Abstract
Background:
Glioblastoma is a particularly common and very aggressive primary brain tumour. One of the main causes of therapy failure is the presence of glioblastoma stem cells that are resistant to chemotherapy and radiotherapy, and that have the potential to form new tumours. This study focuses on validation of eight novel antigens, TRIM28, nucleolin, vimentin, nucleosome assembly protein 1-like 1 (NAP1L1), mitochondrial translation elongation factor (EF-TU) (TUFM), dihydropyrimidinase-related protein 2 (DPYSL2), collapsin response mediator protein 1 (CRMP1) and Aly/REF export factor (ALYREF), as putative glioblastoma targets, using nanobodies.
Methods:
Expression of these eight antigens was analysed at the cellular level by qPCR, ELISA and immunocytochemistry, and in tissues by immunohistochemistry. The cytotoxic effects of the nanobodies were determined using AlamarBlue and water-soluble tetrazolium tests. Annexin V/propidium iodide tests were used to determine apoptotsis/necrosis of the cells in the presence of the nanobodies. Cell migration assays were performed to determine the effects of the nanobodies on cell migration.
Results:
NAP1L1 and CRMP1 were significantly overexpressed in glioblastoma stem cells in comparison with astrocytes and glioblastoma cell lines at the mRNA and protein levels. Vimentin, DPYSL2 and ALYREF were overexpressed in glioblastoma cell lines only at the protein level. The functional part of the study examined the cytotoxic effects of the nanobodies on glioblastoma cell lines. Four of the nanobodies were selected in terms of their specificity towards glioblastoma cells and protein overexpression: anti-vimentin (Nb79), anti-NAP1L1 (Nb179), anti-TUFM (Nb225) and anti-DPYSL2 (Nb314). In further experiments to optimise the nanobody treatment schemes, to increase their effects, and to determine their impact on migration of glioblastoma cells, the anti-TUFM nanobody showed large cytotoxic effects on glioblastoma stem cells, while the anti-vimentin, anti-NAP1L1 and anti-DPYSL2 nanobodies were indicated as agents to target mature glioblastoma cells. The anti-vimentin nanobody also had significant effects on migration of mature glioblastoma cells.
Conclusion:
Nb79 (anti-vimentin), Nb179 (anti-NAP1L1), Nb225 (anti-TUFM) and Nb314 (anti-DPYSL2) nanobodies are indicated for further examination for cell targeting. The anti-TUFM nanobody, Nb225, is particularly potent for inhibition of cell growth after long-term exposure of glioblastoma stem cells, with minor effects seen for astrocytes. The anti-vimentin nanobody represents an agent for inhibition of cell migration.
Keywords: cell migration, cytotoxicity, DPYSL2, glioblastoma, glioblastoma stem cells, nanobodies, NAP1L1, TUFM, vimentin
Introduction
Glioblastoma is an astrocytoma-type brain tumour that represents 60% of all brain tumours in adults, with an incidence 2–3 cases per 100,000 people in Europe and North America.1 According to the World Health Organisation, glioblastoma belongs to the most malignant, grade IV group of brain tumours.2 It is also one of the deadliest cancers, as most patients with glioblastoma die within 14 months of diagnosis, despite receiving standard therapies.3 The fatal nature of glioblastoma arises for several reasons. The boundaries of these tumours are poorly defined, which makes it difficult to surgically remove the entire mass of the tumour.3 Furthermore, groups of cells, or even a single cell, can detach from the original tumour and travel to other parts of the brain to form a new tumour.3 In addition, the brain is separated from the rest of the body by the blood-brain barrier, which is hard to penetrate and represents an obstacle in the design of new drugs.4
Glioblastoma is known for high heterogeneity. The cells follow a hierarchical organisation, at the top of which are the most potent glioblastoma stem cells,5 which are cancer stem cells, and are defined as cells that can self-renew and differentiate to other glioblastoma cells.6 Glioblastoma stem cells are also resistant to radiotherapy and temozolomide, the most common chemotherapeutic agent used in standard glioblastoma treatments, and they are therefore believed to be responsible for tumour relapse.7,8 The limited efficacy of glioblastoma treatments due to the high recurrence rates and overall resistance to radiotherapy and chemotherapy therefore dictates the need for the development of better therapeutic approaches, and especially those that target glioblastoma stem cells. One of the established and prospective ways to target cancers is with monoclonal antibodies.9 These, however, have some drawbacks, and have not yet made significant advances for the treatment of glioblastoma.10
This study focused on nanobodies, which are the 15-kDa antigen-recognizing parts of the heavy-chain-only antibodies produced by Camelidae (camelids), and while they retain some specifics of monoclonal antibodies, they also have some unique characteristics.9 Structurally, nanobodies are similar to the heavy chain variable (VH) part of classical antibodies, but with two important exceptions: their CDR3 region is longer, and particular hydrophobic amino acids in the framework-2 region are substituted by hydrophilic amino acids, which makes them water soluble.9 The other advantages of nanobodies over classical monoclonal antibodies are that they are exceptionally stable under harsh conditions, and they can be produced economically in microbial hosts such as Escherichia coli and yeast with high yields.11,12 Nanobodies also penetrate tumours more rapidly and have more favourable tumour distributions in comparison with monoclonal antibodies.13
To translate nanobodies into therapies, however, there are some obstacles that need to be confronted. Nanobodies are eliminated rapidly from the human body because their molecular weight is below the renal cut-off of 60 kDa. However, they can be bound to other protein units to increase their molecular weight, so as not to be rapidly cleared from the serum circulation, and thus to prolong their half-life in the body.14 A very attractive way that has been shown to extend the life span of some drugs is also through the neonatal Fc receptor (FcRn) rescue mechanism.15
An important aspect of nanobodies is that they can potentially be used for glioblastoma treatments, as it appears that there are some mechanisms for their penetration of the blood–brain barrier.16 They can be bound to a functional unit that enables their penetration, such as a protein that binds to α(2,3)-sialoglycoprotein receptors, transferrin receptors or low-density lipoprotein receptor-related protein 1.16 In addition, it has been reported that if the nanobodies have a basic isoelectric point, they may penetrate the blood–brain barrier themselves, and bind to their target.17 However, few such studies have been carried out, and more research is required to characterise more precisely the mechanisms behind the penetration of the blood–brain barrier by different nanobodies. Indeed, to date, there has been only one report of in vivo targeting of glioblastoma with nanobodies, which showed promising results in an experimental mouse model.18 However, naked nanobodies have been used successfully in the intracranial human epidermal growth factor receptor 2 positive breast cancer model for imaging in mice.19
In our previous studies, alpacas were immunised with whole glioblastoma cells enriched in glioblastoma stem cells. Following the protocol of Vincke et al., a phage display nanobody library was constructed.20 Panning of a protein extract from glioblastoma tumour tissue from a patient and of protein extracts from glioblastoma stem cells (i.e. NCH644, NCH421k cells) provided nanobodies against eight antigens that were subsequently identified by mass spectrometry, and were thus proposed as candidates for tumour-class predictive biomarkers.21,22
We report here on the validation of the expression of these antigens at the cellular level, namely in glioblastoma cells and astrocytes, using qPCR, ELISA and immunocytochemistry. We have also performed immunohistochemical analysis of the antigen expression in normal brain, glioblastoma and lower-grade glioma tissues. Furthermore, we have tested the effectiveness of the eight nanobodies directed against these antigens for reduction of cell survival in comparison with temozolomide. The four most effective nanobodies were then selected to examine whether they can be used to induce cell apoptosis/necrosis, and to impair cell migration, and to see if consecutive treatments and/or different nanobody combinations can potentiate their cytotoxicities. The results show that these selected nanobodies can be used to efficiently target glioblastoma cells and glioblastoma stem cells.
Materials and methods
Cell culture
The U251MG and U87MG glioblastoma cell lines were from American Type Culture Collection. They were cultured in Eagle’s minimum essential medium (Sigma-Aldrich) supplemented with 5% foetal bovine serum (Sigma-Aldrich), 1 mM sodium pyruvate (Sigma-Aldrich), 1% non-essential amino-acids solution (Gibco) and 1% antibiotic–antimycotic solution (penicillin/streptomycin/amphotericin B; Gibco). The NCH644 and NCH421k glioblastoma stem cell lines were from CLS Cell Lines Service GmbH, and were cultured separately or as co-cultures (NCH co-cultures) in Neurobasal medium (Gibco) supplemented with 1% GlutaMax (Gibco), B27 supplement (Gibco), 20 ng/mL EGF (Gibco), 20 ng/mL bFGF (Gibco), 1 U/mL heparin (Sigma-Aldrich) and 1% antibiotic–antimycotic solution (penicillin/streptomycin/amphotericin B; Gibco). The human astrocytes were from ScienCell, and were cultured in Astrocyte medium (ScienCell) supplemented with 2% foetal bovine serum (ScienCell), 1% astrocyte growth supplement (ScienCell) and 1% penicillin/streptomycin (ScienCell).
Antibodies
The antibodies used were all from Sigma-Aldrich and are listed in Table 1.
Table 1.
Antibodies used in the study.
| Antibody | Sigma-Aldrich catalogue number | Experimental system |
|---|---|---|
| a-TRIM28 | WH00101551 | ELISA, ICC, IHC |
| a-NUCL | SAB130550 | ELISA, ICC, IHC |
| a-VIM | V6389 | ELISA, ICC, IHC |
| a-NAP1L1 | SAB1400175 | ELISA |
| a-NAP1L1 | SAB1401253 | ICC, IHC |
| a-TUFM | Amab90964 | ELISA, ICC, IHC |
| a-CRMP2 (a-DPYSL2) | C2993 | ELISA |
| a-DPYSL2 | HPA002381 | ICC, IHC |
| a-CRMP1 | HPA035640 | ELISA, ICC, IHC |
| a-ALYREF | A9979 | ELISA, ICC, IHC |
| Goat anti-rabbit AP conjugate | A3687 | ELISA |
| Goat anti-mouse AP conjugate | A3562 | ELISA |
| Anti-mouse CF640R | SAB4600396 | ICC |
| Anti-rabbit CF640R | SAB4600399 | ICC |
ALYREF, Aly/REF export factor; AP, alkaline phosphatase; CF640R, rhodamine-based far-red fluorescent dye; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; ELISA, enzyme-linked immunosorbent assay; ICC, immunocytochemistry; IHC, immunohistochemistry; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Nanobodies
The nanobodies were produced in E. coli and isolated using periplasmic extraction, as described previously.20 All of the nanobodies contained a 6×His-tag and were purified using nickel immobilised metal affinity chromatography and size exclusion chromatography. Their purity was confirmed by SDS-PAGE. The corresponding antigens were determined by mass spectrometry; however, the nanobody affinities and epitopes have not been determined.21,22 For the experiments, the nanobodies were prepared in phosphate-buffered saline (PBS) at the same pH as the nanobody isoelectric point. Eight different nanobodies that were previously characterised by Jovcevska et al.21,22 were initially used: Nb237 [anti-tripartite motif containing 28 protein (TRIM28)], Nb10 (anti-nucleolin), Nb79 (anti-vimentin), Nb179 [anti-nucleosome assembly protein 1-like 1 (NAP1L1)], Nb225 [anti-mitochondrial translation elongation factor (EF-TU) (TUFM)], Nb314 [anti-dihydropyrimidinase-related protein 2 (DPYSL2)], Nb394 [anti-collapsin response mediator protein 1 (CRMP1)] and Nb395 [anti-Aly/REF export factor (ALYREF)].
Quantitative PCR
RNA was isolated from the U251MG and U87MG cells, co-cultures of NCH644 and NCH421K cells (NCH co-cultures) and astrocytes using TRI reagent (Sigma-Aldrich), as instructed by the manufacturer. RNA concentrations were estimated using a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, USA), and their purities determined according to the ratios of absorption 260 nm to 280 nm and 260 nm to 230 nm (A260/A280 and A260/A230). RNA integrity was determined using a bioanalyser (2100; Agilent Technologies, USA).
First, the isolated RNA was treated with recombinant RNAse-free DNAse I for 15 min at 30°C, and then for 10 min at 75°C. Then, 2 µg of each RNA sample was transcribed to cDNA (Transcriptor Universal cDNA Master; settings: 5 min at 25°C, 10 min at 55°C, 5 min at 85°C). Quantitative PCR (qPCR) was performed using the Roche LightCycler 480 platform. The reaction mixture consisted of 0.75 µL cDNA, 2.5 µL 2× LightCycler 480 SYBR Green I Master, 0.3 µL of each 2.5 mM primer, and 1.15 µL distilled H2O. The total volume was 5 µL. qPCR was performed with the following thermal cycling: pre-incubation: 10 s at 95°C; cycling: 20 s at 60°C and 20 s at 72°C for 45 cycles; melting curve: 5 s at 95°C and 1 min at 65°C; continuous: at 97°C; and cooling: 30 s at 4°C. The ribosomal protein L13a (RPL13A) and cytochrome c-1 (CYC1) genes were used for normalisation. The primers used for the normalisation genes were selected from the literature, while the primers for the genes of interest were from PrimerBank PCR primer database for quantitative gene expression analysis (https://pga.mgh.harvard.edu.primerbank).23–28 The primer sequences for all of the genes are presented in Table 2. Analysis of relative mRNA expression was determined as previously described.26 Briefly, the expression of each of the study genes was normalised to the expression of the reference genes.
Table 2.
Primer sequences for the genes analysed in this study.
| Gene | Primer sequence (5‘→3’) | Amplicon length (bp) |
|---|---|---|
| TRIM28 | F: TGA GAC CTG TGT AGA GGC G | 93 |
| R: CGT TCA CCA TCC CGA GAC TT | ||
| NUCL | F: GAA CCG ACT ACG GCT TTC AAT | 93 |
| R: AGC AAA AAC ATC GCT GAT ACC A | ||
| VIM | F: TGC CGT TGA AGC TGC TAA CTA | 248 |
| R: CCA GAG GGA GTG AAT CCA GAT TA | ||
| NAP1L1 | F: AAA GCA CGT CAG CTA ACT GTT | 146 |
| R: TTG AGA GCA TTC ACT CGT CTT TT | ||
| TUFM | F: AAA GAA GGG AGA CGA GTG TGA | 80 |
| R: TGT GGA ACA TCT CAA TGC CTG | ||
| DPYSL2 | F: GTG ACT ACT CTC TGC ATG TGG A | 87 |
| R: TTA CCC CGT GAT CCT TCA CAA | ||
| CRMP1 | F: AGT GAC CGA CTC CTC ATC AAA | 119 |
| R: CCA GGA ACG ATT AAG TTC TCT CC | ||
| ALYREF | F: ACA TTC AGC TTG TCA CGT CAC | 77 |
| R: TCT AGT CAT GCC ACC TCT GTT TA | ||
| RPL13A | F: CCT GGA GGA GAA GAG GAA AGA GA | 126 |
| R: TTG AGG ACC TCT GTG TAT TTG TCA A | ||
| CYC1 | F: GAG GTG GAG GTT CAA GAC GG | 160 |
| R: TAG CTC GCA CGA TGT AGC TG |
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; CYC1, cytochrome c-1; DPYSL2, dihydropyrimidinase-related protein 2; F, forward; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; RPL13A, ribosomal protein L13a; R, reverse; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Enzyme-linked immunosorbent assay
Proteins were isolated from NCH co-cultures, and U251MG and U87MG cells and astrocytes (ProteoExtract Transmembrane Protein Extraction kits; Novagene), following the manufacturer instructions. Protein concentrations were measured using protein assay kits (Pierce BCA; Thermo Scientific).
The isolated cytosolic and membrane protein fractions were diluted to a final concentration of 2 µg/mL in 1 M NaHCO3 (pH 9) and used to coat the enzyme-linked immunosorbent assay (ELISA) plates (NUNC Maxisorp) at 100 µL/well overnight at 4°C. The next day, the wells were washed three times with 0.01% Tween in PBS, and the residual binding sites were blocked with 5% milk in PBS for 1 h at room temperature. Prior to the application of the primary antibodies, the wells were washed again. The primary antibodies were diluted in 1% milk in PBS to a final concentration of 1 µg/mL (Table 1). After a 1-h incubation at room temperature, the wells were washed again, and the secondary goat anti-mouse or goat anti-rabbit alkaline-phosphatase-conjugated antibodies were applied (1:2000 in 1% milk in PBS). After a 1-h incubation at room temperature, the wells were washed and the alkaline phosphatase substrate was applied (Sigma-Aldrich). The signals were measured at 405 nm over different times (30, 90, 150 and 300 min, and overnight). All experiments were performed in triplicate.
Immunocytochemistry
Initially, U251MG cells (4 × 104) were seeded on coverslips, and U87MG cells (1.5 × 105), NCH co-cultures (1.5 × 105) and astrocytes (1 × 105) were seeded on poly-d-lysine (Sigma-Aldrich) pre-treated coverslips, with all cell lines incubated for 24 h. The next day, the cells were washed three times with PBS. The U251MG cells were fixed in ice-cold methanol for 10 min at room temperature, and washed with PBS three times for 10 min. The U87MG cells, NCH co-cultures and astrocytes were fixed in ice-cold 4% formaldehyde for 15 min, washed three times for 10 min with PBS, and then permeabilised with 0.1% Triton X-100 in PBS for 15 min at room temperature. All cell lines were then blocked with 5% bovine serum albumin in PBS for 1 h at room temperature. Then, the primary antibodies were added at the appropriate dilutions (30:1000 mouse anti-TRIM28, 16:1000 mouse anti-nucleolin, 2:1000 mouse anti-vimentin, 12:1000 mouse anti-TUFM, 12:1000 mouse anti-NAP1L1, 8:1000 rabbit anti-DPYSL2, 40:1000 rabbit anti-CRMP1 or 20:1000 mouse anti-ALYREF, Table 1) and incubated overnight at 4°C. The next day, the cells were washed three times with PBS for 10 min, and then incubated with the secondary antibodies (anti-mouse, 3:1000; anti-rabbit, 3:1000) for 1 h at room temperature. Subsequently, the cells were washed three times with PBS for 10 min, incubated with 300 nM 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 10 min, washed again three times with PBS for 10 min, and mounted on slides using ProlongGlass (ThermoFisher Scientific). Images were taken with an imager (Axio Imager M2; Zeiss) equipped with ZEN software. All of the samples were excited by filters at wavelengths of 335–383 nm (DAPI) and 574–599 nm [rhodamine-based far-red fluorescent dye (CF640R)], with filter emission wavelengths of 420–470 nm (DAPI) and 612–682 nm (CF640R). Images of U251MG and U87MG cells and astrocytes were taken under the 40× objective (EC Plan-Neofluar 40×/0.75 M27), and images of the NCH co-cultures were taken under the 63× oil-immersion objective (Plan-Apochromat 63×/1.40 Oil DIC M27), using a digital camera (AxioCamMR3). The images were analysed using ImageJ.29
Immunohistochemistry
Tissues were fixed in paraffin, and slices (4–6 μm) were cut and mounted on adhesion slides (Thermo Scientific). The samples were processed with CC1 buffer (cat. n° 951–124; Ventana) and then incubated with primary antibodies at the appropriate dilutions for 30 min at 38°C (mouse anti-TRIM28, 1:150; mouse anti-nucleolin, 1:200; mouse anti-vimentin, 1:200; mouse anti-NAP1L1, 1:50; mouse anti-TUFM, 1:500; rabbit anti-DPYSL2, 1:200; rabbit anti-CRMP1, 1:100; mouse anti-ALYREF, 1:100) (Table 1). The samples were then stained using an automated slide preparation system (Ventana Discovery). Signal detection was performed using detection kits (iView DAB Ventana). Each tissue sample was ranked as one of five categories (0–4) according to the proportion of positive cells: 0, negative staining; 1, <25% positive; 2, 25–50% positive; 3, 50–75% positive; 4, 75–100% positive. The mean scores were calculated. Student t tests were used to define statistical significance between glioblastoma and lower-grade glioma samples. The samples with corresponding patient information are listed in Table 3 (i.e. sex, average age at operation, isocitrate dehydrogenase status).
Table 3.
Glioblastoma and lower-grade glioma samples.
| Antigen | Glioblastoma |
Lower-grade glioma |
||||||
|---|---|---|---|---|---|---|---|---|
| Sex (n/N) |
Mean age at operation (years) | Isocitrate dehydrogenase status (n/N) | Sex (n/N) |
Mean age at operation (years) | Isocitrate dehydrogenase status (n/N) | |||
| Female | Male | Female | Male | |||||
| TRIM28, TUFM, NUCL | 5/10 | 5/10 | 68 | 10/10 wild type | 4/8 | 4/8 | 48 | 7/8 mutant; 1/8 wild type |
| VIM | 6/11 | 5/11 | 68 | 10/10 wild type | 2/4 | 2/4 | 49 | 4/4 mutant |
| NAP1L1, DPYSL2, CRMP1, ALYREF | 6/11 | 5/11 | 68 | 10/10 wild type | 4/8 | 4/8 | 48 | 7/8 mutant; 1/8 wild type |
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; RPL13A, ribosomal protein L13a; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Cell viability assays
Cell viabilities were determined using the AlamarBlue reagent (Invitrogen). First, the U251MG and U87MG cells (1 × 103), NCH co-cultures (5 × 103) and astrocytes (4 × 103) were seeded in 70 μL medium and incubated for 3 h at 37°C, in a 5% CO2 atmosphere. Then, 10 μg/mL and 100 μg/mL nanobodies or 50 μM temozolomide were added, to a final volume of 100 μL. Non-treated cells were used as the control, and growth medium as the blank. The cells were incubated for 24 h, 48 h and 72 h. Next, 10 μL AlamarBlue was added, followed by a 3-h to 4-h incubation. When the colour had evolved, the absorbance was measured at 570 nm and 600 nm using a microplate reader (Synergy H4; BioTek). Cell survival was determined as described in the manual for the use of the AlamarBlue reagent (Invitrogen). When the cell viabilities were determined in the presence of different nanobody combinations, 4000 cells/well for all cell lines (i.e. U251MG and U87MG cells, NCH co-cultures, astrocytes) were seeded in 70 μL medium and incubated for 3 h. Then, different nanobody combinations (nanobody pairs; 10 or 100 μg/mL each) were added to a final volume of 100 μL. The cells were incubated for 72 h, and then 10 μL water-soluble tetrazolium (Roche) was added. After a 2.5-h incubation with water-soluble tetrazolium, the absorbance was measured at 450 nm and 690 nm using a microplate reader (Synergy H4; BioTek). To determine the cell viabilities after two consecutive treatments, the nanobodies or 50 μM temozolomide were first added to a final volume of 100 μL. After a 72-h incubation at 37°C in a 5% CO2 atmosphere, the growth medium was replaced and the second treatment with nanobodies or temozolomide was administered in the same way. The cells of the NCH co-cultures grow in suspension, and were therefore centrifuged before medium removal. After the second 72-h incubation at 37°C in a 5% CO2 atmosphere, the absorbance was measured at 450 nm and 690 nm using a microplate reader (Synergy H4; BioTek). Cell survival was determined as the ratios between the values for the treated cells and for the control (non-treated) cells. The blank was subtracted from each value.
Apoptosis/necrosis assays
Apoptosis/necrosis assays were performed for U251MG (4 × 105) and U87MG (1.5 × 105) cells, NCH co-cultures (5 × 105) and astrocytes (1 × 105), which were seeded onto coverslips in 24-well plates. After a 3 h incubation at 37°C in a 5% CO2 atmosphere, nanobodies were added to a final concentration of 100 μg/mL. The cells were incubated for 24 h at 37°C in a 5% CO2 atmosphere, and then washed twice with PBS. Subsequently, an Annexin-FITC/propidium iodide kit (Abcam; ab14085) was used as follows: 250 μL binding buffer, 2.5 μL Annexin V and 2.5 μL propidium iodide were added and incubated for 15 min in the dark at room temperature. The coverslips were then mounted on slides using mounting medium (ProlongGlass; ThermoFisher Scientific). Photographs were taken using an imager (Axio Imager M2; Zeiss) equipped with ZEN software. All images were taken with the 40× objective (EC Plan-Neofluar 40×/0.75 M27). Detection of Annexin V-FITC was at the excitation wavelength of 495 nm and the emission wavelength of 519 nm. Propidium iodide was detected under a rhodamine filter with the excitation wavelength of 558 nm and the emission wavelength of 575 nm. Images were analysed using ImageJ.29
Cell migration assays
Silicone inserts with a defined cell-free gap (Ibidi) were put into each well of 12-well plates. Then, the U251MG (1.5 × 104) and U87MG (9 × 104) cells and astrocytes (2 × 104) were seeded into each well of the inserts and incubated for 24 h (U251MG cells) or 48 h (U87MG cells, astrocytes). The inserts were then removed, and nanobodies were added to a final concentration of 100 μg/mL in 1.5 mL total volume. Images for cell migration were taken at two or three different positions every 1–2 h (Axio Observer.Z1, Zeiss) under the 10× objective (EC Plan-Neofluar 10×/0.3 Ph1 M27). The final time point for U251MG cells was 8 h, with 24 h for U87MG cells and 18 h for astrocytes. The NCH co-cultures were not included here due to their buoyancy and inability to attach to the bottom of the wells of the experimental plates. Cell migration was first calculated as distance covered over time (μm/h), and then relative cell migrations were determined as the ratio between the mean speed of cells in the presence of a nanobody and the mean speed of the control cells (without nanobodies).
Statistical analysis
For statistical analysis of the qPCR data, samples that followed Gaussain distribution were analysed using one-way ANOVA, and those that did not follow Gaussian distribution were analysed using Kruskal–Wallis tests. Data from the ELISA and cell migration assays were analysed using Student’s t tests. One-way ANOVA followed by Sidak’s multiple comparison tests were used for analysis of cytotoxicity data. In all cases, p < 0.05 was considered as statistically significant, and significance is indicated as: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Standard deviations were calculated for all data.
Results
mRNA quantification of different antigens by qPCR
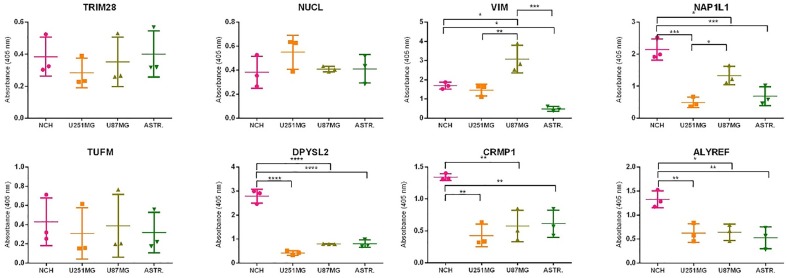
RNA was isolated from all cell lines, with the overall mean concentration of 2481 ng/µL. According to the A260/A280 and A260/A230 ratios, the quality of the RNA was satisfactory, with means of 1.80 and 1.93, respectively. The RNA integrity numbers also showed satisfactory RNA quality, with a mean of 8.60. The qPCR data showed that the relative mRNA expression between the glioblastoma stem cell lines (i.e. NCH co-cultures) and astrocytes was significantly different (higher in the NCH co-cultures) for three genes: NAP1L1 (p < 0.0001), TUFM (p < 0.0001) and CRMP1 (p < 0.0001) (Figure 1, Table 4). Expression of these same genes (i.e. NAP1L1, TUFM, CRMP1) was also significantly higher in the NCH co-cultures in comparison with both the U251MG and U87MG cells. VIM was the only one of the genes examined that significantly distinguished mRNA expression between astrocytes and both mature glioblastoma cell lines (i.e. U251MG, U87MG cells; higher expression in both these cell lines), while TRIM28, NUCL, NAP1L1 and ALYREF significantly distinguished just one or the other of the mature glioblastoma cell lines (i.e. U251MG or U87MG cells) from astrocytes.
Figure 1.
Relative mRNA expression of the selected antigens (as indicated) in the U251MG and U87MG cells, the NCH co-cultures and the astrocytes. The reference genes were RPL13A and CYC1. Data show means ±SD for the relative expression of nine replicates.
**p < 0.01, ***p < 0.001, ****p < 0.0001. Expression of CRMP1 in U87MG cells is not shown, the mRNA was below the detection limit.
Table 4.
Results for the statistical analysis of the relative mRNA expression of the selected antigens in the cell lines examined.
| Gene | Cell comparison | Significance | p-value |
|---|---|---|---|
| TRIM28 | U87MG versus NCH | **** | <0.0001 |
| U87MG versus U251 | **** | <0.0001 | |
| U87MG versus astrocytes | **** | <0.0001 | |
| NUCL | U87MG versus NCH | *** | 0.0003 |
| U87MG versus U251 | **** | <0.0001 | |
| Astrocytes versus U251 | ** | 0.0083 | |
| VIM | U251MG versus NCH | **** | <0.0001 |
| U87MG versus NCH | **** | <0.0001 | |
| U251MG versus U87MG | **** | <0.0001 | |
| U87MG versus astrocytes | **** | <0.0001 | |
| U251MG versus astrocytes | **** | <0.0001 | |
| NAP1L1 | NCH versus U251MG | **** | <0.0001 |
| NCH versus U87MG | *** | 0.0008 | |
| NCH versus astrocytes | **** | <0.0001 | |
| U87MG versus U251 | **** | <0.0001 | |
| U87MG versus astrocytes | **** | <0.0001 | |
| TUFM | NCH versus U251MG | **** | <0.0001 |
| NCH versus U87MG | **** | <0.0001 | |
| NCH versus astrocytes | **** | <0.0001 | |
| U251MG versus U87MG | ** | 0.0013 | |
| DPYSL2 | U87MG versus U251 | *** | 0.0001 |
| NCH versus U251MG | ** | 0.0077 | |
| CRMP1 | NCH versus U251MG | **** | <0.0001 |
| NCH versus astrocytes | **** | <0.0001 | |
| ALYREF | U87MG versus NCH | ** | 0.0056 |
| U87MG versus U251 | ** | 0.0075 | |
| U87MG versus astrocytes | *** | 0.0002 |
Significance for nine replicates for each gene that followed a Gaussian distribution was calculated using one-way ANOVA, and for the values without a Gaussian distribution, by Kruskal-Wallis statistical tests.
**p < 0.01, ***p < 0.001, ****p < 0.0001.
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; RPL13A, ribosomal protein L13a; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
To summarise, based on these findings, VIM, NAP1L1 and TUFM were the most universal biomarkers for the glioblastoma-related cells, and CRMP1 can also be included, as in addition to NAP1L1 and TUFM, CRMP1 showed significant overexpression in the NCH co-cultures.
Quantification of protein expression using ELISA
To quantify the protein expression of the eight antigens, ELISA was performed. The protein was isolated from all cell lines, with a mean concentration of 502 µg/mL. Protein expression in the NCH co-cultures for vimentin, NAP1L1, DPYSL2, CRMP1 and ALYREF was significantly higher than in the astrocytes (Figure 2, Table 5). Expression of NAP1L1, DPYSL2, CRMP1 and ALYREF in the NCH co-cultures was also significantly higher than in both of the mature glioblastoma cell lines, U251MG and U87MG cells (Figure 2, Table 5). Considering the correlation of these findings with the findings from the qPCR, in terms of potential NCH co-culture biomarkers for the further aspects of this study, NAP1L1 and CRMP1 were confirmed, VIM and TUFM were preserved, and DPSYL2 and ALYREF were added as selected molecular targets.
Figure 2.
Protein expression of the selected antigens (as indicated) in the U251MG and U87MG cells, the NCH co-cultures and the astrocytes. Data show means ± SD from three replicates, with absorbance measured at 405 nm.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Table 5.
Statistical analysis of protein expression for the selected antigens in the different cell lines.
| Protein | Cell comparison | Significance | p-value |
|---|---|---|---|
| TRIM28 | All | ns | |
| NUCL | All | ns | |
| VIM | U87MG versus NCH | * | 0.0130 |
| NCH versus astrocytes | * | 0.0263 | |
| U87MG versus U251 | ** | 0.0053 | |
| U87MG versus astrocytes | *** | 0.0002 | |
| NAP1L1 | NCH versus U251MG | *** | 0.0004 |
| NCH versus U87MG | * | 0.0286 | |
| NCH versus astrocytes | *** | 0.0009 | |
| TUFM | All | ns | |
| DPYSL2 | NCH versus U251MG | **** | <0.0001 |
| NCH versus U87MG | **** | <0.0001 | |
| NCH versus astrocytes | **** | <0.0001 | |
| CRMP1 | NCH versus U251MG | ** | 0.0015 |
| NCH versus U87MG | ** | 0.0044 | |
| NCH versus astrocytes | ** | 0.0060 | |
| ALYREF | NCH versus U251MG | ** | 0.0093 |
| NCH versus U87MG | * | 0.0108 | |
| NCH versus astrocytes | ** | 0.0043 |
Significance of three samples was calculated using Student’s t-tests. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, non-significant.
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
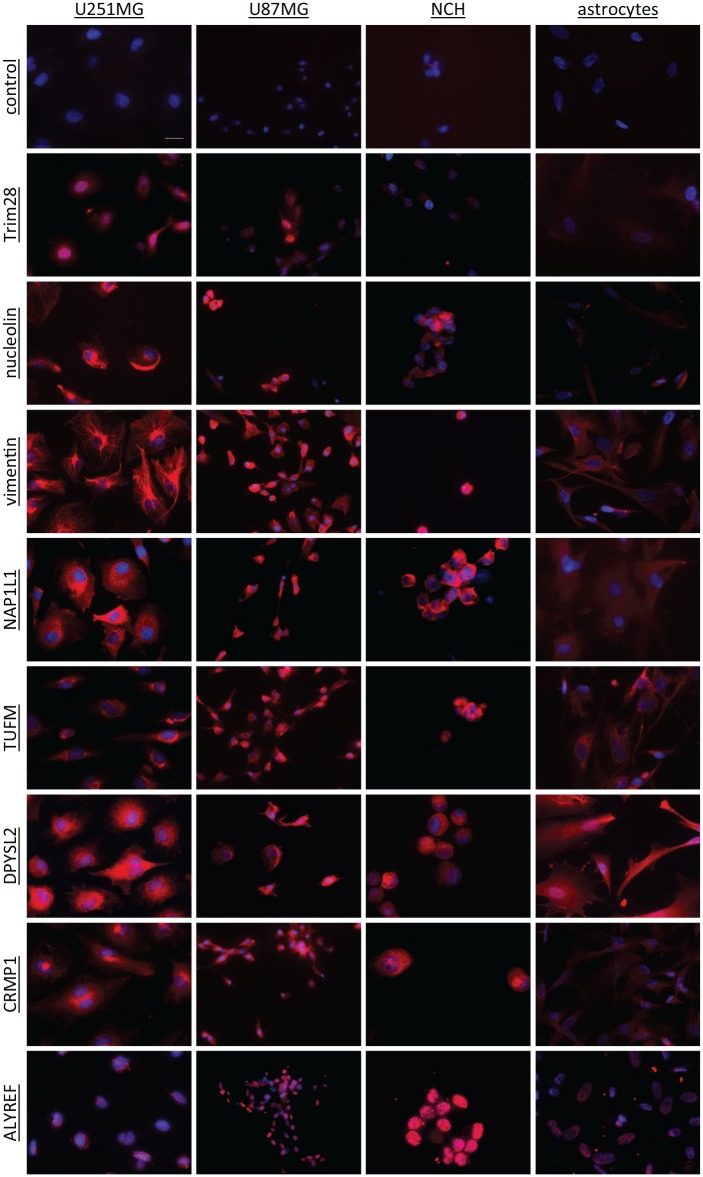
Immunofluorescent staining
To further confirm the protein expression of these eight antigens in the cell lines, and to examine their cellular locations, immunocytochemistry was performed using commercial antibodies (Table 1) and with staining of the nuclei with DAPI (blue), as shown in Figure 3. These data showed that the nuclear protein TRIM28 was expressed in all three glioblastoma cell lines (i.e. U251MG, U87MG cells, NCH co-cultures), and was almost absent in astrocytes. Nucleolin was expressed mostly in the nuclei of U87MG cells, and on the membranes and in the cytoplasm of U251MG and U87MG cells, and the NCH co-cultures. The signal intensities were strong for all three glioblastoma-related cell lines, and were weaker in astrocytes. Vimentin showed a cytoplasmic location, with very strong signal intensity in all three glioblastoma-related cell lines, and again weaker in astrocytes. The location of NAP1L1 was also cytoplasmic, with similar strong signals to vimentin for all three glioblastoma-related cell lines, and weak again for astrocytes. TUFM was located around the nuclei, with high signal intensity, not only in all three glioblastoma-related cell lines, but also in astrocytes. The location of DPYSL2 and CRMP1 was cytoplasmic, although they were also seen in nuclei. Expression of DPYSL2 was seen in all four cell lines, including astrocytes, while, for CRMP1, expression was high in glioblastoma-related cells, and very low in astrocytes. ALYREF was a nuclear protein, and was expressed in all four cell lines.
Figure 3.
Representative immunocytochemical images of the selected antigens (as indicated) in the U251MG and U87MG cells, the NCH co-cultures and the astrocytes. The antigens were stained with a commercial antibody (CF640R fluorophore; red) and the nuclei with DAPI (blue). The control had no primary antibody included. Scale bar, 20μm (top left image; applicable to all images).
DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
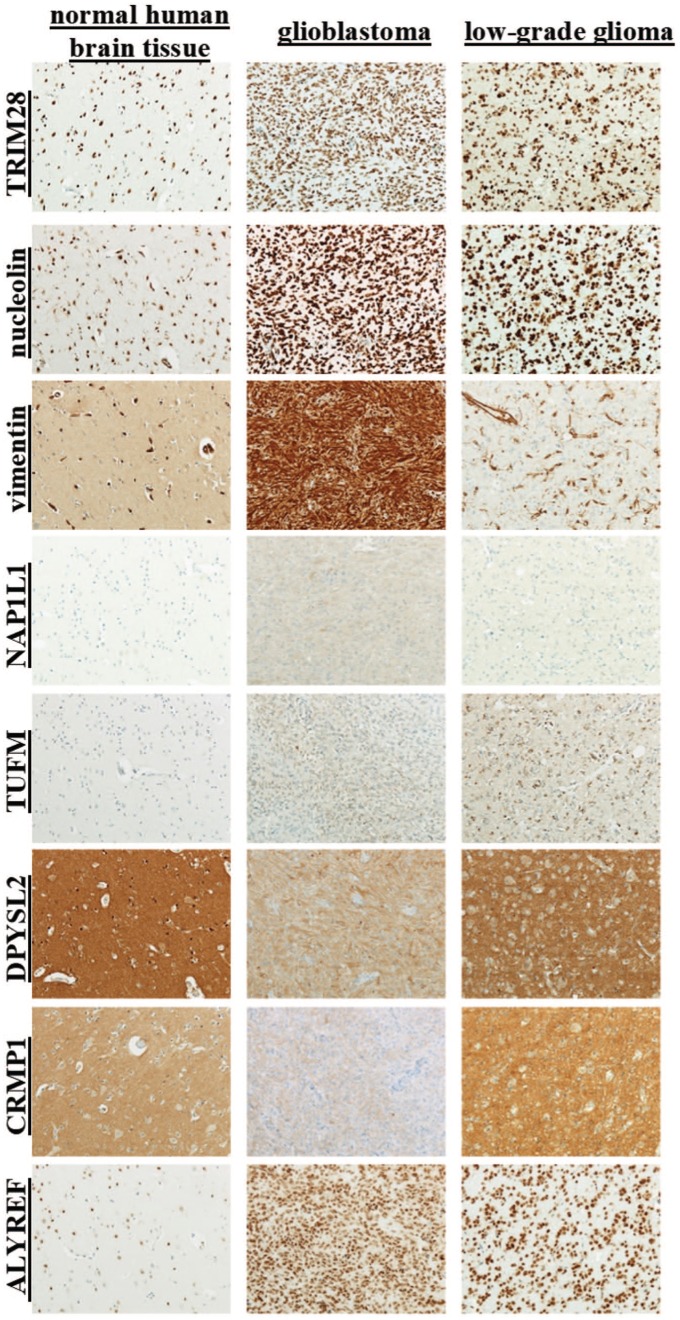
Antigen detection in normal brain, glioblastoma and lower-grade glioma using immunohistochemistry
To evaluate the clinical value of these eight antigens as potential glioblastoma markers at the tissue level, immunohistochemical analysis was performed to determine their expression in glioblastoma and lower-grade glioma (oligodendroglioma and diffuse astrocytoma) cancerous tissues versus normal brain tissue. TRIM28, nucleolin and ALYREF are nuclear proteins, and they were expressed in the normal brain tissue, and highly expressed in the glioblastoma and lower-grade glioma (Figure 4). DPYSL2 and CRMP1 showed a cytoplasmic location, and their expression was negative in glia cells and neurons, but positive in neuropils. Thus, DPYSL2 and CRMP1 showed scores in glioblastoma of 2.33 and 1.64, respectively, with no statistical differences in their expression between glioblastoma and lower-grade glioma (Table 6). Vimentin, NAP1L1 and TUFM were not expressed in the normal brain tissue. Vimentin had strong ubiquitous cytoplasmic expression in glioblastoma (score, 3.91), which was also significantly stronger than in the lower-grade glioma (score, 1.5; p-value, 4.3 × 10–6). NAP1L1 expression in glioblastoma and lower-grade glioma was not uniform (25.0%, 37.5% negative samples, respectively). TUFM was expressed in all of the glioblastoma and lower-grade glioma cases, with no difference between glioblastoma and lower-grade glioma.
Figure 4.
Representative immunohistochemical images for the selected proteins (as indicated) in normal brain, glioblastoma and low-grade glioma tissues.
Table 6.
Statistical analysis from the immunohistochemical detection of the selected proteins in normal brain, and in GB and LGG tissue.
| Antigen | Expression in normal brain tissue |
Positive tissues (%) |
Score |
Statistical analysis (GB versus LGG) |
|||
|---|---|---|---|---|---|---|---|
| GB | LGG | GB | LGG | Significance | p-value | ||
| TRIM28 | Yes | 100 | 100 | 3.8 | 3.875 | No | |
| NUCL | Yes | 100 | 100 | 3.91 | 4 | No | |
| VIM | No † | 100 | 100 | 3.91 | 1.5 | Yes | 4.3 × 10–6 |
| NAP1L1 | No | 75 | 62.5 | 1.42 | 0.875 | No | |
| TUFM | No | 100 | 100 | 2.3 | 1.875 | No | |
| DPYSL2 | Yes‡ | 100 | 100 | 2.33 | 1.875 | No | |
| CRMP1 | Yes‡ | 100 | 100 | 1.64 | 2.125 | No | |
| ALYREF | Yes | 100 | 100 | 4 | 3.875 | No | |
Significance of samples (for numbers see Table 3) was calculated using Student’s t tests.
VIM expression in glia cells and neurons was negative, but positive in blood vessels.
DPYSL2 and CRMP1 expression was positive only in neuropils.
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; GB, glioblastoma; LGG, lower-grade glioma; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Expression of the antigens in the different cell lines and tissues according to each of the methods used in this study (i.e., qPCR, ELISA, immunocytochemistry; tissue immunohistochemistry) is shown in Table 7.
Table 7.
Expression of the antigens in the different cell lines and tissues according to each of the methods used in this study [i.e. qPCR, ELISA, (ICC); tissue].
| Antigen | Cell lines |
Tissue |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U251MG cells |
U87MG cells |
NCH co-culture |
Astrocytes |
IHC |
|||||||||||
| qPCR | ELISA | ICC | qPCR | ELISA | ICC | qPCR | ELISA | ICC | qPCR | ELISA | ICC | Normal | GB | LGG | |
| TRIM28 | + | + | + | + | + | + | + | + | + | + | + | – | + | + | + |
| Nucleolin | + | + | + | + | + | + | + | + | + | + | + | – | + | + | + |
| Vimentin | + | + | + | + | + | + | + | + | + | + | + | – | – | + | + |
| NAP1L1 | + | + | + | + | + | + | + | + | + | + | + | – | – | +/– | +/– |
| TUFM | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + |
| DPYSL2 | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + |
| CRMP1 | + | + | + | + | + | + | + | + | + | + | + | – | – | + | + |
| ALYREF | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Data are presented as either positive (+) or negative (–) for expression.
NAP1L1 showed positive expression in some tissues and negative expression in others (+/–).
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; ELISA, enzyme-linked immunosorbent assay; GB, glioblastoma; ICC, immunocytochemistry; IHC, immunohistochemistry; LGG, lower-grade glioma; NAP1L1, nucleosome assembly protein 1-like 1; Normal, normal human brain; NUCL, nucleolin; qPCR, quantitative polymerase chain reaction; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Cytotoxicity of temozolomide and the nanobodies
In the second stage of this study, viability tests for cell survival were carried out to determine the cytotoxicities of 50 μM temozolomide and the different nanobodies on the different cell lines, using AlamarBlue reagent. The nanobodies corresponding to the eight antigens included in this study were Nb237 (anti-TRIM28), Nb10 (anti-nucleolin), Nb79 (anti-vimentin), Nb179 (anti-NAP1L1), Nb225 (anti-TUFM), Nb314 (anti-DPYSL2), Nb394 (anti-CRMP1) and Nb395 (anti-ALYREF). The cytotoxicities were determined on the four different cell lines, as before, as U251MG and U87MG cells, NCH co-cultures and astrocytes, at three different times: 24 h, 48 h and 72 h.
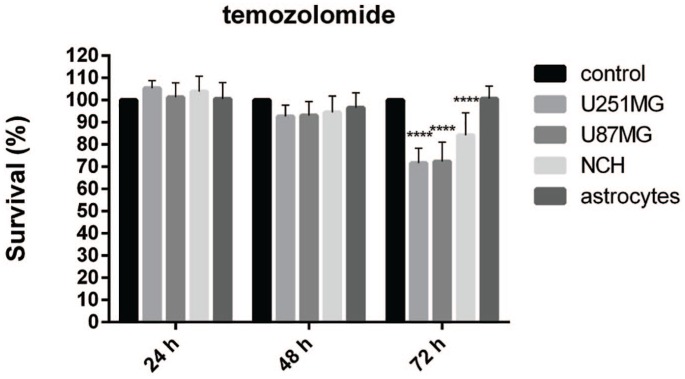
The effects of temozolomide are presented in Figure 5. No negative impacts on cell growth were seen after 24 h and 48 h of incubation. However, late cytotoxic effects were observed after 72 h for the U251MG and U87MG cells, where the cell viability was reduced to 72% in both cases, and for the survival of the NCH co-cultures of the glioblastoma stem cells, which was decreased to 84%. Temozolomide did not affect the survival of the astrocytes.
Figure 5.
Effects of 50 μM temozolomide on cell survival for the cell lines (as indicated). The AlamarBlue viability assay was used.
Data are means of three samples with three replicates (n = 9). **** p < 0.0001.
Following the comparative data on the effects of temozolomide on the cells included in this study, the cytotoxicities of the selected nanobodies were determined. All of the nanobodies were tested at two different concentrations: 10 μg/mL and 100 μg/mL. The cytotoxic effects were determined after 24 h, 48 h and 72 h of incubation (Figure 6). These data were also compared with the effects of temozolomide.
Figure 6.
Effects of the selected nanobodies (as indicated; 10 μg/mL, 100 μg/mL) on cell survival of the cell lines (as indicated). Treatments were for 24 h, 48 h and 72 h.
Data are means of three samples with three replicates (n = 9). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The anti-TRIM28 nanobody, Nb237, did not affect the survival of any of the cell lines at any of the times or concentrations. In contrast, the anti-nucleolin nanobody, Nb10, decreased the survival of all of the cell lines. At the higher concentration, Nb10 affected the U251MG and U87MG cells, and the astrocytes, as was already apparent after 24 h of incubation, when the survival of these three cells types had decreased by 20–30%. The survival of the astrocytes decreased gradually with time of treatment down to 46% after 72 h of incubation, while the survival of the U251MG and U87MG cells remained at ~75%. The low Nb10 concentration did not eliminate the cytotoxic effects on the astrocytes, as their survival continued to decrease over time, finally by 16% here. At the higher concentration of the anti-vimentin nanobody, Nb79, there was decreased survival of the U251MG cells and astrocytes after 72 h of treatment, by 40%, and of the U87MG cells by 19%. The low concentration of Nb79 gradually reduced the survival for the NCH co-cultures (by 15%), but had no apparent effects on the astrocytes. The anti-NAP1L1 nanobody, Nb179, at the higher concentration gradually decreased the survival of the U251MG and U87MG cells and astrocytes, which, after 72 h of incubation, had dropped by 45%, 32% and 55%, respectively. At its low concentration, Nb179 had no apparent impacts on the U251MG cells and astrocytes, but decreased the survival of the U87MG cells and NCH co-cultures by 14% and 16%, respectively. The anti-TUFM nanobody, Nb225, at both the low and higher concentrations did not have any significant effects on the U251MG and U87MG cells and astrocytes, although it had significant effects at the higher concentration on the NCH co-cultures, when after 72 h the survival dropped to 71%. The cytotoxic effects in this case were significantly stronger than the effects of 50 μM temozolomide alone (71% versus 84% survival after 72 h; p = 0.0151), as shown in Table 8. The anti-DPYSL2 nanobody, Nb314, and the anti-CRMP1 nanobody, Nb394, showed similar cell survival profiles. These low nanobody concentrations significantly decreased the survival of the NCH co-cultures by 18% (Nb314) and 21% (Nb394) after 72 h of exposure. Neither of these nanobodies had any impact on the U251MG and U87MG cells and astrocytes. The anti-ALYREF nanobody, Nb395, had no effects on the U251MG and U87MG cells or astrocytes. On the other hand, after 72 h, Nb395 significantly decreased the survival of the NCH co-cultures at the low and higher concentrations by 21% and 23%, respectively.
Table 8.
Relative cell survivals after the nanobody treatments, in comparison with the temozolomide treatments, all after 72 h.
| Nanobody | Concentration |
U251MG |
U87MG |
NCH |
Astrocytes |
||||
|---|---|---|---|---|---|---|---|---|---|
| (μg/mL) | Sign. | p-value | Sign. | p-value | Sign. | p-value | Sign. | p-value | |
| Nb10 | 10 | ns | ns | ns | **** | p < 0.0001 | |||
| 100 | ns | ns | ns | **** | p < 0.0001 | ||||
| Nb79 | 100 | * | 0.0106 | ns | ns | **** | p < 0.0001 | ||
| Nb179 | 100 | ** | 0.0018 | ns | ns | **** | p < 0.0001 | ||
| Nb225 | 100 | ns | ns | * | 0.0151 | ns | |||
Data only shown for the nanobodies with significantly stronger effects than 50 μM temozolomide Significance of three samples with three replicates was calculated by one-way ANOVA.
p < 0.05, **p < 0.01, ****p < 0.0001, ns, not significant.
Sign., significance.
Cytotoxicity of different nanobody combinations
According to qPCR, ELISA, immunocytochemistry and immunohistochemistry, VIM, NAP1L1 and TUFM were the most versatile glioblastoma-related cell biomarkers, which were not apparently, or were weakly, expressed in astrocytes, and were not expressed at all in normal brain tissue. DPYSL2 was also included in the selection of the nanobodies here as the expression difference between the NCH co-cultures and the astrocytes was higher in comparison with CRMP1. ALYREF was excluded due to its expression in normal brain tissue. In addition, the anti-VIM and anti-NAP1L1 nanobodies gradually decreased the survival of the NCH co-cultures and/or mature glioblastoma cells, whereas the anti-TUFM and anti-DPSYL2 nanobodies had relatively strong effects on NCH co-cultures after 72 h of exposure, with no effects on astrocytes. Therefore, these four nanobodies were chosen for further experiments, as Nb79 (anti-VIM), Nb179 (anti-NAP1L1), Nb225 (anti-TUFM) and Nb314 (anti-DPSYL2), in terms of their specificities towards glioblastoma cells and their effectiveness towards glioblastoma stem cells. The concentrations selected were 10 μg/mL for Nb79, Nb179 and Nb314, and 100 μg/mL for Nb225. First, the cell lines were treated with five different nanobody combinations for 72 h: Nb79+Nb179, Nb179+Nb225, Nb225+Nb314, Nb179+Nb314 and Nb225+Nb79. Then the absorbances were measured and cell survivals determined (Figure 7). Instead of the NCH co-culture used in the previous part of this study, here, NCH644 and NCH421k cells were tested separately. These data show that all combinations of nanobodies except Nb179+Nb314 reduced survival of these glioblastoma cells. The Nb79 and Nb179 combination reduced the survival of U251MG, U87MG and NCH644 cells by 20–25%. The Nb179+Nb225 combination significantly reduced the survival of U87MG cells to 66%, of U251 cells to 83%, and the survival of U87MG cells at the same nanobody concentrations was also lower than the cytotoxic effects of each of Nb179 and Nb225 separately (Figure 6). The Nb225+Nb314 combination significantly reduced survival of U251MG and U87MG cells by 24% and 25%, respectively, and of NCH421k cells by 14%. The survival of both mature glioblastoma cell lines was also considerably lower than the effects of both of these nanobodies separately (Figure 6). The Nb225+Nb79 combination significantly reduced the survival of U87MG cells to 78%. These effects were greater than the effects of Nb225 and Nb79 separately (Figure 6).
Figure 7.
Effect of the concurrent nanobody treatments (as indicated) on cell survival of the cell lines (as indicated). Treatments were for 72 h.
Data are means ± SD of three samples with three replicates (n = 9). *p < 0.05, **p < 0.01, ****p < 0.0001.
Both of the initial trials with isolated nanobodies (Figure 6) and nanobody combinations (Figure 7) showed that the nanobodies had mainly negative effects on glioblastoma-related cells. To avoid potential long-term silencing of the cytotoxic effect after single nanobody applications, we studied the impacts of consecutive treatments of the cells with different nanobodies in relation to consecutive treatment with 50 μM temozolomide. These data for temozolomide showed that it had no apparent impact on either of the glioblastoma stem cells (NCH644, NCH421k cells) or astrocytes. On the other hand, two 50 μM treatments in a 72-h period decreased the survival of U251MG cells by 60%, while there was increased survival of U87MG cells (>3-fold) (Figure 8).
Figure 8.
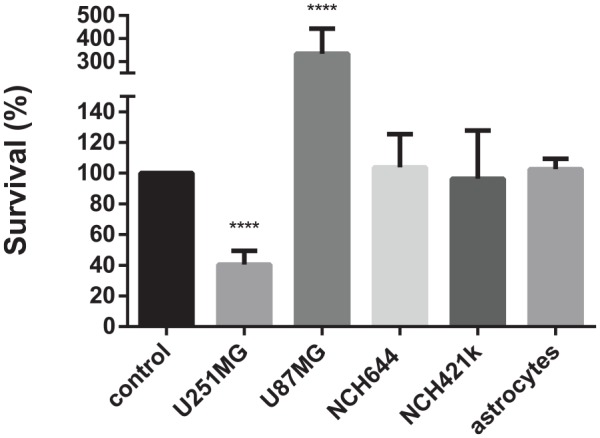
Effects of consecutive 50 μM temozolomide treatments (2 × 72 h) on cell survival of the cell lines (as indicated).
Data are means ± SD of three samples with three replicates (n = 9). ****p < 0.0001.
To test the effects of these consecutive treatments with nanobodies, seven combinations were tested: Nb79+Nb79, Nb179+Nb179, Nb225+Nb225, Nb314+Nb314, Nb79+Nb179, Nb179+Nb225 and Nb225+Nb314, as presented in Figure 9. These combinations were chosen based on the results of the previous single (Figure 6) and concurrent (Figure 7) treatments with nanobody pairs. Glioblastoma-related cells and astrocytes were exposed to these nanobodies for 72 h, and then the second nanobody was administrated and the incubations prolonged for another 72 h. These consecutive treatments with Nb79 decreased the survival of all the glioblastoma cell lines. U87MG cell survival decreased to 46%, and the survival of NCH644 and NCH421k cells decreased to 60%. The effects on U87MG, NCH644 and NCH421k cells were also greater than the effects of the separate and consecutive treatments with temozolomide (Table 9) and the single treatment with Nb79 and concurrent treatment with nanobody pairs with Nb79. Consecutive treatments with Nb225 also greatly decreased survival of all the cell lines. The survival of U251MG and U87MG cells was reduced to 76% and 50%, respectively. The survival of both glioblastoma stem cell lines was reduced significantly, to 22% (NCH644 cells) and 55% (NCH421k cells), and these effects on the cells were significantly higher than the effects of single or consecutive temozolomide treatments (Table 9). They were also again higher in NCH421k cells in comparison with the single treatments with Nb225 and concurrent treatments with nanobody pairs with Nb225. The consecutive treatments with Nb179 and Nb225 decreased the survival of the U87MG cells significantly, by 46%, and that of NCH644 cells by 45%. The effects on both these cell lines were significantly greater in comparison with those of temozolomide (Table 9), as well as of the separate Nb179 and Nb225 treatments, and the concurrent treatments with the Nb179+Nb225 pair. The Nb225 and Nb314 consecutive treatments significantly reduced survival of U251MG, U87MG and NCH644 cells by 30%, 32% and 42%, respectively. The suppressive effects on NCH644 cells were stronger than the effects of temozolomide alone, while the effects on both glioblastoma cell lines (U251MG, U87MG cells) were more pronounced than those of the separate applications of Nb225 or Nb314, and competitive with those of concurrent Nb225+Nb314 applications. All the consecutive nanobody combinations were cytotoxic for astrocytes to some degree. The least cytotoxic effects on astrocytes were seen when these cells were consecutively treated with Nb79, which reduced astrocyte survival by only 10%; conversely, the strongest suppressing effects were observed with the combination of Nb179 and Nb225, which reduced cell survival by 28%.
Figure 9.
Effects of consecutive treatment with the selected nanobodies (as indicated; 2 × 72 h) on cell survival of the cell lines (as indicated).
Data are means ± SD of three samples with three replicates (n = 9). *p < 0.05, **p < 0.01, *** p < 0.001, ****p < 0.0001.
Table 9.
Relative cell survivals after consecutive treatments with the nanobodies, in comparison with consecutive treatments with temozolomide.
| Consecutive nanobody pairs | U251MG |
U87MG |
NCH644 |
NCH421k |
Astrocytes |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sign. | p-value | Sign. | p-value | Sign. | p-value | Sign. | p-value | Sign. | p-value | |
| Nb79+Nb79 | * | 0.0229 | **** | 0.0001 | **** | 0.0001 | ** | 0.005 | *** | 0.0003 |
| Nb179+Nb179 | ** | 0.0023 | **** | 0.0001 | ** | 0.0031 | ns | 0.9699 | **** | 0.0001 |
| Nb225+Nb225 | ** | 0.0047 | **** | 0.0001 | **** | 0.0001 | *** | 0.0007 | **** | 0.0001 |
| Nb314+Nb314 | *** | 0.0001 | **** | 0.0001 | * | 0.0337 | ns | 0.2453 | **** | 0.0001 |
| Nb79+Nb179 | *** | 0.0007 | **** | 0.0001 | * | 0.0246 | ns | 0.968 | **** | 0.0001 |
| Nb179+Nb225 | ** | 0.0034 | **** | 0.0001 | **** | 0.0001 | ns | 0.9998 | **** | 0.0001 |
| Nb225+Nb314 | * | 0.0308 | **** | 0.0001 | **** | 0.0001 | ns | 0.9999 | **** | 0.0001 |
Significances of three samples with three replicates were calculated by one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
ANOVA, analysis of variance; Sign., significance.
Nanobodies induce cell apoptosis and necrosis
To gain some insight into the destructive effects of the selected nanobodies (i.e. Nb79, Nb179, Nb225, Nb314) on the cells (i.e. do the nanobodies induce apoptosis and/or necrosis?), apoptosis/necrosis assays were performed using Annexin V and propidium iodide staining. Annexin V-FITC (green) was used to detect the apoptotic cells, as it can bind to phosphatidylserine, and propidium iodide was used to detect necrotic cells, as it can bind to DNA (Abcam, ab14085).
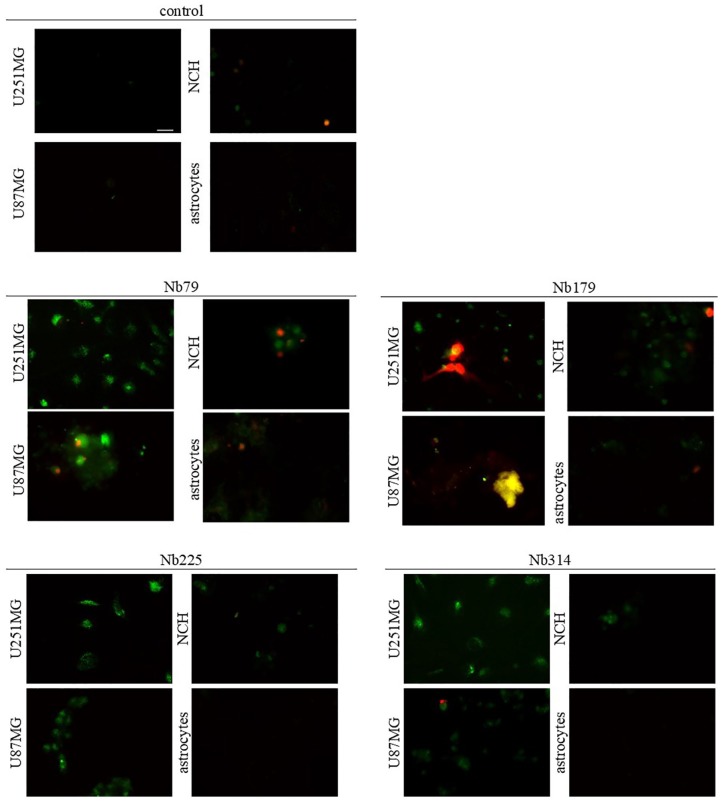
The cells were incubated with the nanobodies at 100 μg/mL for 24 h before analysis. These data show that Nb79 provoked apoptosis of the U251MG and U87MG cells, NCH co-cultures and astrocytes (Figure 10). Nb79 also induced necrosis of the NCH co-cultures and astrocytes. Nb179 induced apoptosis of the U251MG and U87MG cells and NCH co-cultures, and also caused extensive necrosis of the U251MG cells. Additionally, some apoptotic astrocytes were observed. Nb225 and Nb314 both induced apoptosis of the U251MG and U87MG cells and NCH co-cultures, while no necrosis was observed. None of the nanobodies induced apoptosis of the astrocytes after 24 h of incubation.
Figure 10.
Representative images for apoptosis/necrosis assays of treatments with the selected nanobodies (as indicated; 100 μg/mL, 24 h) of the cell lines (as indicated). Annexin V (green) shows apoptotic cells; propidium iodide was used to show necrotic cells. Scale bar, 20 μm (top left image; applicable to all images).
Nanobodies and cell migration
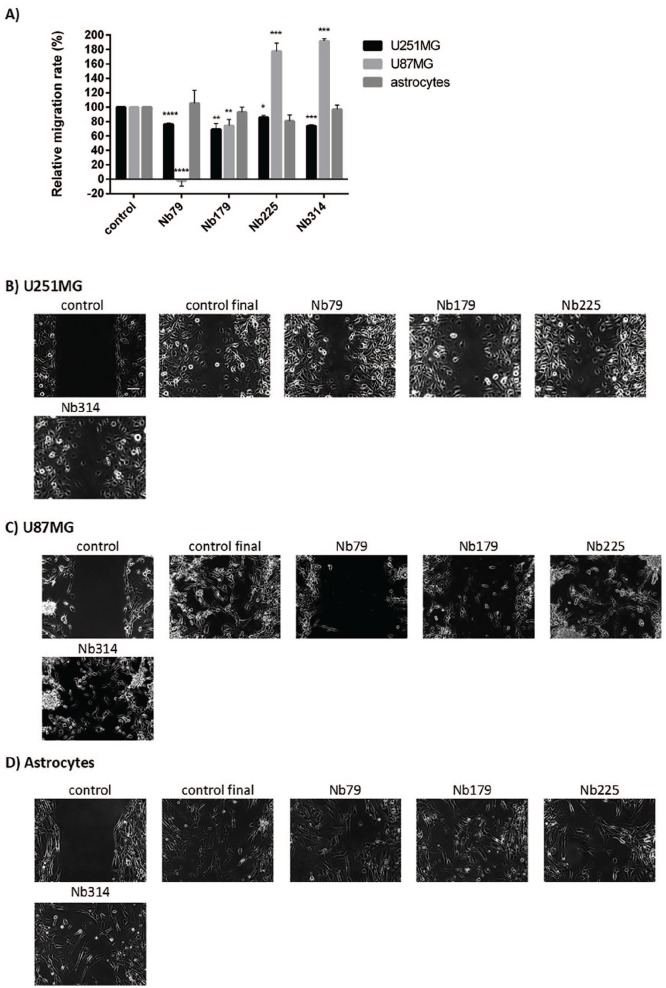
Cell migration is one of the essential characteristics of glioblastoma cells. To determine the possible effects of the nanobodies on cell migration, cell migration assays were carried out for the U251MG and U87MG cells and astrocytes in the presence of the four nanobodies Nb79, Nb179, Nb225 and Nb314. Silicone inserts with two wells (Ibidi) were used, and the cells were seeded into each well to form a wound. The inserts were then removed, and the nanobodies were added. Figure 11B–D presents the final stages of the cell migration, once the control wound of the untreated cells had overgrown. Figure 11A shows the relative rates of cell migration. All four of these nanobodies, Nb79, Nb179, Nb225 and Nb314, reduced the relative migration speed of the U251MG cells, with Nb179 having the greatest effects (31% reduction; p = 0.003); Nb79 reduced the migration speed of U251MG cells by 24% (p = 0.00002), Nb225 by 14% (p = 0.02), and Nb314 by 26% (p = 0.00002). The most prominent effects were observed on the U87MG cells with Nb79, which completely inhibited cell migration (p = 3.4 × 10–6). The other nanobody that decreased the migration speed of U87MG cells was Nb179 (22% reduction; p = 0.0097). Nb225, however, reduced the migration speed of astrocytes by 20%, although this did not reach statistical significance.
Figure 11.
Quantification (A) and representative images (B–D) for the effects of treatments with the selected nanobodies (as indicated; 100 μg/mL) on the migration speed of the cell lines (as indicated). (A) Single samples were analysed from images captured for two or three different positions. Data are means ± SD of three measurements.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (B–D) Scale bar, 100 μm (top left image; applicable to all images). Nanobody treatments were 7 h for U251MG cells, 24 h for U87MG cells and 18 h for astrocytes.
SD, standard deviation.
Discussion
Glioblastoma is one of the deadliest forms of cancer, as most of these patients die within 14 months of diagnosis.3 New therapies are therefore urgently required. In our previous studies, eight different antigens were identified as potential glioblastoma biomarkers/biomarker candidates: TRIM28, nucleolin, vimentin, NAP1L1, TUFM, DPYSL2, CRMP1 and ALYREF.21,22 The roles of these antigens in their molecular and cellular pathways under normal physiological conditions, as well as in cancer and glioma, are summarised in Table 10. All these antigens have previously been linked to various cancers, and some also to glioblastoma, usually with positive association with malignancy. The present study investigated their promise as targets for the design of new approaches to glioblastoma therapy. The first part of the present study focused on evaluation of the expression of novel antigens in glioblastoma cells and tissues: TRIM28, nucleolin, vimentin, NAP1L1, TUFM, DPYSL2, CRMP1 and ALYREF. These data show that NAP1L1 and CRMP1 were significantly overexpressed in glioblastoma stem cells versus astrocytes and differentiated glioblastoma cells (U251MG, U87MG cells) at the mRNA and protein levels, as was shown by qPCR and ELISA. Protein expression levels of DPYSL2 and ALYREF were also significantly higher in the glioblastoma stem cells versus the astrocytes and the U251MG and U87MG cells. Protein expression of vimentin in glioblastoma stem cells is also significantly higher than in astrocytes, as confirmed by ELISA and immunocytochemistry. TUFM has previously been linked to glioblastoma as TUFM protein expression was up-regulated in glioblastoma stem cells in relation to U251MG and U87MG cells, although in a study that did not include astrocytes as a reference cell line.38 Our data show that, in contrast to mRNA expression, in glioblastoma stem cells, TUFM protein expression is not higher than in any of the glioblastoma cell lines, compared with astrocytes.
Table 10.
Roles of the eight antigens investigated in this study. Literature review of the physiological roles of the antigens, and their roles in cancer and glioma.
| Antigen | Physiological role | Role in cancer | Role in glioma |
|---|---|---|---|
| TRIM28 | – E3 ubiquitin ligase30
- Gene repressor31 - Sumo E3 ligase32 - DNA repair31 |
– Cooperation with MDM2 to promote p53 ubiquitylation and degradation30
- Overexpression linked to growth promotion of non–small-cell lung cancer33 |
– Positively correlated with malignancy, and overexpression linked to poor survival34 |
| TUFM | – Mitochondrial elongation factor35
- Promotes autophagy35 |
– Overexpression in colorectal cancer36
- Potential prognostic marker in colorectal cancer37 |
– Up-regulated in glioblastoma cells38 |
| NUCL | – Regulation of Pol l transcription39
- Correct folding of pre-rRNA39 - Histone chaperone39 |
– Blocks apoptosis in cancer cells40
- Overexpression related to disease progression for ErbB2-positive breast cancer patients41 |
– Up-regulated in glioma cell line42
- Knockdown of NUCL gene in glioblastoma cell lines linked do decreased cell proliferation42 |
| VIM | – Type III intermediate filament43
- Role in early stages of brain development44 |
– Marker of epithelial to mesenchymal transition43
- Linked to invasive type of gastric carcinoma43 |
– High expression linked to poor survival45 |
| DPYSL2 | – Role in microtubule formation46
- Regulates dendrite and axon growth and retraction47 |
– Decreased expression in breast cancer48 | – Expression of CRMP2 and its phosphorylation promotes glioblastoma proliferation49 |
| CRMP1 | – Axon growth and branching during neuronal development50
- Microtubule dynamics51 |
– Decreases cell proliferation, migration and apoptosis in gastric cancer cells52
- Metastasis suppressor of prostate cancer50 |
– Loss of CRMP1 in EGFRvIII glioblastoma is linked to more aggressive phenotype53 |
| NAP1L1 | – Nucleosome assembly54
- Histone chaperone55 - Regulator of neuronal differentiation54 |
– Overexpression in small intestinal carcinoid56 | – Up-regulated in glioblastoma57 |
| ALYREF | – mRNA export (part of exon junction complex)58 | – Down-regulated in high grade tumours (thyroid, bladder, stomach)59
- Up-regulated in high grade serous ovarian carcinoma cancer cells and cancer stem cells60 |
– Up-regulated in glioblastoma22,61
- Overexpression linked to reduced survival61 - siRNA knockdown in U251 cells does not affect survival and proliferation61 |
ALYREF, Aly/REF export factor; CRMP1, collapsin response mediator protein 1; DPYSL2, dihydropyrimidinase-related protein 2; EGFRvIII, epidermal growth factor receptor variant III; MDM2, human murine double minute 2; NAP1L1, nucleosome assembly protein 1-like 1; NUCL, nucleolin; siRNA, small interfering RNA; TRIM28, tripartite motif containing 28 protein; TUFM, mitochondrial translation elongation factor (EF-TU); VIM, vimentin.
Knowledge of the cellular site of an antigen is important for the planning of any strategy of targeted treatment of oncological diseases. To detect the presence and cellular location of the selected antigens in various glioblastoma-related cells, we used immunofluorescence assays. The primary location of nucleolin was the nucleoli, as also determined by UNIPROT,62 while other locations, such as cytoplasmic (U251MG, U87MG cells, NCH co-cultures) and distinct membranes (U251MG cells), were also observed in the present study as seen in Figure 3. The unusual nucleolin localisation on the plasma membrane and in the cytoplasm, which was reported as characteristic for cancer cells, is associated with higher malignancy.63 The location of the second antigen, NAP1L1, was cytoplasmic, which is in agreement with the COMPARTMENT and PROTEINATLAS databases.64,65 The primary location of the next antigen, DPYSL2, was in the cytoplasm, as reported by UNIPROT; however, nuclear localisation was also seen in the present study (Figure 3).62 This finding correlates with previous observations of an unusual nuclear location of DPYSL2 in glioblastoma cells.49 Similarly, CRMP1 was located both in the cytoplasm and the nucleus. This unusual nuclear location of CRMP1 was already predicted by the COMPARTMENTS database, although with low confidence64; however, we have confirmed this for the first time experimentally.
To evaluate the clinical value of these antigens, we performed immunohistochemistry analysis. These data determined and compared protein expression of these novel biomarker candidates in normal brain, glioblastoma and lower-grade glioma tissues. Vimentin was not expressed in glia cells and neurons of normal brain, while on the other hand, blood vessels were positive (Figure 4). Vimentin expression in glioblastoma cells was significantly higher than in lower-grade glioma cells. Similar data were reported from Western blotting in a previous study by Jovčevska et al.22 It was also previously reported that vimentin is expressed in early brain development, and is replaced later by other cytoskeletal proteins.44 We thus propose that vimentin is an antigen that can differentiate between normal brain tissue, glioblastoma and lower-grade glioma. TUFM was similarly reported not to be expressed in normal brain tissue, which was confirmed in the present study; on the other hand, it was expressed in all glioblastoma tissues,38 which defines TUFM as an antigen that can also differentiate between normal human brain tissue and glioblastoma. DPYSL2 and CRMP1 were also not expressed in the glia cells and neurons of the normal brain tissue, but are on the other hand, expressed in neuropils (Figure 4). However, it should be noted that a relatively small number of samples were tested in the present study. For more accurate and further confirmation of antigen expression in any specific tumour, it will be necessary to use a larger number of tissue samples in a follow-up study.
Functional tests were performed to evaluate the possibility of specific targeting of the selected antigens, which would lay the foundation for the creation of new therapeutic approaches for glioblastoma. To this end, we used camelid nanobodies that were directed against these antigens that were produced in our previous studies.21,22 In addition to numerous advantages over conventional antibodies,11,13,14 nanobodies might cross the blood–brain barrier,17 penetrate into a tumour more rapidly, and have a more favorable tumour distribution.14 Also, due to their small size (15 kDa), they can easily reach hidden or cryptic targets; however, excess nanobodies are eliminated rapidly from the body through renal excretion.66 Due to their favorable characteristics, nanobodies have gained significant attention in both cancer imaging and therapies, especially nanobodies against human epidermal growth factor receptor 2 and epidermal growth factor, as well as nanobodies applied in immuno-oncology.15 The glioblastoma-associated cells (i.e. U251MG and U87MG cells, NCH co-cultures, and astrocytes) were treated with these nanobodies against the selected antigens at two concentrations. Cell survival was compared with the effects of temozolomide, a chemotherapeutic that is used widely for treating glioblastoma (as an alkylating agent).67 Temozolomide was used at 50 μM, which is the concentration predicted to enter the brain through an impaired blood–brain barrier.68 These data showed that temozolomide had no impact on astrocytes at any times, and poor effects on the U251MG and U87MG cells and NCH co-cultures. On the contrary, some of the nanobodies (i.e. Nb10, Nb79, Nb179) had detrimental effects on all types of glioblastoma cells tested. Nb79 and Nb179 also had significantly greater effects on U251MG glioblastoma cells than temozolomide alone (Table 8). However, at the higher concentrations, Nb10, Nb79 and Nb179 also significantly reduced survival of astrocytes. Therefore, a specific delivery system would be required to circumvent these effects on astrocytes, such as the use of a specifically designed carrier, or direct application to a tumour. For glioblastoma stem cells, the most promising data here were seen for Nb225, which, after prolonged exposure, significantly decreased survival of glioblastoma stem cells (i.e. NCH co-cultures). Thus Nb225 is a more potent cytotoxic agent than temozolomide, while also not affecting the survival of astrocytes.
In this study, for nanobodies tested for their effects on cell migration, none significantly reduced the migration rate of astrocytes, in contrast to their impact on glioblastoma cells (Figure 11). The most prominent effect was seen for the anti-vimentin nanobody Nb79, which completely inhibited the migration of U87MG cells, and decreased the migration rate of U251MG cells. Vimentin is one of the markers of epithelial-to-mesenchymal transition, a process commonly observed in cancers, where cells lose their epithelial phenotype and acquire migratory, mesenchymal characteristics.43 Withaferin A, a natural compound from the Withania somnifera plant, has been described as a vimentin-targeting compound that is cytotoxic and prevents metastasis of breast cancer.69 This study showed that the benefits of targeting vimentin might not only be reduced cell survival, but also inhibition of migration of glioblastoma cells.
Based on our initial findings qPCR, ELISA, immunocytochemistry and immunohistochemistry findings, four nanobodies were chosen for further studies: Nb79 (anti-VIM), Nb179 (anti-NAP1L1), Nb225 (anti TUFM) and Nb314 (anti-DPSYL2). At 10 μg/mL (Nb79, Nb179, Nb314) or 100 μg/mL (Nb225), these nanobodies did not show any significant effects on the survival of astrocytes, while all had cytotoxic effects on glioblastoma stem cells. Therefore, the cytotoxic effects of six different combinations of these nanobodies on the survival of the cells were determined after concurrent treatments: the cells were treated with two different, either co-operative or competing, nanobodies, and incubated for 72 h. None of the combinations had any significant cytotoxic effects on astrocyte survival. The combination of Nb79 and Nb179 had the most pronounced effects on the NCH644 and NCH421k glioblastoma stem cell lines, although this combination did not decrease the survival of the cells by more than 20%. In addition, Nb79 and Nb179 also reduced the survival of mature glioblastoma cell lines U87MG (by 25%) and U251MG (by 18%). The greatest destructive effects on the U87MG mature glioblastoma cell line were seen for the combination of Nb179 and Nb225, which decreased survival of U87MG cells by >30%, and U251 cells by ~20%, while the combination of Nb225 and Nb314 decreased the survival of both of these glioblastoma cell lines by ~25%. From these data, we can conclude that co-administration of nanobodies has, at least in most cases, better effects for the reduction of the survival of glioblastoma cells than separate use of these individual nanobodies. No differences were seen between glioblastoma stem cells and astrocytes. In this respect, the most effective co-administration was seen for the following nanobody pairs: Nb79 (anti-VIM)/Nb179 (anti NAP1L1); Nb179 (anti-NAP1L1)/Nb225 (anti-TUFM); and NB225 (anti-TUFM)/Nb314 (anti-DPSYL2). Good effects were also achieved with co-administration of Nb225 (anti-TUFM)/Nb79 (anti-VIM), with survival of U87MG cells decreased by >20%.
In the continuation of this study, we were interested in whether re-addition of the same or different nanobodies after long-term treatments can increase the cytotoxic effects of these nanobodies on glioblastoma cells (Figure 9). First, in this respect, a reference trial with temozolomide was performed. We found that two rounds of treatment with 50 μM temozolomide had no effects on glioblastoma stem cells (NCH421k, NCH644 cells) and astrocytes. It is interesting that temozolomide significantly reduced the survival of U251MG cells by 60%, while survival of U87MG cells was increased three-fold. This difference in sensitivities towards temozolomide can be attributed to differences between the individual cell lines that reflect the highly heterogeneous nature of glioblastoma tumours.70 However, it should be noted that survival detection was determined indirectly by measurement of the metabolic activities of the cells.71 Therefore, the higher metabolic activity of U87MG cells might explain higher signals when these cells were exposed to two rounds of temozolomide treatment.
Finally, the cytotoxic effects on cell survival of six different combinations of the four nanobodies (Nb79, Nb179, Nb225, Nb314) as consecutive treatments were determined. Thus, after the first incubation of the cells with the nanobodies at 10 μg/mL (Nb79, Nb179, Nb314) or 100 μg/mL (Nb225) for 72 h, a second incubation followed for a further 72 h with the same or other nanobody added. In general, the cytotoxic effects in all of these cases were stronger than for single nanobodies or concurrent treatments with two nanobodies. The most prominent effects here were from consecutive treatment with 10 μg/mL Nb79 (anti-VIM) and 100 μg/mL Nb225 (anti-TUFM). The first of these treatments (Nb79) reduced survival of U87MG cells by >50%, of U251MG cells by 40%, and of the two glioblastoma stem cell lines by 40%. The second, consecutive, treatment (Nb225) then reduced survival of the glioblastoma stem cell line by >50% for NCH421k cells, and by ~80% for NCH644 cells. With these stem cells, this combination treatment thus provided significantly higher destructive effects than temozolomide. This also confirmed the significant cytotoxic effects of the anti-TUFM nanobodies (Nb225) on the survival of glioblastoma cells, which has already been reported for single-dose treatment with an anti-TUFM nanobody on glioblastoma stem cells.38 Similar data have been reported for other sequential applications of nanobodies, where survival levels of the glioblastoma cells ranged from 50% to 80%, and of glioblastoma stem cells from 55% to 85%. To some extent, these consecutive treatments with nanobodies also affected astrocytes, with survival mainly at 80–90%. However, the destructive effects on glioblastoma cells, especially glioblastoma stem cells, was considerably higher than the cytotoxic effects seen with astrocytes.
Full-length immunoglobulin antibodies that bind specifically to the target molecule with high affinity have been widely developed as a research tool, as well as for diagnostic and therapeutic purposes, but, because of their large size and hydrophilicity, they usually cannot cross intact cell or subcellular membranes in living cells. The vast majority of disease-related molecules and protein–protein interactions occur mainly in the cytosol and/or internal compartments of cells, which is also true of all target antigens in this study. Unlike standard antibodies, nanobodies, because of their small size, can better penetrate tissues and inhibit molecular targets, such as enzymes, that are much less addressed by standard antibodies. Numerous studies have reported single-domain antibodies that could transmigrate across human brain endothelial cells in vitro,72–75 but no cell penetration mechanism has been described. In our previous study we demonstrated efficient cross-membrane cellular uptake of the His-tagged anti-TUFM nanobody; however, here again, the mechanism itself remained unanswered.38 Proteins taken up by endocytosis may be released from the endosomes by so-called ‘endosomal escape’.76 Studies of several groups have shown that histidines can provide additional pH-buffering by protonation of imidazole groups, facilitating endosomal escape.77–80 The nanobody, when bound to the target protein, can either act as an antagonist to prevent binding of a ligand, or as an allosteric inhibitor that alters the enzymatic activity of the protein.81 In this study, most nanobodies showed a greater or lesser cytotoxic effect depending on the type of target cell, while anti-TRIM28 nanobody Nb237 showed no effect on glioblastoma cells, which could rule out the potential impact of contaminants in nanobody preparations obtained for all Nbs in the same way. We did not use the other reference nanobody in this study, but further detailed validation of the results will certainly require a more convincing control nanobody, such as anti-GFP.
Any interpretation of the findings of the present study needs also to take into consideration the heterogenous nature of glioblastoma. Indeed, individual glioblastoma stem cells and mature glioblastoma cells differ in their sensitivities to temozolomide, and the same appears to be true when they are treated with the nanobodies. It should also be noted that the cell lines tested in the present study were commercially available cell lines. To provide better understanding of the impact of nanobodies on glioblastoma cells, primary cell lines isolated from patients would be a more appropriate model, because of their closer resemblance to the ‘real’ environment. The preparation of such cells is foreseen for further studies. In addition, this study will be further enhanced by testing the effects of these nanobodies on three-dimensional cell models, as these better resemble the true state of a tumour, and allow for a more ‘pathophysiological’ study of the penetration and anti-migration effects of these nanobodies. Also, the blood–brain barrier could be applied to study the potential penetration of nanobodies.
To translate nanobody therapies into glioblastoma treatments, and to circumvent astrocyte cytotoxicity, optimal treatment schemes will be required. One possibility would be the construction of tumour-stem-cell-specific delivery systems that take into consideration tumour-stem-cell-specific membrane markers.82–85 Alternatively, nanobody administration directly to the location of a tumour can be considered.86 Membrane-specific biomarkers in the field of glioblastoma research have been the subject of numerous studies in recent years, and efforts to discover new and even more specific biomarkers have been increasing.84,85 Therefore, expectations of great progress of their use in the development of efficient delivery systems appear to be justified.
Conclusion
In this study, we have shown that vimentin, NAP1L1, DPYSL2, CRMP1 and ALYREF have significantly higher expression in glioblastoma stem cells compared with astrocytes. The small-scale immunohistochemical study performed here has shown that, as a biomarker, vimentin can be used to differentiate between normal brain, glioblastoma and lower-grade glioma tissues, while TUFM, DPYSL2 and CRMP1 can be used to differentiate between glioblastoma and normal brain tissue. We suggest that nanobodies Nb79 (anti-vimentin), Nb179 (anti-NAP1L1), Nb225 (anti-TUFM) and Nb314 (anti-DPYSL2) are examined further for cell-targeting purposes. The anti-TUFM nanobody (Nb225), in particular, potently inhibited cell growth after long-term exposure of glioblastoma stem cells, with only minor effects on astrocytes. Further, the anti-vimentin nanobody (Nb79) can be considered for inhibition of cell migration. In addition, Nb79 was also potent in the reduction of glioblastoma and glioblastoma stem cell survival after long-term consecutive treatments, again with minor effects on astrocytes. However, to take these nanobodies forward into preclinical trials, and then ultimately to use them as therapies, optimal treatment schemes remain to be designed, or specific delivery systems to be constructed, to circumvent any cytotoxic effects on astrocytes.
Acknowledgments
The authors would like to thank Daniel Velkavrh (Institute of Pathology, Faculty of Medicine, University of Ljubljana, Slovenia) for technical support with the immunohistochemistry, Helena Klavžar (CFGBC, Faculty of Medicine, University of Ljubljana, Slovenia) for technical support with Agilent (RIN analysis), and Tea Tomšič (National Institute of Chemistry, Ljubljana) for technical support with the SEC analysis. The authors would also like to thank Christopher Berrie for scientific English editing of the manuscript.
Footnotes
Author contributions: A.Z., I.J., N.Š. were involved in the planning of the experimental work, and performed most of the experiments; R.K. was involved in the planning and supervision of the study; J.M. performed the immunohistochemistry; J.Š. and I.K. performed the cell migration assays; M.S.V. provided the chemotherapeutic agent; A.Z. drafted the manuscript. All of the authors have read, discussed and approved the final version of the manuscript.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Ethical statement: The tissue samples used in this study were anonymous formalin-fixed, paraffin-embedded (FFPE) archival tissues from which the patient identity was not apparent. In addition, the study was approved by the National Committee for Medical Ethics of the Republic of Slovenia in the form of Consents Nos. 0120-40/2018/4 and 0120-190/2018/4, part of which is a condition for informed patient consent prior to surgery and the study.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was financially supported by the P1-0104, P1-0207 and P1-0390 Research Programme Grants and a Junior Researcher Grant to A. Z. from the Slovenian Research Agency (ARRS) and the EC Interreg TRANS-GLIOMA project. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iDs: Alja Zottel  https://orcid.org/0000-0002-8587-3390
https://orcid.org/0000-0002-8587-3390
Ivana Jovčevska  https://orcid.org/0000-0002-0418-2986
https://orcid.org/0000-0002-0418-2986
Contributor Information
Alja Zottel, Medical Centre for Molecular Biology, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.
Ivana Jovčevska, Medical Centre for Molecular Biology, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.
Neja Šamec, Medical Centre for Molecular Biology, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.
Jernej Mlakar, Institute of Pathology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia.
Jernej Šribar, Department of Molecular and Biomedical Sciences, Jožef Stefan Institute, Ljubljana, Slovenia.
Igor Križaj, Department of Molecular and Biomedical Sciences, Jožef Stefan Institute, Ljubljana, Slovenia.
Marija Skoblar Vidmar, Department of Radiotherapy, Institute of Oncology Ljubljana, Ljubljana, Slovenia.
Radovan Komel, Medical Centre for Molecular Biology, Institute of Biochemistry, Faculty of Medicine, University of Ljubljana, Vrazov trg 2, 1000, Ljubljana, Slovenia.
References
- 1. Rock K, McArdle O, Forde P, et al. A clinical review of treatment outcomes in glioblastoma multiforme– the validation in a non-trial population of the results of a randomised phase III clinical trial: has a more radical approach improved survival? Br J Radiol. 2012; 85: e729–e733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 2010; 60: 166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miranda A, Blanco-Prieto M, Sousa J, et al. Breaching barriers in glioblastoma. Part I: molecular pathways and novel treatment approaches. Int J Pharm 2017; 531: 372–388. [DOI] [PubMed] [Google Scholar]
- 5. Jovcevska I, Kocevar N, Komel R. Glioma and glioblastoma - how much do we (not) know? Mol Clin Oncol 2013; 1: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lathia JD, Mack SC, Mulkearns-Hubert EE, et al. Cancer stem cells in glioblastoma. Genes Dev 2015; 29: 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012; 488: 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Revets H, De Baetselier P, Muyldermans S. Nanobodies as novel agents for cancer therapy. Expert Opin Biol Ther 2005; 5: 111–124. [DOI] [PubMed] [Google Scholar]
- 10. Chinot OL, Wick W, Mason W, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 2014; 370: 709–722. [DOI] [PubMed] [Google Scholar]
- 11. Arbabi Ghahroudi M, Desmyter A, Wyns L, et al. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett 1997; 414: 521–526. [DOI] [PubMed] [Google Scholar]
- 12. Dumoulin M, Conrath K, Van Meirhaeghe A, et al. Single-domain antibody fragments with high conformational stability. Protein Sci 2002; 11: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Debie P, Lafont C, Defrise M, et al. Size and affinity kinetics of nanobodies influence targeting and penetration of solid tumours. J Control Release 2020; 317: 34–42. [DOI] [PubMed] [Google Scholar]
- 14. Kijanka M, Dorresteijn B, Oliveira S, et al. Nanobody-based cancer therapy of solid tumors. Nanomedicine (Lond) 2015; 10: 161–174. [DOI] [PubMed] [Google Scholar]
- 15. Lecocq Q, De Vlaeminck Y, Hanssens H, et al. Theranostics in immuno-oncology using nanobody derivatives. Theranostics 2019; 9: 7772–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rissiek B, Koch-Nolte F, Magnus T. Nanobodies as modulators of inflammation: potential applications for acute brain injury. Front Cell Neurosci 2014; 8: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Bourgeois JP, Celli S, et al. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J 2012; 26: 3969–3979. [DOI] [PubMed] [Google Scholar]
- 18. Vosjan MJ, Vercammen J, Kolkman JA, et al. Nanobodies targeting the hepatocyte growth factor: potential new drugs for molecular cancer therapy. Mol Cancer Ther 2012; 11: 1017–1025. [DOI] [PubMed] [Google Scholar]
- 19. Zhou Z, Vaidyanathan G, McDougald D, et al. Fluorine-18 labeling of the HER2-targeting single-domain antibody 2Rs15d using a residualizing label and preclinical evaluation. Mol Imaging Biol 2017; 19: 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vincke C, Gutierrez C, Wernery U, et al. Generation of single domain antibody fragments derived from camelids and generation of manifold constructs. Methods Mol Biol 2012; 907: 145–176. [DOI] [PubMed] [Google Scholar]
- 21. Jovcevska I, Zupanec N, Kocevar N, et al. TRIM28 and beta-actin identified via nanobody-based reverse proteomics approach as possible human glioblastoma biomarkers. PLoS One 2014; 9: e113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jovcevska I, Zupanec N, Urlep Z, et al. Differentially expressed proteins in glioblastoma multiforme identified with a nanobody-based anti-proteome approach and confirmed by OncoFinder as possible tumor-class predictive biomarker candidates. Oncotarget 2017; 8: 44141–44158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004; 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 24. Grube S, Gottig T, Freitag D, et al. Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR. J Neurooncol 2015; 123: 35–42. [DOI] [PubMed] [Google Scholar]
- 25. Valente V, Teixeira SA, Neder L, et al. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol 2009; 10: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q, Ishikawa T, Michiue T, et al. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med 2012; 126: 943–952. [DOI] [PubMed] [Google Scholar]
- 28. Wang X, Spandidos A, Wang H, et al. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 2012; 40: D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011; 11: 792–804. [DOI] [PubMed] [Google Scholar]
- 31. Bunch H. Role of genome guardian proteins in transcriptional elongation. FEBS Lett 2016; 590: 1064–1075. [DOI] [PubMed] [Google Scholar]
- 32. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci 2017; 42: 297–311. [DOI] [PubMed] [Google Scholar]
- 33. Liu L, Zhao E, Li C, et al. TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 2013; 37: 71–78. [DOI] [PubMed] [Google Scholar]
- 34. Qi ZX, Cai JJ, Chen LC, et al. TRIM28 as an independent prognostic marker plays critical roles in glioma progression. J Neurooncol 2016; 126: 19–26. [DOI] [PubMed] [Google Scholar]
- 35. Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 2012; 13: 780–788. [DOI] [PubMed] [Google Scholar]
- 36. Shi H, Hood KA, Hayes MT, et al. Proteomic analysis of advanced colorectal cancer by laser capture microdissection and two-dimensional difference gel electrophoresis. J Proteomics 2011; 75: 339–351. [DOI] [PubMed] [Google Scholar]
- 37. Shi H, Hayes M, Kirana C, et al. TUFM is a potential new prognostic indicator for colorectal carcinoma. Pathology 2012; 44: 506–512. [DOI] [PubMed] [Google Scholar]
- 38. Samec N, Jovcevska I, Stojan J, et al. Glioblastoma-specific anti-TUFM nanobody for in-vitro immunoimaging and cancer stem cell targeting. Oncotarget 2018; 9: 17282–17299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol 2007; 17: 80–86. [DOI] [PubMed] [Google Scholar]
- 40. Abdelmohsen K, Gorospe M. RNA-binding protein nucleolin in disease. RNA Biol 2012; 9: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregorio AC, Lacerda M, Figueiredo P, et al. Meeting the needs of breast cancer: a nucleolin’s perspective. Crit Rev Oncol Hematol 2018; 125: 89–101. [DOI] [PubMed] [Google Scholar]
- 42. Xu Z, Joshi N, Agarwal A, et al. Knocking down nucleolin expression in gliomas inhibits tumor growth and induces cell cycle arrest. J Neurooncol 2012; 108: 59–67. [DOI] [PubMed] [Google Scholar]
- 43. Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011; 68: 3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quick Q, Paul M, Skalli O. Roles and potential clinical applications of intermediate filament proteins in brain tumors. Semin Pediatr Neurol 2015; 22: 40–48. [DOI] [PubMed] [Google Scholar]
- 45. Zhao J, Zhang L, Dong X, et al. High expression of vimentin is associated with progression and a poor outcome in glioblastoma. Appl Immunohistochem Mol Morphol 2018; 26: 337–344. [DOI] [PubMed] [Google Scholar]
- 46. Quach TT, Honnorat J, Kolattukudy PE, et al. CRMPs: critical molecules for neurite morphogenesis and neuropsychiatric diseases. Mol Psychiatry 2015; 20: 1037–1045. [DOI] [PubMed] [Google Scholar]
- 47. Quach TT AN, Lerch J, Honnorat J, et al. Cytoskeleton and CRMPs in neuronal morphogenesis and neurological diseases: potential targets for new therapies. J Neurol Disord 2016; 4: 1–8. [Google Scholar]
- 48. Shimada K, Ishikawa T, Nakamura F, et al. Collapsin response mediator protein 2 is involved in regulating breast cancer progression. Breast Cancer 2014; 21: 715–723. [DOI] [PubMed] [Google Scholar]
- 49. Moutal A, Villa LS, Yeon SK, et al. CRMP2 phosphorylation drives glioblastoma cell proliferation. Mol Neurobiol 2018; 55: 4403–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cai G, Wu D, Wang Z, et al. Collapsin response mediator protein-1 (CRMP1) acts as an invasion and metastasis suppressor of prostate cancer via its suppression of epithelial-mesenchymal transition and remodeling of actin cytoskeleton organization. Oncogene 2017; 36: 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neufeld G, Kessler O. The semaphorins: versatile regulators of tumour progression and tumour angiogenesis. Nat Rev Cancer 2008; 8: 632–645. [DOI] [PubMed] [Google Scholar]
- 52. Ren L, Li F, Di M, et al. MicroRNA-187 regulates gastric cancer progression by targeting the tumor suppressor CRMP1. Biochem Biophys Res Commun 2017; 482: 597–603. [DOI] [PubMed] [Google Scholar]
- 53. Mukherjee J, DeSouza LV, Micallef J, et al. Loss of collapsin response mediator Protein1, as detected by iTRAQ analysis, promotes invasion of human gliomas expressing mutant EGFRvIII. Cancer Res 2009; 69: 8545–8554. [DOI] [PubMed] [Google Scholar]
- 54. Qiao HM, Li YX, Feng C, et al. Nap1l1 Controls embryonic neural progenitor cell proliferation and differentiation in the developing brain. Cell Rep 2018; 22: 2279–2293. [DOI] [PubMed] [Google Scholar]
- 55. Marteijn JA, Lans H, Vermeulen W, et al. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014; 15: 465–481. [DOI] [PubMed] [Google Scholar]
- 56. Oberg K. Genetics and molecular pathology of neuroendocrine gastrointestinal and pancreatic tumors (gastroenteropancreatic neuroendocrine tumors). Curr Opin Endocrinol Diabetes Obes 2009; 16: 72–78. [DOI] [PubMed] [Google Scholar]
- 57. Heroux MS, Chesnik MA, Halligan BD, et al. Comprehensive characterization of glioblastoma tumor tissues for biomarker identification using mass spectrometry-based label-free quantitative proteomics. Physiol Genomics 2014; 46: 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Le Hir H, Sauliere J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol 2016; 17: 41–54. [DOI] [PubMed] [Google Scholar]
- 59. Dominguez-Sanchez MS, Saez C, Japon MA, et al. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer 2011; 11: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parida S, Chakraborty S, Maji RK, et al. Elucidating the gene regulatory networks modulating cancer stem cells and non-stem cancer cells in high grade serous ovarian cancer. Genomics. Epub ahead of print 6 February 2018. DOI: 10.1016/j.ygeno.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 61. Correa BR, de Araujo PR, Qiao M, et al. Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol 2016; 17: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017; 45: D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Berger CM, Gaume X, Bouvet P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015; 113: 78–85. [DOI] [PubMed] [Google Scholar]
- 64. Binder JX, Pletscher-Frankild S, Tsafou K, et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014; 2014: bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015; 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 66. Chakravarty R, Goel S, Cai W. Nanobody: the “magic bullet” for molecular imaging? Theranostics 2014; 4: 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol 2012; 5: 102–114. [DOI] [PubMed] [Google Scholar]
- 68. Beier D, Rohrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res 2008; 68: 5706–5715. [DOI] [PubMed] [Google Scholar]
- 69. Thaiparambil JT, Bender L, Ganesh T, et al. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int J Cancer 2011; 129: 2744–2755. [DOI] [PubMed] [Google Scholar]
- 70. Motaln H, Koren A, Gruden K, et al. Heterogeneous glioblastoma cell cross-talk promotes phenotype alterations and enhanced drug resistance. Oncotarget 2015; 6: 40998–41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Prabst K, Engelhardt H, Ringgeler S, et al. Basic colorimetric proliferation assays: MTT, WST, and resazurin. Methods Mol Biol. 2017; 1601: 1–17. [DOI] [PubMed] [Google Scholar]
- 72. Abulrob A, Sprong H, Van Bergen en, Henegouwen P, et al. The blood-brain barrier transmigrating single domain antibody: mechanisms of transport and antigenic epitopes in human brain endothelial cells. J Neurochem 2005; 95: 1201–1214. [DOI] [PubMed] [Google Scholar]
- 73. Muruganandam A, Tanha J, Narang S, et al. Selection of phage-displayed llama single-domain antibodies that transmigrate across human blood-brain barrier endothelium. FASEB J 2002; 16: 240–242. [DOI] [PubMed] [Google Scholar]
- 74. Jones DR, Taylor WA, Bate C, et al. A camelid anti-PrP antibody abrogates PrP replication in prion-permissive neuroblastoma cell lines. PLoS One 2010; 5: e9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rutgers KS, Nabuurs RJ, van den Berg SA, et al. Transmigration of beta amyloid specific heavy chain antibody fragments across the in vitro blood-brain barrier. Neuroscience 2011; 190: 37–42. [DOI] [PubMed] [Google Scholar]
- 76. Pei D, Buyanova M. Overcoming endosomal entrapment in drug delivery. Bioconjug Chem 2019; 30: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lachelt U, Kos P, Mickler FM, et al. Fine-tuning of proton sponges by precise diaminoethanes and histidines in pDNA polyplexes. Nanomedicine 2014; 10: 35–44. [DOI] [PubMed] [Google Scholar]
- 78. Kos P, Lachelt U, Herrmann A, et al. Histidine-rich stabilized polyplexes for cMet-directed tumor-targeted gene transfer. Nanoscale 2015; 7: 5350–5362. [DOI] [PubMed] [Google Scholar]
- 79. Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug Chem 1999; 10: 406–411. [DOI] [PubMed] [Google Scholar]
- 80. Bertrand E, Goncalves C, Billiet L, et al. Histidinylated linear PEI: a new efficient non-toxic polymer for gene transfer. Chem Commun (Camb) 2011; 47: 12547–12549. [DOI] [PubMed] [Google Scholar]
- 81. Jovcevska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs 2020; 34: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fakhoury M. Drug delivery approaches for the treatment of glioblastoma multiforme. Artif Cells Nanomed Biotechnol 2016; 44: 1365–1373. [DOI] [PubMed] [Google Scholar]
- 83. Kang YJ, Cutler EG, Cho H. Therapeutic nanoplatforms and delivery strategies for neurological disorders. Nano Converg 2018; 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Podergajs N, Motaln H, Rajcevic U, et al. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget 2016; 7: 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vidak M, Jovcevska I, Samec N, et al. Meta-analysis and experimental validation identified FREM2 and SPRY1 as new glioblastoma marker candidates. Int J Mol Sci 2018; 19: pii: E1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bastiancich C, Danhier P, Preat V, et al. Anticancer drug-loaded hydrogels as drug delivery systems for the local treatment of glioblastoma. J Control Release 2016; 243: 29–42. [DOI] [PubMed] [Google Scholar]