Abstract
Oxidative stress is an imbalance between production and accumulation of oxygen reactive species and/or reactive nitrogen species in cells and tissues, and the capacity of detoxifying these products, using enzymatic and non-enzymatic components, such as glutathione. Oxidative stress plays roles in several pathological processes in the nervous system, such as neurotoxicity, neuroinflammation, ischemic stroke, and neurodegeneration. The concepts of oxidative stress and rare diseases were formulated in the eighties, and since then, the link between them has not stopped growing. The present review aims to expand knowledge in the pathological processes associated with oxidative stress underlying some groups of rare diseases: Friedreich’s ataxia, diseases with neurodegeneration with brain iron accumulation, Charcot-Marie-Tooth as an example of rare neuromuscular disorders, inherited retinal dystrophies, progressive myoclonus epilepsies, and pediatric drug-resistant epilepsies. Despite the discrimination between cause and effect may not be easy on many occasions, all these conditions are Mendelian rare diseases that share oxidative stress as a common factor, and this may represent a potential target for therapies.
Keywords: Friedreich’s ataxia, neurodegenerative disorders with brain iron accumulation (NBIA), Charcot-Marie-Tooth disease (CMT), inherited retinal dystrophy (IRD), progressive myoclonus epilepsy (PME), Unverricht–Lundborg disease (ULD), Lafora disease (LD), Dravet syndrome
1. Introduction
Balanced levels of reactive oxygen species (ROS) and/or reactive nitrogen species (RNS) and antioxidant defense mechanisms are present in normal physiological processes. However, when this equilibrium is disturbed, a process called oxidative stress is observed. As a result of this imbalance cellular, damage is produced by excessive ROS/RNS [1].
Eukaryotic organisms need oxygen to normal physiological functions. Nevertheless, incomplete metabolic reduction of oxygen to water produces most cellular ROS. Reduction of oxygen forms radical superoxide (O2•−), which itself can produce hydrogen peroxide (H2O2). This molecule, in turn, may be fully reduced to inert H2O or may be partially reduced to hydroxyl radical (OH•) and hydroxide ion. One example of this partial reduction is Fenton reaction, where reduced metal ions, such as ferrous iron (Fe2+), act as a catalyst. ROS present elevated reactivity, which produces interactions with biological molecules and hence, changing normal cell functions that may lead to cell death.
Sources of free radical species in humans can be divided based on their original source as internal or external. The main internal sources are mitochondria, peroxisomes, endoplasmic reticulum (ER), NADPH, and xanthine oxidases, ischemia-reperfusion mechanisms, and neutrophils, eosinophils, and macrophages activity. The main external factors are smoking and alcohol, infections, inflammatory processes, UV irradiation, photooxidation, electromagnetic radiation, and xenobiotics. In order to balance ROS content, cells promote antioxidant defenses mechanisms, which can be divided into enzymatic and non-enzymatic. The main non-enzymatic antioxidant in mammalian cells is glutathione (GSH), which has an essential role as a cofactor of different antioxidant enzymes, including glutathione peroxidase enzyme-4 (GPX4). This enzyme has a phospholipid hydroperoxidase activity that reduces lipid hydroperoxides (R–OOH) to lipid alcohols (R–OH). The activity of GPX4 needs two GSH molecules, which are converted to GSSG. GSSG can be recycled back to GSH in an NADPH-dependent manner by GSH reductase. In susceptible cells, inhibition of GPX4 [2,3] or GSH unavailability [4,5] triggers a lethal lipid peroxidation process.
Oxidative stress has been related to different pathological processes such as neurotoxicity, neuroinflammation, ischemic stroke, and neurodegenerative diseases, such as Parkinson’s disease, Alzheimer’s disease, Huntington’s disease, or Friedreich’s ataxia [6,7,8,9]. In this review, we focus in some groups of rare diseases: Friedreich’s ataxia, diseases with neurodegeneration with brain iron accumulation (NBIA), Charcot-Marie-Tooth as an example of rare neuromuscular disorders, inherited retinal dystrophies (IRDs), and progressive myoclonus epilepsies (PMEs) and pediatric drug-resistant epilepsies.
2. Friedreich’s Ataxia
Friedreich’s ataxia (FRDA, MIM 229300) is the most prevalent hereditary ataxia [10,11]. FRDA is a rare childhood-onset disease characterized by gait and limb ataxia, lower limb areflexia, and dysarthria. A mixed origin of ataxia results from spinocerebellar degeneration, peripheral sensory neuropathy, cerebellar, and vestibular pathology, and the posterior adding of the pyramidal disabilities [12]. Other non-neurological features of FRDA are scoliosis, diabetes, and cardiac symptoms [13,14,15]. The majority of FRDA patients are homozygous for an unstable guanine-adenine-adenine (GAA) expansion in the first intron of the FXN gene that localizes in chromosome 9q21.11 producing decreased protein levels of frataxin [16,17]. The principal function of frataxin is not clear; however, the early lethality in embryos of Fxn knockout mice underscores the importance of frataxin function in cell survival [18]. Previous studies have reported the involvement of the FXN protein in mitochondrial biogenesis [19] and the synthesis of iron-sulfur clusters (ISC) [20]. Mitochondrial respiratory chain dysfunction [21], mitochondrial iron accumulation [22], decreased mitochondrial DNA levels, oxidative stress [23,24], reduced generation of ATP [23], and altered lipid metabolism are molecular and pathological features of FRDA. One of the main pathways related to oxidative stress that is altered in FRDA is the Nfr2-pathway [25,26]. Human and mouse models of this disease showed a defective activation of phase II enzymes by Nfr2. Furthermore, different studies with Nfr2-inducers counteracts oxidative stress, cell death, and mitochondrial defects in different human and mouse models of FRDA [27,28].
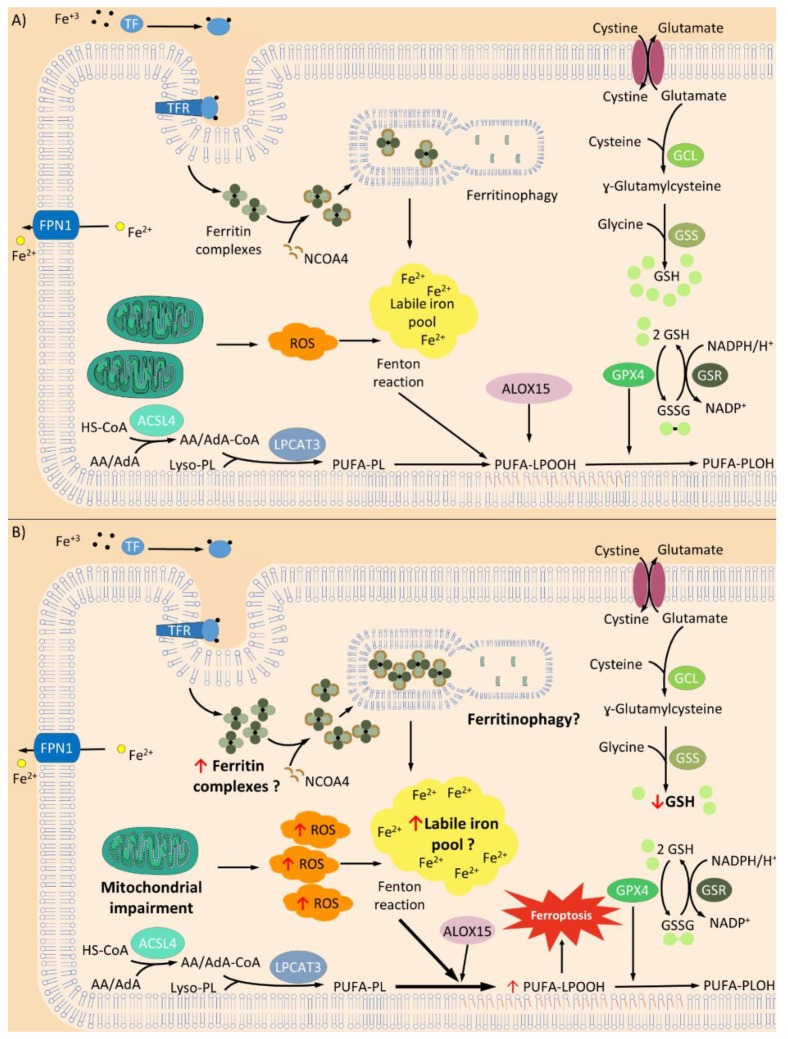
Ferroptosis is a new term for regulated cell death pathways (RCD) that is remarkably distinct at morphological, biochemical, and genetic levels from other RCD, such as apoptosis, classical autophagy, and necrosis [5]. This pathway is characterized by the overwhelming, iron-dependent accumulation of lethal lipid hydroperoxides [5]. It is noteworthy that this RCD is not a novel concept and it has been described several times before [29,30]. Diverse players of the ferroptosis pathway have been reported in the literature as isolated effects, but it is still unknown their connections in the whole landscape of this RCD. Some of these players are shown in Figure 1.
Figure 1.
Main mechanisms in ferroptosis. (A) General process of ferroptosis. Balanced levels of reactive oxygen species (ROS) generation and antioxidant activity maintains an inactive ferroptosis cell death pathway; however, some disturbance in the players of this equilibrium could produce a ferroptosis activation. Two important components of ferroptosis are the process of transferrin (TF)-iron import and ferroportin (FPN1) export. Changes in iron uptake or iron export produce a suppression or activation of ferroptosis since the free iron pool is fundamental [31,32]. Ferroptosis may be enhanced through the process of ferritinophagy, by which ferritin is targeted by nuclear receptor coactivator 4 (NCOA4) and delivered to the lysosome for degradation in autophagosome [33,34,35]. Consequently, a large quantity of iron can be rapidly released. Currently, it has been suggested that ferroptosis initiation might be triggered by an increase in free iron levels itself [36]. Iron increase/or accumulation induces the Fenton reaction which increases the production of ROS. Furthermore, the action of iron-dependent oxidases [37], specifically lipoxygenase activity of 15-LOX (ALOX15) that oxidizes polyunsaturated fatty acids (PUFAs) phospholipids (PUFA-PLs) containing arachidonate or adrenate moieties, triggers the ferroptosis pathway [38,39]. In addition, increased activity of acyl-CoA synthetase long-chain family member 4 (ACSL4), an enzyme that preferentially activates arachidonic (AA) and adrenoic acids (AdA), has been reported to increase ferroptosis [38,40]. Inhibition of glutathione peroxidase enzyme 4 (GPX4) [2,3] or glutathione (GSH) unavailability [4,5] produces lipid hydroperoxide accumulation that triggers ferroptosis. (B) Suggested mechanisms of ferroptosis in Friedreich’s ataxia (FRDA). It has been observed increased levels of lipoperoxides and ROS in FRDA neurons. Furthermore, in these neurons, it has also been reported lower reduced GSH concentration and GSH homeostasis disturbance, respectively [41,42,43]. Controversial studies are reporting either normal or increased iron levels in the nervous system of FRDA patients (reviewed in [44]). Recently a study in FRDA patients, using magnetic resonance imaging, showed increased iron concentration in the extrapyramidal motor system compared with controls [45]. However, there is no information about distribution, location and redox state of the iron in cells from FRDA (indicated with “?”). Increased autophagic processes are related to FRDA in different models of disease [46,47], but there is no information about ferritinophagic activity. Further studies should be performed in order to unravel if this mechanism increases the iron content in FRDA cells. All these ferroptotic characteristics observed in FRDA may indicate that this kind of cell death may have a role in the physiopathology of this disease. Transferrin receptor (TFR) Lysophosphatidylcholine Acyltransferase 3 (LPCAT3); Glutamate Cysteine Ligase (GCL); Glutathione synthetase (GSS); Glutathione reductase (GR). ↑ indicates increased levels and ↓ indicates decreased levels.
The discovery of ferroptosis as a possible way of neuronal death in FRDA is a major achievement since it explains many of the already known cellular, metabolic, and pathophysiological characteristics of neuronal degeneration in FRDA, like oxidative stress, changes in calcium levels, neuronal degeneration, and mitochondrial and membrane damage [48]. FRDA neurons showed higher lipoperoxide levels, increased ROS and lower reduced GSH concentration, and enhanced sensitivity to oxidants compared with control neurons [41,42]. Furthermore, patients with FRDA suffer a disturbance of GSH homeostasis (Figure 1) [43].
In recent years, a few studies have started to show the implication of miRNAs regulation in ferroptosis [49,50,51]. Additionally, small RNA analysis performed in FRDA revealed altered levels of miRNAs [52,53,54]. The plasma of FRDA patients showed higher levels of miR-130b-5p and miR-142-3p. Since these miRNAs target the mRNAs coding for proteins involved in fatty acid metabolism and β-oxidation, they could affect ferroptosis regulation [52]. All these results indicate that ferroptosis may be an important pathway linked with FRDA pathophysiology and hence, a potential therapeutic target.
3. Neurodegenerative Disorders with Brain Iron Accumulation
Syndromes of neurodegeneration with brain iron accumulation (NBIA) are a group of movement disorders, which share an abnormal deposit of iron in the brain, predominantly in the basal ganglia. NBIA disorders start at any age, from early childhood to adulthood, and the course is relentlessly progressive causing early death or severe motor, cognitive, and sensorial disabilities in survival patients. The clinical course includes progressive speech difficulties, dysphagia, dystonia, spasticity, parkinsonism, pure akinesia, ocular motor dysfunction, vision loss, and neuropsychiatric disturbances. NBIA forms are rare diseases with an estimated prevalence of less than 1/1,000,000.
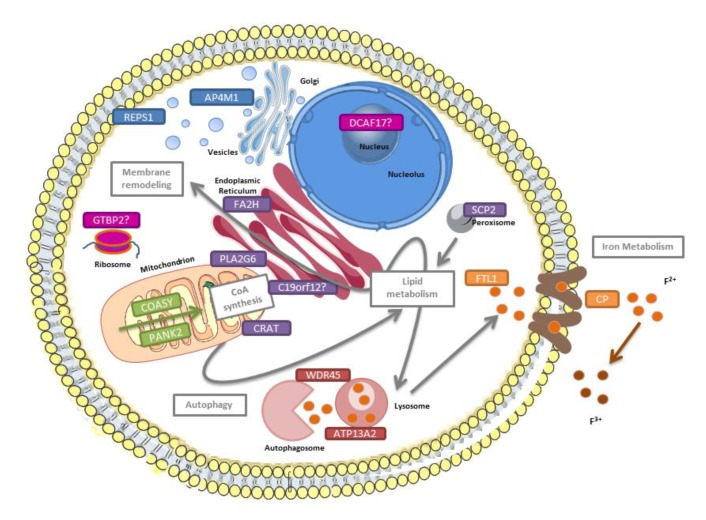
Ten NBIA genes are widely accepted, although, with the description of some cases, five additional genes could also be considered NBIA genes (Table 1) [55,56]. Little is known about the underlying disease mechanisms. Several pathways must be considered: iron and lipid metabolism, membrane remodeling, coenzyme A (CoA) synthesis, and autophagy (Figure 2). However, most of the genes play directly or indirectly a role in mitochondria and cause ion imbalance of Ca2+, mitochondrial-ER altered communication, and impaired oxidative phosphorylation (OXPHOS). In this review, the focus is on the NBIA forms in which altered ROS plays an essential role in the pathomechanism.
Table 1.
NBIA forms: genes and pathways.
| NBIA form | MIM # | Frequency Inheritance |
Gene MIM * |
Protein Location | Pathway |
|---|---|---|---|---|---|
| Pantothenate kinase-associated neurodegeneration (PKAN) | 234200 | 35%–50% AR |
PANK2 606157 |
Mitochondria | CoA synthesis (fatty acid metabolism) |
| Phospholipase 2, group VI-associated neurodegeneration (PLAN) | 610217 | 20% AR |
PLA2G6 603604 |
Mitochondria, ER, cytosol | Membrane phospholipids turnover |
| Mitochondrial membrane protein-associated neurodegeneration (MPAN) | 614298 | 6%–10% AR |
C19ORF12 614297 |
Mitochondria, ER, MAM | Lipid metabolism ? Membrane remodeling ? |
| β-propeller-associated neurodegeneration (BPAN) | 300894 | 1%–2% XD |
WDR45 300526 |
ER | Autophagy |
| Fatty acid hydroxylase-associated neurodegeneration (FA2H) | 612319 | Rare AR |
FA2H 611026 |
ER | Lipid metabolism Membrane remodeling |
| Neuroferritinopathy (NF) | 606159 | Rare AD |
FTL1 134790 |
Cytosol | Iron homeostasis |
| Aceruloplasminemia | 604290 | Rare AR |
CP 117700 |
Plasma membrane | Iron homeostasis |
| Woodhouse-Sakati syndrome | 241080 | Rare AR |
DCAF17 612515 |
Nucleolus | Unknown |
| Kufor-Rakeb syndrome | 606693 | 2 probands AR |
ATP13A2 610513 |
Lysosome, mitochondria | Autophagy |
| COASY protein-associated neurodegeneration (CoPAN) | 615643 | 4 probands AR |
COASY 609855 |
Mitochondria, cytosol | CoA synthesis (fatty acid metabolism) |
| Jaberi-Elahi syndrome (JABELS) + NBIA | 617988 | 1 family AR |
GTPBP2 607434 |
Cytoplasm | Unknown |
| Leukoencephalopathy with dystonia and motor neuropathy + NBIA | 613724 | 1 proband AR |
SCP2 184755 |
Peroxisomes | Lipid metabolism Membrane remodeling |
| NBIA7 | 617916 | 1 family AR |
REPS1 614825 |
Cytoplasm, endosome | Endocytosis Vesicle transport |
| Hereditary spastic paraplegia + NBIA | 612936 | 1 family AR |
AP4M1 602296 |
Endosome | Vesicle formation |
| NBIA8 | 617917 | 1 proband AR |
CRAT 600184 |
Mitochondria | Lipid metabolism |
MIM: Mendelian Inheritance in Man database, # refer to diseases, * refer to genes, ? refer to not known with absolute certainty; NBIA: Neurodegeneration with brain iron accumulation; MIM: Mendelian inheritance in man; AR: autosomal recessive; XD: X-linked dominant; AD: autosomal dominant; ER: endoplasmic reticulum; MAM: Mitochondrial-associated membrane.
Figure 2.
Pathways underlying NBIA disorders. PANK2 and COASY (in green) are involved in the coenzyme A (CoA) synthesis. PLA2G6, C19orf12, FA2H, SCP2, and CRAT (in purple) are related to lipid metabolism and membrane remodeling. CP and FTL1 (in orange) are implicated in iron metabolism; WDR45 and ATP13A2 (in maroon) play a role in autophagy; and finally, REPS1 and AP4M1 (in blue) are associated with vesicle trafficking. The function of DCAF17 and GTPBP2 (in fuchsia) remains elusive.
PANK2 and COASY are implicated in the biosynthesis of CoA. Mutations in PANK2 cause PKAN (Pantothenate Kinase-Associated Neurodegeneration, MIM 234200), which is the most frequent NBIA form (35–50% of NBIA patients), and deficiency of COASY leads to COPAN (COasy Protein-Associated Neurodegeneration, MIM 615643), which is an ultra-rare form of NBIA (Table 1). PANK2 catalyzes the ATP-dependent phosphorylation of pantothenate, the first step of the CoA synthesis, whereas COASY regulates the last two steps in the CoA synthesis. In Drosophila, the dissection of the CoA synthesis route revealed that this pathway is important for maintaining DNA and cellular integrity [57]. Moreover, fly mutants for Pank2 (dPANK/fmbl) and Coasy (dPPAT-DPCK), display locomotor dysfunction, increased sensitivity to oxidative stress, and altered lipid homeostasis [57].
The absence of PANK2 causes the accumulation of substrates in the CoA synthesis pathway, mainly N-pantothenyl cysteine and free cysteine. Free cysteine suffers spontaneous autoxidation due to the deposits of iron, which produces free radicals, and finally, oxidative damage and cell death [58,59]. The null fly model fumble, which is the orthologue of the human PANK2 gene in Drosophila melanogaster, shows decreased CoA levels, mitochondrial dysfunction, increased protein oxidation and impairment of lipid homeostasis [60]. Neurons of the Pank2 knockout mouse present altered mitochondrial membrane potential and defective respiration that would contribute to ROS generation [61]. In the murine Pank2 model, signs such as motor dysfunction, neurological impairment, and premature death, are only observed when the mouse is fed with a ketogenic diet [62]. Fibroblasts and induced Pluripotent Stem Cells (iPSCs)-derived neurons, exhibit elevated levels of ROS production, together with premature death, and mitochondrial dysfunction, including impairment of mitochondrial iron-dependent biosynthesis, probably due to iron mishandling [63,64,65,66,67].
Mutations in PLA2G6 cause PLAN (PLA2G6-associated neurodegeneration, MIM 615643). The encoded protein by PLA2G6, iPLA2-VI, hydrolyzes at the sn-2 position of glycerophospholipids to produce free fatty acids and lysophospholipids, and it can play a key function in lipid homeostasis, which is essential for membrane remodeling. PLA2G6 is also linked to mitochondrial dynamics since high levels of ROS cause the accumulation of iPLA2-VI in mitochondria, which gives apoptosis protection [68]. In fact, the Pla2g6 knockout mice are characterized by progressive cerebellar atrophy, Purkinje cell loss, and neuroinflammation, but no iron accumulation is observed. Moreover, this murine model displays degeneration of mitochondria and axonal termini in the spinal cord due to altered lipid profile, suggesting that the deficit of Pla2g6 may lead to anomalies in membrane remodeling, and reduced mitochondrial potential in neurons, highlighting that Pla2g6 participates in mitochondrial homeostasis.
Mutations in C19ORF12 are responsible for a relatively frequently NBIA form named MPAN (Mitochondrial membrane Protein-Associated Neurodegeneration, MIM 614298) (Table 1). The C19ORF12 gene encodes for a transmembrane glycine zipper present into mitochondria, ER and MAMs (Mitochondria Associated Membranes) [69]. According to the findings described in fibroblasts from one MPAN patient, the loss of C19ORF12 leads to high mitochondrial Ca2+ levels and increased H2O2 inducing oxidative damage [69]. The function of C19ORF12 remains elusive, although a role as regulatory protein for magnesium transporters is suggested given its homology to the magnesium transporters E (MgtE) [69].
Two NBIA forms are due to iron-related genes, aceruloplasminemia (MIM 604290) and neuropherritinopathy (MIM 256600), caused by mutations in the CP and FTL1 genes, respectively (Table 1). CP is responsible for the peroxidation of ferrous transferrin to ferric transferrin and due to its ferroxidase activity, is a strong antioxidant [70]. In cell systems, the absence of CP elicits that the ferrous iron entering the central nervous system cannot be oxidized and this is accumulated in cells as non-transferrin-bound iron that represents a toxic iron form, which induces oxidative stress, and in the end, causes neuronal death [71]. Several Cp knockout mice are available and no all of them have neurological phenotype; however, they share the abnormal iron accumulation and lipids peroxidation in the brain [72,73]. FTL1 encodes the L-ferritin subunit, one of the two subunits of the ferritin, which is the major iron storage protein. The overexpression of mutated FTL1 in cell models yields an increase of ferritin chains and a decrease of transferrin receptor 1 (TfR1) expression together with a production of ROS after treatment with H2O2, suggesting that the pathomechanism is likely due to deregulation of cellular iron homeostasis and oxidative damage, which would be primary cause of cell death [74,75]. Transgenic mice models, expressing mutated Ftl1 show iron deposition and oxidative stress in brain, and a neurological deterioration with reduced lifespan and motor dysfunction [76,77,78].
The loss of ATP13A2 function, which codifies an ATPase that participates in the maintaining of lysosomal iron stores, causes Kufor-Rakeb syndrome (MIM 606693) (Table 1) characterized by α-synuclein accumulation and mitochondrial stress-induced neurotoxicity [79,80]. Using Chinese Hamster Ovary (CHO) cells that stably express the ATP13A2 protein, the cytotoxicity induced by iron was reduced, which may suggest that the ATP13A2 overexpression has a protective role against the cell damage caused by iron [81].
4. Neuromuscular Diseases
Charcot-Marie-Tooth (CMT) disease is one of the most clinically important inheritable peripheral neuropathies. CMT affects the structure and function of nerves, both motor and sensitive. There are at least 80 different genes affected in the disease. The genes affected may cause degeneration of either the axons or the myelin sheath in the peripheral nervous system. This degenerative process impairs action potential conduction, a decrement in the velocity of nerve transmission causing muscle atrophy and alterations in the sensitivity. It is currently incurable.
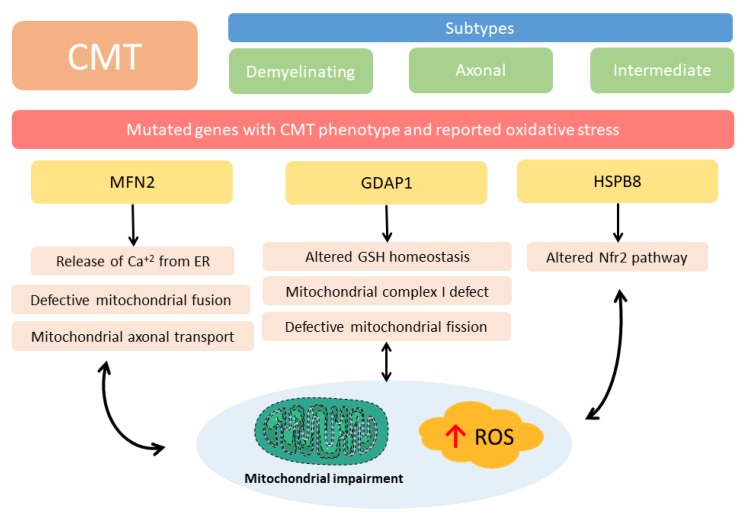
Classification of CMT is rather complex and is based on clinical and genetic criteria. CMT disease is clinically divided into three main types CMT1: the demyelinating form, characterized by a decrease in the nervous conduction velocity. CMT2 or axonal CMT, where there is a loss in the number of axons and a decrease in the number of Schwann cells; and an intermediate clinical form with some decrease in the velocity of conduction and a decrease in the number of axons. An important number of the mutated genes that produce CMT are related to mitochondrial proteins than develop an oxidative stress associated phenotype. However, not all the different types of CMT develop oxidative stress or mitochondrial impairment as far as we know. We will briefly review here the pathophysiological role of two of the most common causative genes for axonal CMT2, mitofusin 2 (MFN2) and ganglioside induced differentiation-associated protein 1 (GDAP1), where oxidative stress takes place when these proteins are mutated (Figure 3), but not all the other types of CMT where oxidative stress has not been demonstrated.
Figure 3.
Charcot-Marie-Tooth disease (CMT) pathways related to oxidative stress. CMT is clinically divided into three main types, the demyelinating form, characterized by a decrease in the nervous conduction velocity; axonal CMT, where there is a loss in the number of axons and an intermediate clinical form with some decrease in the velocity of conduction and a decrease in the number of axons. At least 80 different genes are affected in the disease. Some of them produce oxidative stress or mitochondrial impairment. MFN2 and GDAP1 are the best known. HSPB8 has been related to the Nrf2 pathway. ER: Endoplasmic reticulum, GDAP1: ganglioside induced differentiation associated protein 1, GSH: glutathione, HSPB8: Small Heat shock protein 8, MFN2: mitofusin 2, Nrf2: Nuclear factor erythroid 2-related factor 2, ROS: reactive oxygen species.
Mitofusin 2 protein is located mainly in the outer mitochondrial membrane where it contributes to the process of mitochondrial fusion by homo- and hetero-dimerization of MFN2 with mitofusin 1 (MFN1) [82]. MFN2 is also present in ER membrane-bound to mitochondrial MFN1 and MFN2, allowing the release of Ca2+ from ER into mitochondria [83,84]. MFN2 is also involved, possibly due to Ca2+ influx, in increasing outer membrane permeability, and consequently in the regulation of apoptosis [85]. It has been related to mitochondrial dynamics and in the control of the oxidative phosphorylation machinery [86]. MFN2 is present in many cell types. Nonetheless, the presence of MFN2 mutations is only able to produce dysfunctions in peripheral nerves; this is the case of CMT. It seems that MFN1 is sufficient to accomplish the functions of MFN2 in the majority of tissues, except in peripheral nerves, due to the low physiological levels of MFN1 compared to other tissues [87]. It is still unclear why mutations in MFN2 lead to CMT2A (MIM 609260) and what are the potential mechanisms that chief the pathological process, but clearly two are the most obvious: firstly, a defective mitochondrial fusion (for a review see [88]), and secondly, oxidative stress. In fact, Han et al. [89] have recently demonstrated in mouse adult hippocampus and cortex neurons that downregulation of MFN2 produces mitochondrial fragmentation and subsequent neurodegeneration induced by oxidative stress giving rise to neuroinflammation in vivo [89].
Among the main characteristics that are modified when MFN2 is mutated, mitochondrial transport is of particular interest. An abnormal axonal transport of mitochondria could explain why mutations in MFN2, induce distal axonal degeneration giving rise to the clinical features of CMT2A neuropathy, where the longest axons are mostly affected. An important body of literature underscores the role of MFN2 in mitochondrial axonal transport (for a review see [87]).
GDAP1 is the other major protein mutated in CMT that gives rise to oxidative stress in CMT. It causes different clinical phenotypes of the disease (CMT2K and AR-CMT2K, MIM 607831; RI-CMTA, MIM 608340; CMT4A, MIM 214400). GDAP1 is ubiquitously expressed, but its presence is especially high in neurons [90]. GDAP1 is part of the glutathione S-transferase (GST) family [91], but it is unclear if the GST activity present in GDAP1 has any physiological relevance. However, inhibition of GDAP1 expression increases the susceptibility of the NSC34 motor neuron-like cells to GSH depletion [92]. Similarly, fibroblasts from CMT4A patients that also have reduced GDAP1 levels, show decreased GSH levels and mitochondrial membrane potential impairment, pointing oxidative stress as the main player in the pathogenesis of this type of CMT [92]. Very recently, it has been reported that mutations in the GST domain of GDAP1 are associated with a mitochondrial complex I defect and worsening of the clinical phenotype of the disease. This looks very interesting since other reported mutations that are located outside the GST domain show mild clinical phenotypes [93].
Decreased expression of GDAP1 levels in CMT produces mitochondrial morphological abnormalities impairing mitochondrial function, increasing the production of free radical species and altering the cytosolic calcium handling by mitochondria [94]. Decreased levels of GDAP1 produce disarrangement of Ca2+ metabolism in neurons and mitochondrial damage in a knockout mouse model of CMT. These mitochondrial changes give rise to axonal degeneration, neuronal death and neuroinflammation [95,96] producing a vicious circle of oxidative stress that contributes to the clinical features of the disease. However, we should remember that the correlation genotype/phenotype is complicated and CMT is a good example. In fact, Junctophilin-1 was described as a modifier gene of GDAP1 and that this gives rise to phenotypic variability [97].
Missense mutations, such as p.K141N, in the heat shock protein HSPB8 cause CMT2L. According to Yang et al. [98], this mutation induced mitochondrial aggregation and caused increased oxidative stress injury in SH-SY5Y cells. L-3-n-butylphthalide (NBP), extracted from seeds of celery (Apium), was able to prevent mitochondrial aggregation and oxidative stress activating the Nrf2 pathway. Nrf2 is considered a master regulator that induces antioxidant gene expression through binding to antioxidant response elements (AREs) located in the promoters of genes encoding antioxidants enzymes.
5. Inherited Retinal Dystrophies
Inherited retinal dystrophies (IRDs) are considered a group of heterogeneous retinal diseases that cause progressive and severe loss of vision by altering the retinal structure or/and retinal function [99]. IRDs are generally characterized by a permanent loss of light-sensitive retinal neurons, or their support cells leading eventually to legal blindness [99]. Although each disorder has individually a low prevalence, as a whole, they affect 1/3000–1/4000 individuals [100,101].
The features that best define IRDs are its high clinical variability and high genetic heterogeneity. Currently, 271 genes and 36 loci responsible for some type of IRD are known (RetNet, https://sph.uth.edu/retnet/; last accessed March 2020). Additionally, different mutations in the same gene may lead to different clinical conditions and to the same clinical condition but with different inheritance patterns [102].
Apart from inherited mutations, the retina is susceptible to several environmental insults and stress, including damage by light, and oxidative stress that can alter its structure and/or function [103]. The retina is especially vulnerable to changes in oxygen tension and oxidative stress because this is rich in polyunsaturated lipid membranes. Oxygen supply to the photoreceptors in the outer retina (main oxygen consumers) comes primarily from the choroid, while the retinal vasculature supplies oxygen to the inner retina. Elevated oxygen levels or hyperoxia are well known that results in photoreceptor cell death. Hyperoxia increases the production of ROS and RNS, which have been related to several IRD [104,105,106,107,108]. In particular, hyperoxia has been described in the outer retina of different models of Retinitis Pigmentosa (RP), the most prevalent IRD, such as RCS (Royal College of Surgeons) rats or P23H mutant transgenic rats [105,109]. Several studies support an oxygen toxicity hypothesis for RP. This hypothesis proposes that cone death (secondary photoreceptor loss) is due to hyperoxia, which is the consequence of the reduction in oxygen uptake following the initial loss of rod (due to various rod-related mutations) [110]. Some pieces of evidence of the gradual loss of oxygen metabolism in animal models of RP or patients include the altered distribution of intraretinal oxygen at different stages of the disease [109], the decreased content of oxygen-sensitive hypoxia-inducible factor-1α (HIF-1α) [111], or the thinning of inner retinal vessels [105,112,113]. Evidence for oxidative damage in RP are the increase of ROS or RNS, increase oxidation of macromolecules (lipids, proteins, and nucleic acids) and decrease in antioxidants in the retina, aqueous humor, and plasma of RP animal models and patients [107,114,115,116,117]. For instance, 8-oxo-7,8-dihydroguanine (8-oxoG), a major oxidized base in DNA, is increased in retinas of RP models and the vitreous of RP patients [118,119,120]. Accumulation of superoxide radicals and subsequent increase of peroxynitrite is present in the outer retinas of rd1 mice, a model of RP, contributing to cone death [121]. Recently, Perdices et al. [122] also showed hepatic oxidative stress in P23H rhodopsin transgenic rats [105,122]. It is necessary to neutralize free radicals by the antioxidant defense system, otherwise, free radicals will interact with lipids, proteins, DNA, etc., affecting the cone survival. However, collected data from patients suggest that the antioxidant defense system is affected during RP [115,117,123].
Photoreceptors contain a high density of mitochondria in the inner segments that supply the ATP for the phototransduction processes, mainly through oxidative phosphorylation [124]. Most of the oxygen (~90%) is used within mitochondria. Then, mitochondrial oxygen metabolism is the main source of ROS production (e.g., superoxide and hydrogen peroxide) [125]. Mitochondrial oxidative stress and energy failure is a significant early event of IRD [126]. It was observed that retinal degeneration is accompanied by mitochondrial failure and metabolic imbalance, including downregulation of oxidative phosphorylation pathway and alterations in energetic pathways (glycolysis, β-oxidation), and changes in mitochondrial proteins [114,126,127,128].
When mitochondria are damaged, they can be selectively removed by autophagy, called mitophagy. Some genes associated with autosomal dominant RP, including PRPF6, PRPF31, SNRNP200, and PRPF8 (core spliceosomal components) [129,130,131], are suggested to be responsible for ULK1 mRNA mis-splicing (an important protein for mitophagy initiation) and subsequent mitophagy defects [132].
Among genes involved in IRD, some mutations in mitochondrial genes are found such IDH3A, which generates NADH used for mitochondrial ATP production and is associated to early-onset retinal degeneration [133]; and aconitase (similar to IDH3A), that results in severe neurological disease apart from retinal degeneration [134,135].
The transcription factor Nrf2 exhibits antioxidant and anti-inflammatory properties in several tissues including the retina. Activation of Nrf2 through carnosic acid prevents retinal degeneration in Mef2d mice exposed to light damage [136]. Analogs of progesterone, which regulate Nrf2, also prevent retinal degeneration in inherited or light-damage models of retinal degeneration. For instance, Norgestrel activates and increases Nrf2 expression and its target, superoxide dismutase 2 reducing mitochondrial oxidative stress in a light damage model [137]. In RP, activation of Nrf2 and upregulation of its targets slow retinal degeneration in rabbits [138] and, mice by preserving photoreceptor, reducing inflammation and visual loss [139].
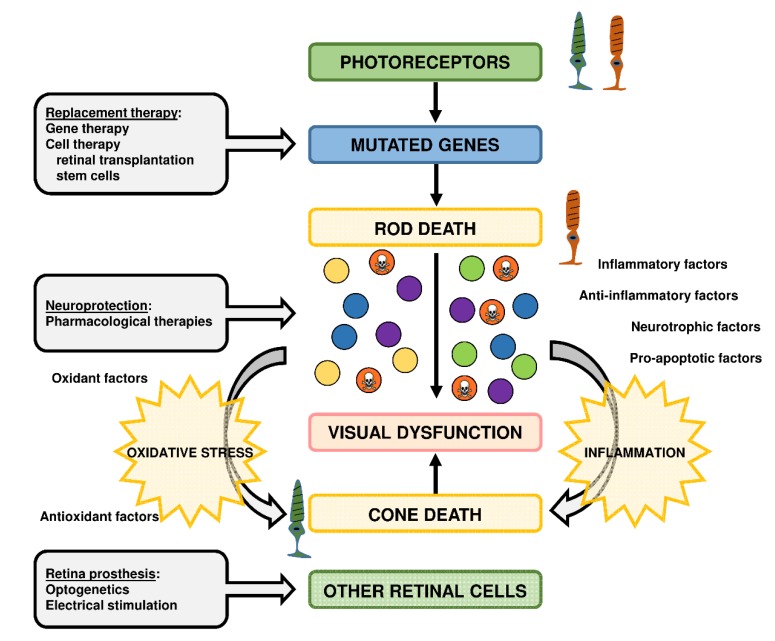
To date, there are no commercially available treatments to prevent photoreceptor cell loss and preserve vision in IRDs, but the development of several therapeutic approaches are ongoing, including nutritional supplements, neurotrophic factors, antioxidants supplementation, gene therapy, stem cell-based therapy, optogenetics, or retinal prostheses (Figure 4) [140,141,142,143]. These approaches depend on the subtype of IRD, its severity, and stage, the therapeutic target molecule, the knowledge of the mutated gene, etc. Some of these approaches try to correct or replace the genetic defect (gene or stem cell-based therapies) or to slow down the IRD progression (nutritional supplements, neurotrophic factors, antioxidants, etc.). In particular, several antioxidant formulations including curcumin, Tauroursodeoxycholic Acid (TUDCA), vitamin A, vitamin A in combination with docosahexaenoic acid (DHA), lutein, etc., have been used to mitigate the degenerative process in RP models and patients [116,144,145,146,147,148,149,150].
Figure 4.
Therapeutic approaches for retinitis pigmentosa (RP), the most prevalent inherited retinal dystrophy. RP is characterized by progressive rod photoreceptor degeneration (mutated genes) in the initial stage and eventual cone photoreceptor degeneration in later stages. It is highly probable that cone degeneration is influenced by the release of inflammatory molecules, oxidant radicals, pro-apoptotic factors, etc. from rods and other cells, independently of the gene defect. Several therapeutic approaches have been developed to preserve retinal function or restore vision depending on the stage of the disease. They can be led to repair mutated photoreceptors (gene therapy), to replace damaged photoreceptors (cell therapy), or to protect photoreceptors and other retinal cells to the “toxic environment” (neuroprotective compounds including antioxidants). At later stages of RP, therapeutic approaches can be led to other retinal cells to restore visual activity with the remaining retinal circuits (optogenetics, retinal prostheses).
6. Rare Epilepsies
Epilepsy is a neurological disorder characterized by a predisposition to generate epileptic seizures and the associated cognitive, psychological, and social consequences [151]. The first-line treatment for epilepsy is anti-seizure drugs (ASDs). However, despite the availability of many ASDs, approximately one-third of patients fail to achieve seizure control or soon become resistant to the effects of the ASDs [151,152]. Consequently, there is a critical need for the development of innovative anti-epileptogenic treatment strategies to either ameliorate the progression or/and limit the detrimental consequences of the disease. Neuroinflammation, oxidative stress, and epilepsy are interconnected. It is becoming clear that brain inflammation promotes neuronal hyperexcitability and seizures, and that dysregulation in the glia immune-inflammatory function is a common factor that predisposes or contributes to the generation of seizures. At the same time, acute seizures upregulate the production of pro-inflammatory cytokines in microglia and astrocytes, triggering a downstream cascade of inflammatory mediators. Therefore, epileptic seizures, oxidative stress, and inflammatory mediators form a positive feedback loop, reinforcing each other [153]. It has also been shown that seizures can alter the redox state of the mitochondria [154]. As neuroinflammation and oxidative stress are intimately related hallmarks of many epileptic syndromes, this offers a window of opportunity for the use of anti-inflammatory and antioxidant drugs in these conditions. This possibility is supported by recent reports that clearly state that neuroinflammatory pathways may serve as treatment targets and biomarkers in different forms of epilepsy [153,155].
In this section, we will focus our attention on two different types of rare epilepsies: progressive myoclonus epilepsies (PMEs) and pediatric drug-resistant epilepsies. We describe examples of both types of pathology to illustrate the involvement of oxidative stress and neuroinflammation despite their clinical differences, and the window of opportunity that this information offers in the development of new treatments.
6.1. Progressive Myoclonus Epilepsies (PMEs)
The PMEs are a group of neurological disorders characterized by the occurrence of focal and generalized seizures, myoclonus, and progressive neurological deterioration, accompanied by cerebellar signs and dementia [156,157,158,159,160]. PMEs consist of more than a dozen different diseases and they are classified as rare diseases since each of them affects less than 1/2000 individuals. The most common PME conditions are: (1) Unverricht–Lundborg disease (ULD) or epilepsy progressive myoclonus 1 (EPM1), due to mutations in the CSTB gene encoding cystatin B, a lysosomal cysteine protease inhibitor. (2) Lafora disease (LD) or epilepsy progressive myoclonus 2 (EPM2), due to mutations in either EPM2A gene, encoding the glucan phosphatase laforin, or EPM2B gene, encoding the E3-ubiquitin ligase malin. (3) Neuronal ceroid lipofuscinosis (NCL), a collection of disorders due to mutations in more than ten different CLN genes. (4) Sialidosis, a lysosomal storage disease due to mutations in the NEU1 gene encoding the lysosomal enzyme alpha-N-acetylneuraminidase; and (5) mitochondrial myopathies, such as myoclonic epilepsy with ragged fibers (MERRF) due to mutations in the mitochondrial gene MT-TK encoding the tRNALys, among others [158].
Although the primary cause of PMEs is different in each case, recent reports suggest that oxidative stress and neuroinflammation are common traits in all these conditions [161,162,163]. This reinforces the idea that oxidative stress and neuroinflammation are at the crossroad of rare neurodegenerative disorders. Among PMEs, we will focus on the two most characterized forms: ULD and LD.
6.1.1. Unverricht–Lundborg Disease (ULD)
The onset of ULD (MIM 254800) is around late childhood and early adolescence. It is characterized by the appearance of generalized tonic-clonic seizures that may occur without prior myoclonic jerks. As the disease progresses, myoclonus increases in intensity and frequency culminating in generalized tonic-clonic seizures. ULD also progresses with associated neurological symptoms, such as ataxia, impaired walking, and cognitive impairment. On the contrary, to other PMEs, early death is not common in ULD and the outcome of adult patients ranges from minimal impairment with an independent active life to severe disability [164,165]. ULD is an autosomal recessive disorder due to mutations in the gene encoding cystatin B/Stefin B (CSTB), an 11 kDa lysosomal cysteine protease inhibitor. The most common mutation leads to the expansion of minisatellite sequence repeats (CCCCGCCCCGCG) in the 5′-untranslated region of the CSTB gene, to reach a number of 30–80 repeats in the causative disease range, leading to its reduced expression [165]. In addition, frameshift mutations and deletions are also found among ULD patients, although they are less common [164].
There is a mouse model of ULD that lacks the CSTB gene (Cstb−/−), which reproduces the pathophysiology of the disease: they show myoclonic seizures, ataxia, and progressive neuronal loss. In these mice, it has been proposed that early-onset neuroinflammation was key in the pathogenesis of ULD and that glial-derived pro-inflammatory chemokines and cytokines contributed to recurrent excitation and epilepsy [161]. In the absence of Cstb, there is also a clear mitochondrial dysfunction, with decreased membrane potential and increased ROS production, which could stimulate an inflammatory response. The authors suggest that this mitochondrial dysfunction could be the primary cause of the pathophysiology of ULD [166].
6.1.2. Lafora Disease (LD)
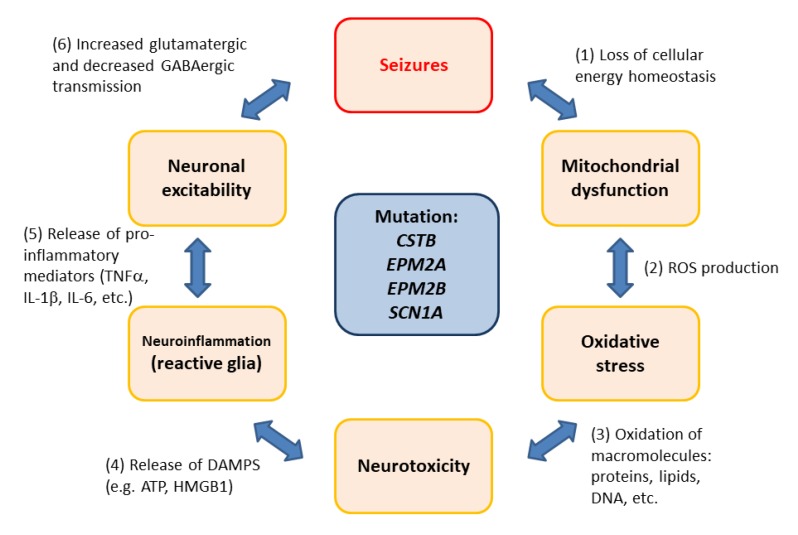
LD (MIM 254780) is a rare, fatal form of PME characterized by the accumulation of insoluble poorly branched glycogen inclusions (named Lafora bodies, LBs) in the brain and peripheral tissues. The onset of LD occurs around late childhood and early adolescence and it is characterized by the appearance of generalized tonic-clonic seizures, myoclonus, absences, and visual hallucinations. The disease progresses rapidly with a worsening of seizures and dementia, leading to the death of the patient after a decade from the onset of the first symptoms [167]. LD is caused by mutations in the EPM2A gene, which encodes the glucan phosphatase laforin [168], and the EPM2B gene, encoding the E3-ubiquitin ligase malin [169]. Laforin and malin form a functional complex that regulates glycogen synthesis, the homeostasis of glucose transporters, the maintenance of proteostasis and the response to oxidative stress, among other physiological pathways [167] (Figure 5).
Figure 5.
The vicious circle of oxidative stress and neuroinflammation in rare epilepsies with a genetic origin. (1) There is an inverse relationship between mitochondria functionality and the appearance of seizures. Alterations in cellular energy homeostasis result in neuronal pathology. (2) Mitochondrial dysfunction leads to oxidative stress due to the accumulation of reactive oxygen species (ROS). (3) Oxidized macromolecules (e.g., proteins, lipids, DNA) affect neuronal viability, leading to neurotoxicity. (4) Neuronal release of danger associated molecular patterns (DAMPs; e.g., HMGB1, ATP) triggers a neuroinflammatory response, leading to the activation of glia (microglia and astrocytes). (5) The production of pro-inflammatory mediators (TNFα, IL-1β, IL-6, etc.) affects neuronal excitability. (6) Increased glutamatergic transmission and decreased GABAergic inhibition leads to seizures.
LBs contain, in addition to carbohydrate moieties, up to 6% of proteinaceous material, which includes glycogen metabolizing enzymes, laforin, chaperones, autophagic components, proteasome subunits, advanced glycation end products, etc. [170,171,172]. Most of these products are post-translationally modified by ubiquitination, suggesting a cellular attempt to eliminate LBs by the usual protein clearance mechanisms, namely proteasomes and autophagy. However, these mechanisms are unsuccessful in disposing of the LBs and become deteriorated. In fact, it has been suggested that in the absence of laforin or malin there is a decrease in the activity of proteasomes [173,174] and an impairment in autophagy, which likely occurs at the initial step of autophagosome formation [134,170,175,176]. As a consequence, activation of the unfolded protein response pathway proceeds, which eventually leads to increased endoplasmic reticulum stress [171,174]. In this regard, it has also been described that laforin interacts physically with the chaperone Hsp70 and helps to decrease the toxicity of the unfolded proteins [177]. Therefore, all these results suggest a possible role of the laforin-malin complex in regulating cellular proteostasis [134,170,173,174,175,176].
Since the autophagy machinery is impaired, mechanisms that require this process are also affected in cellular and animal models of the disease. In this sense, we have recently described a defect in mitophagy, most likely due to defects in the initial steps of autophagosome formation [178]. Finally, we have also reported that oxidative stress is affected in the absence of a functional laforin-malin complex [179]. We demonstrated that in cellular and animal models of LD, there were higher levels of ROS and oxidative stress products. This defect was probably due to an altered mitochondrial function and also, to alterations in the levels of antioxidant enzymes and to decreased activity of enzymes involved in the detoxification of ROS [179].
Recently, we have analyzed by RNA-Seq technology the genes that were differentially expressed in the brain of Epm2a−/− and Epm2b−/− mice in comparison to control animals and observed that Epm2a−/− and Epm2−/− mouse brains overexpress a common set of genes mostly related to inflammation [162]. We also defined that reactive glia was responsible for the expression of these genes. These results are in agreement with the presence of pro-inflammatory markers (e.g., IL-1beta, TNF-alpha, and IL-6) in the brain of Epm2a−/− and Epm2−/− mice [180], and suggest that neuroinflammation should be considered as one of the most important traits in LD.
6.2. Dravet Syndrome
Pediatric drug-resistant epilepsies include a group of epileptic encephalopathies that are characterized by being caused by monogenic mutations, an early onset before or around the first year of age, the presence of associated developmental and cognitive impairments and resistance to common anticonvulsant treatments [181]. These pathologies include the Otahara, West, atypical Rett, and Dravet syndrome (DS) or severe myoclonic epilepsy of infancy (SMEI). The DS (MIM 607208) can be considered as a paradigmatic example in this type of epilepsy, since the involved gene was one of the first to be identified, and has well-known genetics, etiology, and pathophysiology [182,183,184]. DS has an early onset in the first few months of life, usually triggered by fever, with a clonic or hemiclonic seizure, which is followed by further episodes that can be prolonged and even result in status epilepticus. With time, patients usually suffer more febrile or afebrile seizures that can be of different types from the initial ones: tonic-clonic, myoclonic, absence. In addition to seizures, they can also suffer cognitive delay, movement disorders and, more seriously, sudden death in a relatively high proportion (>10%).
When the genetic basis of DS was uncovered [185], it came as a surprise that it was caused by dominant mutations on the SCN1A gene. This gene codes for the α subunit of the type 1 voltage-gated sodium channel, and these channels are usually involved in the excitation of neuronal and muscular cells. The conundrum was resolved when it was found that the mutations prevented the activation of GABAergic inhibitory interneurons, not the excitatory neurons, disrupting the excitation/inhibition balance [186,187]. There are no efficient medications to control the seizures and the neurological damage, although some first-line ASDs such as valproate, stiripentol, or benzodiazepines can help to control the seizures [188]; the most recent therapeutic development has been the successful clinical trials of cannabidiol (CBD) [189], which was approved to treat DS in America and Europe in 2019.
In patients suffering from severe DS, there are evident features of encephalopathy [190,191], probably due to an exacerbated inflammatory state. Although oxidative stress is not the primary cause of DS, the available evidence strongly indicates that oxidative stress and neuroinflammation are involved in the pathology pediatric epilepsy, mainly in the ensuing encephalopathy and deterioration of cognitive capabilities. Nuclear Metabolic Resonance (NMR) metabolomic analysis of pediatric drug-resistant epilepsies revealed changes in lactate, creatine, citrate, and lipids consistent with an increase in oxidative stress [192]. Regarding DS, GABAergic neurons differentiated from patient-derived iPSCs displayed a defective response to oxidative stress [193]. In this study, the authors also found reduced levels of the GSTM1 gene, which is also associated with epilepsy and encodes a glutathione-S-transferase, partly explaining the susceptibility to oxidative stress.
6.3. Oxidative Stress and Neuroinflammation as Therapeutic Targets in Rare Epilepsies
Neuroinflammation and oxidative stress emerge as important traits to be considered in the pathophysiology of rare epilepsies, and therefore, they constitute pharmacological targets for this group of diseases. We could hypothesize that mitochondria dysfunction accompanied by ROS production could be the trigger of the initial inflammatory response, which will eventually lead to epilepsy.
Regarding oxidative stress, some of the current treatments could already be addressing this problem. In addition to DS, cannabidiol (CBD) is being used or in clinical trials for several types of epilepsy [194], and it has a neuroprotective effect by inhibiting nitric oxide production, one of the main neurotoxic effectors [195]. The ketogenic diet, high in fats and low in carbohydrates and proteins, is also widely used to treat refractory epilepsies [196]. The β-oxidation of fatty acids produces ketone bodies, acetoacetate, and β-hydroxybutyrate, a metabolic state similar to starvation known as ketosis. In a rat model of fever induced by lipopolysaccharides, the ketogenic diet produced an anti-inflammatory state, with reduced levels of pro-inflammatory cytokines [197]. β-hydroxybutyrate acts as an epigenetic factor through histone deacetylases and increases the expression of the antioxidant genes FOXO3A and MT2 [198].
The initial oxidative insult could activate astrocytes and/or microglia to prime a general activation of these cells in the brain, which would then lead to neuronal degeneration. This is an important issue since it enhances the importance of astrocytes and microglia in this pathology and identifies these cells as the primary cause of the disease instead of the neurocentric idea that they are produced by initial neuronal problems. In addition, it has been demonstrated that anti-inflammatory interventions in animal models of epilepsy have both anti-epileptogenic and disease-modifying therapeutic effects [153,155]. However, it has also been stated that general anti-inflammatory drugs should not be used due to their wide central and peripheral effects [199] and that the anti-inflammatory strategy should be based on the signaling pathways that are altered in each epileptic condition. Some of these specific compounds are already in clinical use for the treatment of autoimmune diseases, so the use of specific brain-penetrant anti-inflammatory compounds that are used in other pathologies could be repurposed for drug-resistant epilepsies [153]. Therefore, the use of specific anti-inflammatory compounds is an alternative therapeutic strategy that should be explored for the treatment of PMEs.
7. Conclusions
As it is well known, free radical species are physiological cell signaling molecules, but when there is an imbalance in their levels, pathological oxidative stress can induce paramount changes in cell function even inducing cell death. In this scenario, the role of mitochondria, one of the main cellular sources of ROS, plays a pivotal role. The division between cause and effect is not always easy to establish because mitochondrial dysfunction and oxidative stress are tightly dependent on each other in the pathogenesis of neurodegenerative disorders. Moreover, oxidative stress and inflammation feed on each other and contribute to neurodegeneration. The NADPH oxidase and hydrogen peroxide release from activated inflammatory cells are also important factors. Here, we have reviewed the disease mechanism in disparate groups of neurodegenerative rare disorders that comprise retinopathies, epilepsies, movement disorders, and neuromuscular diseases. All of them are monogenic rare disorders that share oxidative stress as a common factor and may represent potential targets for therapies. Many neurodegenerative polygenic diseases (e.g., Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s disease, etc.) also show oxidative stress-related damage. Therefore, to unravel the disease mechanisms of Mendelian conditions results of great interest because this should be easier than for common disorders, and the findings may be also applied in the understanding of the pathophysiological mechanisms underlying common entities.
Funding
Generalitat Valenciana [Grant PROMETEO/2018/135].
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sies H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaschler M.M., Stockwell B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017;482:419–425. doi: 10.1016/j.bbrc.2016.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W.S., Kim K.J., Gaschler M.M., Patel M., Shchepinov M.S., Stockwell B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA. 2016;113:E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao J.Y., Poddar A., Magtanong L., Lumb J.H., Mileur T.R., Reid M.A., Dovey C.M., Wang J., Locasale J.W., Stone E., et al. A Genome-wide Haploid Genetic Screen Identifies Regulators of Glutathione Abundance and Ferroptosis Sensitivity. Cell Rep. 2019;26:1544–1556. doi: 10.1016/j.celrep.2019.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewerenz J., Ates G., Methner A., Conrad M., Maher P. Oxytosis/Ferroptosis-(Re-) Emerging Roles for Oxidative Stress-Dependent Non-apoptotic Cell Death in Diseases of the Central Nervous System. Front. Neurosci. 2018;12:214. doi: 10.3389/fnins.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guiney S.J., Adlard P.A., Bush A.I., Finkelstein D.I., Ayton S. Ferroptosis and cell death mechanisms in Parkinson’s disease. Neurochem. Int. 2017;104:34–48. doi: 10.1016/j.neuint.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Masaldan S., Bush A.I., Devos D., Rolland A.S., Moreau C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free Radic. Biol. Med. 2019;133:221–233. doi: 10.1016/j.freeradbiomed.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Neitemeier S., Jelinek A., Laino V., Hoffmann L., Eisenbach I., Eying R., Ganjam G.K., Dolga A.M., Oppermann S., Culmsee C. BID links ferroptosis to mitochondrial cell death pathways. Redox Biol. 2017;12:558–570. doi: 10.1016/j.redox.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidichandani S.I., Delatycki M.B. Friedreich Ataxia. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. GeneReviews®[Internet] University of Washington; Seattle, WA, USA: 1998. [updated 2017 June 1] [PubMed] [Google Scholar]
- 11.Musselman K.E., Stoyanov C.T., Marasigan R., Jenkins M.E., Konczak J., Morton S.M., Bastian A.J. Prevalence of ataxia in children: A systematic review. Neurology. 2014;82:80–89. doi: 10.1212/01.wnl.0000438224.25600.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delatycki M.B., Corben L.A. Clinical features of Friedreich ataxia. J. Child. Neurol. 2012;27:1133–1137. doi: 10.1177/0883073812448230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cady R.B., Bobechko W.P. Incidence, natural history, and treatment of scoliosis in Friedreich’s ataxia. J. Pediatr. Orthop. 1984;4:673–676. doi: 10.1097/01241398-198411000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Harding A.E., Hewer R.L. The heart disease of Friedreich’s ataxia: A clinical and electrocardiographic study of 115 patients, with an analysis of serial electrocardiographic changes in 30 cases. Q. J. Med. 1983;52:489–502. [PubMed] [Google Scholar]
- 15.Korner A., Barta L. Association of diabetes mellitus with Friedreich’s ataxia. Orv. Hetil. 1983;124:1391–1392. [PubMed] [Google Scholar]
- 16.Pandolfo M. Friedreich ataxia: The clinical picture. J. Neurol. 2009;256:3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- 17.Sharma R., De Biase I., Gomez M., Delatycki M.B., Ashizawa T., Bidichandani S.I. Friedreich ataxia in carriers of unstable borderline GAA triplet-repeat alleles. Ann. Neurol. 2004;56:898–901. doi: 10.1002/ana.20333. [DOI] [PubMed] [Google Scholar]
- 18.Cossee M., Puccio H., Gansmuller A., Koutnikova H., Dierich A., LeMeur M., Fischbeck K., Dolle P., Koenig M. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet. 2000;9:1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- 19.Jasoliya M.J., McMackin M.Z., Henderson C.K., Perlman S.L., Cortopassi G.A. Frataxin deficiency impairs mitochondrial biogenesis in cells, mice and humans. Hum. Mol. Genet. 2017;26:2627–2633. doi: 10.1093/hmg/ddx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaubel R.A., Isaya G. Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Mol. Cell Neurosci. 2013;55:50–61. doi: 10.1016/j.mcn.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 22.Martelli A., Puccio H. Dysregulation of cellular iron metabolism in Friedreich ataxia: From primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front. Pharmacol. 2014;5:130. doi: 10.3389/fphar.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Gimenez J.L., Gimeno A., Gonzalez-Cabo P., Dasi F., Bolinches-Amoros A., Molla B., Palau F., Pallardo F.V. Differential expression of PGC-1alpha and metabolic sensors suggest age-dependent induction of mitochondrial biogenesis in Friedreich ataxia fibroblasts. PLoS ONE. 2011;6:e20666. doi: 10.1371/journal.pone.0020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chantrel-Groussard K., Geromel V., Puccio H., Koenig M., Munnich A., Rotig A., Rustin P. Disabled early recruitment of antioxidant defenses in Friedreich’s ataxia. Hum. Mol. Genet. 2001;10:2061–2067. doi: 10.1093/hmg/10.19.2061. [DOI] [PubMed] [Google Scholar]
- 25.Paupe V., Dassa E.P., Goncalves S., Auchere F., Lonn M., Holmgren A., Rustin P. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PLoS ONE. 2009;4:e4253. doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Rosa P., Bertini E.S., Piemonte F. The NRF2 Signaling Network Defines Clinical Biomarkers and Therapeutic Opportunity in Friedreich’s Ataxia. Int. J. Mol. Sci. 2020;21:916. doi: 10.3390/ijms21030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Rosa P., Russo M., D’Amico J., Petrillo S., Aquilano K., Lettieri-Barbato D., Turchi R., Bertini E.S., Piemonte F. Nrf2 Induction Re-establishes a Proper Neuronal Differentiation Program in Friedreich’s Ataxia Neural Stem Cells. Front. Cell Neurosci. 2019;13:356. doi: 10.3389/fncel.2019.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrillo S., Piermarini E., Pastore A., Vasco G., Schirinzi T., Carrozzo R., Bertini E., Piemonte F. Nrf2-Inducers Counteract Neurodegeneration in Frataxin-Silenced Motor Neurons: Disclosing New Therapeutic Targets for Friedreich’s Ataxia. Int. J. Mol. Sci. 2017;18:2173. doi: 10.3390/ijms18102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn T., Stockwell B.R. The development of the concept of ferroptosis. Free Radic. Biol. Med. 2019;133:130–143. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell Mol. Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng N., Shi B.J., Li S.L., Zhong Z.Y., Li Y.C., Xua W.L., Zhou H., Cai J.H. Knockdown of ferroportin accelerates erastin-induced ferroptosis in neuroblastoma cells. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3826–3836. doi: 10.26355/eurrev_201806_15267. [DOI] [PubMed] [Google Scholar]
- 33.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santana-Codina N., Mancias J.D. The Role of NCOA4-Mediated Ferritinophagy in Health and Disease. Pharmaceuticals (Basel) 2018;11:114. doi: 10.3390/ph11040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 38.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S., Dar H.H., Liu B., Tyurin V.A., Ritov V.B., et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah R., Shchepinov M.S., Pratt D.A. Resolving the Role of Lipoxygenases in the Initiation and Execution of Ferroptosis. ACS Cent. Sci. 2018;4:387–396. doi: 10.1021/acscentsci.7b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan H., Li X., Zhang X., Kang R., Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem. Biophys. Res. Commun. 2016;478:1338–1343. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 41.Codazzi F., Hu A., Rai M., Donatello S., Salerno Scarzella F., Mangiameli E., Pelizzoni I., Grohovaz F., Pandolfo M. Friedreich ataxia-induced pluripotent stem cell-derived neurons show a cellular phenotype that is corrected by a benzamide HDAC inhibitor. Hum. Mol. Genet. 2016;25:4847–4855. doi: 10.1093/hmg/ddw308. [DOI] [PubMed] [Google Scholar]
- 42.Lupoli F., Vannocci T., Longo G., Niccolai N., Pastore A. The role of oxidative stress in Friedreich’s ataxia. FEBS Lett. 2018;592:718–727. doi: 10.1002/1873-3468.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bulteau A.L., Planamente S., Jornea L., Dur A., Lesuisse E., Camadro J.M., Auchere F. Changes in mitochondrial glutathione levels and protein thiol oxidation in yfh1 yeast cells and the lymphoblasts of patients with Friedreich’s ataxia. Biochim. Biophys. Acta. 2012;1822:212–225. doi: 10.1016/j.bbadis.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Llorens J.V., Soriano S., Calap-Quintana P., Gonzalez-Cabo P., Molto M.D. The Role of Iron in Friedreich’s Ataxia: Insights from Studies in Human Tissues and Cellular and Animal Models. Front. Neurosci. 2019;13:75. doi: 10.3389/fnins.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding I.H., Raniga P., Delatycki M.B., Stagnitti M.R., Corben L.A., Storey E., Georgiou-Karistianis N., Egan G.F. Tissue atrophy and elevated iron concentration in the extrapyramidal motor system in Friedreich ataxia: The IMAGE-FRDA study. J. Neurol. Neurosurg. Psychiatry. 2016;87:1261–1263. doi: 10.1136/jnnp-2015-312665. [DOI] [PubMed] [Google Scholar]
- 46.Bolinches-Amoros A., Molla B., Pla-Martin D., Palau F., Gonzalez-Cabo P. Mitochondrial dysfunction induced by frataxin deficiency is associated with cellular senescence and abnormal calcium metabolism. Front. Cell Neurosci. 2014;8:124. doi: 10.3389/fncel.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edenharter O., Schneuwly S., Navarro J.A. Mitofusin-Dependent ER Stress Triggers Glial Dysfunction and Nervous System Degeneration in a Drosophila Model of Friedreich’s Ataxia. Front. Mol. Neurosci. 2018;11:38. doi: 10.3389/fnmol.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cotticelli M.G., Xia S., Lin D., Lee T., Terrab L., Wipf P., Huryn D.M., Wilson R.B. Ferroptosis as a Novel Therapeutic Target for Friedreich’s Ataxia. J. Pharmacol Exp. Ther. 2019;369:47–54. doi: 10.1124/jpet.118.252759. [DOI] [PubMed] [Google Scholar]
- 49.Bai T., Liang R., Zhu R., Wang W., Zhou L., Sun Y. MicroRNA-214-3p enhances erastin-induced ferroptosis by targeting ATF4 in hepatoma cells. J. Cell Physiol. 2020 doi: 10.1002/jcp.29496. [DOI] [PubMed] [Google Scholar]
- 50.Luo M., Wu L., Zhang K., Wang H., Zhang T., Gutierrez L., O’Connell D., Zhang P., Li Y., Gao T., et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ. 2018;25:1457–1472. doi: 10.1038/s41418-017-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K., Wu L., Zhang P., Luo M., Du J., Gao T., O’Connell D., Wang G., Wang H., Yang Y. miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol. Carcinog. 2018;57:1566–1576. doi: 10.1002/mc.22878. [DOI] [PubMed] [Google Scholar]
- 52.Seco-Cervera M., Gonzalez-Rodriguez D., Ibanez-Cabellos J.S., Peiro-Chova L., Gonzalez-Cabo P., Garcia-Lopez E., Vilchez J.J., Sanz-Gallego I., Pallardo F.V., Garcia-Gimenez J.L. Circulating miR-323-3p is a biomarker for cardiomyopathy and an indicator of phenotypic variability in Friedreich’s ataxia patients. Sci. Rep. 2017;7:5237. doi: 10.1038/s41598-017-04996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seco-Cervera M., Gonzalez-Rodriguez D., Ibanez-Cabellos J.S., Peiro-Chova L., Pallardo F.V., Garcia-Gimenez J.L. Small RNA-seq analysis of circulating miRNAs to identify phenotypic variability in Friedreich’s ataxia patients. Sci. Data. 2018;5:180021. doi: 10.1038/sdata.2018.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dantham S., Srivastava A.K., Gulati S., Rajeswari M.R. Differentially Regulated Cell-Free MicroRNAs in the Plasma of Friedreich’s Ataxia Patients and Their Association with Disease Pathology. Neuropediatrics. 2018;49:35–43. doi: 10.1055/s-0037-1607279. [DOI] [PubMed] [Google Scholar]
- 55.Tello C., Darling A., Lupo V., Pérez-Dueñas B., Espinós C. On the complexity of clinical and molecular bases of neurodegeneration with brain iron accumulation. Clin. Genet. 2018;93:731–740. doi: 10.1111/cge.13057. [DOI] [PubMed] [Google Scholar]
- 56.Levi S., Tiranti V. Neurodegeneration with Brain Iron Accumulation Disorders: Valuable Models Aimed at Understanding the Pathogenesis of Iron Deposition. Pharmaceuticals (Basel) 2019;12:27. doi: 10.3390/ph12010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosveld F., Rana A., van der Wouden P.E., Lemstra W., Ritsema M., Kampinga H.H., Sibon O.C. De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum. Mol. Genet. 2008;17:2058–2069. doi: 10.1093/hmg/ddn105. [DOI] [PubMed] [Google Scholar]
- 58.Perry T.L., Norman M.G., Yong V.W., Whiting S., Crichton J.U., Hansen S., Kish S.J. Hallervorden-Spatz disease: Cysteine accumulation and cysteine dioxygenase deficiency in the globus pallidus. Ann. Neurol. 1985;18:482–489. doi: 10.1002/ana.410180411. [DOI] [PubMed] [Google Scholar]
- 59.Zhou B., Westaway S.K., Levinson B., Johnson M.A., Gitschier J., Hayflick S.J. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat. Genet. 2001;28:345–349. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- 60.Rana A., Seinen E., Siudeja K., Muntendam R., Srinivasan B., van der Want J.J., Hayflick S., Reijngoud D.J., Kayser O., Sibon O.C. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc. Natl. Acad. Sci. USA. 2010;107:6988–6993. doi: 10.1073/pnas.0912105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunetti D., Dusi S., Morbin M., Uggetti A., Moda F., D’Amato I., Giordano C., d’Amati G., Cozzi A., Levi S., et al. Pantothenate kinase-associated neurodegeneration: Altered mitochondria membrane potential and defective respiration in Pank2 knock-out mouse model. Hum. Mol. Genet. 2012;21:5294–5305. doi: 10.1093/hmg/dds380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunetti D., Dusi S., Giordano C., Lamperti C., Morbin M., Fugnanesi V., Marchet S., Fagiolari G., Sibon O., Moggio M., et al. Pantethine treatment is effective in recovering the disease phenotype induced by ketogenic diet in a pantothenate kinase-associated neurodegeneration mouse model. Brain. 2014;137:57–68. doi: 10.1093/brain/awt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez-Cordoba M., Fernandez Khoury A., Villanueva-Paz M., Gomez-Navarro C., Villalon-Garcia I., Suarez-Rivero J.M., Povea-Cabello S., de la Mata M., Cotan D., Talaveron-Rey M., et al. Pantothenate Rescues Iron Accumulation in Pantothenate Kinase-Associated Neurodegeneration Depending on the Type of Mutation. Mol. Neurobiol. 2019;56:3638–3656. doi: 10.1007/s12035-018-1333-0. [DOI] [PubMed] [Google Scholar]
- 64.Santambrogio P., Dusi S., Guaraldo M., Rotundo L.I., Broccoli V., Garavaglia B., Tiranti V., Levi S. Mitochondrial iron and energetic dysfunction distinguish fibroblasts and induced neurons from pantothenate kinase-associated neurodegeneration patients. Neurobiol. Dis. 2015;81:144–153. doi: 10.1016/j.nbd.2015.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campanella A., Privitera D., Guaraldo M., Rovelli E., Barzaghi C., Garavaglia B., Santambrogio P., Cozzi A., Levi S. Skin fibroblasts from pantothenate kinase-associated neurodegeneration patients show altered cellular oxidative status and have defective iron-handling properties. Hum. Mol. Genet. 2012;21:4049–4059. doi: 10.1093/hmg/dds229. [DOI] [PubMed] [Google Scholar]
- 66.Orellana D.I., Santambrogio P., Rubio A., Yekhlef L., Cancellieri C., Dusi S., Giannelli S.G., Venco P., Mazzara P.G., Cozzi A., et al. Coenzyme A corrects pathological defects in human neurons of PANK2-associated neurodegeneration. EMBO Mol. Med. 2016;8:1197–1211. doi: 10.15252/emmm.201606391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arber C., Angelova P.R., Wiethoff S., Tsuchiya Y., Mazzacuva F., Preza E., Bhatia K.P., Mills K., Gout I., Abramov A.Y., et al. iPSC-derived neuronal models of PANK2-associated neurodegeneration reveal mitochondrial dysfunction contributing to early disease. PLoS ONE. 2017;12:e0184104. doi: 10.1371/journal.pone.0184104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Z., Zhang X., Zhao C., Choi J., Shi J., Song K., Turk J., Ma Z.A. Protection of pancreatic beta-cells by group VIA phospholipase A(2)-mediated repair of mitochondrial membrane peroxidation. Endocrinology. 2010;151:3038–3048. doi: 10.1210/en.2010-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venco P., Bonora M., Giorgi C., Papaleo E., Iuso A., Prokisch H., Pinton P., Tiranti V. Mutations of C19orf12, coding for a transmembrane glycine zipper containing mitochondrial protein, cause mis-localization of the protein, inability to respond to oxidative stress and increased mitochondrial Ca(2)(+) Front. Genet. 2015;6:185. doi: 10.3389/fgene.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gutteridge J.M. Lipid peroxidation and antioxidants as biomarkers of tissue damage. Clin. Chem. 1995;41:1819–1828. doi: 10.1093/clinchem/41.12.1819. [DOI] [PubMed] [Google Scholar]
- 71.Brissot P., Ropert M., Le Lan C., Loreal O. Non-transferrin bound iron: A key role in iron overload and iron toxicity. Biochim. Biophys. Acta. 2012;1820:403–410. doi: 10.1016/j.bbagen.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 72.Harris Z.L., Durley A.P., Man T.K., Gitlin J.D. Targeted gene disruption reveals an essential role for ceruloplasmin in cellular iron efflux. Proc. Natl. Acad. Sci. USA. 1999;96:10812–10817. doi: 10.1073/pnas.96.19.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel B.N., Dunn R.J., Jeong S.Y., Zhu Q., Julien J.P., David S. Ceruloplasmin regulates iron levels in the CNS and prevents free radical injury. J. Neurosci. 2002;22:6578–6586. doi: 10.1523/JNEUROSCI.22-15-06578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cozzi A., Rovelli E., Frizzale G., Campanella A., Amendola M., Arosio P., Levi S. Oxidative stress and cell death in cells expressing L-ferritin variants causing neuroferritinopathy. Neurobiol. Dis. 2010;37:77–85. doi: 10.1016/j.nbd.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 75.Cozzi A., Santambrogio P., Corsi B., Campanella A., Arosio P., Levi S. Characterization of the l-ferritin variant 460InsA responsible of a hereditary ferritinopathy disorder. Neurobiol. Dis. 2006;23:644–652. doi: 10.1016/j.nbd.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Deng X., Vidal R., Englander E.W. Accumulation of oxidative DNA damage in brain mitochondria in mouse model of hereditary ferritinopathy. Neurosci. Lett. 2010;479:44–48. doi: 10.1016/j.neulet.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Capoccia S., Maccarinelli F., Buffoli B., Rodella L.F., Cremona O., Arosio P., Cirulli F. Behavioral characterization of mouse models of neuroferritinopathy. PLoS ONE. 2015;10:e0118990. doi: 10.1371/journal.pone.0118990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maccarinelli F., Pagani A., Cozzi A., Codazzi F., Di Giacomo G., Capoccia S., Rapino S., Finazzi D., Politi L.S., Cirulli F., et al. A novel neuroferritinopathy mouse model (FTL 498InsTC) shows progressive brain iron dysregulation, morphological signs of early neurodegeneration and motor coordination deficits. Neurobiol. Dis. 2015;81:119–133. doi: 10.1016/j.nbd.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Usenovic M., Tresse E., Mazzulli J.R., Taylor J.P., Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, alpha-synuclein accumulation, and neurotoxicity. J. Neurosci. 2012;32:4240–4246. doi: 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajagopalan S., Rane A., Chinta S.J., Andersen J.K. Regulation of ATP13A2 via PHD2-HIF1alpha Signaling Is Critical for Cellular Iron Homeostasis: Implications for Parkinson’s Disease. J. Neurosci. 2016;36:1086–1095. doi: 10.1523/JNEUROSCI.3117-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rinaldi D.E., Corradi G.R., Cuesta L.M., Adamo H.P., de Tezanos Pinto F. The Parkinson-associated human P5B-ATPase ATP13A2 protects against the iron-induced cytotoxicity. Biochim. Biophys. Acta. 2015;1848:1646–1655. doi: 10.1016/j.bbamem.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Cartoni R., Martinou J.C. Role of mitofusin 2 mutations in the physiopathology of Charcot-Marie-Tooth disease type 2A. Exp. Neurol. 2009;218:268–273. doi: 10.1016/j.expneurol.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 83.De Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 84.Merkwirth C., Langer T. Mitofusin 2 builds a bridge between ER and mitochondria. Cell. 2008;135:1165–1167. doi: 10.1016/j.cell.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 85.Suen D.F., Norris K.L., Youle R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pich S., Bach D., Briones P., Liesa M., Camps M., Testar X., Palacin M., Zorzano A. The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum. Mol. Genet. 2005;14:1405–1415. doi: 10.1093/hmg/ddi149. [DOI] [PubMed] [Google Scholar]
- 87.Filadi R., Pendin D., Pizzo P. Mitofusin 2: From functions to disease. Cell Death Dis. 2018;9:330. doi: 10.1038/s41419-017-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pareyson D., Saveri P., Sagnelli A., Piscosquito G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci. Lett. 2015;596:66–77. doi: 10.1016/j.neulet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 89.Han S., Nandy P., Austria Q., Siedlak S.L., Torres S., Fujioka H., Wang W., Zhu X. Mfn2 Ablation in the Adult Mouse Hippocampus and Cortex Causes Neuronal Death. Cells. 2020;9:116. doi: 10.3390/cells9010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pedrola L., Espert A., Wu X., Claramunt R., Shy M.E., Palau F. GDAP1, the protein causing Charcot-Marie-Tooth disease type 4A, is expressed in neurons and is associated with mitochondria. Hum. Mol. Genet. 2005;14:1087–1094. doi: 10.1093/hmg/ddi121. [DOI] [PubMed] [Google Scholar]
- 91.Marco A., Cuesta A., Pedrola L., Palau F., Marin I. Evolutionary and structural analyses of GDAP1, involved in Charcot-Marie-Tooth disease, characterize a novel class of glutathione transferase-related genes. Mol. Biol. Evol. 2004;21:176–187. doi: 10.1093/molbev/msh013. [DOI] [PubMed] [Google Scholar]
- 92.Noack R., Frede S., Albrecht P., Henke N., Pfeiffer A., Knoll K., Dehmel T., Meyer Zu Horste G., Stettner M., Kieseier B.C., et al. Charcot-Marie-Tooth disease CMT4A: GDAP1 increases cellular glutathione and the mitochondrial membrane potential. Hum. Mol. Genet. 2012;21:150–162. doi: 10.1093/hmg/ddr450. [DOI] [PubMed] [Google Scholar]
- 93.Cassereau J., Chevrollier A., Codron P., Goizet C., Gueguen N., Verny C., Reynier P., Bonneau D., Lenaers G., Procaccio V. Oxidative stress contributes differentially to the pathophysiology of Charcot-Marie-Tooth disease type 2K. Exp. Neurol. 2020;323:113069. doi: 10.1016/j.expneurol.2019.113069. [DOI] [PubMed] [Google Scholar]
- 94.Barneo-Munoz M., Juarez P., Civera-Tregon A., Yndriago L., Pla-Martin D., Zenker J., Cuevas-Martin C., Estela A., Sanchez-Arago M., Forteza-Vila J., et al. Lack of GDAP1 induces neuronal calcium and mitochondrial defects in a knockout mouse model of charcot-marie-tooth neuropathy. PLoS Genet. 2015;11:e1005115. doi: 10.1371/journal.pgen.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernandez-Lizarbe S., Civera-Tregon A., Cantarero L., Herrer I., Juarez P., Hoenicka J., Palau F. Neuroinflammation in the pathogenesis of axonal Charcot-Marie-Tooth disease caused by lack of GDAP1. Exp. Neurol. 2019;320:113004. doi: 10.1016/j.expneurol.2019.113004. [DOI] [PubMed] [Google Scholar]
- 96.Lopez Del Amo V., Seco-Cervera M., Garcia-Gimenez J.L., Whitworth A.J., Pallardo F.V., Galindo M.I. Mitochondrial defects and neuromuscular degeneration caused by altered expression of Drosophila Gdap1: Implications for the Charcot-Marie-Tooth neuropathy. Hum. Mol. Genet. 2015;24:21–36. doi: 10.1093/hmg/ddu416. [DOI] [PubMed] [Google Scholar]
- 97.Pla-Martin D., Calpena E., Lupo V., Marquez C., Rivas E., Sivera R., Sevilla T., Palau F., Espinos C. Junctophilin-1 is a modifier gene of GDAP1-related Charcot-Marie-Tooth disease. Hum. Mol. Genet. 2015;24:213–229. doi: 10.1093/hmg/ddu440. [DOI] [PubMed] [Google Scholar]
- 98.Yang X.D., Cen Z.D., Cheng H.P., Shi K., Bai J., Xie F., Wu H.W., Li B.B., Luo W. L-3-n-Butylphthalide Protects HSPB8 K141N Mutation-Induced Oxidative Stress by Modulating the Mitochondrial Apoptotic and Nrf2 Pathways. Front. Neurosci. 2017;11:402. doi: 10.3389/fnins.2017.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cohen J. Diseases of the retina: 1975-76 review. Am. J. Optom. Physiol. Opt. 1977;54:485–494. doi: 10.1097/00006324-197707000-00010. [DOI] [PubMed] [Google Scholar]
- 100.Bunker C.H., Berson E.L., Bromley W.C., Hayes R.P., Roderick T.H. Prevalence of retinitis pigmentosa in Maine. Am. J. Ophthalmol. 1984;97:357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 101.Fishman G.A. Retinitis pigmentosa. Genetic percentages. Arch. Ophthalmol. 1978;96:822–826. doi: 10.1001/archopht.1978.03910050428005. [DOI] [PubMed] [Google Scholar]
- 102.den Hollander A.I., Black A., Bennett J., Cremers F.P. Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Investig. 2010;120:3042–3053. doi: 10.1172/JCI42258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Athanasiou D., Aguila M., Bevilacqua D., Novoselov S.S., Parfitt D.A., Cheetham M.E. The cell stress machinery and retinal degeneration. FEBS Lett. 2013;587:2008–2017. doi: 10.1016/j.febslet.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gupta N., Brown K.E., Milam A.H. Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp. Eye Res. 2003;76:463–471. doi: 10.1016/S0014-4835(02)00332-9. [DOI] [PubMed] [Google Scholar]
- 105.Yu D.Y., Cringle S., Valter K., Walsh N., Lee D., Stone J. Photoreceptor death, trophic factor expression, retinal oxygen status, and photoreceptor function in the P23H rat. Investig. Ophthalmol. Vis. Sci. 2004;45:2013–2019. doi: 10.1167/iovs.03-0845. [DOI] [PubMed] [Google Scholar]
- 106.Stone J., Maslim J., Valter-Kocsi K., Mervin K., Bowers F., Chu Y., Barnett N., Provis J., Lewis G., Fisher S.K., et al. Mechanisms of photoreceptor death and survival in mammalian retina. Prog. Retin. Eye Res. 1999;18:689–735. doi: 10.1016/S1350-9462(98)00032-9. [DOI] [PubMed] [Google Scholar]
- 107.Shen J., Yang X., Dong A., Petters R.M., Peng Y.W., Wong F., Campochiaro P.A. Oxidative damage is a potential cause of cone cell death in retinitis pigmentosa. J. Cell Physiol. 2005;203:457–464. doi: 10.1002/jcp.20346. [DOI] [PubMed] [Google Scholar]
- 108.Stringham J.M., Stringham N.T. Nitric Oxide and Lutein: Function, Performance, and Protection of Neural Tissue. Foods. 2015;4:678–689. doi: 10.3390/foods4040678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu D.Y., Cringle S.J., Su E.N., Yu P.K. Intraretinal oxygen levels before and after photoreceptor loss in the RCS rat. Investig. Ophthalmol. Vis. Sci. 2000;41:3999–4006. [PubMed] [Google Scholar]
- 110.Yu D.Y., Cringle S.J. Retinal degeneration and local oxygen metabolism. Exp. Eye Res. 2005;80:745–751. doi: 10.1016/j.exer.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 111.Olivares-Gonzalez L., Martinez-Fernandez de la Camara C., Hervas D., Millan J.M., Rodrigo R. HIF-1alpha stabilization reduces retinal degeneration in a mouse model of retinitis pigmentosa. FASEB J. 2018;32:2438–2451. doi: 10.1096/fj.201700985R. [DOI] [PubMed] [Google Scholar]
- 112.Eysteinsson T., Hardarson S.H., Bragason D., Stefansson E. Retinal vessel oxygen saturation and vessel diameter in retinitis pigmentosa. Acta Ophthalmol. 2014;92:449–453. doi: 10.1111/aos.12359. [DOI] [PubMed] [Google Scholar]
- 113.Zong Y., Lin L., Yi C., Huang X., Fu Y., Dong Y., Qian X., Li Y., Gao Q. Retinal vessel oxygen saturation and vessel diameter in retinitis pigmentosa at various ages. Graefes. Arch. Clin. Exp. Ophthalmol. 2016;254:243–252. doi: 10.1007/s00417-015-3039-6. [DOI] [PubMed] [Google Scholar]
- 114.Martinez-Fernandez de la Camara C., Hernandez-Pinto A.M., Olivares-Gonzalez L., Cuevas-Martin C., Sanchez-Arago M., Hervas D., Salom D., Cuezva J.M., de la Rosa E.J., Millan J.M., et al. Adalimumab Reduces Photoreceptor Cell Death in A Mouse Model of Retinal Degeneration. Sci. Rep. 2015;5:11764. doi: 10.1038/srep11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martinez-Fernandez de la Camara C., Salom D., Sequedo M.D., Hervas D., Marin-Lambies C., Aller E., Jaijo T., Diaz-Llopis M., Millan J.M., Rodrigo R. Altered antioxidant-oxidant status in the aqueous humor and peripheral blood of patients with retinitis pigmentosa. PLoS ONE. 2013;8:e74223. doi: 10.1371/journal.pone.0074223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Komeima K., Rogers B.S., Campochiaro P.A. Antioxidants slow photoreceptor cell death in mouse models of retinitis pigmentosa. J. Cell Physiol. 2007;213:809–815. doi: 10.1002/jcp.21152. [DOI] [PubMed] [Google Scholar]
- 117.Campochiaro P.A., Strauss R.W., Lu L., Hafiz G., Wolfson Y., Shah S.M., Sophie R., Mir T.A., Scholl H.P. Is There Excess Oxidative Stress and Damage in Eyes of Patients with Retinitis Pigmentosa? Antioxid. Redox Signal. 2015;23:643–648. doi: 10.1089/ars.2015.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murakami Y., Ikeda Y., Yoshida N., Notomi S., Hisatomi T., Oka S., De Luca G., Yonemitsu Y., Bignami M., Nakabeppu Y., et al. MutT homolog-1 attenuates oxidative DNA damage and delays photoreceptor cell death in inherited retinal degeneration. Am. J. Pathol. 2012;181:1378–1386. doi: 10.1016/j.ajpath.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 119.Narayan D.S., Wood J.P., Chidlow G., Casson R.J. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016;94:748–754. doi: 10.1111/aos.13141. [DOI] [PubMed] [Google Scholar]
- 120.Bramall A.N., Wright A.F., Jacobson S.G., McInnes R.R. The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu. Rev. Neurosci. 2010;33:441–472. doi: 10.1146/annurev-neuro-060909-153227. [DOI] [PubMed] [Google Scholar]
- 121.Komeima K., Usui S., Shen J., Rogers B.S., Campochiaro P.A. Blockade of neuronal nitric oxide synthase reduces cone cell death in a model of retinitis pigmentosa. Free Radic. Biol. Med. 2008;45:905–912. doi: 10.1016/j.freeradbiomed.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perdices L., Fuentes-Broto L., Segura F., Ben Gdara N., Sanchez-Cano A.I., Insa G., Orduna E., Pinilla I. Hepatic oxidative stress in pigmented P23H rhodopsin transgenic rats with progressive retinal degeneration. Free Radic. Biol. Med. 2018;124:550–557. doi: 10.1016/j.freeradbiomed.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 123.Campochiaro P.A., Mir T.A. The mechanism of cone cell death in Retinitis Pigmentosa. Prog. Retin. Eye Res. 2018;62:24–37. doi: 10.1016/j.preteyeres.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 124.Ames A., III, Li Y.Y., Heher E.C., Kimble C.R. Energy metabolism of rabbit retina as related to function: High cost of Na+ transport. J. Neurosci. 1992;12:840–853. doi: 10.1523/JNEUROSCI.12-03-00840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orrenius S., Gogvadze V., Zhivotovsky B. Mitochondrial oxidative stress: Implications for cell death. Annu. Rev. Pharmacol. Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 126.Vlachantoni D., Bramall A.N., Murphy M.P., Taylor R.W., Shu X., Tulloch B., Van Veen T., Turnbull D.M., McInnes R.R., Wright A.F. Evidence of severe mitochondrial oxidative stress and a protective effect of low oxygen in mouse models of inherited photoreceptor degeneration. Hum. Mol. Genet. 2011;20:322–335. doi: 10.1093/hmg/ddq467. [DOI] [PubMed] [Google Scholar]
- 127.Maleki S., Gopalakrishnan S., Ghanian Z., Sepehr R., Schmitt H., Eells J., Ranji M. Optical imaging of mitochondrial redox state in rodent model of retinitis pigmentosa. J. Biomed. Opt. 2013;18:16004. doi: 10.1117/1.JBO.18.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Griciuc A., Roux M.J., Merl J., Giangrande A., Hauck S.M., Aron L., Ueffing M. Proteomic survey reveals altered energetic patterns and metabolic failure prior to retinal degeneration. J. Neurosci. 2014;34:2797–2812. doi: 10.1523/JNEUROSCI.2982-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McKie A.B., McHale J.C., Keen T.J., Tarttelin E.E., Goliath R., van Lith-Verhoeven J.J., Greenberg J., Ramesar R.S., Hoyng C.B., Cremers F.P., et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum. Mol. Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 131.Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T., et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/S1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 132.Xu G., Li T., Chen J., Li C., Zhao H., Yao C., Dong H., Wen K., Wang K., Zhao J., et al. Autosomal dominant retinitis pigmentosa-associated gene PRPF8 is essential for hypoxia-induced mitophagy through regulating ULK1 mRNA splicing. Autophagy. 2018;14:1818–1830. doi: 10.1080/15548627.2018.1501251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peter V.G., Nikopoulos K., Quinodoz M., Granse L., Farinelli P., Superti-Furga A., Andreasson S., Rivolta C. A novel missense variant in IDH3A causes autosomal recessive retinitis pigmentosa. Ophthalmic Genet. 2019;40:177–181. doi: 10.1080/13816810.2019.1605391. [DOI] [PubMed] [Google Scholar]
- 134.Aguado C., Sarkar S., Korolchuk V.I., Criado O., Vernia S., Boya P., Sanz P., de Cordoba S.R., Knecht E., Rubinsztein D.C. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum. Mol. Genet. 2010;19:2867–2876. doi: 10.1093/hmg/ddq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Metodiev M.D., Gerber S., Hubert L., Delahodde A., Chretien D., Gerard X., Amati-Bonneau P., Giacomotto M.C., Boddaert N., Kaminska A., et al. Mutations in the tricarboxylic acid cycle enzyme, aconitase 2, cause either isolated or syndromic optic neuropathy with encephalopathy and cerebellar atrophy. J. Med. Genet. 2014;51:834–838. doi: 10.1136/jmedgenet-2014-102532. [DOI] [PubMed] [Google Scholar]
- 136.Nagar S., Noveral S.M., Trudler D., Lopez K.M., McKercher S.R., Han X., Yates J.R., III, Pina-Crespo J.C., Nakanishi N., Satoh T., et al. MEF2D haploinsufficiency downregulates the NRF2 pathway and renders photoreceptors susceptible to light-induced oxidative stress. Proc. Natl. Acad. Sci. USA. 2017;114:E4048–E4056. doi: 10.1073/pnas.1613067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Byrne A.M., Ruiz-Lopez A.M., Roche S.L., Moloney J.N., Wyse-Jackson A.C., Cotter T.G. The synthetic progestin norgestrel modulates Nrf2 signaling and acts as an antioxidant in a model of retinal degeneration. Redox Biol. 2016;10:128–139. doi: 10.1016/j.redox.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakagami Y., Hatano E., Inoue T., Yoshida K., Kondo M., Terasaki H. Cytoprotective Effects of a Novel Nrf2 Activator, RS9, in Rhodopsin Pro347Leu Rabbits. Curr. Eye Res. 2016;41:1123–1126. doi: 10.3109/02713683.2015.1078362. [DOI] [PubMed] [Google Scholar]
- 139.Campello L., Kutsyr O., Noailles A., Michalska P., Fernandez-Sanchez L., Martinez-Gil N., Ortuno-Lizaran I., Sanchez-Saez X., de Juan E., Lax P., et al. New Nrf2-Inducer Compound ITH12674 Slows the Progression of Retinitis Pigmentosa in the Mouse Model rd10. Cell Physiol. Biochem. 2020;54:142–159. doi: 10.33594/000000210. [DOI] [PubMed] [Google Scholar]
- 140.Wubben T.J., Zacks D.N., Besirli C.G. Retinal neuroprotection: Current strategies and future directions. Curr. Opin. Ophthalmol. 2019;30:199–205. doi: 10.1097/ICU.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 141.Benati D., Patrizi C., Recchia A. Gene editing prospects for treating inherited retinal diseases. J. Med. Genet. 2019 doi: 10.1136/jmedgenet-2019-106473. [DOI] [PubMed] [Google Scholar]
- 142.Ben M’Barek K., Habeler W., Regent F., Monville C. Developing Cell-Based Therapies for RPE-Associated Degenerative Eye Diseases. Adv. Exp. Med. Biol. 2019;1186:55–97. doi: 10.1007/978-3-030-28471-8_3. [DOI] [PubMed] [Google Scholar]
- 143.Sahaboglu A., Miranda M., Canjuga D., Avci-Adali M., Savytska N., Secer E., Feria-Pliego J.A., Kayik G., Durdagi S. Drug repurposing studies of PARP inhibitors as a new therapy for inherited retinal degeneration. Cell Mol. Life Sci. 2019 doi: 10.1007/s00018-019-03283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee S.Y., Usui S., Zafar A.B., Oveson B.C., Jo Y.J., Lu L., Masoudi S., Campochiaro P.A. N-Acetylcysteine promotes long-term survival of cones in a model of retinitis pigmentosa. J. Cell Physiol. 2011;226:1843–1849. doi: 10.1002/jcp.22508. [DOI] [PubMed] [Google Scholar]
- 145.Berson E.L., Rosner B., Sandberg M.A., Weigel-DiFranco C., Brockhurst R.J., Hayes K.C., Johnson E.J., Anderson E.J., Johnson C.A., Gaudio A.R., et al. Clinical trial of lutein in patients with retinitis pigmentosa receiving vitamin A. Arch. Ophthalmol. 2010;128:403–411. doi: 10.1001/archophthalmol.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Berson E.L. Long-term visual prognoses in patients with retinitis pigmentosa: The Ludwig von Sallmann lecture. Exp. Eye Res. 2007;85:7–14. doi: 10.1016/j.exer.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sanz M.M., Johnson L.E., Ahuja S., Ekstrom P.A., Romero J., van Veen T. Significant photoreceptor rescue by treatment with a combination of antioxidants in an animal model for retinal degeneration. Neuroscience. 2007;145:1120–1129. doi: 10.1016/j.neuroscience.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 148.Shintani K., Shechtman D.L., Gurwood A.S. Review and update: Current treatment trends for patients with retinitis pigmentosa. Optometry. 2009;80:384–401. doi: 10.1016/j.optm.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 149.Emoto Y., Yoshizawa K., Uehara N., Kinoshita Y., Yuri T., Shikata N., Tsubura A. Curcumin suppresses N-methyl-N-nitrosourea-induced photoreceptor apoptosis in Sprague-Dawley rats. In Vivo. 2013;27:583–590. [PubMed] [Google Scholar]
- 150.Piano I., D’Antongiovanni V., Testai L., Calderone V., Gargini C. A Nutraceutical Strategy to Slowing Down the Progression of Cone Death in an Animal Model of Retinitis Pigmentosa. Front. Neurosci. 2019;13:461. doi: 10.3389/fnins.2019.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Devinsky O., Vezzani A., O’Brien T.J., Jette N., Scheffer I.E., de Curtis M., Perucca P. Epilepsy. Nat. Rev. Dis. Primers. 2018;4:18024. doi: 10.1038/nrdp.2018.24. [DOI] [PubMed] [Google Scholar]
- 152.Patel D.C., Tewari B.P., Chaunsali L., Sontheimer H. Neuron-glia interactions in the pathophysiology of epilepsy. Nat. Rev. Neurosci. 2019;20:282–297. doi: 10.1038/s41583-019-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vezzani A., Balosso S., Ravizza T. Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat. Rev. Neurol. 2019;15:459–472. doi: 10.1038/s41582-019-0217-x. [DOI] [PubMed] [Google Scholar]
- 154.Liang L.P., Patel M. Seizure-induced changes in mitochondrial redox status. Free Radic. Biol. Med. 2006;40:316–322. doi: 10.1016/j.freeradbiomed.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 155.Ravizza T., Vezzani A. Pharmacological targeting of brain inflammation in epilepsy: Therapeutic perspectives from experimental and clinical studies. Epilepsia Open. 2018;3:133–142. doi: 10.1002/epi4.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kalviainen R. Progressive Myoclonus Epilepsies. Semin Neurol. 2015;35:293–299. doi: 10.1055/s-0035-1552620. [DOI] [PubMed] [Google Scholar]
- 157.Knupp K., Wirrell E. Progressive myoclonic epilepsies: It takes a village to make a diagnosis. Neurology. 2014;82:378–379. doi: 10.1212/WNL.0000000000000091. [DOI] [PubMed] [Google Scholar]
- 158.Orsini A., Valetto A., Bertini V., Esposito M., Carli N., Minassian B.A., Bonuccelli A., Peroni D., Michelucci R., Striano P. The best evidence for progressive myoclonic epilepsy: A pathway to precision therapy. Seizure. 2019;71:247–257. doi: 10.1016/j.seizure.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shahwan A., Farrell M., Delanty N. Progressive myoclonic epilepsies: A review of genetic and therapeutic aspects. Lancet Neurol. 2005;4:239–248. doi: 10.1016/S1474-4422(05)70043-0. [DOI] [PubMed] [Google Scholar]
- 160.Zupanc M.L., Legros B. Progressive myoclonic epilepsy. Cerebellum. 2004;3:156–171. doi: 10.1080/14734220410035356. [DOI] [PubMed] [Google Scholar]
- 161.Joensuu T., Tegelberg S., Reinmaa E., Segerstrale M., Hakala P., Pehkonen H., Korpi E.R., Tyynela J., Taira T., Hovatta I., et al. Gene expression alterations in the cerebellum and granule neurons of Cstb−/− mouse are associated with early synaptic changes and inflammation. PLoS ONE. 2014;9:e89321. doi: 10.1371/journal.pone.0089321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lahuerta M., Gonzalez D., Aguado C., Fathinajafabadi A., Garcia-Gimenez J.L., Moreno-Estelles M., Roma-Mateo C., Knecht E., Pallardo F.V., Sanz P. Reactive Glia-Derived Neuroinflammation: A Novel Hallmark in Lafora Progressive Myoclonus Epilepsy That Progresses with Age. Mol. Neurobiol. 2020;57:1607–1621. doi: 10.1007/s12035-019-01842-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parviainen L., Dihanich S., Anderson G.W., Wong A.M., Brooks H.R., Abeti R., Rezaie P., Lalli G., Pope S., Heales S.J., et al. Glial cells are functionally impaired in juvenile neuronal ceroid lipofuscinosis and detrimental to neurons. Acta Neuropathol. Commun. 2017;5:74. doi: 10.1186/s40478-017-0476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Crespel A., Ferlazzo E., Franceschetti S., Genton P., Gouider R., Kalviainen R., Korja M., Lehtinen M.K., Mervaala E., Simonato M., et al. Unverricht-Lundborg disease. Epileptic Disord. 2016;18:28–37. doi: 10.1684/epd.2016.0841. [DOI] [PubMed] [Google Scholar]
- 165.Lasek-Bal A., Lukasik M., Zak A., Sulek A., Bosak M. Unverricht-Lundborg disease: Clinical course and seizure management based on the experience of polish centers. Seizure. 2019;69:87–91. doi: 10.1016/j.seizure.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 166.Kopitar-Jerala N. Innate Immune Response in Brain, NF-Kappa B Signaling and Cystatins. Front. Mol. Neurosci. 2015;8:73. doi: 10.3389/fnmol.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Garcia-Gimeno M.A., Knecht E., Sanz P. Lafora Disease: A Ubiquitination-Related Pathology. Cells. 2018;7:87. doi: 10.3390/cells7080087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Minassian B.A., Lee J.R., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 169.Chan E.M., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 170.Criado O., Aguado C., Gayarre J., Duran-Trio L., Garcia-Cabrero A.M., Vernia S., San Millan B., Heredia M., Roma-Mateo C., Mouron S., et al. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum. Mol. Genet. 2012;21:1521–1533. doi: 10.1093/hmg/ddr590. [DOI] [PubMed] [Google Scholar]
- 171.Rao S.N., Maity R., Sharma J., Dey P., Shankar S.K., Satishchandra P., Jana N.R. Sequestration of chaperones and proteasome into Lafora bodies and proteasomal dysfunction induced by Lafora disease-associated mutations of malin. Hum. Mol. Genet. 2010;19:4726–4734. doi: 10.1093/hmg/ddq407. [DOI] [PubMed] [Google Scholar]
- 172.Sinadinos C., Valles-Ortega J., Boulan L., Solsona E., Tevy M.F., Marquez M., Duran J., Lopez-Iglesias C., Calbo J., Blasco E., et al. Neuronal glycogen synthesis contributes to physiological aging. Aging Cell. 2014;13:935–945. doi: 10.1111/acel.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Garcia-Gimenez J.L., Seco-Cervera M., Aguado C., Roma-Mateo C., Dasi F., Priego S., Markovic J., Knecht E., Sanz P., Pallardo F.V. Lafora disease fibroblasts exemplify the molecular interdependence between thioredoxin 1 and the proteasome in mammalian cells. Free Radic. Biol. Med. 2013;65:347–359. doi: 10.1016/j.freeradbiomed.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 174.Vernia S., Rubio T., Heredia M., Rodriguez de Cordoba S., Sanz P. Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PLoS ONE. 2009;4:e5907. doi: 10.1371/journal.pone.0005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Puri R., Suzuki T., Yamakawa K., Ganesh S. Dysfunctions in endosomal-lysosomal and autophagy pathways underlie neuropathology in a mouse model for Lafora disease. Hum. Mol. Genet. 2012;21:175–184. doi: 10.1093/hmg/ddr452. [DOI] [PubMed] [Google Scholar]
- 176.Sanchez-Martin P., Lahuerta M., Viana R., Knecht E., Sanz P. Regulation of the autophagic PI3KC3 complex by laforin/malin E3-ubiquitin ligase, two proteins involved in Lafora disease. Biochim. Biophys. Acta Mol. Cell Res. 2019;1867:118613. doi: 10.1016/j.bbamcr.2019.118613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Garyali P., Siwach P., Singh P.K., Puri R., Mittal S., Sengupta S., Parihar R., Ganesh S. The malin-laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system. Hum. Mol. Genet. 2009;18:688–700. doi: 10.1093/hmg/ddn398. [DOI] [PubMed] [Google Scholar]
- 178.Lahuerta M., Aguado C., Sanchez-Martin P., Sanz P., Knecht E. Degradation of altered mitochondria by autophagy is impaired in Lafora disease. FEBS J. 2018;285:2071–2090. doi: 10.1111/febs.14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Roma-Mateo C., Aguado C., Garcia-Gimenez J.L., Ibanez-Cabellos J.S., Seco-Cervera M., Pallardo F.V., Knecht E., Sanz P. Increased oxidative stress and impaired antioxidant response in Lafora disease. Mol. Neurobiol. 2015;51:932–946. doi: 10.1007/s12035-014-8747-0. [DOI] [PubMed] [Google Scholar]
- 180.Lopez-Gonzalez I., Viana R., Sanz P., Ferrer I. Inflammation in Lafora Disease: Evolution with Disease Progression in Laforin and Malin Knock-out Mouse Models. Mol. Neurobiol. 2017;54:3119–3130. doi: 10.1007/s12035-016-9884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jain P., Sharma S., Tripathi M. Diagnosis and management of epileptic encephalopathies in children. Epilepsy Res. Treat. 2013;2013:501981. doi: 10.1155/2013/501981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Dravet C. The core Dravet syndrome phenotype. Epilepsia. 2011;52:3–9. doi: 10.1111/j.1528-1167.2011.02994.x. [DOI] [PubMed] [Google Scholar]
- 183.Gataullina S., Dulac O. From genotype to phenotype in Dravet disease. Seizure. 2017;44:58–64. doi: 10.1016/j.seizure.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 184.Guerrini R. Dravet syndrome: The main issues. Eur. J. Paediatr. Neurol. 2012;16:S1–S4. doi: 10.1016/j.ejpn.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 185.Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ogiwara I., Miyamoto H., Morita N., Atapour N., Mazaki E., Inoue I., Takeuchi T., Itohara S., Yanagawa Y., Obata K., et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: A circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J. Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yu F.H., Mantegazza M., Westenbroek R.E., Robbins C.A., Kalume F., Burton K.A., Spain W.J., McKnight G.S., Scheuer T., Catterall W.A. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat. Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- 188.Auvin S., Avbersek A., Bast T., Chiron C., Guerrini R., Kaminski R.M., Lagae L., Muglia P., Cross J.H. Drug Development for Rare Paediatric Epilepsies: Current State and Future Directions. Drugs. 2019;79:1917–1935. doi: 10.1007/s40265-019-01223-9. [DOI] [PubMed] [Google Scholar]
- 189.Devinsky O., Cross J.H., Laux L., Marsh E., Miller I., Nabbout R., Scheffer I.E., Thiele E.A., Wright S. Trial of Cannabidiol for Drug-Resistant Seizures in the Dravet Syndrome. N. Engl. J. Med. 2017;376:2011–2020. doi: 10.1056/NEJMoa1611618. [DOI] [PubMed] [Google Scholar]
- 190.Okumura A., Uematsu M., Imataka G., Tanaka M., Okanishi T., Kubota T., Sudo A., Tohyama J., Tsuji M., Ohmori I., et al. Acute encephalopathy in children with Dravet syndrome. Epilepsia. 2012;53:79–86. doi: 10.1111/j.1528-1167.2011.03311.x. [DOI] [PubMed] [Google Scholar]
- 191.Tang S., Lin J.P., Hughes E., Siddiqui A., Lim M., Lascelles K. Encephalopathy and SCN1A mutations. Epilepsia. 2011;52:e26–e30. doi: 10.1111/j.1528-1167.2011.03019.x. [DOI] [PubMed] [Google Scholar]
- 192.Boguszewicz L., Jamroz E., Ciszek M., Emich-Widera E., Kijonka M., Banasik T., Skorupa A., Sokol M. NMR-based metabolomics in pediatric drug resistant epilepsy—Preliminary results. Sci. Rep. 2019;9:15035. doi: 10.1038/s41598-019-51337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schuster J., Laan L., Klar J., Jin Z., Huss M., Korol S., Noraddin F.H., Sobol M., Birnir B., Dahl N. Transcriptomes of Dravet syndrome iPSC derived GABAergic cells reveal dysregulated pathways for chromatin remodeling and neurodevelopment. Neurobiol. Dis. 2019;132:104583. doi: 10.1016/j.nbd.2019.104583. [DOI] [PubMed] [Google Scholar]
- 194.Silvestro S., Mammana S., Cavalli E., Bramanti P., Mazzon E. Use of Cannabidiol in the Treatment of Epilepsy: Efficacy and Security in Clinical Trials. Molecules. 2019;24:1459. doi: 10.3390/molecules24081459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Esposito G., De Filippis D., Maiuri M.C., De Stefano D., Carnuccio R., Iuvone T. Cannabidiol inhibits inducible nitric oxide synthase protein expression and nitric oxide production in beta-amyloid stimulated PC12 neurons through p38 MAP kinase and NF-kappaB involvement. Neurosci. Lett. 2006;399:91–95. doi: 10.1016/j.neulet.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 196.D’Andrea Meira I., Romao T.T., Pires do Prado H.J., Kruger L.T., Pires M.E.P., da Conceiçao P.O. Ketogenic Diet and Epilepsy: What We Know So Far. Front. Neurosci. 2019;13:5. doi: 10.3389/fnins.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dupuis N., Curatolo N., Benoist J.F., Auvin S. Ketogenic diet exhibits anti-inflammatory properties. Epilepsia. 2015;56:e95–e98. doi: 10.1111/epi.13038. [DOI] [PubMed] [Google Scholar]
- 198.Shimazu T., Hirschey M.D., Newman J., He W., Shirakawa K., Le Moan N., Grueter C.A., Lim H., Saunders L.R., Stevens R.D., et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Andreasson K.I., Bachstetter A.D., Colonna M., Ginhoux F., Holmes C., Lamb B., Landreth G., Lee D.C., Low D., Lynch M.A., et al. Targeting innate immunity for neurodegenerative disorders of the central nervous system. J. Neurochem. 2016;138:653–693. doi: 10.1111/jnc.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]