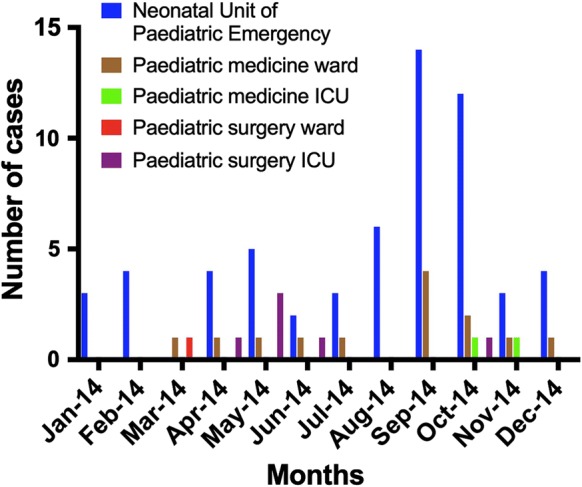
Ward-wise distribution of paediatric candidemia cases by C. krusei
Case–Control Study
The clinical characteristics and risk factor analysis are given in Table 1. Significant risk factors observed in C. krusei candidemia cases included gastrointestinal disease (p = 0.018), prior use of antibiotics (p = 0.021), especially exposure to carbapenems (p = 0.039) when compared to C. pelliculosa candidemia cases. Other risk factors such as the presence of congenital malformations (p = 0.002), ICU admission (p = 0.009) and mechanical ventilation (p = 0.003), significantly observed among C. tropicalis candidemia, did not have any association with C. krusei candidemia. The in-hospital mortality rate was significantly lower among C. krusei candidemia patients as compared to C. tropicalis candidemia patients [23% (17/74) vs. 42.9% (15/35); p = 0.033]. Time to blood culture positivity for C. krusei was observed to be much shorter as compared to C. tropicalis [median 2 days (IQR = 1–4) vs. 5 days (IQR = 1–13.75); p value 0.007].
Table 1.
Evaluation of clinical characteristics of C. krusei candidemia cases in paediatric ward
| C. krusei | C. tropicalis | C. pelliculosa | C. krusei versus C. tropicalisp value | C. krusei versus C. pelliculosap value | |
|---|---|---|---|---|---|
| (n = 82) | (n = 36) | (n = 27) | |||
| No. (%) | No. (%) | No. (%) | |||
| Neonates | |||||
| Total | 65 (79.3) | 25 (69.4) | 22 (81.5) | 0.248 | 0.804 |
| Males | 47 (72.4) | 17 (68) | 16 (72.7) | ||
| Females | 18 (27.6) | 8 (32) | 6 (27.3) | ||
| Children | |||||
| Total | 17 (20.7) | 11 (30.6) | 5 (18.5) | 0.248 | 0.804 |
| Males | 11 (64.7) | 6 (54.5) | 5 (100) | ||
| Females | 6 (35.3) | 5 (45.5) | 0 (0) | ||
| Age [median (IQR)] | 13 days (6–28 days) | 14 days (4–112 days) | 13 days (2–28 days) | 0.865 | 0.296 |
| Days to positivity to blood culture (median, IQR) | 2 (1–4) days | 5 (1–13.75) days | 1 (1–3) days | 0.007* | 0.794 |
| Underlying disease | |||||
| Congenital malformations | 14 (17.1) | 16 (44.4) | 1 (3.7) | 0.002* | 0.248 |
| Gastrointestinal | 20 (24.4) | 15 (41.7) | 1 (3.7) | 0.059 | 0.018* |
| Late onset neonatal sepsis | 28 (34.1) | 11 (30.6) | 8 (29.6) | 0.703 | 0.665 |
| Early onset neonatal sepsis | 10 (2.8) | 1 (2.8) | 5 (18.5) | 0.095 | 0.297 |
| Neurological | 13 (15.9) | 4 (11.1) | 4 (14.8) | 0.499 | 0.584 |
| Jaundice | 4 (4.9) | 2 (5.6) | 0 (0) | 0.596 | 0.314 |
| Cardiac | 3 (3.7) | 2 (5.6) | 1 (3.7) | 0.484 | 0.686 |
| Renal | 1 (1.2) | 0 (0) | 2 (7.4) | 0.695 | 0.151 |
| Infection | 5 (6.1) | 3 (8.3) | 3 (11.1) | 0.463 | 0.312 |
| Gastroenteritis | 5 (6.1) | 1 (2.8) | 1 (3.7) | 0.404 | 0.537 |
| Tetanus | 0 (0) | 1 (2.8) | 1 (3.7) | 0.305 | 0.248 |
| Risk factors | |||||
| Prematurity | 9 (11) | 3 (8.3) | 3 (11) | 0.754 | 0.613 |
| ALL | 0 (0) | 1 (2.8) | 0 (0) | 0.305 | - |
| ICU admission | 19 (23.2) | 17 (47.2) | 2 (7.4) | 0.009* | 0.072 |
| Iatrogenic risk factors | |||||
| Urinary catheter | 5 (6.1) | 2 (5.6) | 0 (0) | 0.637 | 0.234 |
| Central line catheter | 1 (1.2) | 3 (8.3) | 0 (0) | 0.084 | 0.752 |
| Nasogastric/orogastric tube | 9 (11) | 7 (19.4) | 4 (14.8) | 0.171 | 0.407 |
| Endotracheal tube | 6 (7.3) | 3 (8.3) | 0 (0) | 0.556 | 0.173 |
| Mechanical ventilation | 15 (18.3) | 16 (44.4) | 2 (7.4) | 0.003* | 0.147 |
| Antibiotics | 24 (29.3) | 13 (36.1) | 2 (7.4) | 0.461 | 0.021* |
| Vancomycin | 15 (18.3) | 8 (22.2) | 1 (3.7) | 0.620 | 0.052 |
| Piperacillin–tazobactam | 3 (3.7) | 1 (2.8) | 1 (3.7) | 0.643 | 0.686 |
| Aminoglycosides | 15 (18.3) | 5 (13.9) | 2 (7.4) | 0.557 | 0.147 |
| Carbapenems | 16 (19.5) | 7 (19.4) | 1 (3.7) | 0.993 | 0.039* |
| Cephalosporins | 8 (9.8) | 1 (2.8) | 1 (3.7) | 0.176 | 0.294 |
| Metronidazole | 2 (2.4) | 4 (11.1) | 1 (3.7) | 0.069 | 0.578 |
| Erythromycin | 2 (2.4) | 0 (0) | 0 (0) | 0.481 | 0.564 |
| Mortality | 17 (23) | 15 (42.9) | 4 (14.8) | 0.033* | 0.371 |
ALL acute lymphoblastic leukaemia, ICU intensive care unit, IQR interquartile range, italicized words in the first column indicates the group under which different factors/parametes were analysed
*Statistically significant
Environmental Surveillance
Of 40 environmental samples collected from different sites in paediatric wards, C. krusei was isolated from washbasin of paediatric emergency ward. Among hand wash samples, C. krusei was present in only one HCW in NUPE. Other major Candida species such as C. tropicalis (n = 11), C. albicans (n = 6), C. parapsilosis (n = 3) and C. glabrata (2) were also isolated from environment and hands of HCWs from paediatric emergency ward.
Infection Control and Preventive Intervention
The routine surveillance data of hand hygiene compliance in paediatric unit of our hospital revealed lower rate in 2014 (58.51%, 433/740) which corroborated with increased C. krusei candidemia cases in that year. After intensifying hand hygiene practices, compliance increased from 58.51% in 2014 to 73.2% (1058/1445) in 2016 and a concurrent decline in number of C. krusei candidemia cases was observed (Fig. 4).
Fig. 4.
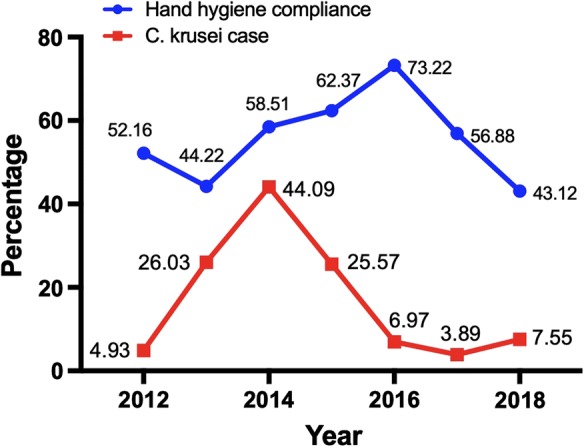
Year-wise trend in paediatric candidemia cases caused by C. krusei and hand hygiene compliance in paediatric units
Antifungal Susceptibility Testing (AFST)
MIC, geometric mean, MIC50 and MIC90 of 82 C. krusei isolates against various antifungal agents are provided in Table 2. Amphotericin B MIC’s of ≥ 1 mg/L was noted in 70.7% (58/82) isolates. Only 8.6% (7/82), 4.8% (4/82) and 6.1%, (5/82) of the isolates tested had the MIC of ≥ 1 mg/L against caspofungin, micafungin and anidulafungin, respectively. Among the azole group of antifungals, all isolates had MIC of ≤ 0.5 mg/L against itraconazole, voriconazole and posaconazole except for two isolates which showed MICs of ≥ 2 mg/L against voriconazole.
Table 2.
In-vitro antifungal susceptibility data (MIC, mg/L) of 82 C. krusei isolates against seven antifungal drugs
| Antifungals | GM | Range | MIC 50 | MIC 90 |
|---|---|---|---|---|
| Amphotericin B | 0.896341 | 0.25–2 | 1 | 1 |
| Voriconazole | 0.413049 | 0.05–8 | 0.25 | 0.25 |
| Itraconazole | 0.305132 | 0.12–0.5 | 0.25 | 0.5 |
| Posaconazole | 0.24122 | 0.06–0.5 | 0.25 | 0.5 |
| Caspofungin | 0.350802 | 0.12–2 | 0.12 | 0.5 |
| Anidulafungin | 0.275976 | 0.03–4 | 0.12 | 0.5 |
| Micafungin | 0.447805 | 0.06–12 | 0.12 | 0.5 |
GM geometric mean, MIC minimum inhibitory concentration
Molecular Typing [Fluorescent Amplified Fragment Length Polymorphism (FAFLP)]
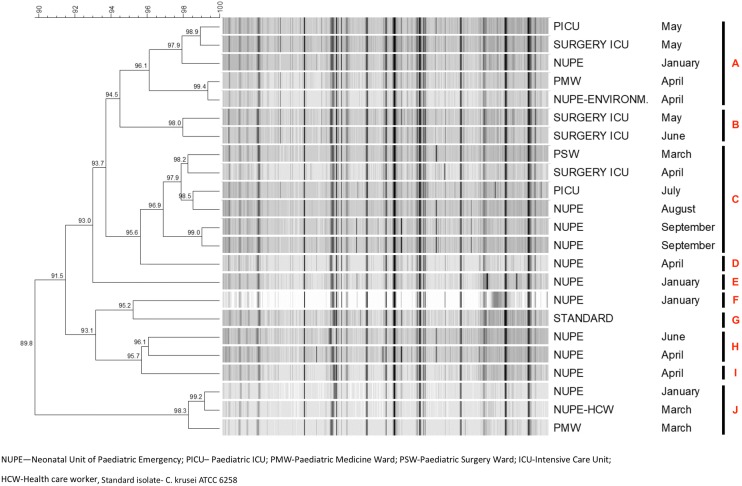
FAFLP analysis of all C. krusei isolates from 2014 showed a similarity coefficient of > 89.8% and formed ten different clusters (Fig. 5). C. krusei isolates from blood (n = 20) along with an isolate from environment (isolate = NUPE-ENVIRONM, washbasin), hand isolate from HCW (NUPE-HCW) and control strain (C. krusei ATCC 6258, STANDARD) were tested. Cluster ‘A’ consisted of four blood culture isolates and one environmental isolate obtained during January to May. Cluster ‘B’ consists of two isolates obtained from surgical ICU. Cluster ‘C’ consisted of six blood isolates, of which three belonged to NUPE and one isolate each belonged to PICU, surgical ICU and PSW. Cluster ‘J’ consists of three isolates of which two were from blood and one from hands of HCW. Clusters ‘D’, ‘E’, ‘F’ and ‘I’ consist of one isolate each. C. krusei ATCC 6258 formed a separate cluster (Cluster ‘G’).
Fig. 5.
Fingerprint profile generated by FAFLP analysis of C. krusei isolated from blood and environment in the year 2014
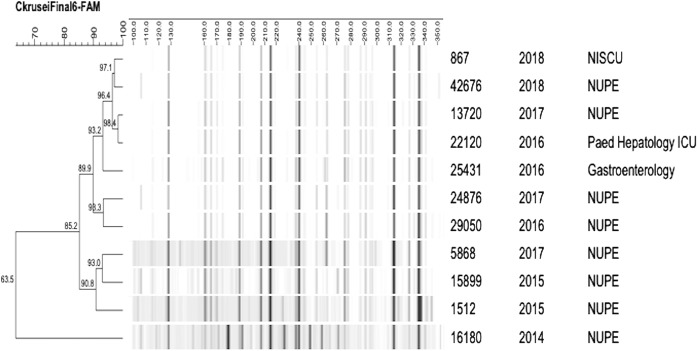
FAFLP analysis of representative C. krusei isolates over different years (2014–2018) showed that isolates from 2014 were not genetically related to C. krusei from other years (Fig. 6). Major cluster containing four isolates belonging to year 2016, 2017 and 2018 showed similarity of > 96.4%. None of the other isolates tested showed any clonality with isolates of other years. The majority of the isolates tested belonged to NUPE ward, and one each from paediatric gastroenterology ward and paediatric hepatology ICU.
Fig. 6.
Fingerprint profile generated by FAFLP analysis of C. krusei isolated over different years
Discussion
Candida krusei candidemia outbreak is reported very infrequently both in adults and paediatric population across the globe. The present study represents the largest cluster (n = 82) of C. krusei candidemia cases reported till date. A sudden upsurge in the number of C. krusei candidemia cases to 44.09% in the year 2014 as compared to 4.93% in 2012 prompted us to perform epidemiological investigation. The majority of the cases were neonates (79.3%) who are an established risk group for developing candidemia due to their weak immune system and fragile skin/gut barrier [18]. The previously described outbreaks of C. krusei candidemia are either from neonates or from haematology oncology wards (Table 3). We noticed the presence of gastrointestinal disease and prior exposure to antibiotics, especially to carbapenems as significant risk factors for acquiring C. krusei candidemia compared to C. pelliculosa candidemia patients. These risk factors along with total parenteral nutrition and central venous catheter are previously reported in neonatal C. krusei candidemia cases [19, 20]. The use of fluconazole prophylaxis is reported as a risk factor for the majority of C. krusei outbreaks though this practice is not prevalent in our hospital [18, 19]. The overall in-hospital mortality rate (22.9%, n = 17) among C. krusei infected patients was lower than C. tropicalis (42.9%) candidemia patients which may be attributed to its lower virulence potential as described previously [21].
Table 3.
Literature review of outbreaks due to Candida krusei worldwide
| Sl no | Year | Country | Hospital area | Risk factor | Patients/HCW/environment | Duration | Origin | Typing method | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1996 | USA | Haemato-oncology | Fluconazole prophylaxis | 4/–/– | 1 month | Genotype A/A1 | REA | Noskin et al. |
| 2 | 2006 | Netherlands | Haemato-oncology | Fluconazole prophylaxis | 13/–/– | 2.5 years | Genotype 1/6/8 | CKTNR/CKRS1 assay | Vos et al. |
| 3 | 2007 | Finland | Haemato-oncology | Fluconazole prophylaxis | 6/–/2 (sink trap) | 1 month | Same cluster | CHEF PFGE | Hautala et al. |
| 4 | 2011 | Portugal | Neutropenia unit of haematology | Fluconazole prophylaxis (n = 2) | 3/–/2 (surfaces) | 2 months | Related strains | REA | Ricardo et al. |
| 5 | 2012 | Brazil | NICU | Fluconazole prophylaxis (n = 1) | 3/–/– | 3 years | NA | NA | Selma Amaral-Lopes et al. |
| Fluconazole therapy (n = 1) | |||||||||
| Premature | |||||||||
| 6 | 2014 | India | NICU | Prematurity | 7/–/1 (multi-electrolyte dextrose infusate) | 11 days | Clonal | RAPD | Rongpharpi et al. |
| 7 | 2015 | India (2 outbreaks) | NICU | Prematurity, low birth weight | (a) 2/–/1 (multi-electrolyte dextrose infusate) | (a) 2 days | Related | RAPD | Duggal et al. |
| (b) 4/–/1 (suction apparatus) | (b) 10 days | ||||||||
| 8 | 2018 | South Africa (2 outbreaks) | Neonatal unit | Neonates | (a) 48/–/– | Both 4 months | NA | NA | van Schalkwyk et al. |
| (b) 41/–/– | |||||||||
| 9 | 2018 | India | Paediatric unit | Paediatrics including Neonates | 82/1/1 (hands of healthcare worker and washbasin) | 1 year | Clonal | FAFLP | Present study |
FAFLP fluorescent amplified fragment length polymorphism, HCW health care workers, NA not available, NICU neonatal intensive care unit, PFGE pulse field gel electrophoresis, RAPD random amplified polymorphic DNA, REA restriction endonuclease assay, USA United States of America
We conducted an environmental surveillance of the wards as well as hand yeast carriage of HCWs to find the possible source of outbreak. C. krusei was isolated from washbasin of NUPE consistently being used by the HCWs after attending the patients. The FAFLP results suggest that three major clones of C. krusei were circulating throughout the year in these paediatric units. The majority of the isolates revealed genetic similarity with the environmental isolates. NUPE ward was common among all the four clusters of C. krusei isolates. This is probably because NUPE ward is an emergency ward where the first physical examination of all the paediatric patients is done before being transferred to different units. Due to huge rush of neonatal patients in NUPE, high patient-to-bed ratio may compromise the infection control practices and contribute to higher risk of cross-transmission. Probability of patients acquiring C. krusei during their stay in NUPE cannot be neglected.
Exogenous acquisition of Candida species is common among several Candida species including C. krusei, C. tropicalis, C. parapsilosis, C. pelliculosa, C. lusitaniae and recently C. auris [22–29]. Table 3 shows the previous C. krusei candidemia outbreaks across the world [11–13, 18, 22, 30–32]. Few of them could localize the environmental source of transmission. Hautala et al. [13] and Ricardo et al. [30] from Europe could isolate the related strains of outbreak from sink traps and surfaces, respectively. Several recurrent outbreaks of C. krusei candidemia have been reported in neonatal ICUs due to contaminated environmental sources from India and South Africa [11, 12, 18]. Rongpharpi et al. and Duggal et al. reported three C. krusei outbreaks over a period of 4 years (September 2011, May 2013 and October 2014). In their study from a single centre, they found contaminated intravenous multi-electrolyte dextrose infusate bottle (different brands) as the source in first two outbreaks and a suction apparatus in the third outbreak [11, 12]. A large outbreak of C. krusei candidemia (n = 48, 8.2% incidence) was reported by van Schalkwyk et al. [18] among infants of Gauteng Province in South Africa over a period of 4 months in 2014. Subsequently, another large outbreak of 41 cases of C. krusei was reported in April–July 2015 although source could not be traced in both these outbreaks. None of the previous studies have reported isolation of C. krusei from hands of HCWs. We could track down the possible sources of transmission of C. krusei in our study to a washbasin in the NUPE ward and hands of one of the HCWs who was posted in different wards at the same time. Moist surface like washbasins tend to provide a niche for the survival of yeasts and may get into the hands of HCWs by consistently using this washbasin. Nevertheless, overcrowding, limited infrastructural capacity, compromise in preventive hygiene measures and non-compliance with infection control practices may also contribute in causing outbreaks.
Rapid identification of aetiologic agent is a prerequisite in any outbreak situation. All the yeasts isolated in this study from the blood and environment were accurately identified by MALDI-TOF MS as per our previous protocol [33, 34]. Restriction endonuclease assay (REA), pulse field gel electrophoresis (PFGE) and random amplified polymorphic DNA (RAPD) methods were used in previous studies to find the relatedness among clinical and environmental strains [11–13, 30]. Here, we utilized the most robust and reproducible typing method, FAFLP technique to establish genetic relatedness among blood and environmental isolates. Previously, we reported the effectiveness of AFLP in determining genetic relatedness of isolates due to other fungi from our centre[8, 9, 29, 34–36]. Clustering of environmental isolates with the majority of the blood isolates indicates the hospital environment as the major source of infection. The isolation of C. krusei from washbasin of NUPE increases the probability of transmission to other fomites as well. Additionally, few C. krusei blood isolates clustered with isolate from HCW. This indicates cross-transmission of C. krusei from environmental sources to hands of HCWs and then to the susceptible patients. Moreover, the representative isolates of C. krusei from 2014 analysed did not show genetic relatedness with isolates from other years suggesting frequent introduction of new strains periodically into the hospital [11–13, 30].
Control of the outbreak is a daunting task among overcrowded hospitals but is possible by adhering to strict infection control practices. The overcrowding possibly due to high influx of patients per day, high baby-to-bed ratio and lower staff-to-baby ratio led to an extended outbreak. Cross-transmission can be controlled by HCWs by strictly following hand hygiene protocols as per WHO recommendations [37]. Other crucial practices which involve the removal of catheters, optimal use of antibiotics, rapid transition from parenteral to enteral diet and avoidance of H2 blockers may help restricting acquisition of Candida species [1]. Periodic environmental decontamination and strict adherence to hand hygiene practices post-outbreak and also pinpointing the source helped containing further rise in numbers of C. krusei candidemia patients in our hospital. Decrease in the hand hygiene compliance again in 2018 could be due to the administration protocol in which nursing staff working in non-paediatric units were posted in these areas (Fig. 4).
In conclusion, we report here the largest outbreak of C. krusei candidemia in paediatric unit over a period of 1 year with the isolation of related strains from environment and hands of HCW. The strengthening of infection control practices helped control the outbreak in subsequent years. The molecular typing (FAFLP) of C. krusei strains from same wards from 2014 to 2018 did not show any relatedness signifying the emergence of new strains over the years. Suboptimal adherence to infection control practices probably due to overcrowded paediatric facility led to the huge number of C. krusei candidemia outbreak at our centre. Evaluating the colonization status of the patients may provide much better insight into acquisition of this infection.
Acknowledgements
We would like to acknowledge Indian Council of Medical Research for financial support.
Funding
Research Grant from funding agency—Indian Council of Medical Research with Grant No. AMR/TF/51/13ECDII.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Handling Editor: Takashi Sugita.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Harsimran Kaur, Email: drharsimranpgi@gmail.com.
Shamanth A. Shankarnarayana, Email: shamanth.adekhandi@gmail.com
Vinaykumar Hallur, Email: vinay118@gmail.com.
Jayashree Muralidharan, Email: mjshree64@gmail.com.
Manisha Biswal, Email: manisha.biswal@gmail.com.
Anup K. Ghosh, Email: anupkg3@gmail.com
Pallab Ray, Email: drpallabray@gmail.com.
Arunaloke Chakrabarti, Email: arunaloke@hotmail.com.
Shivaprakash M. Rudramurthy, Email: mrshivprakash@yahoo.com, Email: mrshivprakash@gmail.com
References
- 1.Kaur H, Chakrabarti A. Strategies to reduce mortality in adult and neonatal candidemia in developing countries. J Fungi. 2017;3:41. doi: 10.3390/jof3030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakrabarti A, editor. Fungal infections in Asia: the eastern frontier of mycology. Berlin: Elsevier; 2013. [Google Scholar]
- 3.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yapar N, Pullukcu H, Avkan-Oguz V, Sayin-Kutlu S, Ertugrul B, Sacar S, et al. Evaluation of species distribution and risk factors of candidemia: a multicenter case–control study. Med Mycol. 2011;49:26–31. doi: 10.3109/13693786.2010.501344. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M. Geographic variations in species distribution and echinocandin and azole antifungal resistance rates among Candida bloodstream infection isolates: report from the SENTRY Antimicrobial Surveillance Program (2008 to 2009) J Clin Microbiol. 2011;49:396–399. doi: 10.1128/JCM.01398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinbach WJ, Roilides E, Berman D, Hoffman JA, Groll AH, Bin-Hussain I, et al. Results from a prospective, international, epidemiologic study of invasive candidiasis in children and neonates. Pediatr Infect Dis J. 2012;31:1252–1257. doi: 10.1097/INF.0b013e3182737427. [DOI] [PubMed] [Google Scholar]
- 7.Messer SA, Jones RN, Fritsche TR. International surveillance of Candida spp. and Aspergillus spp.: report from the SENTRY Antimicrobial Surveillance Program (2003) J Clin Microbiol. 2006;44:1782–1787. doi: 10.1128/JCM.44.5.1782-1787.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti A, Singh K, Narang A, Singhi S, Batra R, Rao KLN, et al. Outbreak of Pichia anomala infection in the pediatric service of a tertiary-care center in northern India. J Clin Microbiol. 2001;39:1702–1706. doi: 10.1128/JCM.39.5.1702-1706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti A, Rudramurthy SM, Kale P, Hariprasath P, Dhaliwal M, Singhi S, et al. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care centre. Clin Microbiol Infect. 2014;20:O83–O89. doi: 10.1111/1469-0691.12337. [DOI] [PubMed] [Google Scholar]
- 10.Biswal M, Rudramurthy SM, Jain N, Shamanth AS, Sharma D, Jain K, et al. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect. 2017;97:363–370. doi: 10.1016/j.jhin.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Rongpharpi SR, Gur R, Duggal S, Kumar A, Nayar R, Xess I, et al. Candida krusei fungemia in 7 neonates: clonality tracked to an infusate. Am J Infect Control. 2014;42:1247–1248. doi: 10.1016/j.ajic.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 12.Duggal SD, Jena PP, Gur R, Kumar A, Rongpharpi SR, Pandey M, et al. Recurring events of Candida krusei septicaemia: first report from an ICU. J Mycol. 2015;2015:1–6. doi: 10.1155/2015/721424. [DOI] [Google Scholar]
- 13.Hautala T, Ikäheimo I, Husu H, Säily M, Siitonen T, Koistinen P, et al. A cluster of Candida krusei infections in a haematological unit. BMC Infect Dis. 2007;7:97. doi: 10.1186/1471-2334-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoch CLL, Seifert KA, Huhndorf S, Robert V, Spouge JLL, Levesque CAA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strausbaugh LJ, Sewell DL, Tjoelker RC, Heitzman T, Webster T, Ward TT, et al. Comparison of three methods for recovery of yeasts from hands of health-care workers. J Clin Microbiol. 1996;34:471–473. doi: 10.1128/JCM.34.2.471-473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd edition, Approved Standard M27-A3. Clin. Lab. Stand. Institute, Wayne, PA; 2008.
- 17.Chakrabarti A, Shivaprakash MR, Curfs-Breuker I, Baghela A, Klaassen CH, Meis JF. Apophysomyces elegans: epidemiology, amplified fragment length polymorphism typing, and in vitro antifungal susceptibility pattern. J Clin Microbiol. 2010;48:4580–4585. doi: 10.1128/JCM.01420-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Schalkwyk E, Iyaloo S, Naicker SD, Maphanga TG, Mpembe RS, Zulu TG, et al. Large outbreaks of fungal and bacterial bloodstream infections in a neonatal unit, South Africa, 2012–2016. Emerg Infect Dis. 2018;24:1204–1212. doi: 10.3201/eid2407.171087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patted S, Halkati P, Yavagal S, Patil R. Candida krusei infection presenting as a right ventricular mass in a two month old Infant. Ann Pediatr Cardiol. 2009;2:170. doi: 10.4103/0974-2069.58324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natale F, Castronovo A, Regoli D, De Curtis M, Manzoni P. Successful treatment with caspofungin of refractory Candida krusei candidemia in a very low birth weight preterm infant. Pediatr Infect Dis J. 2009;28:452. doi: 10.1097/INF.0b013e31819d1cf1. [DOI] [PubMed] [Google Scholar]
- 21.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36:288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 22.Noskin GA, Lee J, Hacek DM, Postelnick M, Reisberg BE, Stosor V, et al. Molecular typing for investigating an outbreak of Candida krusei. Diagn Microbiol Infect Dis. 1996;26:117–123. doi: 10.1016/S0732-8893(96)00204-0. [DOI] [PubMed] [Google Scholar]
- 23. da Silva CM, de Carvalho Parahym AMR, Leão MPC, de Oliveira NT, de Jesus Machado Amorim R, Neves RP. Fungemia by Candida pelliculosa (Pichia anomala) in a neonatal intensive care unit: a possible clonal origin. Mycopathologia. 2013;175:175–179. doi: 10.1007/s11046-012-9605-0. [DOI] [PubMed] [Google Scholar]
- 24.Hernández-Castro R, Arroyo-Escalante S, Carrillo-Casas EM, Moncada-Barrón D, Alvarez-Verona E, Hernández-Delgado L, et al. Outbreak of Candida parapsilosis in a neonatal intensive care unit: a health care workers source. Eur J Pediatr. 2010;169:783–787. doi: 10.1007/s00431-009-1109-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin H-C, Lin H-Y, Su B-H, Ho M-W, Ho C-M, Lee C-Y, et al. Reporting an outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. J Microbiol Immunol Infect. 2013;46:456–462. doi: 10.1016/j.jmii.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Sherertz RJ, Gledhill KS, Hampton KD, Pfaller MA, Givner LB, Abramson JS, et al. Outbreak of Candida bloodstream infections associated with retrograde medication administration in a neonatal intensive care unit. J Pediatr Mosby. 1992;120:455–461. doi: 10.1016/S0022-3476(05)80920-5. [DOI] [PubMed] [Google Scholar]
- 27.Welbel SF, McNeil MM, Kuykendall RJ, Lott TJ, Pramanik A, Silberman R, et al. Candida parapsilosis bloodstream infections in neonatal intensive care unit patients: epidemiologic and laboratory confirmation of a common source outbreak. Pediatr Infect Dis J. 1996;15:998–1002. doi: 10.1097/00006454-199611000-00013. [DOI] [PubMed] [Google Scholar]
- 28.Sears D, Schwartz BS. Candida auris: an emerging multidrug-resistant pathogen. Int J Infect Dis. 2017;63:95–8. [DOI] [PubMed]
- 29.Rudramurthy SM, Chakrabarti A, Paul RA, Sood P, Kaur H, Capoor MR, et al. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J Antimicrob Chemother. 2017;72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 30.Ricardo E, Silva AP, Gonçalves T, Costa De Oliveira S, Granato C, Martins J, et al. Candida krusei reservoir in a neutropaenia unit: molecular evidence of a foe. Clin Microbiol Infect. 2011;17:259–263. doi: 10.1111/j.1469-0691.2010.03223.x. [DOI] [PubMed] [Google Scholar]
- 31.Vos MC, Endtz HP, Horst-Kreft D, Doorduijn J, Lugtenburg E, Verbrugh HA, et al. Candida krusei transmission among hematology patients resolved by adapted antifungal prophylaxis and infection control measures. J Clin Microbiol. 2006;44:1111–1114. doi: 10.1128/JCM.44.3.1111-1114.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amaral-Lopes S, Moura A. Neonatal fungal sepsis by Candida krusei: a report of three cases and a literature review. Med Mycol Case Rep. 2012;1(1):24–26. doi: 10.1016/j.mmcr.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect. 2015;21:372–378. doi: 10.1016/j.cmi.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Shankarnarayan SA, Rudramurthy SM, Chakrabarti A, Shaw D, Paul S, Sethuraman N, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Emerg Infect Dis. 2015;24:1956–1958. doi: 10.3201/eid2410.180801. [DOI] [PubMed] [Google Scholar]
- 35.Nagarathnamma T, Chunchanur SK, Rudramurthy SM, Vineetha KR, Ramamurthy K, Joseph J, et al. Outbreak of Pichia kudriavzevii fungaemia in a neonatal intensive care unit. J Med Microbiol. 2017;66:1759–64. doi: 10.1099/jmm.0.000645. [DOI] [PubMed] [Google Scholar]
- 36.Roy U, Jessani LG, Rudramurthy SM, Gopalakrishnan R, Dutta S, Chakravarty C, et al. Seven cases of Saccharomyces fungaemia related to use of probiotics. Mycoses. 2017;60:375–80. doi: 10.1111/myc.12604. [DOI] [PubMed] [Google Scholar]
- 37.Pittet D, Allegranzi B, Boyce J. The World Health Organization guidelines on hand hygiene in health care and their consensus recommendations. Infect Control Hosp Epidemiol. 2009;30:611–22. doi: 10.1086/600379. [DOI] [PubMed] [Google Scholar]