Abstract
Mechanical stimuli are known to modulate biological functions of cells and tissues. Recent studies have suggested that compressive stress alters growth plate cartilage architecture and results in growth modulation of long bones of children. To determine the role of compressive stress in bone growth, we created a microfluidic device actuated by pneumatic pressure, to dynamically (or statically) compress growth plate chondrocytes embedded in alginate hydrogel cylinders. In this article, we describe detailed methods for fabricating and characterizing this device. The advantages of our protocol are: 1) Five different magnitudes of compressive stress can be generated on five technical replicates in a single platform, 2) It is easy to visualize cell morphology via a conventional light microscope, 3) Cells can be rapidly isolated from the device after compression to facilitate downstream assays, and 4) The platform can be applied to study mechanobiology of any cell type that can grow in hydrogels.
Keywords: Bioengineering, Issue 151, Microfluidics, photolithography, soft lithography, growth plate chondrocytes, mechanobiology, cell mechanics, alginate hydrogel, confocal microscopy, image analysis
Introduction
Micro-engineered platforms are valuable tools for studying the molecular, cellular, and tissue level biology because they enable dynamic control of both the physical and chemical microenvironments1,2,3,4,5,6,7,8. Thus, multiple hypotheses can be simultaneously tested in a tightly controlled manner. In the case of growth plate cartilage, there are increasing evidences of an important role of compressive stress in modulating bone growth through action on the growth plate cartilage9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25. However the mechanism of action of compressive stress – in particular, how stress guides the formation of chondrocyte columns in the growth plate – is poorly understood.
The goal of this protocol is to create a pneumatically actuating microfluidic chondrocyte compression device26 to elucidate mechanisms of mechanobiology in growth plate chondrocytes (Figure 1a-c). The device consists of two parts: the pneumatic actuation unit and the alginate gel construct. The microfluidic pneumatic actuation unit is fabricated using polydimethylsiloxane (PDMS) based on the photo- and soft-lithography. This unit contains a 5 × 5 array of thin PDMS membrane balloons which can be inflated differently based on their diameters. The alginate gel construct consists of the chondrocytes embedded in a 5 × 5 array of alginate gel cylinders, and the entire alginate-chondrocyte constructs are assembled with the actuation unit. The alginate gel constructs are compressed by the pneumatically inflated PDMS balloons (Figure 1b). The microfluidic device can generate five different levels of compressive stress simultaneously in a single platform based on differences in the PDMS balloon diameter. Thus, a high-throughput test of chondrocyte mechanobiology under multiple compression conditions is possible.
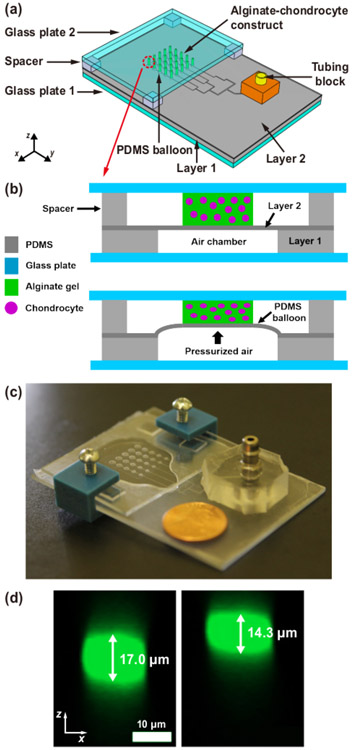
Figure 1. Microfluidic chondrocyte compression device.
(a) Schematic of the assembled device. A 5 × 5 array of alginate–chondrocyte constructs are aligned on PDMS balloons with 5 different diameters (D = 1.2, 1.4, 1.6, 1.8 and 2.0 mm), where D is the diameter of PDMS balloon (or air chamber). (b) Schematic of the device operation. The device is actuated by pneumatic pressure which expands PDMS balloons. (c) Image of an actual device (coin diameter = 19 mm). (d) Vertical cross-sections of a chondrocyte before (left) and under (right) compression on the largest PDMS balloon (D = 2.0 mm) (cell compressive strain, εcell = ∣cell height change/initial cell height∣ × 100 = 16%). This figure is reproduced from 26.
The microfluidic device described in this protocol has many advantages over the conventional compression device such as external fixators14,21,23 and macroscopic compression devices16,19,27,28 for studying chondrocyte mechanobiology: 1) The microfluidic device is cost effective because it consumes smaller volume of samples than the macroscopic compression device, 2) The microfluidic device is time effective because it can test multiple compression conditions simultaneously, 3) The microfluidic device can combine mechanical and chemical stimuli by forming a concentration gradient of chemicals based on the limited mixing in microchannels, and 4) Various microscopy techniques (time-lapse microscopy and fluorescence confocal microscopy) can be applied with the microfluidic device made of transparent PDMS.
We adopted and modified the method of Moraes et al.7,29 to create different compressive stress levels in a single device to enable high-throughput mechanobiology studies of chondrocyte compression. Our approach is appropriate for cells (e.g., chondrocytes) which need three-dimensional (3D) culture environment and for biological assays after compressing cells. Although some microfluidic cell compression devices can compress cells cultured on two-dimensional (2D) substrates30,31,32, they cannot be used for chondrocytes because 2D cultured chondrocytes dedifferentiate. There are microfluidic platforms for compressing 3D cultured cells in photopolymerized hydrogels7,33, but they are limited in isolating cells after compression experiments because isolating cells from photopolymerized hydrogel is not easy. Additionally, the effects of ultraviolet (UV) exposure and photo crosslinking initiators on cells may need to be evaluated. In contrast, our method allows rapid isolation of cells after compression experiments for post biological assays because alginate hydrogels can be depolymerized quickly by calcium chelators. The detailed device fabrication and characterization methods are described in this protocol. A brief procedure for fabricating the microfluidic chondrocyte compression device is shown in Figure 2.
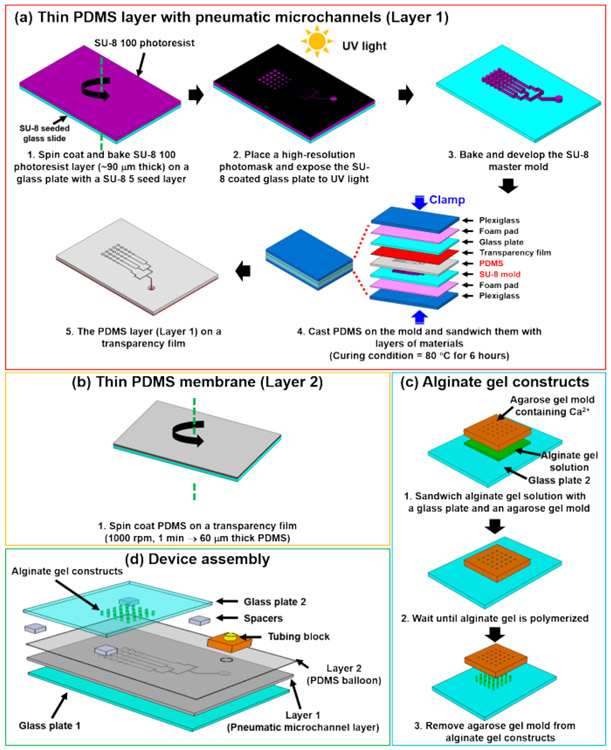
Figure 2. Detailed steps of microfluidic chondrocyte compression device fabrication.
(a) Photolithography for generating a SU-8 master mold and following soft lithography for creating PDMS layer with pneumatic microchannels (Layer 1). (b) Thin PDMS membrane (Layer 2) on a transparency film generated by spin coating. (c) Cylindrical alginate gel casting method on glass (Glass plate 2). (d) Assembly of the microfluidic chondrocyte compression device. This figure is reproduced from 26.
Protocol
NOTE: Wear personal protective equipment (PPE) such as gloves and lab coat for every step in this protocol.
1. Master mold fabrication
NOTE: Perform step 1.1 - 1.3 in a fume hood.
-
Glass treatment
NOTE: Wear a face shield, gloves, and a lab coat for step 1.1.-
Make Piranha solution (60 mL) by mixing sulfuric acid (H2SO4) and hydrogen peroxide (H2O2) with a volume ratio of 3:1.CAUTION: Do not use Piranha solution and acetone in the same fume hood due to the explosion hazard.
- Place a glass plate (50.8 mm × 76.2 mm × 1.2 mm) in Piranha solution for 30 min at 40 °C.
- Rinse the glass plate with deionized water (diH2O).
- Place the glass plate in acetone at room temperature for 10 min.
- Rinse the glass plate with isopropanol and dry it with nitrogen (N2) gas.
- Bake the glass plate at 200 °C for 20 min on a hot plate. All subsequent baking steps use a hot plate.
-
- SU-8 seed layer formation on the glass plate
- Spread SU-8 5 on the glass plate with a disposable pipette to cover around 2/3 of the plate’s surface area.
- Spin the glass plate with SU-8 5 photoresists at 500 rpm for 35 s (initial spinning cycle) and then 2,500 rpm for 40 s (final spinning cycle) using a spin coater. All subsequent spin coating steps include the same initial spinning cycle.
- Bake the SU-8 5 coated glass plate at 65 °C for 2 min and then at 95 °C for 5 min.
-
Expose the SU-8 5 coated glass plate to UV light (60 mW/cm2, distance between UV lamp and photomask is 20 cm, total amount of UV light energy = 60 mJ/cm2) for 1 s.NOTE: The UV exposure time should be adjusted according to the power of a used UV light.
- Bake the SU-8 5 coated glass plate at 65 °C for 2 min and at 95 °C for 5 min.
- Place the baked glass plate in the SU-8 developer for 2 min.
- Bake the SU-8 5 coated glass at 180 °C for 20 min.
- SU-8 channel pattern fabrication using photolithography (Figure 2a Step 1-3)
- Pour SU-8 100 on the SU-8 5 seeded glass plate to cover around 2/3 of the plate’s surface area.
- Spin the glass plate with SU-8 100 at 3,000 rpm for 38 s (≈ 90 μm thick).
- Bake the SU-8 100 coated glass plate at 65 °C for 10 min and then at 95 °C for 30 min. If SU-8 100 is still sticky after this procedure, bake the glass plate for a longer time at 95 °C until SU-8 100 becomes non-sticky.
-
Place a high-resolution microchannel photomask (25,400 dpi, see Supplementary Figure 1) on the SU-8 100 coated glass plate and expose the photomask covered glass plate to UV light for 4 s (total amount of UV light energy = 240 mJ/cm2).NOTE: The UV exposure time should be adjusted according to the power of a used UV light.
- Remove the photomask from the glass plate and bake the glass plate at 65 °C for 2 min and then 95 °C for 20 min.
- Keep the baked glass plate in a container, which is wrapped with aluminum foil to block any light, for overnight curing.
- Develop SU-8 100 channel patterns on the glass plate in the SU-8 developer for 15 min.
- Wash the SU-8 patterned glass plate (master mold) with isopropyl alcohol and dry it with N2 gas. If white particles remain during this process, repeat step 7 for 5 min.
2. Pneumatic actuation unit
-
Microchannel layer (Layer 1)
NOTE: Perform step 2.1.1 - 2.1.2 in a fume hood.- Drop 200 pL of (Tridecafluoro-1,1,2,2-Tetrahydrooctyl)-1-Trichlorosilane on a coverslip and place it in a vacuum chamber with the master mold.
- Apply vacuum for 2 min in the chamber and wait for 6 h (or overnight) for silanization of the mold.
-
Mix PDMS with a weight ratio of 10:1 (prepolymer:curing agent) for 5 min.NOTE: All subsequent PDMS casting step contains the same prepolymer and curing agent weight ratio (10:1).
- Pour PDMS on the master mold and degas PDMS in a vacuum chamber for 30 min.
- Sandwich PDMS with a piece of transparency film.
- Clamp the above sandwiched assembly with a glass plate, foam pads and plexiglass plates (Figure 2a Step 4).
- Cure PDMS in an oven at 80 °C for 6 h.
- Isolate the PDMS layer (Layer 1) with the transparency film from the sandwiched structure (Figure 2a Step 5).
- Activate surfaces of Layer 1 and a clean glass plate (Glass plate 1; 50.8 mm × 76.2 mm × 1.2 mm) using a plasma cleaner for 1 min. NOTE: Plasma cleaning time may vary based on the power of a used plasma cleaner.
- Bond Layer 1 onto Glass plate 1 and place them in the oven at 80 °C for 30 min.
- Remove the transparency film from Layer 1.
- Thin PDMS membrane (Layer 2)
- Spin coat uncured PDMS on a transparency film at 1,000 rpm for 1 min to obtain a 60 μm thick PDMS layer.
- Partially cure spin coated PDMS (Layer 2) in the oven at 80 °C for 20-30 min.
-
Activate Layer 1 on Glass plate 1 and Layer 2 using the plasma cleaner for 1 min.NOTE: Plasma cleaning time may vary based on the power of a used plasma cleaner.
- Bond the Layer 2 onto Layer 1 and place them in the oven at 80 °C overnight.
- Tubing block
- Place metal tubes vertically on a petri dish.
- Gently pour PDMS in the dish to submerge about 3/4 of the metal tubes.
- Cure PDMS in the oven at 60 °C for 6 h (or overnight).
- Cut a piece of PDMS block containing one metal tube.
- Punch a hole on Layer 2 for the inlet.
- Activate the PDMS block and Layer 2 using the plasma cleaner for 1 min.
- Attach the PDMS block onto the inlet part of Layer 2 and place the entire actuation unit in the oven at 80 °C for overnight.
3. Alginate-chondrocyte (or bead) constructs
- Amino-silanized glass plate
- Cut a glass plate (50.8 mm × 76.2 mm × 1.2 mm) into two half-sized glass plates (50.8 mm × 38.1 mm × 1.2 mm) using a diamond scriber.
- Place the glass plates in 0.2 M hydrogen chloride (HCl) and gently shake (e.g., 55 rpm) them overnight.
- Rinse the glass plates with diH2O.
- Shake the glass plates in 0.1 M sodium hydroxide (NaOH) for 1 h at 55 rpm and rinse them with diH2O.
- Shake the glass plates in 1 % (v/v) 3-aminopropyltrimethoxysilane (APTES) for 1 h at 55 rpm and rinse them with diH2O.
- Dry the amino-silanized glass plates in a fume hood overnight.
- Agarose gel mold for alginate gel constructs
- Mix 5% (w/v) agarose and 200 mM calcium chloride (CaCl2) in diH2O.
- Boil the agarose gel solution with a microwave oven (or a hot plate). Boiling time varies based on the volume of the agarose gel solution and the power of the microwave oven (or the temperature of the hot plate).
- Pour the boiled agarose gel solution onto an aluminum mold (see Supplementary Figure S2), and sandwich it with a glass plate.
- Wait for 5 min and unmold the solidified agarose gel from the aluminum mold.
- Growth plate chondrocyte harvest
- Isolate growth plates from the hind limbs of neonatal mice.
- Place the growth plates in 1 mL of 0.25% collagenase for 3 h in an incubator at 37 °C and 8% carbon dioxide (CO2), to remove extracellular matrix (ECM).
- Centrifuge the digested sample for 5 min at 125 x g to make a chondrocyte pellet and remove supernatant from the sample. Here, 1 x g is the acceleration of gravity.
- Resuspend the chondrocyte pellet in 1 mL of Dulbecco's modified Eagle's medium (DMEM).
- Count the number of chondrocytes in DMEM using a cell counter.
- Centrifuge the chondrocytes in DMEM for 5 min at 125 x g to make a chondrocyte pellet again and remove supernatant from the sample.
- Optionally, resuspend the chondrocyte pellet in the chondrocyte culture media (CCM)34 containing 2 μM calcein AM, and incubate the sample at 37 °C for 30 min. Repeat step 3.3.6 and move to step 3.3.8.
- Resuspend the chondrocyte pellet in a desired volume of CCM and keep them in the incubator at 37 °C and 8% C02 before use.
- Alginate-chondrocyte (or bead) constructs (Figure 2c)
- Mix 1.5% (w/v) alginate in phosphate-buffered saline (PBS) with 5 mg/mL of sulfo-NHS, 10 mg/mL of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC).
- Add 8 × 106 chondrocytes into the 1 mL of alginate gel solution [or Add 3 μL of 1 μm-diameter fluorescent beads (542/612 nm) in 1 mL of alginate gel solution (0.3% (v/v))].
- Place 150 μL of the alginate-chondrocyte (or bead) solution on the amino-silanized glass plate fabricated in Step 3.1.
- Cover the alginate gel solution with the agarose gel mold for 3 min.
- Remove excessive alginate gel solution overflowing from the agarose gel mold with a razor blade and remove the agarose gel mold. Then, cylindrical alginate-chondrocyte (or bead) constructs are obtained on the amino-silanized glass plate.
- Place the alginate-chondrocyte constructs in cross-linking solution (50 mM CaCl2 / 140 mM NaCl in diH2O) for 1 min for further polymerization.
4. Device assembly (Figure 2d)
NOTE: PDMS spacers and 3D printed clamps need to be prepared separately.
Locate four 1 mm thick PDMS spacers on the four corners of the Layer 2 of the actuation unit.
Place 700 μL of CCM to cover the air chambers of Layer 2.
Place the alginate-chondrocyte (or bead) constructs on Layer 2 while carefully aligning the constructs with the air chambers.
Clamp the device with 3D printed clamps (Figure 1c).
5. Actuation of the device
Connect the outlet of an air pump (see Table of Materials) with the inlet of a solenoid valve with a silicon tubing.
Connect the outlet of the solenoid valve with the inlet of the assembled device with a silicon tubing.
Connect the solenoid valve with a function generator.
Manipulate the solenoid valve with a square wave (e.g., 1 Hz) generated by the function generator.
Turn on the air pump to actuate the device pneumatically.
6. Imaging of chondrocytes in the device
NOTE: To obtain a good image quality, image chondrocytes (or fluorescent beads) in alginate gel through Glass plate 2 because expanded PDMS balloons and air chambers can distort optical images. If an inverted microscope is used for imaging, the device needs to be setup so that Glass plate 2 faces downward.
Prepare the device with chondrocytes (or fluorescent beads) as shown in previous sections.
Take z-stack images of chondrocytes (or fluorescent beads) with a confocal microscope before and under compression, respectively. Choose a z-step size based on the depth of field of a used confocal imaging system.
The height of chondrocytes (or an alginate-bead construct) can be measured with automatic image processing method shown in previous literatures26,35.
Representative Results
This article shows detailed steps of the microfluidic chondrocyte compression device fabrication (Figure 2). The device contains a 5 × 5 arrays of cylindrical alginate-chondrocyte constructs, and these constructs can be compressed with five different magnitudes of compression (Figure 1, Figure 3 and Figure 4). The height of the pneumatic microchannel is around 90 μm, and the PDMS balloon diameters are 1.2, 1.4, 1.6, 1.8 and 2.0 mm, respectively. The performance of the device was quantified with confocal microscopy with static compression conditions and image processing. Static compression was employed for the microscopic imaging because the z-stack imaging with the confocal microscopy takes a few minutes, so it is too slow for quantitative imaging during the dynamic compression.
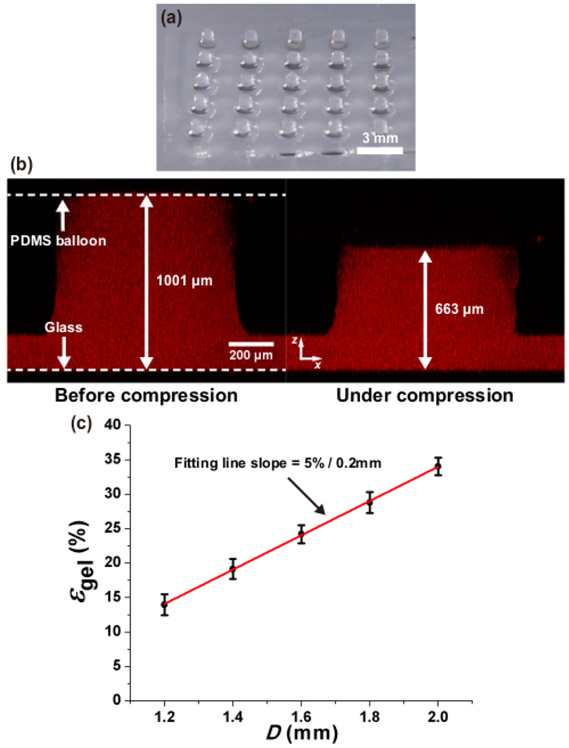
Figure 3. Measurement of alginate gel deformation under static compression.
(a) 5 × 5 arrays of cylindrical alginate gel constructs (diameter: ~800 μm, height: ~1 mm). (b) Alginate gel compressed by the largest PDMS balloon (D = 2.0 mm). The compressive strain of the alginate gel is 33.8%. (c) Compressive strain of alginate gel (εgel) increases around 5% per 0.2 mm increment of PDMS balloon diameter (D). Error bar: standard deviation. Red line: linear fitting line This figure is reproduced from 26.
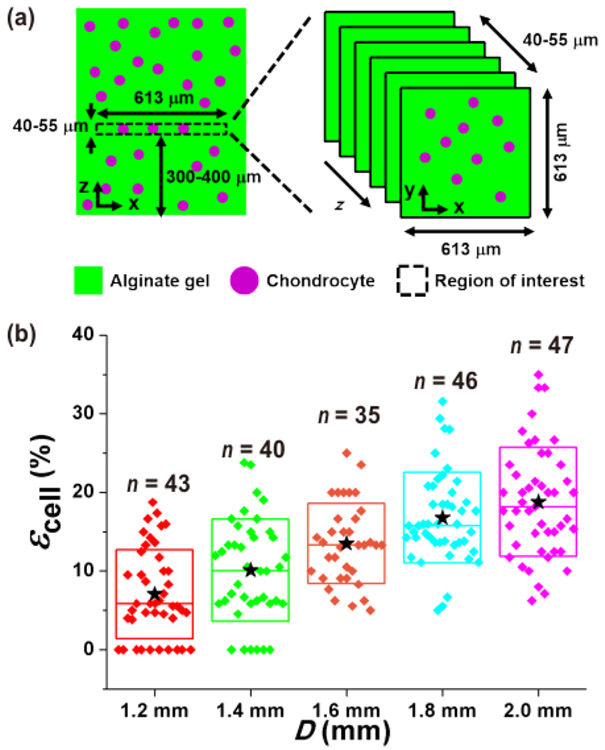
Figure 4. Measurement of chondrocyte deformation under static compression.
(a) A z-stack image [613 μm × 613 μm × 40–55 μm (x × y × z)] was obtained in the middle of the gel construct, 300–400 μm from the gel bottom. (b) Different magnitudes of chondrocyte compressive strain (εcell) resulted as a function of the PDMS balloon diameter (D). ★: mean values. ◆: each data points. Top (or bottom) and middle lines of the box are the standard deviation and median value, respectively. This figure is reproduced from 26.
Figure 3a shows the 5 × 5 arrays of alginate hydrogel columns (diameter: ~0.8 mm, height: ~1 mm) cast on Glass plate 2. These gel constructs were imaged by adding fluorescent beads in the gel. Figure 3b shows an example case that the gel column was compressed by 33.8% in height by the largest PDMS balloon. The resultant compressive strain of the gel constructs increased by approximately 5% per 0.2 mm increment in the PDMS balloon diameter as shown in Figure 3c.
Compressive deformation of chondrocytes was determined by imaging the cells in a 613 μm × 613 μm × 40–55 μm (x × y × z) volume near the gel construct center as shown in Figure 4a. Figure 1d shows an example image of a chondrocyte that was compressed by 16% by the largest PDMS balloon. Figure 4b shows the distribution of the measured cell compression strain values, and overall cells were compressed more by larger PDMS balloons. Therefore, the amount of alginate gel and chondrocyte compression were controlled by the diameter of PDMS balloons (Figure 3 and Figure 4) with a constant pressure of 14 kPa.
Discussion
To test the effects of compressive stress on growth plate chondrocytes, we developed the microfluidic chondrocyte compression device (Figure 1) to apply various levels of compressive stress to the chondrocytes in the alginate hydrogel scaffold for 3D culture in high throughput ways. To assist other researchers to adopt our device or to develop similar devices, we provided details of the device fabrication steps in this protocol article.
The crucial steps in this protocol are 1) fabricating PDMS layer with pneumatic microchannels (Layer 1) without any air bubbles since air bubbles in Layer 1 may damage pneumatic microchannels while the backing transparency film is peeled off, 2) maintaining a constant temperature (e.g., 80 °C) for curing PDMS balloons (Layer 2) because the elasticity of PDMS is known to depend on the curing temperature36, 3) aligning the alginate gel constructs with the PDMS balloons, and 4) using fresh amino-silanized glass plate (Glass plate 2) for bonding the alginate gel columns on Glass plate 2 within two days after the salinization treatment.
The major limitation of this protocol is that it is relatively labor intensive to fabricate the device because the process involves photolithography and multiple steps of soft lithography. Additionally, the performance of microfluidic cell compression devices fabricated based on our protocol needs to be evaluated whenever different types of hydrogels and cells are used. This is because any differences in the mechanical properties of hydrogels and cells will affect device performance.
Although our microfluidic cell compression device is for applying dynamic compression to chondrocytes, its performance was evaluated by imaging statically compressed alginate gels and cells. This is because it was hard to image gels and cells under dynamic compression with the conventional confocal microscopy. We compared static (14 kPa, 1 h) and dynamic compression (14 kPa, 1 Hz, 1 h) in terms of the permanent deformation of alginate gel and found that the permanent deformation of the gel under the dynamic compression was negligible compared to the static compression (see Supplementary Figure S3).
One advantage of our method is that it can be used for other cell types which need 3D culture environment. The resultant compression of the device can be modulated depending on applications by changing the diameter and thickness of the PDMS balloons and/or the pressure in the balloon. It is also possible to modify the elasticity of the PDMS balloon by adjusting the mixing ratio between the prepolymer and the curing agent. Cells in this device can be imaged in real time using light/fluorescence microscopy, and the device can be rapidly disassembled for cell harvest to enable downstream analysis. Another advantage is the ability to generate five distinct mechanical stress levels with five technical replicates per each stress level using a single device. Combining replication and a dose-response analysis ensures a high degree of rigor and reproducibility in the results.
Supplementary Material
Figure S1. MicroChannel photomask design for step 1.3.4 (unit = mm).
Figure S2. Aluminum mold design for step 3.2.3 (unit = mm).
Figure S3. Permanent deformation of the alginate gel (1.5%, w/v) under 1 h-long dynamic (1 Hz) and static compression. This figure is reproduced from 26.
Acknowledgments
We thank Drs. Christopher Moraes and Stephen A. Morin for their support for device design and fabrication. This study was supported by Bioengineering for Human Health grant from the University of Nebraska-Lincoln (UNL) and the University of Nebraska Medical Center (UNMC), and grant AR070242 from the NIH/NIAMS. We thank Janice A. Taylor and James R. Talaska of the Advanced Microscopy Core Facility at the University of Nebraska Medical Center for providing assistance with confocal microscopy.
Footnotes
Disclosures
The authors have nothing to disclose.
Video Link
The video component of this article can be found at https://www.jove.com/video/59676/
References
- 1.Lu H et al. Microfluidic shear devices for quantitative analysis of cell adhesion. Analytical Chemistry. 76 (18), 5257–5264, (2004). [DOI] [PubMed] [Google Scholar]
- 2.Malek AM, Izumo S Mechanism of endothelial cell shape change and cytoskeletal remodeling in response to fluid shear stress. Journal of Cell Science. 109 (4), 713–726, (1996). [DOI] [PubMed] [Google Scholar]
- 3.Paguirigan AL, Beebe DJ Microfluidics meet cell biology: bridging the gap by validation and application of microscale techniques for cell biological assays. BioEssays. 30 (9), 811–821, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Cardeña G, Comander J, Anderson KR, Blackman BR, Gimbrone MA Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proceedings of the National Academy of Sciences. 98 (8), 4478–4485, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh D et al. Reconstituting organ-level lung functions on a chip. Science. 328 (5986), 1662–1668, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moraes C, Chen J-H, Sun Y, Simmons CA Microfabricated arrays for high-throughput screening of cellular response to cyclic substrate deformation. Lab on a Chip. 10 (2), 227–234, (2010). [DOI] [PubMed] [Google Scholar]
- 7.Moraes C, Wang G, Sun Y, Simmons CA A microfabricated platform for high-throughput unconfined compression of micropatterned biomaterial arrays. Biomaterials. 31 (3), 577–584, (2010). [DOI] [PubMed] [Google Scholar]
- 8.Sundararaghavan HG, Monteiro GA, Firestein BL, Shreiber DI Neurite growth in 3D collagen gels with gradients of mechanical properties. Biotechnology and Bioengineering. 102 (2), 632–643, (2009). [DOI] [PubMed] [Google Scholar]
- 9.Bougault C, Paumier A, Aubert-Foucher E, Mallein-Gerin F Molecular analysis of chondrocytes cultured in agarose in response to dynamic compression. BMC Biotechnology. 8 (1), 71, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaviani R, Londono I, Parent S, Moldovan F, Villemure I Compressive mechanical modulation alters the viability of growth plate chondrocytes in vitro. Journal of Orthopaedic Research. 33 (11), 1587–1593, (2015). [DOI] [PubMed] [Google Scholar]
- 11.Ménard A-L et al. In vivo dynamic loading reduces bone growth without histomorphometric changes of the growth plate. Journal of Orthopaedic Research. 32 (9), 1129–1136, (2014). [DOI] [PubMed] [Google Scholar]
- 12.Robling AG, Duijvelaar KM, Geevers JV, Ohashi N, Turner CH Modulation of appositional and longitudinal bone growth in the rat ulna by applied static and dynamic force. Bone. 29 (2), 105–113, (2001). [DOI] [PubMed] [Google Scholar]
- 13.Sergerie K et al. Growth plate explants respond differently to in vitro static and dynamic loadings. Journal of Orthopaedic Research. 29 (4), 473–480, (2011). [DOI] [PubMed] [Google Scholar]
- 14.Valteau B, Grimard G, Londono I, Moldovan F, Villemure I In vivo dynamic bone growth modulation is less detrimental but as effective as static growth modulation. Bone. 49 (5), 996–1004, (2011). [DOI] [PubMed] [Google Scholar]
- 15.Walsh AJL, Lotz JC Biological response of the intervertebral disc to dynamic loading. Journal of Biomechanics. 37 (3), 329–337, (2004). [DOI] [PubMed] [Google Scholar]
- 16.Zimmermann EA et al. In situ deformation of growth plate chondrocytes in stress-controlled static vs dynamic compression. Journal of Biomechanics. 56, 76–82, (2017). [DOI] [PubMed] [Google Scholar]
- 17.Akyuz E, Braun JT, Brown NAT, Bachus KN Static versus dynamic loading in the mechanical modulation of vertebral growth. Spine. 31 (25), E952–E958, (2006). [DOI] [PubMed] [Google Scholar]
- 18.Alberty A, Peltonen J, Ritsilä V Effects of distraction and compression on proliferation of growth plate chondrocytes: A study in rabbits. Acta Orthopaedica Scandinavica. 64 (4), 449–455, (1993). [DOI] [PubMed] [Google Scholar]
- 19.Amini S, Veilleux D, Villemure I Tissue and cellular morphological changes in growth plate explants under compression. Journal of Biomechanics. 43 (13), 2582–2588, (2010). [DOI] [PubMed] [Google Scholar]
- 20.Aronsson DD, Stokes IAF, Rosovsky J, Spence H Mechanical modulation of calf tail vertebral growth: implications for scoliosis progression. Journal of Spinal Disorders. 12 (2), 141–146, (1999). [PubMed] [Google Scholar]
- 21.Cancel M, Grimard G, Thuillard-Crisinel D, Moldovan F, Villemure I Effects of in vivo static compressive loading on aggrecan and type II and X collagens in the rat growth plate extracellular matrix. Bone. 44 (2), 306–315, (2009). [DOI] [PubMed] [Google Scholar]
- 22.Reich A et al. Weight loading young chicks inhibits bone elongation and promotes growth plate ossification and vascularization. Journal of Applied Physiology. 98 (6), 2381–2389, (2005). [DOI] [PubMed] [Google Scholar]
- 23.Stokes IA, Mente R L., latridis JC, Farnum CE, Aronsson DD Enlargement of growth plate chondrocytes modulated by sustained mechanical loading. Journal of Bone and Joint Surgery. 84 (10), 1842–1848, (2002). [DOI] [PubMed] [Google Scholar]
- 24.Stokes IAF, Clark KC, Farnum CE, Aronsson DD Alterations in the growth plate associated with growth modulation by sustained compression or distraction. Bone. 41 (2), 197–205, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee D, Erickson A, Dudley AT, Ryu S Mechanical stimulation of growth plate chondrocytes: previous approaches and future directions. Experimental Mechanics. 10.1007/s11340-018-0424-1, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D, Erickson A, You T, Dudley AT, Ryu S Pneumatic microfluidic cell compression device for high-throughput study of chondrocyte mechanobiology. Lab on a Chip. 18 (14), 2077–2086, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilak F Compression-induced changes in the shape and volume of the chondrocyte nucleus. Journal of Biomechanics. 28 (12), 1529–1541, (1995). [DOI] [PubMed] [Google Scholar]
- 28.Knight MM, Ghori SA, Lee DA, Bader DL Measurement of the deformation of isolated chondrocytes in agarose subjected to cyclic compression. Medical Engineering & Physics. 20 (9), 684–688, (1998). [DOI] [PubMed] [Google Scholar]
- 29.Moraes C, Sun Y, Simmons CA Microfabricated platforms for mechanically dynamic cell culture. Journal of Visualized Experiments. doi: 10.3791/2224 (46), e2224, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim WY et al. A pneumatic micro cell chip for the differentiation of human mesenchymal stem cells under mechanical stimulation. Lab on a Chip. 7 (12), 1775–1782, (2007). [DOI] [PubMed] [Google Scholar]
- 31.Hosmane S et al. Valve-based microfluidic compression platform: single axon injury and regrowth. Lab on a Chip. 11 (22), 3888–3895, (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ho KKY, Wang YL, Wu J, Liu AP Advanced microfluidic device designed for cyclic compression of single adherent cells. Frontiers in Bioengineering and Biotechnology. 6 (148), (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo J et al. Interconnectable dynamic compression bioreactors for combinatorial screening of cell mechanobiology in three dimensions. ACS Applied Materials & Interfaces. 10 (16), 13293–13303, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erickson AG et al. A tunable, three-dimensional in Vitro culture model of growth plate cartilage using alginate hydrogel acaffolds. Tissue Engineering Part A. 24 (1-2), 94–105, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee D, Rahman MM, Zhou Y, Ryu S Three-dimensional confocal microscopy indentation method for hydrogel elasticity measurement. Langmuir. 31 (35), 9684–9693, (2015). [DOI] [PubMed] [Google Scholar]
- 36.Johnston ID, McCluskey DK, Tan CKL, Tracey MC Mechanical characterization of bulk Sylgard 184 for microfluidics and microengineering. Journal of Micromechanics and Microengineering. 24 (3), 035017, (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. MicroChannel photomask design for step 1.3.4 (unit = mm).
Figure S2. Aluminum mold design for step 3.2.3 (unit = mm).
Figure S3. Permanent deformation of the alginate gel (1.5%, w/v) under 1 h-long dynamic (1 Hz) and static compression. This figure is reproduced from 26.