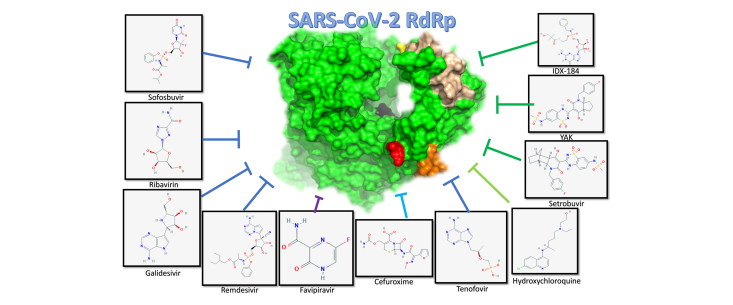
Abstract
New treatment against SARS-CoV-2 now is a must. Nowadays, the world encounters a huge health crisis by the COVID-19 viral infection. Nucleotide inhibitors gave a lot of promising results in terms of its efficacy against different viral infections. In this work, molecular modeling, docking, and dynamics simulations are used to build a model for the viral protein RNA-dependent RNA polymerase (RdRp) and test its binding affinity to some clinically approved drugs and drug candidates. Molecular dynamics is used to equilibrate the system upon binding calculations to ensure the successful reproduction of previous results, to include the dynamics of the RdRp, and to understand how it affects the binding. The results show the effectiveness of Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, Cefuroxime, Tenofovir, and Hydroxychloroquine, in binding to SARS-CoV-2 RdRp. Additionally, Setrobuvir, YAK, and IDX-184, show better results, while four novel IDX-184 derivatives show promising results in attaching to the SARS-CoV-2 RdRp. There is an urgent need to specify drugs that can selectively bind and subsequently inhibit SARS-CoV-2 proteins. The availability of a punch of FDA-approved anti-viral drugs can help us in this mission, aiming to reduce the danger of COVID-19. The compounds 2 and 3 may tightly bind to the SARS-CoV-2 RdRp and so may be successful in the treatment of COVID-19.
Keywords: COVID-19, SARS-CoV-2, RdRp, molecular docking, molecular dynamics simulation, drug repurposing
Graphical Abstract
Communicated by Ramaswamy H. Sarma
Introduction
Late in December 2019, a rapid outbreak of a mysterious coronavirus, identified later as SARS-CoV-2, emerged in the city of Wuhan, China (Bogoch et al., 2020; Hui et al., 2020; Rothan & Byrareddy, 2020). Any traveler to Wuhan City in Hubei Province 2 weeks before the onset of the symptoms of pneumonia (termed COVID-19) is suspected to be infected with the novel coronavirus (Bogoch et al., 2020; Organization, 2020a, 2020b, 2020c). The SARS-CoV-2 is suggested to be originated in the wildlife when bat translates the virus to a secondary host that, by his role, transmits the virus to the human being by direct contact in the Wuhan market (Hui et al., 2020). The National Health Commission of China confirmed, for the first time, the human-to-human transmission of the COVID-19 in late January 2020 (Yang, 2020). WHO declared COVID-19 as a Public Health Emergency of International Concern (PHEIC), then as a pandemic in March 2020 (Ibrahim et al., 2020). There is now a total lockdown in many countries, attempting to reduce the spread of the new mysterious virus (Parr, 2020; Yang, 2020). The total infections reported until today reached two million people worldwide with the majority in Europe and North America. The first million infections were reported from the beginning of the outbreak and until April 2, 2020, while the second million was just reached in two weeks. More than 170,000 deaths are reported worldwide, with at least 40,000 in the united states. The common COVID-19 symptoms are fever, dry cough, malaise, shortness of breath, and respiratory distress, while the loss of olfactory and taste perceptions are reported in many cases (Hui et al., 2020).
SARS-CoV-2 is a Betacoronavirus like the SARS and MERS human coronaviruses (Chan et al., 2015; Elfiky et al., 2017; Ibrahim et al., 2020). Until today, seven different strains of human coronaviruses (HCoVs) have been detected (229E and NL63 (Alphacoronaviruses), and OC43, HKU1, SARS, MERS, and SARS-CoV-2 (Betacoronaviruses)) (Elfiky et al., 2017; Hui et al., 2020; WHO., 2016).
Human coronaviruses are positive-sense RNA (30 kb) viruses. Two types of proteins characterize HCoVs, structural (Spike (S), Nucleocapsid (N), Matrix (M), and Envelope (E)) and non-structural proteins (nsp1 up to nsp16) including the RNA dependent RNA polymerase (RdRp) (nsp12) (Elfiky et al., 2017; Hasan et al., 2020). RdRp is a vital enzyme for the life cycle of RNA viruses. It has been targeted in various viral infections (HCV, Zika virus (ZIKV), and HCoVs) (Elfiky, 2016; 2017, 2019; Elfiky et al., 2013; Elfiky & Elshemey, 2016; 2018; Elfiky & Ismail, 2017; 2019; Ganesan & Barakat, 2017). RdRp active site is conserved among different organisms, while two successive, surface-exposed aspartate residues are protruding from a beta-turn motif. (Doublie & Ellenberger, 1998; Elfiky, 2020a; Elfiky & Ismail, 2018). Several studies are suggesting the effectiveness of different anti-viral drugs and compounds against the coronavirus proteins (Aanouz et al., 2020; Boopathi et al., 2020; Elfiky & Azzam, 2020; Elmezayen et al., 2020; Enayatkhani et al., 2020; Gupta et al., 2020; Khan et al., 2020a, 2020b; Muralidharan et al., 2020; Pant et al., 2020; Sarma et al., 2020).
In this study, the SARS-CoV-2 RdRp model built in previous studies is subjected to Molecular Dynamics Simulation (MDS) for up to 51 ns (Elfiky, 2020a, 2020b). After cluster analysis of the MDS trajectories, molecular docking was performed to test more than 30 compounds against the SARS-CoV-2 RdRp. These compounds include some FDA-approved drugs that are used to treat Hepatitis C Virus (HCV), the human immunodeficiency virus (HIV), Ebola virus (EBOV), and Influenza viruses in addition to an antibiotic compound Zinacef. The drugs are Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, Cefuroxime, Tenofovir, and Hydroxychloroquine. Additionally, compounds currently under clinical trials against different viruses are tested in this study against the new strain of human coronavirus RdRp after exploring its dynamics.
Materials and methods
Structural retrieval
PubChem database is used to retrieve the 3 D structures of the small molecules used in this study (Kim et al., 2015). SCIGRESS 3.4 software is used to optimize the compounds and to activate (addition of triphosphate group at 5′ position) the small molecules to be ready for the docking experiments (Elfiky & Elshemey, 2016; Summers et al., 2012). Geometry optimization for the ligands is performed through the classical MM3 force field, followed by the semiempirical parameterization method 6 (PM6) (Lii & Allinger, 1989; Stewart, 2007). After that, the density functional theory (B3LYP functional) is used to optimize the molecules further and to ensure that the molecules in its minimum energy (Elfiky et al., 2017; Leach, 2001; Stewart, 2007).
RdRp modeling
Swiss Model web server is utilized to build the all atoms 3 D RdRp model for SARS-CoV-2 using the gene (NC_045512.2) retrieved from the National Center for Biotechnology Information (NCBI) as in previous studies (Biasini et al., 2014; Elfiky, 2020a, 2020b; NCBI., 2020). SARS HCoV solved structure (PDB ID: 6NUR, chain A) is used as a template (97.08% identity) (Kirchdoerfer & Ward, 2019). The model is valid based on the Structure Analysis and Verification Server (SAVES) of the University of California, Los Angeles (UCLA). ( Eisenberg et al., 1997; Hooft et al., 1996; Joan Pontius, 1996; Laskowski et al., 1996; SAVES, 2020). After validation, the model is subjected to Molecular Dynamics Simulation (MDS) for 51 ns. NAMD software is used to perform the MDS calculations utilizing the CHARMM 36 force field (Phillips et al., 2005). The TIP3P water model is used as the solvent with added NaCl (0.154 M) to the protein-water system (Mark & Nilsson, 2001). Water is minimized (conjugate gradient), followed by the minimization of the protein system for 10000 steps each. The temperature is adjusted slowly to reach 310 K, and then an equilibration run is performed with the NPT ensemble (constant number of molecules, pressure, and temperature) for 1 ns. This is followed by the production run at the NVT ensemble (constant number of molecules, volume, and temperature) for 51 ns (Elfiky & Elshemey, 2018; Elfiky & Ismail, 2019; Ismail et al., 2020). The chimera software package is used to perform clustering analysis of the production run trajectories leading to 8 different clusters (Pettersen et al., 2004). A representative protein model is selected from each cluster for use in the docking experiments.
Molecular docking
AutoDock Vina software was used in all docking experiments, with the eight representative SARS-CoV-2 RdRp models as the docking targets (Trott & Olson, 2010). A total of 31 compounds were tested against the RdRp models for SARS-CoV-2. The four physiological nucleotides (GTP, UTP, CTP, and ATP), six drugs approved against different viruses (Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, and Tenofovir), the anti-malaria drug Hydroxychloroquine, the antibiotic drug Cefuroxime (Zinacef), thirteen different compounds (IDX-184, YAK (PAEBVIJMNXTTAT-AEFFLSMTSA-N), Setrobuvir, 2’C-methylcytidine, Valopectibine, Uprifosbuvir, Balaprevir, BMS-986094, PSI-6206, PSI-6130, R1479, MK0608, and R7128) were (or currently) in clinical trials against other viral RdRp. Four compounds derived from IDX-184 have proved its potency against Zika and Hepatitis C Viruses, and two compounds possess a low binding affinity to RdRp (Cinnamaldehyde and Thymoquinone) are used as negative controls (Elfiky, 2017, 2019; 2020b; Elfiky & Ismail, 2017; 2018; 2019).
Protein-Ligand Interaction Profiler (PLIP) web server (Technical University of Dresden) is used to mine the docking complexes (Salentin et al., 2015).
Results and discussion
A previous study by the same author pointed out the effectiveness of some in-market drugs against the COVID-19-causing coronavirus strain, SARS-CoV-2 RdRp (Elfiky, 2020b). It is crucial to test the binding affinity of the compounds in different dynamic states of the protein. Few questions need to be addressed; are the compounds still able to bind the RdRp tightly? Is the active site of the RdRp is always available to receive the small molecule? Can IDX-184 give some potent derivative molecules that can specifically target SARS-CoV-2 RdRp?
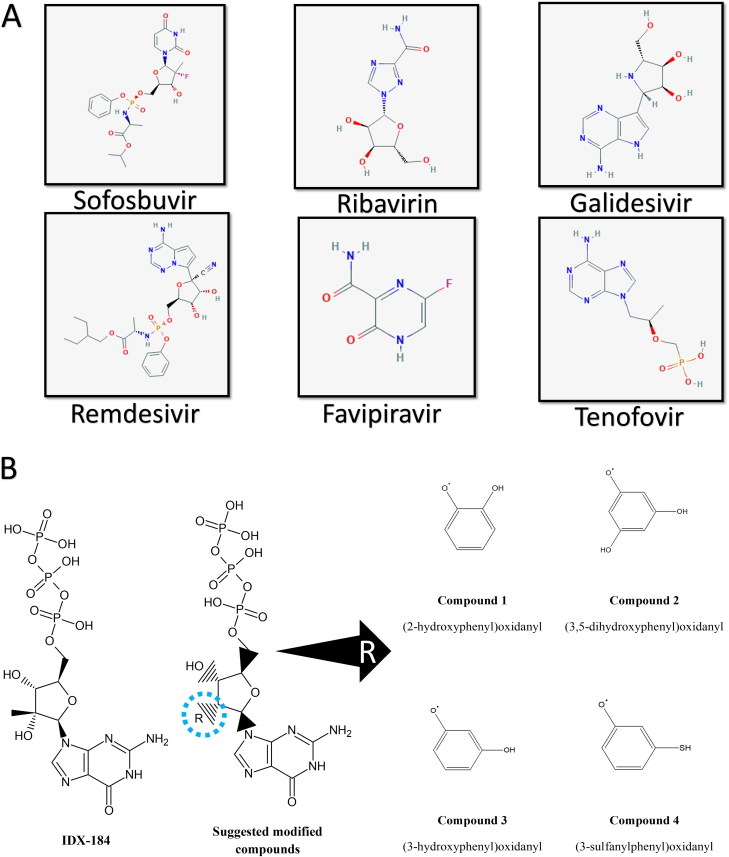
Figure 1A shows the 2 D structures of the anti-viral drugs Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, and Tenofovir. These drugs are subjected to the activation process inside the human cells (phosphorylation). Additionally, the four modified compounds (2-hydroxyphenyl)oxidanyl (compound 1), (3,5-dihydroxyphenyl)oxidanyl (compound 2), (3-hydroxyphenyl)oxidanyl (compound 3), and (3-sulfanylphenyl)oxidanyl (compound 4) are shown in Figure 1B in its active form (triphosphate). These four compounds are modified IDX-184 (at position 2′ in the ribose ring). IDX-184, in turn, is a modified form of the nucleotide guanosine triphosphate (GTP) (2′ methylated GTP).
Figure 1.
(A) 2 D structures of the FDA approved anti-viral drugs; Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, and Tenofovir. (B) 2 D structures of the guanosine derivative IDX-184 and its modifications; compound 1 ((2-hydroxyphenyl)oxidanyl), compound 2 ((3,5-dihydroxyphenyl)oxidanyl), compound 3 ((3-hydroxyphenyl)oxidanyl), and compound 4 ((3-sulfanylphenyl)oxidanyl).
RdRp dynamics
Figure 2 shows the dynamics of the SARS-CoV-2 RdRp during the 51 ns production run. Figure 2A presents the Root Mean Square Deviation (RMSD) in Å (blue line), Radius of Gyration (RoG) in Å (orange line), and Surface Accessible Surface Area (SASA) in Å2 (gray line) versus time in ns. As implicated from the RMSD, RoG, and SASA, the RdRp system is equilibrated during the first ten nanoseconds (ns) of the simulation, reaching 2.6 Å, 29 Å, and 38500 Å2, respectively. Figure 2B shows the per residues Root Mean Square Fluctuations (RMSF) in Å versus time in ns. The structure of the RdRp is depicted in cartoon and surface representations showing the active site aspartates D651 and D652 (black) and the most movable regions including the N (blue balls) and C (red balls) terminals, the K712-D716 region (orange), A731-V739 region (yellow), and Y775-M790 region (wheat). As shown in the surface representation in the left-hand side of the figure, the active site (black surface) is located in the NTP tunnel of the RdRp. The two aspartic acids D651 and D652 are surface accessible, while the most movable (RMSF of up to 5 Å) regions (orange, yellow, and wheat surfaces) are present in the NTP tunnel. One has to test whether the dynamics of these regions will affect small molecule fitting to the active site or not. Different conformations of the RdRp are selected at different time steps. Chimera software is utilized to perform cluster analysis of the 51 ns MDS data. Eight different clusters are obtained; from each, one representative structure is selected for the binding affinity calculation. The structures chosen for use in the docking study are the RdRp conformations at 30.2, 31.6, 34.4, 35.5, 36.9, 39.5, 46.3, and 49.7 ns.
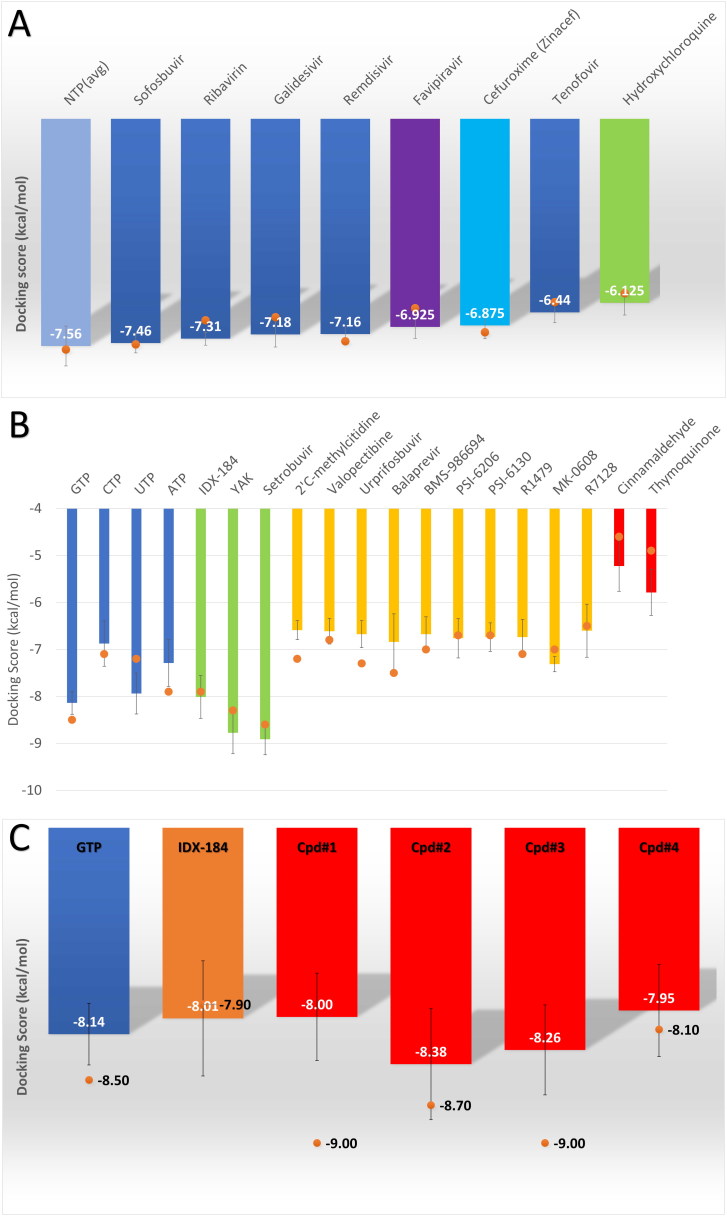
Figure 3.
(A) Bar graph representing the average binding energies (in kcal/mol) calculated by AutoDock Vina software for NTPs (sky blue column), Sofosbuvir (blue), Ribavirin (blue), Galidesivir (blue), Remdesivir (blue), Favipiravir (violet), Cefuroxime (cyan), Tenofovir (blue), and Hydroxychloroquine (green). (B) Bar graph representing the average binding energies for the nucleotides (GTP, CTP, UTP, and ATP), and other compounds. The compounds IDX-184, YAK, Setrobuvir (green column) are the best based on its binding affinities to SARS-CoV-2 RdRp. The negative control compounds (Cinnamaldehyde and Thymoquinone) are depicted in red columns, while other compounds with moderate affinities in yellow columns. (C) A bar graph shows the average binding energies for the IDX-184 (orange column) and its derivative compounds (1-4) (red columns). Error bars represent the standard deviation, while orange circles represent the binding affinities of the compounds to the solved SARS-CoV-2 RdRp structure (PDB ID: 7BTF).
Figure 2.
MDS analysis (A) Root Mean Square Deviation (RMSD) (blue line), Radius of Gyration (RoG) (orange line), and Surface Accessible Surface Area (SASA) (gray line) versus time of the simulation. (B) The per residue Root Mean Square Fluctuation (RMSF) (blue line) during the 51 ns of the MDS. The structure of SARS-CoV-2 RdRp shown by surface (left) and cartoon (right) representations. The N (blue) and C (red) terminals are shown in the balls representations, while the active site aspartates are depicted in black. The most movable parts of the protein are colored in orange (K712-D716), yellow (A731-V739), and wheat (Y775-M790), while the entire protein is in green.
Molecular docking
The eight different conformations of the SARS-CoV-2 RdRp are used as a target for the small molecule compounds using AutoDock Vina software. A flexible ligand-flexible binding site approach is used in the current study. The two active site aspartates D651 and D652 (D760 and D761 in the PDB 7BTF) are declared as flexible during the search for the possible binding mode for the flexible small molecules utilizing the vina scoring function. The grid box used for the search was 30 × 30 × 30 Å and centered at (142.5, 139.2, 149.3) Å with minimal differences in the centers between the different RdRp conformations.
Figure 3A shows the average binding affinities (in kcal/mol) calculated for the docking of the drugs (Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir (violet), Cefuroxime (cyan), Tenofovir, and Hydroxychloroquine (green)) into the SARS-CoV-2 RdRp different conformations, error bars represent the standard deviation. The average binding energy for the four nucleotides (GTP, CTP, UTP, and ATP) (sky blue column) is added to the figure for comparison. As implicated from the value, the average binding affinities for all the drugs are in the same range (-6.13 (Hydroxychloroquine) and down to -7.46 (Sofosbuvir) kcal/mol) as that of the NTP average binding affinity (-7.56 kcal/mol). These drugs can compete with NTP for the binding site of RdRp and hence can induce RdRp inhibition. Due to the safety profile for these drugs (approved by the FDA before for other viruses or microbes), it can be successful candidates as anti-COVID-19 possible drugs as reported earlier (Al-Tawfiq et al., 2020; Dong et al., 2020; Elfiky, 2020a, 2020b; Lythgoe & Middleton, 2020).
Figure 3B shows the average binding energies for the docking of the four nucleotides (blue columns), the compounds (green and yellow columns), and the negative control molecules (red columns) into SARS-CoV-2 RdRp different conformations. IDX-184, YAK, and Setrobuvir (green columns) show excellent binding affinities (-8.01, −8.78, and -8.91 kcal/mol, respectively) to RdRp even better than the physiological NTPs (blue columns) (-7.56 kcal/mol as an average value for the four NTPs). For the other compounds (yellow columns), moderate average binding energies are shown to RdRp ranging from -6.59 kcal/mol (2’C-methylcytidine) down to -7.31 kcal/mol (MK-0608). These values of the average binding energies still can compete for RdRp active site with that of CTP (-6.88 kcal/mol) and ATP (-7.29 kcal/mol). For the negative controls (Cinnamaldehyde and Thymoquinone), the average binding energies (-5.23 and -5.79 kcal/mol, respectively) are significantly different from that of the NTPs.
Figure 3C shows the average binding energies for the binding of the four IDX-184 derived compounds (compound 1 – 4) to SARS-CoV-2 RdRp different conformations. Compounds 1 and 4 show average binding energies (-8.00 and -7.95 kcal/mol, respectively) almost the same as the parent compound IDX-184 (-8.01 kcal/mol). On the other hand, compounds 2 and 3 show better average binding affinities (-8.38 and -8.26 kcal/mol, respectively) even better than the competing physiological nucleotide, GTP (-8.14 kcal/mol). These two compounds are up-and-coming anti-SARS-CoV-2 RdRp candidates that are suggested to be excellent binders and hence possible inhibitors of the RdRp.
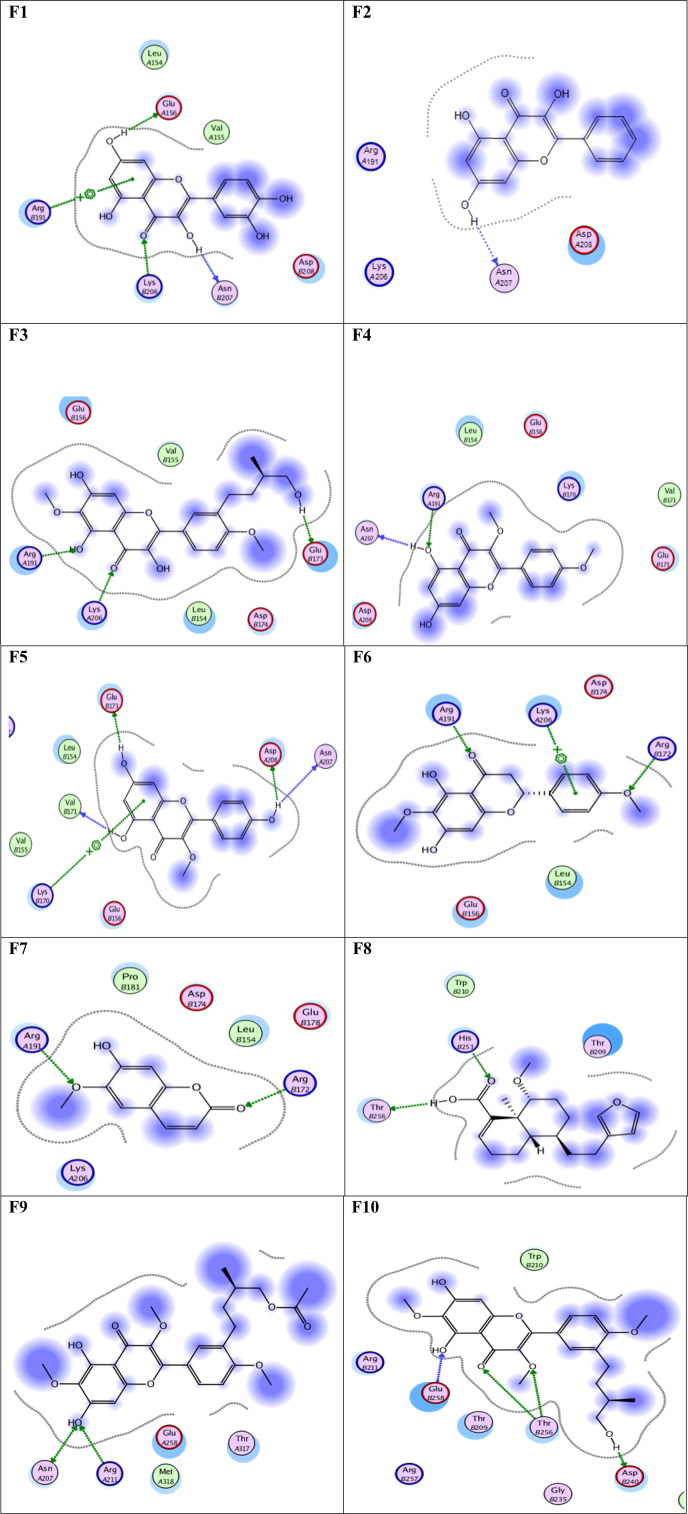
Tables 1 and 2 show the interaction details after docking the compound 2 ((3,5-dihydroxyphenyl)oxidanyl) and compound 3 ((3-hydroxyphenyl)oxidanyl) into the eight different conformations of the SARS-CoV-2 RdRp, respectively. The AutoDock Vina scores (in kcal/mol) are listed for each docking complex in addition to the number of interactions for each type of communication established. The main driving force for the binding is the H-bonds, followed by the created salt bridges. A few π-cation interactions are found, while one complex only is able to form two hydrophobic interactions. The central residues that form H-bonds are, R444, Y510, D514, T571, N582, and D651, while K289, D514, and D651 are the most three contributing residues in salt bridge formation. Noticeably, compound 2 form hydrophobic interactions only with the RdRp conformation at 46.3 ns (through Y510 and C513), and this is the lowest binding energy reported (-9.3 kcal/mol).
Table 1.
The interactions formed between compounds 2 ((3,5-dihydroxyphenyl)oxidanyl) and SARS-CoV-2 RdRp upon docking.
| RdRp conformation at | AutoDock score (kcal/mol) | H-bonding |
Salt
bridges |
other
interactions |
|||
|---|---|---|---|---|---|---|---|
| number | Amino acids involved | number | Amino acids involved | number | Amino acids involved | ||
| 30.2 ns | −8.4 | 9 | C513, T571(2),N582, D651, E702, C704, S705, Q706 | 2 | D514, D651 | ||
| 31.6 ns | −8.7 | 7 | D514(2), T571(2), T578, N582, D651 | 2 | D514, D651 | ||
| 34.4 ns | −8.3 | 6 | K436, Y510, K512, C513, D514, D651 | 3 | R444, K512, K689 | ||
| 35.5 ns | −7.8 | 12 | K442, N443, R444(3), A445, Y510, P511, K512(2), C513, K689 | 2 | K442, R444 | 2 | K689(2) |
| 36.9 ns | −7.9 | 6 | I439, R446, D514, T571, N582, S650 | 2 | R446(2) | ||
| 39.5 ns | −8.4 | 7 | K391, D514, T571(2), N582, D651(2) | ||||
| 46.3 ns | −9.3 | 8 | D509, Y510, K512(2), C513, T571, N582, D651 | 8 | K442, R444(2), K512(3), D651, K689 | 2 | Y510, C513 |
| 49.7 ns | −8.2 | 7 | Y510, C513, D651, D652, F703, S705, Q706 | 2 | D509, D651 | ||
Bold residues are interacting through π-cation, while underlined residues are creating hydrophobic contacts.
Table 2.
The interactions formed between compounds 3 ((3-hydroxyphenyl)oxidanyl) and SARS-CoV-2 RdRp upon docking.
| RdRp conformation at | AutoDock score (kcal/mol) | H-bonding |
Salt
bridges |
other
interactions |
|||
|---|---|---|---|---|---|---|---|
| number | Amino acids involved | number | Amino acids involved | number | Amino acids involved | ||
| 30.2 ns | −8.3 | 7 | N582, S650, D652(2), W691, C704, S705 | 1 | D651 | ||
| 31.6 ns | −8.5 | 8 | R444, D514, T571(2), N582, S650(2), D651 | 2 | D514, D651 | ||
| 34.4 ns | −7.7 | 6 | A449, D514, T571, S573(2), N582 | 3 | K391, D514, D651 | ||
| 35.5 ns | −8.4 | 11 | Y346, K442, N443, R444, D509, Y510, P511, K512(2), C513, D651 | 2 | K512, K689 | ||
| 36.9 ns | −7.8 | 10 | R444, C513, D514, T571(2), N582, S650(2), D651(2) | ||||
| 39.5 ns | −8.2 | 2 | R444, K512 | 9 | K442, R444, K512(3), D514, D651, K689(2) | ||
| 46.3 ns | −8.9 | 7 | R444, W508(2), Y510(2), N586, A653 | 1 | R444 | 1 | Y510 |
| 49.7 ns | −8.3 | 6 | Y510, D651, D652(2), S705, Q706 | 2 | D509, D651 | ||
Bold residues are interacting through π-cation.
A few days ago, two structures for SARS-CoV-2 RdRp were released in the protein data bank (PDB ID: 6M71 and 7BTF) (Gao et al., 2020). The RMSDs between the different conformations of the RdRp model after the MDS and the solved structures are less than 2 Å. Additionally, the solved structure 7BTF is docked by all the 31 compounds used in this study. The values of the binding affinity with the solved structure are shown in orange circles in Figures 3A-3C. As implicated from these docking experiments, the data is in an excellent agreement with presented RdRp model (Elfiky, 2020b).
Conclusions
COVID-19 pandemic is a public health crisis that needs to be treated quickly and effectively. More than 120000 deaths from the 2 million infections are reported until the writing of this manuscript. The present study attempts to test anti-RdRp drugs and suggest possible derivative molecules that may inhibit the SARS-CoV-2 RdRp. Sofosbuvir, Ribavirin, Galidesivir, Remdesivir, Favipiravir, Cefuroxime, Tenofovir, and Hydroxychloroquine can bind to the RdRp active site tightly and supposed to be good candidates for clinical trials. Moreover, the compounds Setrobuvir, YAK, and IDX-184 can tightly wrap to the SARS-CoV-2 RdRp, hence contradicting its function leading to viral eradication. Additionally, IDX-184 derived compounds (3,5-dihydroxyphenyl)oxidanyl and (3-hydroxyphenyl)oxidanyl may be used to target SARS-CoV-2 RdRp effectively, after the binding assays confirmation, in vitro, and in vivo studies.
Supplementary Material
Acknowledgements
MDS calculations are conducted on the supercomputing facility of the Bibliotheca Alexandrina, Alexandria, Egypt. This work is done during the Junior associate award granted to the author for the period 2016–2021 from the Abdus Salam International Center for Theoretical Physics (ICTP), Trieste, Italy.
Competing interest
The author declares that there is no competing interest in this work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J. A., Al-Homoud A. H., & Memish Z. A. (2020). Remdesivir as a possible therapeutic option for the COVID-19. Travel Medicine and Infectious Disease, 101615 10.1016/j.tmaid.2020.101615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., & Schwede T. (2014). SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42(W1), W252–W258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I. I., Watts A., Thomas-Bachli A., Huber C., Kraemer M. U. G., & Khan K. (2020). Pneumonia of unknown etiology in Wuhan, China: Potential for international spread via commercial air travel. Journal of Travel Medicine. 10.1093/jtm/taaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 Coronavirus Structure, Mechanism of Action, Anti-viral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F., Lau S. K., To K. K., Cheng V. C., Woo P. C., & Yuen K.-Y. (2015). Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews, 28(2), 465–522. 10.1128/CMR.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Hu S., & Gao J. (2020). Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discoveries & Therapeutics, 14(1), 58–60. 10.5582/ddt.2020.01012 [DOI] [PubMed] [Google Scholar]
- Doublie S., & Ellenberger T. (1998). The mechanism of action of T7 DNA polymerase. Current Opinion in Structural Biology, 8(6), 704–712. 10.1016/S0959-440X(98)80089-4 [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Lüthy R., & Bowie J. U. (1997). VERIFY3D: Assessment of protein models with three-dimensional profiles In Charles W. Carter Jr., and Robert M. Sweet (Eds.), Methods in enzymology (Vol. 277, pp. 396–404). Elsevier. [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2016). Zika viral polymerase inhibition using anti-HCV drugs both in market and under clinical trials. Journal of Medical Virology, 88(12), 2044–2051. 10.1002/jmv.24678 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2017). Zika virus: Novel Guanosine Derivatives revealed strong binding and possible inhibition of the polymerase [Research article]. Future Virology, 12(12), 721–728. 10.2217/fvl-2017-0081 [DOI] [Google Scholar]
- Elfiky A. A. (2019). Novel guanosine derivatives as anti-HCV NS5b polymerase: A QSAR and molecular docking study. Medicinal Chemistry, 15(2), 130–137. 10.2174/1573406414666181015152511 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel Guanosine Derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Elshemey W. M. (2016). IDX-184 is a superior HCV direct-acting anti-viral drug: A QSAR study. Medicinal Chemistry Research, 25(5), 1005–1008. 10.1007/s00044-016-1533-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Elshemey W. M. (2018). Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. Journal of Medical Virology, 90(1), 13–18. 10.1002/jmv.24934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., Elshemey W. M., Gawad W. A., & Desoky O. S. (2013). Molecular modeling comparison of the performance of NS5b polymerase inhibitor (PSI-7977) on prevalent HCV genotypes. Protein Journal, 32(1), 75–80. 10.1007/s10930-013-9462-9 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A., & Ismail A. (2019). Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sciences, 238, 116958 10.1016/j.lfs.2019.116958 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A., & Ismail A. M. (2017). Molecular Modeling and Docking revealed superiority of IDX-184 as HCV polymerase Inhibitor. Future Virology, 12(7), 339–347. 10.2217/fvl-2017-0027 [DOI] [Google Scholar]
- Elfiky A. A., & Ismail A. M. (2018). Molecular docking revealed the binding of nucleotide/side inhibitors to Zika viral polymerase solved structures. SAR and QSAR in Environmental Research, 29(5), 409–418. 10.1080/1062936X.2018.1454981 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A., Mahdy S. M., & Elshemey W. M. (2017). Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. Journal of Medical Virology, 89(6), 1040–1047. 10.1002/jmv.24736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A., & Barakat K. (2017). Applications of Computer-Aided Approaches in The Development of Hepatitis C Antiviral Agents. Expert Opinion on Drug Discovery, 12(4), 407–425. 10.1080/17460441.2017.1291628 [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., … Rao Z. (2020). Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major anti-viral drug target. BioRxiv. 10.1101/2020.03.16.993386 [DOI] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft R. W., Vriend G., Sander C., & Abola E. E. (1996). Errors in protein structures. Nature, 381(6580), 272 10.1038/381272a0 [DOI] [PubMed] [Google Scholar]
- Hui D. S., I Azhar E., Madani T. A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T. D., Memish Z. A., Drosten C., Zumla A., & Petersen E. (2020). The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases, 91, 264–266. 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I. M., Abdelmalek D. H., Elshahat M. E., & Elfiky A. A. (2020). COVID-19 Spike-host cell receptor GRP78 binding site prediction. Journal of Infection, 80(5), 554–562. 10.1016/j.jinf.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A. M., Elfiky A. A., & Elshemey W. M. (2020). Recognition of the gluconeogenic enzyme, Pck1, via the Gid4 E3 ligase: An in silico perspective. Journal of Molecular Recognition, 33(3), e2821 10.1002/jmr.2821 [DOI] [PubMed] [Google Scholar]
- Joan Pontius J. R. a S. J. W. (1996). Deviations from standard atomic volumes as a quality measure for protein crystal structures. Journal of Molecular Biology, 264, 121–136. 10.1006/jmbi.1996.0628 [DOI] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020. a). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020. b). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kim S., Thiessen P. A., Bolton E. E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., & Shoemaker B. A. (2015). PubChem substance and compound databases. Nucleic Acids Research, 44(D1), D1202–D1213. 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R. N., & Ward A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Communications, 10(1), 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., & Thornton J. M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular Nmr, 8(4), 477–486. 10.1007/BF00228148 [DOI] [PubMed] [Google Scholar]
- Leach A. (2001). Molecular modelling: Principles and applications (2nd ed.). England: Prentice Hall; https://doi.org/citeulike-article-id:571146 [Google Scholar]
- Lii J. H., & Allinger N. L. (1989). Molecular mechanics. The MM3 force field for hydrocarbons. 3. The van der Waals’ potentials and crystal data for aliphatic and aromatic hydrocarbons. Journal of the American Chemical Society, 111(23), 8576–8582. 10.1021/ja00205a003 [DOI] [Google Scholar]
- Lythgoe M., & Middleton P. (2020). Ongoing clinical trials for the management of the COVID-19 pandemic. Trends in pharmaceutical Sciences [In press]. [DOI] [PMC free article] [PubMed]
- Mark P., & Nilsson L. (2001). Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. The Journal of Physical Chemistry A, 105(43), 9954–9960. 10.1021/jp003020w [DOI] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- NCBI. (2020). National Center of Biotechnology Informatics (NCBI) database website http://www.ncbi.nlm.nih.gov/http://www.ncbi.nlm.nih.gov/
- Organization W. H. (2020. a). Infection prevention and control during health care when novel coronavirus (nCoV) infection is suspected: Interim guidance.
- Organization W. H. (2020. b, January 10). Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection: Interim guidance.
- Organization W. H. (2020. c, January). Surveillance case definitions for human infection with novel coronavirus (nCoV): Interim guidance v1.
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr J. (2020). Pneumonia in China: Lack of information raises concerns among Hong Kong health workers. England: British Medical Journal Publishing Group. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., & Ferrin T. E. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., & Schulten K. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26(16), 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H. A., & Byrareddy S. N. (2020). The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. Journal of Autoimmunity, 109, 102433 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentin S., Schreiber S., Haupt V. J., Adasme M. F., & Schroeder M. (2015). PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAVES. (2020). Structural analysis and verification server website http://nihserver.mbi.ucla.edu/SAVES/
- Stewart J. J. P. (2007). Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. Journal of Molecular Modeling, 13(12), 1173–1213. 10.1007/s00894-007-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers K. L., Mahrok A. K., Dryden M. D., & Stillman M. J. (2012). Structural properties of metal-free apometallothioneins. Biochemical and Biophysical Research Communications, 425(2), 485–492. 10.1016/j.bbrc.2012.07.141 [DOI] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2016, June 23). Middle East respiratory syndrome coronavirus (MERS-CoV). Retrieved October 8 2016 from https://www.who.int/csr/don/24-february-2020-mers-saudi-arabia/en/.
- Yang L. (2020). China confirms human-to-human transmission of coronavirus [Web page]. https://www.theguardian.com/world/2020/jan/20/coronavirus-spreads-to-beijing-as-china-confirms-new-cases
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.