Abstract
Induction of broadly neutralizing monoclonal antibodies (bNAbs) that bind to the viral envelope glycoproteins is a major goal of hepatitis C virus (HCV) vaccine research. The study of bNAbs arising in natural infection is essential in this endeavor. We generated a human antibody, 8D6, recognizing the E2 protein of HCV isolated from a chronic hepatitis C patient. This antibody shows broadly neutralizing activity, which covers a pan-genotypic panel of cell culture-derived HCV virions (HCVcc). Functional and epitope analyses demonstrated that 8D6 can block the interaction between E2 and CD81 by targeting a highly conserved epitope on E2. We describe how the 8D6 lineage evolved via somatic hypermutation to achieve broad neutralization. We found that the V(D)J recombination-generated junctional and somatic hypermutation-induced disulfide bridge (C-C) motif in the CDRH3 is critical for the broad neutralization and binding activity of 8D6. This motif is conserved among a series of broadly neutralizing HCV antibodies, indicating a common binding model. Next, the 8D6 inferred germline (iGL) was reconstructed and tested for its binding affinity and neutralization activity. Interestingly, 8D6 iGL-mediated relatively strong inhibition of the 1b genotype PR79L9 strain, suggesting that PR79L9 may serve as a potential natural viral strain that provides E2 sequences that induce bNAbs. Overall, our detailed epitope mapping and genetic studies of the HCV E2-specific mAb 8D6 have allowed for further refinement of antigenic sites on E2 and reveal a new mechanism to generate a functional CDRH3, while its iGL can serve as a probe to identify potential HCV vaccine strains.
Keywords: Hepatitis C virus, neutralizing antibody, inferred germline, CDRH3, disulfide bridge motif
Subject terms: Infection, Adaptive immunity
Introduction
Hepatitis C virus (HCV) infection is a global health problem. More than 71 million people worldwide have been infected.1 In addition, 50–80% of infected individuals develop a chronic infection that increases the risk of serious liver diseases, such as liver cirrhosis and hepatocellular carcinoma.2,3
Notably, HCV remains the sole human hepatitis virus for which a vaccine is not yet commercially available.4 Highly effective direct-acting antivirals (DAAs) for the treatment of chronic hepatitis C have been developed recently; however, resistance-associated variants have been identified during DAA therapy and can lead to therapy failure.5 In addition, some HCV-infected populations, especially in developing countries, do not have access to DAAs owing to their high cost. Furthermore, reinfection can occur after treatment.6 Therefore, an effective prophylactic vaccine is essential for combating the global HCV epidemic.
A major longstanding barrier to the development of HCV vaccines is the great antigenic variability of circulating virus strains, particularly within the E1 and E2 envelope glycoproteins, which are the primary targets of neutralizing antibodies. HCV can be classified into seven genotypes, with up to 30% divergence, and each genotype can be further divided into numerous subtypes, with 15–20% divergence at the nucleotide level.7 A major goal of HCV vaccine research is to elicit broadly neutralizing monoclonal antibodies (bNAbs) that bind E2 or E1/E2 from diverse HCV strains. The origin and development of bNAbs have not been clearly defined and are poorly understood in natural human HCV infection, which is an urgent problem to solve in HCV vaccine development.8
In this study, we report a new natively paired monoclonal antibody, 8D6, which was isolated from a chronic hepatitis C patient by single B-cell technology. This antibody exhibits broadly neutralizing activity against genetically diverse HCV isolates. Epitope mapping showed that 8D6 targets a highly conserved epitope, which partially overlaps but is distinct from the epitopes recognized by AR3A and AR3C. A CX4C motif in the CDRH3 region of 8D6 was found to be critical for the neutralization activity of multiple bNAbs. Specific HCV strains that bind to inferred germline (iGL) Abs were identified. We hold the opinion that 8D6 not only has potential for prophylaxis of HCV infections but also represents a potential probe for developing a prophylactic HCV vaccine.
Results
The fully human mAb 8D6 was isolated from an HCV-infected patient and showed broad neutralizing activity against different isolates of HCV
Using single-cell PCR to generate fully human monoclonal antibodies specific for a viral antigen,9 we isolated HCV E2 antigen-specific memory B cells (CD19+ IgG+ E2+) from the peripheral blood of an HCV 2a chronically infected patient. This donor showed potent broad plasma neutralizing activity against different subtypes of cell culture-derived HCV virions (HCVcc) (Supplementary Fig. 1a, b). Paired IgH and Igκ variable genes were amplified by RT-PCR and nested PCRs and cloned into a human IgG1 scaffold. Seven genetically distinct mAbs were obtained. Subsequent screening of mAbs for binding activity with soluble E2 and for HCVcc neutralizing potency led to the identification of one mAb, named 8D6, which performed best among the seven mAbs (Supplementary Fig. 1c, d).
8D6 was able to bind to the E2 protein of HCV, covering a panel of 12 different HCV strains from 6 distinct genotypes (Fig. 1a). Immunofluorescence analysis indicated that 8D6 binds to E2 proteins of different HCV genotypes expressed within HCVcc-infected Huh7.5.1 cells (Supplementary Fig. 2). In addition, the binding affinity of 8D6 to the purified recombinant Con1 (genotype 1b) E2 protein was determined by biolayer interferometry, with KD (dissociation constant) = 4.25 nM (Supplementary Fig. 3). These data showed that 8D6 binds E2 in vitro and in the context of HCV infection from widely divergent strains of genotypes 1–6.
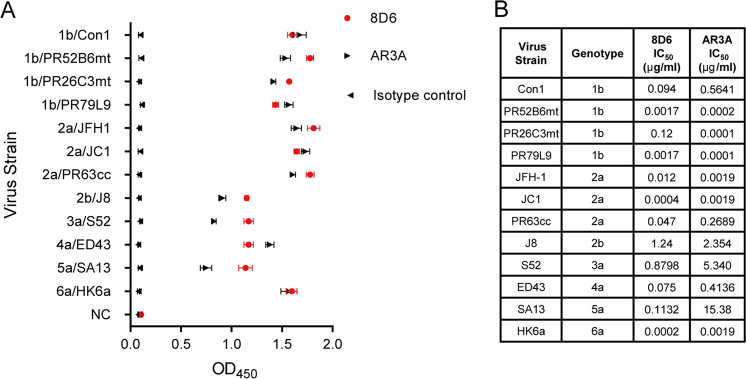
Fig. 1.
In vitro binding and neutralizing activities of 8D6. a Binding (OD450 ELISA values) of antibody to representative soluble E2 from genotypes 1–6. AR3A served as a positive control,9 the anti-influenza virus mAb 3E1 served as the isotype control [8], and the values are representative of three independent experiments. b Fifty percent inhibitory concentration (IC50 (micrograms per milliliter)) of the indicated antibodies against a panel of 12 cell culture-produced HCV strains (HCVCC) expressing envelope glycoproteins from genotypes 1–6 was performed. One experiment representative of two independent experiments is shown
To analyze the neutralizing potency of 8D6, 12 HCVcc strains expressing envelope glycoproteins from genotypes 1–6 were constructed to perform a neutralization assay. For comparison, a previously described bNAb AR3A was tested against the same viral panel under the same conditions.10 As shown in Fig. 1b, consistent with the binding assay, 8D6 exhibited broad cross-neutralizing activity against all 12 strains, with IC50 values ranging from 0.2 ng/ml to 1.24 μg/ml. We observed that 8D6 could also neutralize four clinical isolates, PR52B6mt (genotype 1b), PR26C3mt (genotype 1b), PR79L9 (genotype 1b) and PR63cc (genotype 2a), that were isolated from Chinese patients.11,12 Notably, 8D6 was highly active against PR63cc, which has been reported to be more resistant to an NS5A inhibitor (daclatasvir) than JFH1.12 AR3A also cross-neutralized all isolates evaluated in the assays. The neutralizing profiles of 8D6 and AR3A were differently dependent on the E1E2 context (Fig. 1b, Supplementary Fig. 4). Compared with 8D6, another isolated mAb, 8F6, exhibited lower breath and potency, as it failed to neutralize virus strains Con1 (genotype 1b) and S52 (genotype 3a) and weakly neutralized PR79, SA13, and HK6a with a relatively higher IC50 value, although it exhibited cross-binding activity with E2 (Supplementary Fig. 1c, d).
8D6 blocks E2 binding to CD81 to inhibit HCV entry
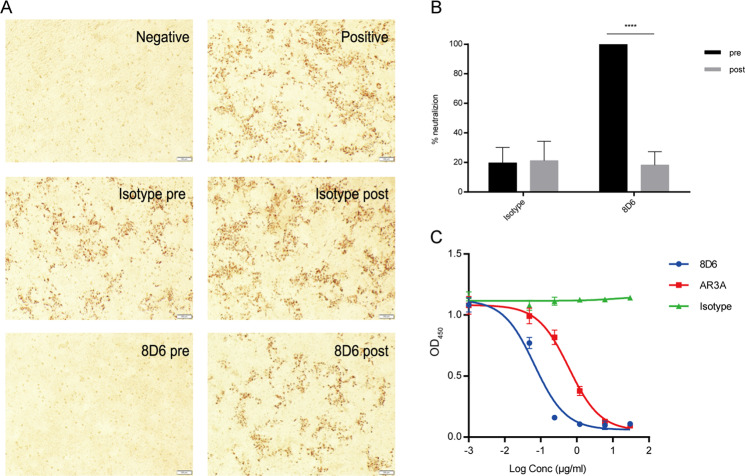
Next, we explored the mechanism of 8D6 neutralization and investigated whether its function was related to pre- or post-attachment steps during HCV infection. The data showed that 8D6 significantly blocked HCV entry into hepatoma cells (Huh7.5.1) when 8D6 was incubated with HCV (JC1) for 1 h before infection. When added at 4 h after infection, 8D6 did not inhibit HCV infection efficiently and showed progressively decreasing activity compared with that observed for the pre-attachment step (Fig. 2a, b). These results suggested that the efficiency of 8D6 was dependent on the time of the intervention. Several cell membrane proteins are involved in HCV entry, of which CD81 binds to HCV E2 protein directly to mediate HCV entry. 13 As shown in Fig. 2c, both 8D6 and the positive control mAb AR3A could efficiently block E2 binding to CD81 in a dose-dependent manner.7 The data demonstrated that 8D6 mainly inhibits HCV entry by blocking E2 binding to the CD81 receptor.
Fig. 2.
Mechanism of the neutralizing activity of 8D6. a Immunohistochemistry results indicate that 8D6 neutralized HCV during the re-attachment steps. For the pre-attachment assay, 10 µg/ml mAbs were incubated with HCVcc for 1 h before the mixture was added to the cells. For the post-attachment assay, HCVcc was first incubated with Huh7.5.1 cells for 4 h at 37 °C to allow virus entry. Cells were washed to remove unattached virus followed by adding 8D6 to the medium. The anti-influenza virus mAb 3E1 was used as an isotype control. The cells were incubated at 37 °C for 72 h, followed by methyl alcohol fixation and NS5A immunostaining. pre, pre-attachment; post, post-attachment. b Statistical analysis and a graph were used to quantify the virus plaques that escaped 8D6 neutralization. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS, not significant (two-tailed Student’s t test). c The effect of 8D6 on the binding of E2 to CD81 was studied by ELISA. AR3A served as a positive control, and 3E1 served as the isotype control. The data are representative of at least three independent experiments
Epitope mapping revealed that 8D6 recognizes a highly conserved epitope which overlaps with but is distinct from the epitopes recognized by AR3A and AR3C
To provide insight into the binding interface between 8D6 and the E2 protein, we performed epitope mapping by several methods. The previously described six bNAbs targeting five distinct clusters of epitopes on E1E2 were selected to serve as reference antibodies.4,7,10,14 First, we analyzed the binding activity of 8D6 with native and denatured recombinant E2 protein. 8D6 bound native E2 but did not bind the denatured form, indicating that it recognized a conformational epitope (Supplementary Fig. 5a). As bNAbs are more likely to target the conserved E2 core (E2c) rather than the highly variable regions (HVRs), which were reported to modulate the exposure of antibody epitopes on E2 and their ability to prevent E2-CD81 interactions, 15 we tested the ability of the 8D6 mAb to bind HVR-deleted E2c proteins. As shown in Supplementary Fig. 5b, the binding activity of 8D6 to E2 was not affected when the three HVRs were deleted, indicating that 8D6 recognized the core conserved domain within the E2 protein. To determine the binding sites of 8D6, we further performed competition enzyme-linked immunosorbent assay (ELISA) between 8D6 and a panel of previously described bNAbs. The data showed that 8D6, AR3A, and AR3C cross competed with each other, indicating that their epitopes are in close proximity or overlap on the surface of E2. The 8D6 and AR3 antibodies could block the binding of HC84.27 specifically directed against a short peptide located in the E2 protein between residues 427 and 446 (AS434).16 However, 8D6 and AR3 antibodies could not be competed by HC84.27, which might be owing to the binding interface of 8D6 and AR3 antibodies being wider than that of HC84.27. In addition, 8D6 did not cross-compete with AP33, AR4A or AR5A. (Fig. 3a). Next, we determined the amino-acid residues of E2 that were critical for 8D6 binding by alanine-mutagenesis scanning (Fig. 3b). In this experiment, the critical residues L413, N415, G418, and W420 were correctly identified as the epitope targeted by the well-characterized mAb AP33, indicating that the assay was reliable.17 According to the alanine-mutagenesis scanning results, some residues located on the back layer of E2 and the Ig β sandwich core were critical for E2 binding to all three conformation-dependent bNAbs. These mutations also influenced the binding activities of other conformation-sensitive mAbs targeting E2, suggesting that they may affect global folding of E1E2, not directly involved in mAb interactions.18 Thus, the discontinuous epitope targeted by 8D6 was likely to be formed by at least two segments, which were composed of a serpentine stretch of residues 421–448 in the front layer and a portion of the CD81 receptor-binding loop, and these segments are also involved in the interaction between E2 and CD81.14,18 We demonstrated that residues T425, L427, C429, G436, L438, N448, G530, and D535 were probably involved in the binding of 8D6 to E2. Notably, the binding activity of these E2 mutants to CD81 was significantly reduced, except for the N448A mutation (Fig. 3c), suggesting that most of the identified residues targeted by 8D6 are critical for the interaction between the E2 protein and CD81.
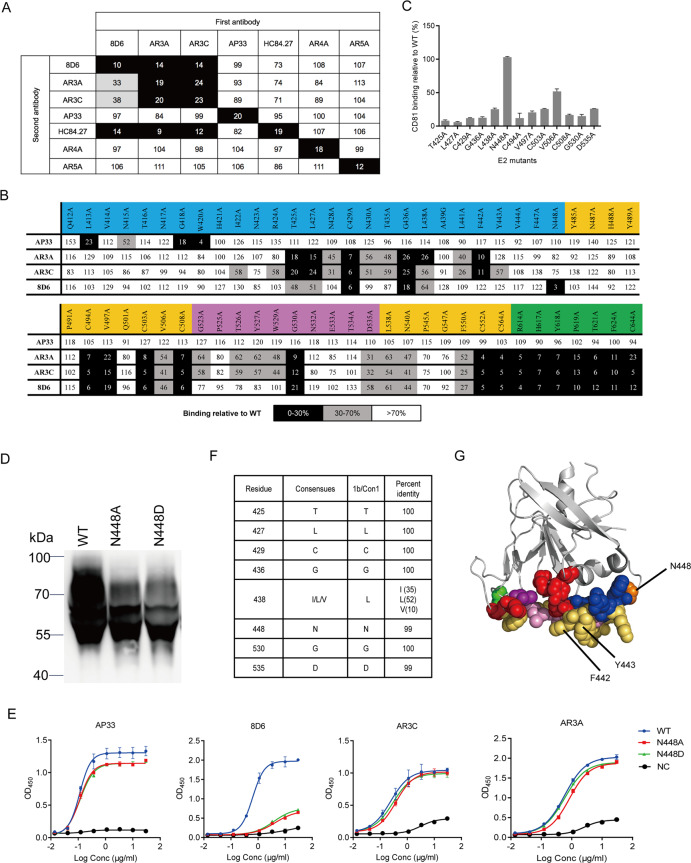
Fig. 3.
8D6 recognizes a conserved site on E2. a Competitive binding between 8D6 and the reference mAbs. For epitope binding, the data indicate the percentage of binding of the secondary antibody in the presence of the primary antibody compared with that of the secondary antibody alone. b Alanine-scanning mutagenesis study. The results are expressed as the percentage of binding of WT E2. The binding of previously described AP33, AR3A, and AP3C was tested as a control. For a, b, residual binding level: <30% presented in black, 30–70% presented in gray, and >70% presented in white. Sequences for regions are colored by structural components: front layer (blue), β-sandwich core (yellow), CD81 binding loop (purple), and back layer (green). c Binding of E2 mutants to CD81. Each value is calculated as the binding relative to WT E2 (%). The mean±SEM of duplicate wells is shown in two independent experiments. d Western blot analysis under reducing conditions of WT (Con1 E2 384–661), N448A, and N448D mutants. The reduced protein was detected with HRP-conjugated mouse anti-His antibody. Markers are shown on the left (kDa). e The E2 glycovariants were verified by binding to 8D6 by ELISA. AP33, AR3A, and AR3C were used as controls. f Conservation of the 8D6 core epitope in ~700 strains. Nonredundant HCV polyprotein sequences downloaded from public databases were aligned using ClustalX. Shown at each position is the frequency of the most common residue. g Critical binding residues of 8D6, AR3C, and HEPC3. The crystallized structure of HCV E2 protein (PDB accession: 4MWF) is shown in cartoon (gray), and critical binding residues of mAbs are marked in spheres with different colors. Green, binding residues unique to 8D6; red, binding residues unique to AR3C; blue, binding residues unique to HEPC3; purple, shared binding residue of 8D6 and AR3C; orange, shared binding residue of 8D6 and HEPC3; yellow orange, shared binding residues of AR3C and HEPC3; and pink, shared binding residues of 8D6, AR3C, and HEPC3. Contact residues for AR3C and HEPC3 were identified by the crystal structure of the antibody–E2 complex (Supplementary Table 1)
The majority of the 8D6-binding residues were found to be highly conserved among 700 virus strains (>90% conservation) from genotypes 1–6 (Fig. 3f and Supplementary Fig. 6). Although the binding epitope of 8D6 overlapped with those of AR3A and AR3C, there were differences in specificity for antibody recognition, particularly in the front layer. We showed that a highly conserved N-linked glycosylation site, N448, contributed to the E2c-8D6 interaction but did not affect AR3A and AR3C binding. We also generated the N448D mutation to knock out the glycan at N448 in Con1 soluble E2. As shown in Fig. 3d, the WT sample migrated as a diffuse band owing to glycan modification heterogeneity. The N448 substitution resulted in a band below that of WT, suggesting possible glycosylation at N448. Both N448A and N448D did not bind 8D6 but showed binding with AR3A and AR3C, indicating that the glycan at N448 may be involved in 8D6 recognition (Fig. 3e). Resistance to 8D6 was conferred by the introduction of the N448A/N448D mutation into the JC1 clone (Supplementary Fig. 7). The bNAbs HEPC3 and HEPC74 isolated from individuals who spontaneously cleared HCV infection were also demonstrated to recognize the glycan at N448 by structural analysis.19
Interestingly, AR3A- and AR3C-binding activities were moderately impaired by N428A, N430A, T435A, and L441A and significantly impaired by F442A, but none of the mutants affected 8D6 binding. Residues 442 and 443, which form a hydrophobic protrusion, are critical binding residues for domain D HMAbs and AR3 clusters.20,21 F442 is less conserved, and its mutations have been reported to confer high-level AR3A resistance to H77/JFH1ΔHVR.22 As shown in Supplementary Fig. 5c, the binding activity of AR3C, AR3A, and HC84.22 to the E2 F442Y mutant was greatly impaired, whereas we did not observe decreased 8D6 binding. High ambiguity-driven docking (HADDOCK) docking was performed to generate a model of the E2-8D6 Fab complex (Supplementary Fig. 8). We then compared the differences between the epitopes recognized by 8D6 and two structurally well-characterized antibodies AR3C and HEPC3.14,19 The epitope that 8D6 recognized overlapped with but was distinct from the AR3C and HEPC3 epitopes (Fig. 3g).
8D6 gene sequence analysis revealed a common CX4C motif in CDRH3, which is critical for broad neutralizing activity against HCV infection
To better understand how 8D6 acquired broad neutralizing activity, we performed gene sequence analysis of 8D6 using IMGT and IgBLAST (Supplementary Fig. 9a, b). To date, several antibodies targeting the discontinuous epitope that covers the front layer and a portion of the CD81 receptor-binding loop have been reported to derive from the VH1-69 family.14,23 8D6 was found to utilize the VH7-4-1*02 and VL1–39*01 germline genes, which are unique among the reported human HCV bNAbs. 8D6-binding activity was compromised significantly when its heavy chain or light chain was replaced with that of anti-influenza mAb 3E1. In addition, loss of binding was also observed with the 8D6 VH single-domain antibody. The result suggests that 8D6 binding requires both HC and LC (Supplementary Fig. 10a). As shown in Supplementary Fig. 9a, the 8D6 antibody has a 20-amino-acid heavy chain complementarity-determining region (CDR3) comprising IGHD5-18*01 and IGHJ3*01 and carries 15 and 11 amino-acid substitutions in VH and VL, respectively, compared with the iGL.
To investigate the relative contributions of somatic hypermutations in the heavy and light chains of 8D6, we produced recombinant 8D6 variants that were completely or partially reverted to the germline sequence, which were designated VH rev, VL rev, and VH VL rev. We screened the binding ability of the reverted mutants of the 8D6 mAb (Fig. 4a and Supplementary Fig. 10b) and measured their neutralizing activity against different HCVcc strains (Fig. 4b). The data showed that the cross-binding and neutralizing activity of 8D6 VH rev were greatly abolished compared with those of mature 8D6, indicating that somatic hypermutations in the heavy chain are essential for 8D6 function. In addition, the reverted light chain moderately affected binding, indicating that somatic hypermutation in the light chain is also needed but makes a smaller contribution to 8D6 binding than mutations in the heavy chain.
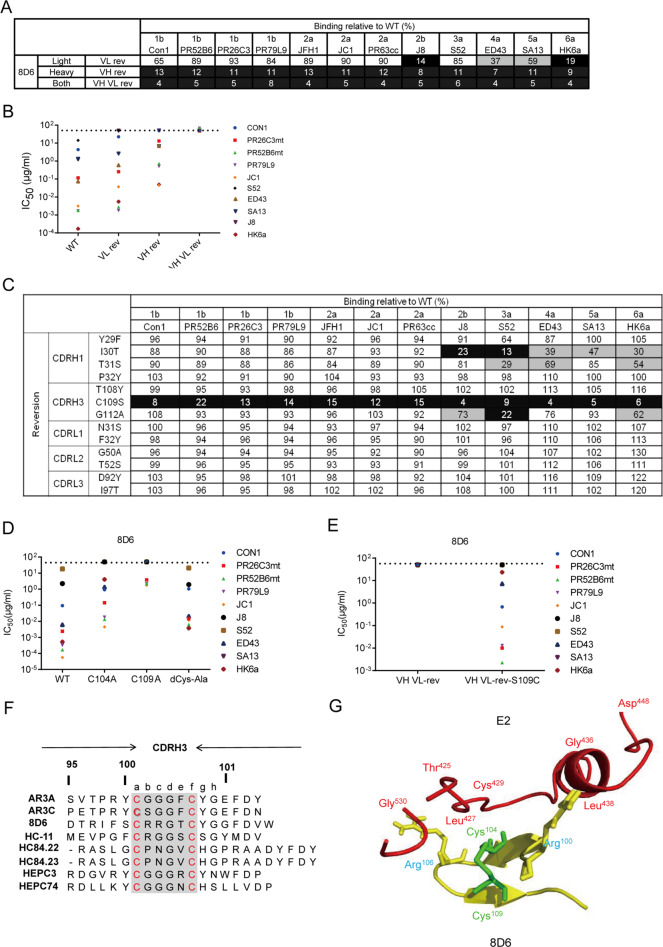
Fig. 4.
Role of somatic mutations in the neutralization and binding breadth of 8D6. a Binding to a diverse panel of E2 proteins from genotypes 1–6 by 8D6 variants that were completely or partially reverted to the germline sequence (designated VH rev, VL rev, and VH VL rev). b VH rev, VL rev, and VH VL rev neutralization IC50 values were determined against a panel of 10 HCVcc strains. Values above 50 mg/ml were scored as negative (dashed line). c Binding to a panel of diverse E2 proteins in the cell supernatant by different versions of 8D6 in which individual somatic mutations in the CDR of the VH or VL chain were reverted to the germline. For a and b, the binding activity relative to the wild type is shown, with <25% presented in black, 25–75% presented in gray, and >75% presented in white. d Neutralization of 8D6 mutants carrying one or two cysteine to alanine mutations in CDRH3. IC50 values were determined against a panel of 10 HCVcc strains. Values above 50 mg/ml were scored as negative (dashed line). For b, d, average IC50 values were obtained from at least two independent experiments. e Neutralizing activity against a panel of HCVcc by 8D6 VH VL rev (iGL) and VH VL rev variant S109C. f Sequence alignment of CDRH3 sequences of a panel of bNAbs against HCV shows conservation of the CX4C motif (highlighted in gray) in the 8D6, HC-11, HC84.22, HC84.23, AR3A, and AR3C antibodies. g A close-up view of the interaction between the CDRH3 loop of the 8D6 Fab (yellow) and HCV E2 (red) from the HADDOCK model. Green: disulfide bonds
Next, we sought to determine the individual somatic mutations in the CDRs of the 8D6 heavy and light chains that play essential roles in its binding activity. We produced a series of 8D6 variants with each somatic mutation individually reverted to the germline-encoded mAb sequence and examined their binding activities to a panel of E2 proteins (Fig. 4c). Notably, a significant loss of binding for all tested E2 proteins was observed with the substitution of C109 with S in the CDRH3 loop. In addition, an individual reversion of the somatic mutation in all three CDRLs had minor effects. We also performed quantitative kinetic binding analysis of the 8D6 variants using the purified sE2 protein of the Con1 strain (Supplementary Fig. 10c). The results were consistent with those shown in Fig. 4c. These findings indicate that the broad antiviral activity of 8D6 was acquired through only a few somatic hypermutations, in particular, an S to C mutation at position 109 in CDRH3, indicating that 8D6-competing antibodies could be elicited more efficiently with suitable immunogens and strategies. Genetic analyses further showed that the 8D6 VH has a long template-independent addition of nucleotides (N nucleotides) at the VD joint introduced by V-(D)-J rearrangement, resulting in a cysteine at position 104 (Supplementary Fig. 9b, c). We predicted that cysteines 104 and 109 form a CX4C disulfide motif in CDRH3, which is a common strategy for 8D6 neutralization of different genotypes of HCV. To further demonstrate the importance of the CX4C motif for 8D6, we tested the binding and neutralizing activity of single C109A and C104A and double Cys to Ala mutants. All the mutants displayed reduced binding to 1b/Con1 E2 and a panel of E2 proteins by ELISA, with the C109A mutants showing the weakest binding (Supplementary Fig. 11a, b). The mutants also displayed reduced neutralization potencies against a panel of HCVcc strains (Fig. 4d).
To investigate the importance of the CDRH3 disulfide motif in the process of 8D6 evolution, we evaluated the effects on the binding and neutralization potencies of the substitution of S109 with C in the CDRH3 of 8D6 iGL. The iGL S109C mutant acquired improved binding affinity to Con1, whereas 8D6 iGL showed no detectable binding signal (Supplementary Fig. 10d). The iGL S109C mutant also partially restored iGL neutralizing activity against a panel of HCVcc strains (Fig. 4e). It is possible that the single cysteine in CDRH3 drives 8D6 iGL somatic hypermutation, which contributes to a gradual increase in affinity for E2. Somatic hypermutation of Ig variable regions requires the activity of activation-induced cytidine deaminase, which favors canonical WRC (W = A or T, R = A or G) hot spot motifs.24 Interestingly, there are two WRC hot spots in germline IGHD5-18*01, where four somatic mutations were coincidentally introduced into 8D6 (Supplementary Fig. 9c). Thus, the codon AGC encoding serine is biased to be mutated to TGC encoding cysteine with only one nucleotide change through somatic hypermutation.
To determine whether the CX4C motif in CDRH3 is a common phenomenon employed by other bNAbs against E2, we analyzed the CDRH3 sequences of published bNAbs (Fig. 4f) and found that a panel of antibodies share a C-X-X-X-G-C motif in the CDRH3 region.10,19,25,26 These human mAbs have been isolated from different subjects with chronic or acute HCV infection with different genotypes and could neutralize multiple viral strains from many different genotypes (Supplementary Table 1). We constructed single and double amino-acid substitutions in the CX4C motif in AR3A, AR3C, and HC84.22, and tested the binding and neutralizing activity of the mutants. The results showed that the binding and neutralizing activity of single and double Cys mutants decreased compared with those of their original antibodies, indicating the important contribution of this motif to the interaction of this series of bNAbs with E2. Furthermore, 8D6, AR3A, and AR3C carrying two Cys mutations in CDRH3 displayed higher binding and neutralizing activity than single Cys mutants. The mutation of only one residue of the proposed cysteine pair may result in a conformational change of CDRH3, which might be responsible for their poor binding activity. In contrast, HC84.22 carrying double Cys mutants completely lost its neutralizing activity and ability to bind E2, suggesting that it might use a different conformation of disulfide-stabilized CDRH3 to recognize E2 (Supplementary Fig. 11a–c).
Interestingly, as shown in Supplementary Table 1, all of the bNAbs were found to have overlapping epitopes with multiple shared binding residues on E2, generally containing at least two segments, most of the front layer and the CD81 receptor-binding loop, suggesting a common binding model. We observed that three highly conserved residues, L427, C429, and G436, in the front layer are the common requisites for all bNAbs, which are also critical for the binding of E2 to CD81 (Fig. 3c). AR3C structural studies and recently published crystal structures of full-length E2 ectodomain complexes with HEPC3 and HEPC74 have shown that the CDRH3 disulfide motif stabilizes a β-hairpin fold to recognize the front layer in the CD81 receptor-binding loop and that the first cysteine residue makes hydrogen bonds with C429.14,19 The HADDOCK model reinforced the importance of the CDRH3 of the 8D6 Fab and the 425–438 loop of E2 for the antibody–antigen interaction (Fig. 4g).
A suitable HCV vaccine strain was selected by the germline version of the 8D6 mAb
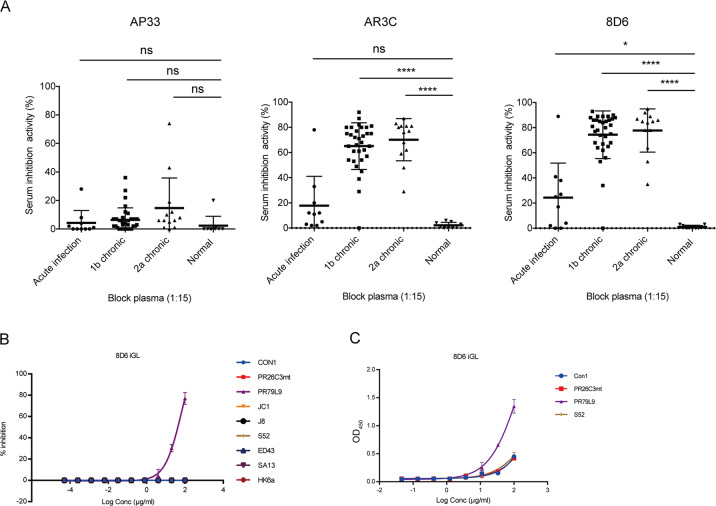
We next evaluated the predominance of 8D6-competing antibodies in the antibody repertoires induced in patients with HCV infection (Fig. 5a). In these experiments, competition ELISA was used to detect plasma-derived 8D6 and AR3-competing Abs. We found that the plasma from donors chronically infected with 1b or 2a significantly suppressed 8D6 and AR3C-binding activity with E2 compared to plasma from normal healthy controls. In contrast, the binding of the antigenic site comprising amino acids 412–423 (AS412)-specific antibody AP33 could not be blocked by either chronically or acutely infected patient plasma. Antibody binding to AS412 has been reported to be rare in HCV-infected patients.27 The plasma from patients with acute infection contained limited 8D6- and AR3C-competing-Ab responses, which may be owing to low antibody titers (Supplementary Fig. 12) or low affinity without long-term maturation. However, one out of 10 subjects who spontaneously cleared HCV infection could significantly suppress 8D6-binding activity (82%), indicating that certain patients with spontaneously cleared infection could produce 8D6- and AR3-competing bNAbs. HEPC3 or HEPC74 targeting overlapping epitopes have been isolated from individuals who spontaneously cleared HCV.23,28 Therefore, our study together with previous studies indicated that 8D6-competing antibodies might be induced in many persistently HCV-infected patients and certain patients who spontaneously cleared HCV infection, thus representing a reproducible class of neutralizing antibodies that provide theoretical support for human HCV vaccine research.
Fig. 5.
The PR79L9 virus is sensitive to 8D6 iGL. a Blocking of AP33, AR3C, and 8D6 by human plasma. Data are presented as the percentage inhibition. Samples included acute plasma samples from 10 patients who were able to spontaneously clear HCV infection, plasma samples from 32 patients with chronic 1b infection, samples from 13 patients with chronic 2a infection, and nine samples from healthy subjects participating as controls. b HCVcc neutralizing activity scanning of 8D6 iGL. c Binding of 8D6 iGL to various purified E2 proteins by ELISA. All data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS, not significant (two-tailed Student’s t test)
It has generally been accepted that germline Ab-targeting immunogens are promising for initiating bNAb induction in HIV studies.29 We hypothesized that the 8D6 mAb inferred germline (8D6 iGL) could be used as a probe to identify suitable HCV vaccine strains. To address this possibility, we used the germline sequence of the 8D6 mAb to generate 8D6 iGL mAbs. The inhibitory effects of 8D6 iGL mAbs on HCVcc isolates from different HCV genotypes were tested (Fig. 5b). One neutralization-sensitive isolate was selected, which is a chimeric genotype 1b HCVcc (PR79L9) constructed from clinical isolates. All other HCVcc strains were resistant to neutralization by 8D6 iGL. The neutralization pattern of 8D6 iGL was confirmed by its ability to bind to purified recombinant soluble PR79L9 E2 (Fig. 5c). These results suggest that PR79L9 E2 might serve as a priming immunogen for activating B-cell precursors and priming an 8D6-competing bNAb response.
Discussion
The identification and characterization of bNAbs against HCV has been a long-sought goal for immunotherapy and vaccine development.30 In this study, we isolated bNAb 8D6 from an individual who experienced chronic HCV 2a infection by using single B-cell clone technology. The 8D6 antibody reacted with E2 proteins across genotypes 1–6 and neutralized diverse strains from numerous genotypes of HCV. Moreover, 8D6 efficiently inhibited HCV infection during the early step of the HCV life cycle by blocking the interaction between E2 and its receptor CD81. These results suggest that as highly effective entry inhibitors, 8D6 and other broad neutralizing antibodies may be potential passive antibody-based agents for HCV prevention in the absence of an effective vaccine.
One of the major goals of HCV vaccine research is to identify the mechanism by which bNAbs neutralize the majority of diverse circulating HCV strains.4 Because of the broad neutralization potency of 8D6, identifying its epitopes and functional mechanism will be helpful for vaccine design. In our study, epitope mapping revealed that 8D6 targeted a similar interface as AR3C and HEPC3, but some interactions with amino-acid residues were different. The envelope of HCV is highly N-glycosylated and is involved in complement lectin pathway-mediated cytolytic activity in HCV-infected hepatocytes.31 However, few bNAbs have been reported to interact with glycans. Our work indicated that the highly conserved N-linked glycan at position 448 of E2 is crucial for 8D6 recognition. These observations indicate that specific glycans at N448 might be taken into consideration in terms of designing an E2 protein-based vaccine.
The identification of the highly conserved epitope targeted by 8D6 has allowed for further refinement of antigenic sites on E2. To facilitate rational vaccine design and induce similar bNAbs in human subjects in a reproducible manner, it is necessary to understand how bNAbs, such as 8D6, developed from their unmutated common ancestor during natural infection. We demonstrated that the broad antiviral activity and high affinity of 8D6 was acquired through only a few somatic mutations. In particular, a somatic hypermutation-induced cysteine at position 109 of CDRH3 in 8D6 and a V(D)J recombination-generated cysteine at position 104 formed a CX4C motif that made a key contribution to the 8D6 interaction with E2. Furthermore, based on our findings, we characterized a series of prototype bNAbs that targeted a similar discontinuous surface, and all these antibodies retained a conserved C-X-X-X-G-C motif in CDRH3. The motif may exert two functions: involvement in the direct association with residues on E2 and maintenance of the required intramolecular 3D structure of CDRH3 for epitope detection. Previous structural and functional studies have shown that this motif is critical for the interaction between antibodies and helix α1 in the E2c front layer.14,19 To date, the disulfide motif in CDRH3 is mainly encoded in IGHD2 germline alleles, such as those of AR3C, HEPC3, and HEPC74. Based on our study, the cysteine pair can be produced combinatorially by VDJ recombination and somatic hypermutation of certain DHs with a codon bias towards mutation to cysteine.32,33 Our results suggest that the junctional residues introduced by V-D-J rearrangement could affect subsequent somatic hypermutation and thus determine the maturation pathway of an antibody.34,35 Identifying the mechanism by which bNAbs from different individuals recognize the same antigenic epitope on the virus may provide valuable information for both immunogen design and immunization strategy design in HCV vaccine research.
It is possible to rationally design an epitope-based vaccine that can elicit autologous bNAbs, and targeting the B-cell germline offers a potential strategy to achieve this goal.36,37 Although germline-targeting immunogens have been studied in HIV, iGLs of bNAbs against HCV have rarely been characterized, and very little is known about which tailored immunogens are feasible for priming B-cell precursor stimulation. In this study, we demonstrated that PR79L9 was much more sensitive to 8D6 iGL than other selected HCV strains, including 1b/Con1, in both the recombinant virus neutralizing assay and the soluble E2 ELISA-binding assay (Fig. 5b, c). The interaction between PR79L9 and 8D6 iGL is CDRH3 disulfide motif independent, whereas for the HEPC3/AR3C family of HCV bNAbs, the CDRH3 disulfide motif is essential for initial germline recognition of HCV by naive B cells.19,21 These studies have provided key proof that E2 from certain isolates favorably reacts with germline B-cell receptors of the 8D6 antibody; thus, it is possible to engineer these immunogens to target the 8D6 and other bNAb precursors and expand these specific antibody lineages.
Overall, these findings provide a foundation for the design of potential vaccines that can elicit 8D6-competing bNAbs to combat HCV infection. First, the human immune system may be primed by a related B-cell precursor-stimulating antigen. Second, based on the results of our investigation using a patient plasma competition ELISA (Fig. 5a), the next step would be boosting with more diverse immunogens that resemble other genotype E2 variants to guide the somatic mutations in the VH and VL chains towards increased breadth and potency. These hypotheses should be verified in animal systems expressing human antibody repertoires.38 Collectively, our findings on the potent and broad neutralizing activity achieved by 8D6 provide additional evidence for rational antibody-based HCV vaccine design.
Materials and methods
Ethics statement
Plasma samples and peripheral blood mononuclear cells (PBMCs) were isolated from adult patients who provided written informed consent. This study was approved by the ethics committee of the 1st Hospital of Jilin University and registered with ClinicalTrials.gov (ID: NCT01760148).39 The subjects were assigned to an acute and chronic infection groups based on the infection outcome, as described previously.40
Cells, antibodies, E2 protein, and viruses
HEK293T cells (ATCC CRL-3216) and the Huh7.5-derived subline Huh7.5.1 that is highly permissive for HCV replication41 were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 100 U penicillin per ml, 100 μg streptomycin per ml, and 10% fetal calf serum. ExpiCHO-S mammalian cells (Invitrogen) were cultured in ExpiCHO Expression Medium (Invitrogen) supplemented with 1% penicillin–streptomycin at 37 °C with 8% CO2. PBMCs were isolated from venous blood collected from the donors by using Lympholyte-H density separation medium (GE) according to the manufacturer’s instructions.
A panel of HCVcc strains for the neutralization assay was generated as previously described.42 A mouse anti-E2 mAb (1C9) was generated in our laboratory, and the human anti-E2 mAbs (AR3A, AR4A, and AR5A) were a gift from Mansun Law at the Scripps Research Institute. The AR3C, AP33, HC84.22, and HC84.27 VH and VL sequences were commercially synthesized (Shanghai Generay Biotech Co., Ltd). The soluble E2 proteins of genotype 1b strains PR79, PR26, and Con1, as well as genotype 3 strain S52, were constructed, expressed in Drosophila S2 cells and purified as previously described.11,42,43 The antibodies provided by other groups and prepared in house were not expressed and validated in parallel; thus, any apparent differences in performance should not be overinterpreted.
Generation of HCV E2 sorting probes
The soluble E2 ectodomain encompassing residues 384–661 in HCR6 was synthesized by Generay Biotech Co. (China)44 and cloned into the pBudCE4.1 mammalian expression vector by incorporating an immunoglobulin (Ig) heavy chain (H) signal peptide at the N-terminus for E2 secretion and a 6× His tag at the C-terminus. The construct was transiently expressed in ExpiCHO-S mammalian cells (Invitrogen). The proteins were purified by metal affinity chromatography using a His Trap Excel column (GE, EDTA-resistant). Purified E2 was biotinylated using Thermo Fisher Scientific EZ-Link Sulfo-NHS-LC-Biotin according to the manufacturer’s instructions. Biotinylated proteins were dialyzed against phosphate-buffered saline (PBS) to remove excess biotin.
Isolation of monoclonal antibodies
The generation of human monoclonal antibodies from human memory B cells by single-cell RT-PCR was performed as previously described.9 Peripheral blood mononuclear cells were stained using anti-human IgG (APC), CD19 (FITC), and a Cy3-labeled HCV E2 protein. Single cells were sorted into 96-well PCR plates containing lysis buffer. In brief, IgH and Igκ variable genes were amplified by RT-PCR and nested PCRs using cocktails of specific primers. The VH gene and the Vκ genes were sequenced and cloned into IgG and Igκ expression vectors.
Expression and purification of mAbs
Monoclonal antibodies were produced by transient transfection of ExpiCHO-S cells (Invitrogen). Supernatants from transfected cells were collected after 10 days, and IgG was affinity purified by protein G chromatography (GE Healthcare) and dialyzed against PBS.
Neutralization assay
Antibody neutralization titers were determined by using a previously described focus reduction assay.42 HCVccs were diluted in complete DMEM to approximately 4 × 103 FFU/ml, mixed with an equal volume (50 μl) of diluted antibodies (initial concentration 100 μg/ml), and incubated at 37 °C for 1 h. The virus–plasma mixture was transferred to Huh7.5.1 cells that had been seeded 12 h earlier in 96-well plates (1 × 104 cells/well) and replaced with complete DMEM after 4–6 h of incubation at 37 °C. The cells were incubated at 37 °C for 72 h, followed by methyl alcohol fixation and NS5A immunostaining. The percent neutralization was calculated by comparing the focus numbers of the antibody group to those of the virus-only control.
HCV E2 truncation and site-directed mutagenesis
The DNA sequences encoding the Con1 E2 soluble fragments encompassing amino acids 384–661 (wild type, WT) were fused in frame with an N-terminal human IgE signal peptide and a C-terminal 6× His tag. Deletion of HVR1 was generated by deletion of the sequence encoding amino acids 384–411. Substitution of HVR2 (460–485) and igVR/VR3 (570–580) with a Gly-Ser-Ser-Gly linker was performed by overlap PCR. Residues were selected for alanine-scanning mutagenesis or based on their conservation and previous studies. The alanine-scanning mutants or sited directed mutagenesis mutants were generated with a KOD-Plus mutagenesis kit (TOYOBO) according to the manufacturer’s instructions. All mutations were verified by DNA sequence analysis (Tsingke). To express wild type and mutant E2, HEK293T cells (ATCC) were plated in 12-well plates and transiently transfected with these constructs. At 72 h after transfection, the supernatants were harvested.
Production of HCVcc encoding N448D and N448A mutants
The individual E2 mutations N448D and N448A were introduced into E1E2 in the Jc1 plasmid using a KOD-Plus mutagenesis kit (TOYOBO). The E2 mutations N448D and N448A were confirmed by DNA sequence analysis.45
In vitro transcription and RNA transfection
In vitro transcripts generated with pUC-based plasmids were transfected into cells by electroporation as described previously.46 The concentration of purified RNA was determined by a spectrometer, and the integrity of the transcripts was verified by agarose gel electrophoresis. Transcripts were stored as 10 μg aliquots at −80 °C for further use. For RNA transfection, subconfluent Huh7.5.1 cell monolayers were detached from the culture dish by trypsinization, washed once with PBS, and resuspended at a concentration of 1.5 × 107 cells per ml in Cytomix containing 2 mM ATP and 5 mM glutathione. Ten micrograms of in vitro transcript was mixed with 400 μl of the cell suspension, and the cells were transfected by electroporation at 960 mF and 270 V using a GenePulser system (Bio-Rad) and a cuvette with a gap width of 0.4 cm (Bio-Rad). Immediately after electroporation, the cells were resuspended in complete DMEM and seeded as required.
Immunofluorescence staining
For immunostaining, cells grown on glass coverslips were fixed in formaldehyde (3% [wt/vol] in PBS) after prior washing with PBS and incubated in blocking reagent (3% bovine serum albumin (BSA) in PBS). After overnight incubation at 4 °C, the cells were incubated with monoclonal antibodies specific for NS5A, AR3C, 8D6, 8F6, or 3E1 (negative control) for 1 h at room temperature. The cells were then incubated with Alexa Fluor 488-conjugated secondary antibodies specific for mouse antibodies and Alexa Fluor 555-conjugated secondary antibodies specific for human antibodies (Invitrogen), and cell nuclei were stained with 4,6-diamidino-2-phenylindole. Images were collected and analyzed with an Olympus IX53 confocal microscope.
Biolayer interferometry analysis of mAb-Env binding
Binding affinity (KD) analysis was performed by biolayer interferometry at 25 °C using an Octet Red instrument (ForteBio, Inc.). A 20 μg/ml concentration of the 8D6 or 8F6 mAb was immobilized on an anti-human IgG-Fc (AHC)-coated biosensor surface for 5 min. The baseline interference was then read for 60 s in kinetics buffer (KB: 1× PBS, 0.01% BSA, and 0.02% Tween-20), followed by subsequent immersion of the sensors into wells containing recombinant trimeric E2 diluted in KB for 300 s (association phase). The sensors were then immersed in KB for the indicated times (dissociation phase) for up to 900 s. The mean kon, koff, and apparent KD values were determined from all binding curves that were globally fit to a 1:1 Langmuir binding model with an R2 value ≥ 0.95.
Germline antibody design and antibody engineering
In brief, the closest inferred germline sequences were determined using IMGT, and the inferred germline gene sequences were synthesized (Shanghai Generay Biotech Co., Ltd). The iGL antibodies and VH or VL reverted chimeric antibodies were completely or partially reverted to the germline sequence. Site-directed mutagenesis to revert individual somatic mutations and alanine-scanning mutagenesis were performed using a KOD-Plus mutagenesis kit (TOYOBO). Monoclonal antibodies were produced by transient transfection of ExpiCHO-S cells (Invitrogen).
ELISA
To test mAb binding to E2 in the supernatant without purification, HEK293T cells were transfected with E2 soluble fragment expression constructs encoding amino acids 384–661 from different genotypes. The cell supernatants were harvested after 72 h. Then, 96-well plates were coated with a series of concentrations of antibodies and blocked with PBS containing 2% BSA and 0.5% Tween-20, and E2-containing cell supernatants were added. After a 2-h incubation, the plates were washed, and binding was detected with a horseradish peroxidase (HRP)-conjugated anti-6× His mAb antibody (1:5000; Pierce). After a 1-h incubation, TMB substrate was added, and optical density was measured at 450 nm with a microwell plate autoreader (Thermo Scientific). The human mAb 3E1 against HA2 of influenza H1N1 was used as an isotype control.
To confirm whether the antibodies recognized continuous or discontinuous epitopes, 96-well microwell plates (Nunc) were coated with recombinant soluble E2 or denatured E2 pretreated with 0.1% (wt/vol) SDS/50 mm DTT and were incubated at 100 °C for 5 min, followed by incubation at a concentration of 10 μg/ml in 0.1 m sodium carbonate–bicarbonate buffer (pH 9.6) at 4 °C overnight. Bovine serum albumin (2%) and Tween-20 (0.05%) in PBS were used as the blocking and dilution buffer. After blocking at 37 °C for two h, the ELISA plates were washed, and serially diluted mAbs (twofold dilutions from 10 μg/ml) were added. The mouse mAb 1c9, which is specific for the E2 linear epitope, was used as a positive control. The plates were washed after a 2-h incubation. Next, the bound antibodies were detected with an HRP-conjugated goat anti-human FC antibody (1:5000; Sigma) or HRP-conjugated goat anti-mouse IgG (1:2000; Invitrogen).
For the binding competition assay, excessive amounts of blocking full-length human antibodies (100 μg/ml) or human plasma (1:15 dilution) were added to the E1E2 antigens (Con1 isolate) captured by precoated Galanthus nivalis lectin (20 μg/mL; Sigma)7 or recombinant E2-precoated plates at a concentration of 0.5 μg/ml and incubated for 1 h at 37 °C. The E1E2 antigen was prepared from cell lysates from 293 T cells transfected with a Con1 E1E2 (170–746) expression plasmid. The biotin-labeled monoclonal detection antibodies were then added to the plates. The plates were washed after 2 h of incubation at 37 °C, and binding of the detection antibodies was detected by HRP-conjugated streptavidin (1:200, BD) and TMB substrate. The percentage of inhibition by the blocking antibody was calculated as the percentage reduction in the absorbance values in the presence of blocking antibody.
To investigate the ability of mAbs to block E2 binding to CD81, serially diluted mAbs (fivefold dilution from 30 μg/ml) were incubated with recombinant E2 for 1 h at 37 °C. The mixture was added to plates precoated with 1 μg of the recombinant large extracellular loop from human CD81 (CD81-LEL) containing Phe113-Lys201 and expressed by human cells (purchased from Sino Biological Inc., Beijing, China). After 1 h of incubation at 37 °C, the wells were washed, and E2 bound to CD81-LEL was detected with an HRP-conjugated mouse anti-His monoclonal antibody. The plates were incubated for 1 h, followed by the addition of TMB substrate. The percentage of binding inhibition was calculated as the reduction percentage of E2 binding to CD81 compared with the value in the absence of the antibody. The mAb AR3A was used as a positive control, as it was reported to inhibit E1E2 binding to CD81. The human mAb 3E1 against HA2 of influenza H1N1 was used as an isotype control.
To confirm the important residues that contribute to the binding of E2 to antibodies or sE2 to the CD81 receptor, a sandwich ELISA was performed to measure the E2 concentration in the cell supernatant. In detail, the mouse mAb 1C9, which recognizes a linear epitope in the C-terminal region of E2 (645–661), was used to coat plates, and a tenfold dilution of cell supernatant was then added and captured by 1C9. Serially diluted purified E2 (twofold dilutions from 100 ng/ml) was used as a standard. Subsequently, the antigen was detected by an HRP-conjugated mouse anti-His monoclonal antibody. The concentration of the sample was calculated according to a standard curve generated from the serial dilution data. Another ELISA was performed to analyze the relative binding affinities of these E2S mutants to 8D6. AP33, AR3A, AR3C, and HC84.22 were used as controls. Then, 100 ng/ml concentrations of the E2 mutants were incubated on plates precoated with 500 ng or a serial dilution of mAbs or 500 ng of CD81. After 2 h of incubation, the binding ability of the E2 mutants to human and mouse mAbs or CD81 was detected by an HRP-conjugated mouse anti-His monoclonal antibody. The binding signals of the mutants to the mAbs were compared with those of the wild type.
Immunoblot analysis
Cell supernatants were separated on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gel and transferred onto a nitrocellulose membrane. The membrane was probed with HRP-conjugated anti-His tag mAb (1:5000 dilution; Proteintech) followed by ECL Western Blotting Substrate detection.
Docking
HADDOCK was used to build a structural model of the E2-8D6 Fab complex. The coordinates of HCV E2 were obtained from a previously published crystal structure (PDB accession number: 4mwf; structure of a minimal E2 core domain in complex with the AR3C Fab) [13]. The glycans were removed from the E2 crystal structure for simplicity. Since a high-resolution structure of the 8D6 Fab was not available, its coordinates were adapted from the AR3C Fab of the same protein complex structure (PDB accession number: 4mwf) with modifications. Using PyMOL (The PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger, LLC), residues D30, N31, and Y32 in AR3C Fab HCDR1, L53 and F54 in HCDR2 and 97TPRYCGGGFCYGE in HCDR3 were mutated to the corresponding sequences of the 8D6 Fab (I30, T31, P32, N53, T54, and 100RIFSCRRGTCYGG, respectively). Other sequence differences were not considered. The ambiguous interaction restraints were derived from antigen–antibody interaction competition studies and alanine-scanning mutagenesis, as described above. For E2, the “active” residues involved in the intermolecular interaction were limited to T425, L427, C429, G436, and L438; the “passive” residues involved in the intermolecular interaction were N448, G530, and D535. For the 8D6 Fab, the “active” residues involved in the intermolecular interaction were restricted to 101IFSCRRGTCYGG; the “passive” residues involved were I30, T31, P32, N53, T54, and R100. HADDOCK was run at the following web interface: http://milou.science.uu.nl/services/HADDOCK2.2/haddockserver-easy.html. The best structure with the lowest HADDOCK score is shown in Fig. S4.
Sequence analysis of 8D6 epitope conservation
A total of 700 full-length, nonredundant HCV polyprotein sequences were downloaded from https://euhcvdb.ibcp.fr/euHCVdb/. The sequences were aligned and analyzed using ClustalX, and the percent conservation for each residue is presented as the quality score of each column.
Statistical analyses
All statistical analyses were performed using Graph Pad Prism 6. The P values reported in the figures and figure legends were calculated using unpaired two-tailed Student’s t tests. We accepted the following significance levels: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS, not significant (two-tailed Student’s t test).
Supplementary information
Acknowledgements
We are grateful to Dr. Jens Bukh (University of Copenhagen) for providing chimeric HCVcc plasmids, and to Drs. Dennis Burton and Mansun Law (The Scripps Research Institute) for providing the mAb AR3A. This work was supported by grants from the Chinese National 973 Program (2015CB554302), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000) to B.S., the Chinese National 973 Program (2015CB554300), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29010205) to J.Z., the National Natural Science Foundation of China (31670172), Shanghai Science and Technology Innovation Action (16DZ1910100) to B.S. and Nanjing Galaxy Biopharma C.O.
Author contributions
C. Yi, J. Xia, L. He, and Z. Ling designed and performed the experiments and analyzed the data. X. Wang, Y. Yan, J. Wang, X. Zhao, and S. Ye contributed to reagent and sample preparation and discussed the project. R.-G. Zhang, R.-H. Zhang, H. Zhou, and Z. Huang provided suggestions and reagents and discussed the data. X. Sun, L. Ma prepared cell lines and provided reagents. C. Yi, J. Xia, L., and Z. Ling wrote the manuscript. B. Sun, J. Zhong, J. Lu, G. Long, and J. Niu supervised the project and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Chunyan Yi, Jing Xia, Lan He, Zhiyang Ling
Contributor Information
Gang Long, Email: glong@ips.ac.cn.
Junxia Lu, Email: lujx@shanghaitech.edu.cn.
Jin Zhong, Email: jzhong@ips.ac.cn.
Bing Sun, Email: bsun@sibs.ac.cn.
Supplementary information
The online version of this article (10.1038/s41423-020-0403-1) contains supplementary material.
References
- 1.Mankowski MC, et al. Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc. Natl Acad. Sci. USA. 2018;115:E82–E91. doi: 10.1073/pnas.1718441115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Yang C, et al. Interferon alpha (IFNalpha)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol. Immunol. 2016;13:94–102. doi: 10.1038/cmi.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinchen VJ, Cox AL, Bailey JR. Can broadly neutralizing monoclonal antibodies lead to a hepatitis C virus vaccine? Trends Microbiol. 2018;26:854–864. doi: 10.1016/j.tim.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz J, et al. Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology. 2018;154:976–988. doi: 10.1053/j.gastro.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Hayes CN, Chayama K. Why highly effective drugs are not enough: the need for an affordable solution to eliminating HCV. Expert Rev. Clin. Pharm. 2017;10:583–594. doi: 10.1080/17512433.2017.1313111. [DOI] [PubMed] [Google Scholar]
- 7.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl Acad. Sci. USA. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swadling L, Klenerman P, Barnes E. Ever closer to a prophylactic vaccine for HCV. Expert Opin. Biol. Ther. 2013;13:1109–1124. doi: 10.1517/14712598.2013.791277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu W, et al. Fully human broadly neutralizing monoclonal antibodies against influenza A viruses generated from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology. 2013;435:320–328. doi: 10.1016/j.virol.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, et al. Construction and characterization of infectious hepatitis C virus chimera containing structural proteins directly from genotype 1b clinical isolates. Virology. 2013;443:80–88. doi: 10.1016/j.virol.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, et al. A novel strategy to develop a robust infectious hepatitis C virus cell culture system directly from a clinical isolate. J. Virol. 2014;88:1484–1491. doi: 10.1128/JVI.02929-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feneant L, Levy S, Cocquerel L. CD81 and hepatitis C virus (HCV) infection. Viruses. 2014;6:535–572. doi: 10.3390/v6020535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong L, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alhammad Y, et al. Monoclonal antibodies directed toward the hepatitis C virus glycoprotein E2 detect antigenic differences modulated by the N-terminal hypervariable region 1 (HVR1), HVR2, and intergenotypic variable region. J. Virol. 2015;89:12245–12261. doi: 10.1128/JVI.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krey T, et al. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 2013;9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong L, et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 2012;86:13085–13088. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopal R, et al. Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog. 2017;13:e1006735. doi: 10.1371/journal.ppat.1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flyak AI, et al. HCV broadly neutralizing antibodies use a CDRH3 disulfide motif to recognize an E2 glycoprotein site that can Be targeted for vaccine design. Cell Host Microbe. 2018;24:703–716. doi: 10.1016/j.chom.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fauvelle C, et al. Hepatitis C virus vaccine candidates inducing protective neutralizing antibodies. Expert Rev. Vaccines. 2016;15:1535–1544. doi: 10.1080/14760584.2016.1194759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzarum N, et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci. Adv. 2019;5:eaav1882. doi: 10.1126/sciadv.aav1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velazquez-Moctezuma R., Galli A., Law M., Bukh J., Prentoe J. Hepatitis C virus escape studies of human antibody AR3A reveal a high barrier to resistance and novel insights on viral antibody evasion mechanisms. J. Virol.93, pii: e01909-18 (2019). [DOI] [PMC free article] [PubMed]
- 23.Bailey J. R., et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight2, e92872 (2017). [DOI] [PMC free article] [PubMed]
- 24.MacCarthy T, et al. V-region mutation in vitro, in vivo, and in silico reveal the importance of the enzymatic properties of AID and the sequence environment. Proc. Natl Acad. Sci. USA. 2009;106:8629–8634. doi: 10.1073/pnas.0903803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foung S. & Keck Z. Y. Cluster of neutralizing antibodies of hepatitis C virus. United States patent No: US 2013/0084301 A1 (2013).
- 26.Foung S. & Keck Z. Y. Hepatitis C antibodies and uses. United States patent No: US 8,858,947 B2 (2014).
- 27.Tarr AW, et al. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J. Virol. 2012;86:2739–2749. doi: 10.1128/JVI.06492-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinchen VJ, et al. Broadly neutralizing antibody mediated clearance of human hepatitis C virus infection. Cell Host Microbe. 2018;24:717–730. doi: 10.1016/j.chom.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong PD, Mascola JR. HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity. 2018;48:855–871. doi: 10.1016/j.immuni.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Pierce BG, et al. Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc. Natl Acad. Sci. USA. 2016;113:E6946–E6954. doi: 10.1073/pnas.1614942113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, et al. Specifically binding of L-ficolin to N-glycans of HCV envelope glycoproteins E1 and E2 leads to complement activation. Cell Mol. Immunol. 2009;6:235–244. doi: 10.1038/cmi.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, et al. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deiss TC, et al. Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol. Immunol. 2019;16:53–64. doi: 10.1038/cmi.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furukawa K, Akasako-Furukawa A, Shirai H, Nakamura H, Azuma T. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 1999;11:329–338. doi: 10.1016/s1074-7613(00)80108-9. [DOI] [PubMed] [Google Scholar]
- 35.Ying T, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Ramirez M, Sanders RW, Klasse PJ. Targeting B-cell germlines and focusing affinity maturation: the next hurdles in HIV-1-vaccine development? Expert Rev. vaccines. 2014;13:449–452. doi: 10.1586/14760584.2014.894469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jardine JG, et al. HIV-1 vaccines. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee EC, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 39.Fan W, et al. IFN-lambda4 desensitizes the response to IFN-alpha treatment in chronic hepatitis C through long-term induction of USP18. J. Gen. Virol. 2016;97:2210–2220. doi: 10.1099/jgv.0.000522. [DOI] [PubMed] [Google Scholar]
- 40.Osburn WO, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc. Natl Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D, et al. Altered glycosylation patterns increase immunogenicity of a subunit hepatitis C virus vaccine, inducing neutralizing antibodies which confer protection in mice. J. Virol. 2016;90:10486–10498. doi: 10.1128/JVI.01462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, et al. A trivalent HCV vaccine elicits broad and synergistic polyclonal antibody response in mice and rhesus monkey. Gut. 2019;68:140–149. doi: 10.1136/gutjnl-2017-314870. [DOI] [PubMed] [Google Scholar]
- 44.Broering TJ, et al. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 2009;83:12473–12482. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao F, et al. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog. 2014;10:e1004128. doi: 10.1371/journal.ppat.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long G, et al. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.