Abstract
Objective
Stroke is a severe complication of atrial fibrillation (AF). We aimed to discover key genes and microRNAs related to stroke risk in patients with AF using bioinformatics analysis.
Methods
GSE66724 microarray data, including peripheral blood samples from eight patients with AF and stroke and eight patients with AF without stroke, were downloaded from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) between AF patients with and without stroke were identified using the GEO2R online tool. Functional enrichment analysis was performed using the DAVID database. A protein–protein interaction (PPI) network was obtained using the STRING database. MicroRNAs (miRs) targeting these DEGs were obtained from the miRNet database. A miR–DEG network was constructed using Cytoscape software.
Results
We identified 165 DEGs (141 upregulated and 24 downregulated). Enrichment analysis showed enrichment of certain inflammatory processes. The miR–DEG network revealed key genes, including MEF2A, CAND1, PELI1, and PDCD4, and microRNAs, including miR-1, miR-1-3p, miR-21, miR-21-5p, miR-192, miR-192-5p, miR-155, and miR-155-5p.
Conclusion
Dysregulation of certain genes and microRNAs involved in inflammation may be associated with a higher risk of stroke in patients with AF. Evaluating these biomarkers could improve prediction, prevention, and treatment of stroke in patients with AF.
Keywords: MicroRNAs, stroke, atrial fibrillation, computational biology, microarray analysis, MEF2A, CAND1
Introduction
Persistent or permanent atrial fibrillation (AF) is the most common sustained arrhythmia worldwide. AF is the second most common risk factor for ischemic stroke, the prevention of which is the main goal of treating patients with AF.1–3 In AF patients suffering stroke, a cardiac embolus originating from the left atrial appendage is a common cause.4,5 Patients with AF-related stroke seem to have a worse prognosis, such as more severe disability and greater mortality, than those with stroke in the absence of AF.6,7 Today, an increasing number of studies are focusing on the prevention and intervention of stroke in AF patients. One approach is to find suitable biomarkers by which to identify patients at greatest risk of stroke.
A number of traditional cardiovascular-related biomarkers have been explored to predict or clarify the risk of stroke in patients with AF. Troponin is one of these risk factors. A substudy of the RE-LY trial showed that troponin level is associated with thromboembolic events such as stroke.8 Other research has suggested that cardiac troponin high-sensitivity (cTn-hs) can be a biomarker independently associated with risk of stroke in AF.9 In contrast, a substudy of the ARISTOTLE trial suggested that plasma D-dimer, a marker of fibrin turnover, is a potential predictor of stroke, mortality, and major bleeding.10 The RE-LY substudy showed that the level of N-terminal pro b-type natriuretic peptide (NT-proBNP) is related to the risk of thromboembolic events and cardiovascular mortality.8
Recently, new biomarkers have been reported for prediction and treatment of stroke in AF. For example, phosphodiesterase 4D may be a risk factor for AF and stroke.11 Asymmetrical dimethylarginine may be used to predict a pro-thrombotic state in AF and correlate with the risk of stroke based on the CHADS2/CHA2DS2-VASc score.12 Variations in the gene encoding alanine-glyoxylate aminotransferase 2 may be involved in age-related thromboembolic complications in AF.13 New therapeutic targets have also been studied. Heat-shock protein 70-kDa induction seems to delay thrombus formation with minimal bleeding risk, which is promising for reducing the risk of stroke in AF patients.14
In this study, we downloaded microarray data from the Gene Expression Omnibus (GEO) database and screened differentially expressed genes (DEGs) between AF patients with stroke and those without stroke. MicroRNAs, which are small noncoding RNA molecules (containing about 22 nucleotides), targeting these DEGs were also included in our analysis. We aimed to explore new mechanisms, molecular biomarkers, and potential therapeutic targets for stroke in AF patients.
Methods
Microarray data
All of the data we used were obtained from open access databases on the Internet and did not require ethical permission or patient consent to use.
We downloaded the microarray data of GSE66724 from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with its microarray platform as GPL570 ([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array). The gene expression files were deposited by Allende et al.14 Peripheral blood was used in the microarray analysis. According to the original study,14 16 nonvalvular AF patients were recruited, 8 of whom had suffered a cardioembolic stroke. Stroke and non-stroke patients were matched by CHAD index and sex. All patients were on anticoagulant treatment with acenocoumarol, with an international normalized ratio (INR) between 2 and 3 at the time of blood withdrawal. AF was diagnosed by electrocardiography and lasted more than 3 months. Cardioembolic stroke was diagnosed clinically by imaging techniques (magnetic resonance imaging or X-ray computed tomography). Patients were excluded from the study if they met any of the following criteria: carotid artery lesion occluding more than 50% of the lumen vessel diameter in the side of the infarction, cancer in progress, leukocytosis (>7,000 cells/mL), leukopenia (<3,500 cells/mL), history of venous thromboembolism in the last 3 months, acute coronary syndrome, infection, autoimmune disease, or surgery. Renal failure (creatinine value more than double the normal value), oral contraceptive use, hormonal therapy, and corticoid consumption were also exclusion criteria.
Screening DEGs
DEGs were screened using the online tool GEO2R in the GEO database. GEO2R is an R-programming-based language for analysis of gene expression datasets by t-test or analysis of variance (ANOVA). It can help identify DEGs between two groups of samples under the same condition.15 In the present study, DEGs between stroke and non-stroke AF patients were screened and selected by the cutoff point of P < 0.05 and |logFC| >0.5, where FC is the fold change difference.
Functional enrichment analysis
We uploaded the selected DEGs to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) v6.8 Beta (https://david-d.ncifcrf.gov/) for further analysis. The DAVID database provides useful functional annotation tools to help researchers understand the biological meaning of genes.16,17 In our study, DAVID was used to investigate Gene Ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to the selected DEGs. These biological processes and pathways might be associated with stroke in AF patients. P < 0.05 was chosen as the selection threshold.
Protein–protein interaction analysis
Protein–protein interaction (PPI) pairs of DEGs were obtained from Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/).18 The STRING database is an online database resource containing a collection of comprehensive information of predicted and experimental interactions of proteins. In our study, a PPI confidence score >0.6 was considered the threshold of significance to construct the PPI network. The generated list of PPI pairs was downloaded for further analysis.
MicroRNAs targeting the DEGs
The DEGs were uploaded to the database miRNet (http://www.mirnet.ca/faces/home.xhtml)19 to obtain microRNAs (miRs) targeting these DEGs. miRNet is an online tool with comprehensive support for statistical analysis and functional interpretation of data generated in microRNA studies. It allows researchers to build miR–target interaction networks. The generated list of miR–DEG pairs was downloaded for further analysis.
Construction of miR–DEG network
A miR–DEG network composed of both miR–DEG pairs and PPI pairs was constructed and visualized using Cytoscape software 3.4.0.20 To determine the key functional modules, module clustering analysis for the network was then performed by the Molecular Complex Detection (MCODE)21 plugin. We set the degree cutoff value to 2 and the node score cutoff to 0.2 for the MCODE process.
Results
DEG screening
Data normalization and cross-comparability were assessed (Figure 1), and then DEGs were analyzed. A total of 165 DEGs were selected using the cutoff point of P < 0.05 and |logFC| >0.5, which included 141 upregulated and 24 downregulated DEGs.
Figure 1.
Cross-comparability assessment of microarray data. Dataset GSE66724 included eight peripheral blood samples from AF patients with stroke (Cases) and eight from AF patients without stroke (Controls). The box plot shows the distribution of value data for the samples selected. The box extends from the first to the third quartile; the line inside the box displays the median; and the whiskers extend from the ends of the box to the smallest and largest data values. AF, atrial fibrillation.
Functional enrichment analysis
Twenty-one GO terms (P < 0.05) were significantly enriched by upregulated DEGs, and 29 (P < 0.05) were significantly enriched by downregulated DEGs. Many of these enriched terms were associated with immune or inflammatory processes, which are listed in Table 1. Only one KEGG pathway (hsa04066:HIF-1 signaling pathway) was identified by upregulated DEGs (ANGPT1, EGLN1, IFNGR1) but it was not significant.
Table 1.
| Term | P-value | Genes | |
|---|---|---|---|
| Upregulated | GO:0002764–immune response-regulating signaling pathway | 0.017683 | PELI1, MEF2A, IRS2, LAMTOR3, F2RL1, RASGEF1A, IGKV4-1, ANGPT1, RAPGEF2, FCRL5 |
| GO:0042330–taxis | 0.031543 | ALCAM, NOV, IRS2, LAMTOR3, ST8SIA4, F2RL1, RASGEF1A, ANGPT1, RAPGEF2, EPHB1, SPP1 | |
| GO:0006935–chemotaxis | 0.031543 | ALCAM, NOV, IRS2, LAMTOR3, ST8SIA4, F2RL1, RASGEF1A, ANGPT1, RAPGEF2, EPHB1, SPP1 | |
| GO:0038095–Fc-epsilon receptor signaling pathway | 0.040044 | IRS2, LAMTOR3, RASGEF1A, IGKV4-1, ANGPT1, RAPGEF2 | |
| GO:0034141–positive regulation of toll-like receptor 3 signaling pathway | 0.04084 | PELI1, F2RL1 | |
| GO:0050776–regulation of immune response | 0.041524 | PELI1, MEF2A, IRS2, LAMTOR3, HLX, F2RL1, RASGEF1A, IGKV4-1, ANGPT1, RAPGEF2, FCRL5, IFNGR1 | |
| Downregulated | GO:0002437–inflammatory response to antigenic stimulus | 0.001872 | ELANE, HLA-DRB4, SPN |
| GO:0050728–negative regulation of inflammatory response | 0.006285 | ELANE, HLA-DRB4, SPN | |
| GO:0002862–negative regulation of inflammatory response to antigenic stimulus | 0.012856 | HLA-DRB4, SPN | |
| GO:0002438–acute inflammatory response to antigenic stimulus | 0.022107 | ELANE, SPN | |
| GO:0002684–positive regulation of immune system process | 0.022618 | PSMF1, ELANE, HLA-DRB4, TAB3, SPN | |
| GO:0002861–regulation of inflammatory response to antigenic stimulus | 0.023258 | HLA-DRB4, SPN | |
| GO:0050776–regulation of immune response | 0.029438 | PSMF1, ELANE, HLA-DRB4, TAB3, SPN | |
| GO:0006955–immune response | 0.041101 | APOBEC3B, PSMF1, ELANE, HLA-DRB4, TAB3, SPN | |
| GO:0050778–positive regulation of immune response | 0.043136 | PSMF1, ELANE, HLA-DRB4, TAB3 | |
| GO:0050727–regulation of inflammatory response | 0.045142 | ELANE, HLA-DRB4, SPN |
GO, Gene Ontology; DEG, differentially expressed gene.
Construction of miR–DEG network
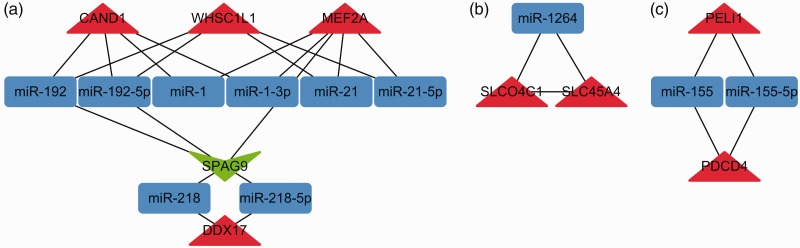
Taking the selected 165 DEGs into account, we identified 38 PPI pairs by STRING, and 4,739 miR–DEG pairs by miRNet. These two pairing pictures were merged and a complex miR–DEG network was generated in Cytoscape. To assess the key functional modules of this network, module clustering was then performed using the MCODE plugin of Cytoscape. Three modules were identified (Figure 2). Two DEGs, MEF2A and PELI1, in these modules also occurred in the GO terms enriched above, and their interacting DEGs, such as CAND1 and PDCD4, and miRs were associated with inflammatory processes or certain cardiovascular diseases.
Figure 2.
The three functional modules. Module A involves MEF2A and CAND1, and module C involves PELI1 and PDCD4, 4 genes reported to be associated with cardiovascular disease or inflammatory processes. Red triangles represent upregulated DEGs, green triangles represent downregulated DEGs, and blue rectangles represent microRNAs. DEG, differentially expressed gene.
Discussion
One of the main goals when treating AF is to prevent stroke. The CHA2DS2-VASc score is a commonly used clinical prediction rule for estimating the risk of stroke in patients with non-rheumatic AF. Risk factors in CHA2DS2-VASc score include congestive heart failure, hypertension, age ≥75 years, diabetes mellitus, prior stroke, transient ischemic attack or thromboembolism, vascular disease, age 65 to 74 years, and sex. The major treatment based on the CHA2DS2-VASc score is the use of anticoagulants to decrease the risk of stroke.22,23 However, despite careful management, some patients still suffer a stroke. In addition, anticoagulants may increase the risk of bleeding in some contraindicated patients. Therefore, new risk factors and targets of therapeutic intervention need to be identified.
Some previous studies proposed that inflammation is an independent risk factor for and contributes to the occurrence of thromboembolism and stroke in AF patients.24–27 Associations between miRNAs and certain cardiovascular diseases have also been uncovered in recent years. Zhang et al.28 suggested a role for miRNAs in the mechanism of inflammation in AF. McManus and Freedman suggested that platelet-derived miRNAs have important roles as biomarkers of susceptibility, prognosis, or treatment of certain cardiovascular diseases, including stroke and AF.29 Llombart et al. suggested that circulating miRNAs may be used in the diagnosis of cardioembolic stroke.30
In the present study, based on the GSE66724 dataset from the GEO database, 165 DEGs were found between AF patients with stroke and those without stroke. Key biological processes were enriched based on these DEGs, many of which were associated with immune or inflammatory process. We constructed three miR–DEG modules, which contained DEGs and miRNAs that were also associated with inflammation or cardiovascular diseases. This implied a mechanism of inflammation precipitating stroke in patients with AF.
Myocyte enhancer factor 2A (MEF2A) is a transcription factor in the Mef2 family. We found that MEF2A was upregulated (P = 0.00495, logFC = 0.624) in AF patients with stroke compared with those without stroke. Its association with stroke or AF has not been well documented. However, certain mutations or polymorphisms in MEF2A were found to be associated with coronary artery disease and myocardial infarction,31–34 which in turn are risk factors for stroke in AF patients. Inflammation is thought to play a role in the pathogenesis of MEF2A-related coronary artery disease.35,36
Cullin associated and neddylation dissociated 1 (CAND1) was another upregulated DEG (P = 0.00985, logFC = 0.555) enriched in this study. CAND1 is a protein-coding gene. Diseases associated with CAND1 include hypertension,37 which is also a risk factor assessed by the CHA2DS2-VASc score.
The miR–DEG module A that we constructed suggested that miR-1, miR-1-3p, miR-21, miR-21-5p, miR-192, and miR-192-5p could be associated with stroke in AF patients via MEF2A or CAND1 pathways. The miR-1 family has important roles in heart diseases such as hypertrophy, myocardial infarction, and arrhythmias.38–40 Calcium signaling is a central regulator of cardiomyocyte growth and function. Ikeda et al.41 suggested that miR-1 regulates cardiomyocyte growth responses by negatively regulating the calcium signaling components calmodulin, MEF2A, and GATA4. miR-21 has been shown to play important roles in the development of heart diseases. Its level was found to increase in fibroblasts of the failing heart, and in vivo silencing of miR-21 was shown to inhibit interstitial fibrosis and improve cardiac function in a pressure-overload cardiac disease mouse model.42 In addition, atrial fibrosis is important for the pathogenesis of atrial fibrillation. miR-21 was found to be overexpressed in atrial tissue from patients with AF and was proposed to play a role in the pathogenesis of atrial fibrosis.43 miR-192 is also upregulated in certain cardiac diseases, including heart failure after acute myocardial infarction and hypertrophic cardiomyopathy.44,45
Pellino E3 ubiquitin protein ligase 1 (PELI1; P = 0.0218, logFC = 0.543) and programmed cell death 4 (PDCD4; P = 0.0138, logFC = 0.66) were two upregulated DEGs discovered in this study. PELI1 is involved in the toll-like receptor (TLR) and interleukin-1 signaling pathways, which are associated with inflammation.46 PELI1 was proposed as a microglia-specific mediator of central nervous system (CNS) inflammation.47 PDCD4 is a protein-coding gene. Diseases associated with PDCD4 include colorectal cancer, in which downregulation of PDCD4 was found to be associated with inflammation and certain miR pathways.48,49 In contrast, activation of PDCD4 was found in coronary microembolization-related cardiac dysfunction that could, in turn, be improved by inhibition of the PDCD4 pathway via miR-21 transfection,50–52 which sheds light upon new therapeutic targets.
The miR–DEG module C that we constructed showed that PELI1 and PDCD4 were co-regulated by miR-155 and miR-155-5p. Previous studies have indicated an intimate relationship between inflammation, innate immunity, and miR-155 expression.53–58 miR-155 is essential for the generation and function of T follicular helper (Tfh) cells through a miR-155-PELI1-c-Rel pathway.59 miR-155 is associated with cancer metastasis through pathways including PDCD4.60–62
As with AF, miR-155 was found to be upregulated in left atrial appendage from nonvalvular AF patients, which indicates a role of miR-155 in electric remodeling of AF.63 In contrast, miR-155 polymorphism was found to be associated with the risk of ischemic stroke.64 miR-155 mediates inflammatory responses in ischemic cerebral tissue and other CNS neuroinflammatory disorders.65 More importantly, inhibition of miR-155 exerts an anti-inflammatory action and promotes recovery after stroke,66,67 indicating that miR-155 may be a potential therapeutic target.
In summary, by bioinformatic approaches, we constructed a stroke-related miR–DEG network in patients with AF. Dysregulation of certain genes and miRNAs in the network involved in inflammation may be associated with a higher risk of stroke in AF patients, which suggests that inflammation might be a potential risk factor of stroke that should be considered when assessing and treating AF patients. Certain genes, including MEF2A, CAND1, PELI1, and PDCD4, as well as miRNAs, including miR-1, miR-1-3p, miR-21, miR-21-5p, miR-192, miR-192-5p, miR-155, and miR-155-5p, were identified and might play key roles in the pathogenesis of stroke in patients with AF. Evaluation of these biomarkers could improve the prediction, prevention, and treatment of stroke in patients with this dysrhythmia. However, further studies are necessary to verify the clinical applications of these findings.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was funded by grants from the National Natural Science Foundation of China (Grant no. 81601711) and Science and Technology Project of Guangzhou (Grant no. 201904010418) to Zhongxing Wang. The funding sources had no involvement in study design; collection, analysis, and interpretation of data; writing the report; or the decision to submit the article for publication.
ORCID iDs
Yingyuan Li https://orcid.org/0000-0002-7160-1474
Fang Ye https://orcid.org/0000-0002-0312-2412
Zhongxing Wang https://orcid.org/0000-0003-4490-6202
References
- 1.Pritchett EL. Management of atrial fibrillation. N Engl J Med 1992; 326: 1264–1271. [DOI] [PubMed] [Google Scholar]
- 2.Lip GY, Metcalfe MJ, Rae AP. Management of paroxysmal atrial fibrillation. Q J Med 1993; 86: 467–472. [DOI] [PubMed] [Google Scholar]
- 3.Disch DL, Greenberg ML, Holzberger PT, et al. Managing chronic atrial fibrillation: a Markov decision analysis comparing warfarin, quinidine, and low-dose amiodarone. Ann Intern Med 1994; 120: 449–457. [DOI] [PubMed] [Google Scholar]
- 4.Anderson DC, Kappelle LJ, Eliasziw M, et al. Occurrence of hemispheric and retinal ischemia in atrial fibrillation compared with carotid stenosis. Stroke 2002; 33: 1963–1967. [DOI] [PubMed] [Google Scholar]
- 5.Harrison MJ, Marshall J. Atrial fibrillation, TIAs and completed strokes. Stroke 1984; 15: 441–442. [DOI] [PubMed] [Google Scholar]
- 6.Lin HJ, Wolf PA, Kelly-Hayes M, et al. Stroke severity in atrial fibrillation. The Framingham Study. Stroke 1996; 27: 1760–1764. [DOI] [PubMed] [Google Scholar]
- 7.Lamassa M, Di Carlo A, Pracucci G, et al. Characteristics, outcome, and care of stroke associated with atrial fibrillation in Europe: data from a multicenter multinational hospital-based registry (The European Community Stroke Project). Stroke 2001; 32: 392–398. [DOI] [PubMed] [Google Scholar]
- 8.Hijazi Z, Oldgren J, Andersson U, et al. Cardiac biomarkers are associated with an increased risk of stroke and death in patients with atrial fibrillation: a Randomized Evaluation of Long-term Anticoagulation Therapy (RE-LY) substudy. Circulation 2012; 125: 1605–1616. [DOI] [PubMed] [Google Scholar]
- 9.Hijazi Z, Ziad H, Johan L, et al. The ABC (age, biomarkers, clinical history) stroke risk score: a biomarker-based risk score for predicting stroke in atrial fibrillation. Eur Heart J 2016; 37: 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christersson C, Wallentin L, Andersson U, et al. D-dimer and risk of thromboembolic and bleeding events in patients with atrial fibrillation–observations from the ARISTOTLE trial. J Thromb Haemost 2014; 12: 1401–1412. [DOI] [PubMed] [Google Scholar]
- 11.Jørgensen C, Yasmeen S, Iversen HK, et al. Phosphodiesterase4D (PDE4D)–A risk factor for atrial fibrillation and stroke? J Neurol Sci 2015; 359: 266–274. [DOI] [PubMed] [Google Scholar]
- 12.Lao MC, Liu LJ, Luo CF, et al. [ Effect of asymmetrical dimethylarginine for predicting pro-thrombotic risk in atrial fibrillation]. Zhonghua Yi Xue Za Zhi 2016; 96: 2059–2063. [DOI] [PubMed] [Google Scholar]
- 13.Seppälä I, Kleber ME, Bevan S, et al. Associations of functional alanine-glyoxylate aminotransferase 2 gene variants with atrial fibrillation and ischemic stroke. Sci Rep 2016; 6: 23207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allende M, Molina E, Guruceaga E, et al. Hsp70 protects from stroke in atrial fibrillation patients by preventing thrombosis without increased bleeding risk. Cardiovasc Res 2016; 110: 309–318. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. limma: Linear Models for Microarray Data. In: Statistics for Biology and Health. pp.397–420.
- 16.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011; 39: D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan Y, Siklenka K, Arora SK, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 2016; 44: W135–W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito R, Smoot ME, Ono K, et al. A travel guide to Cytoscape plugins. Nat Methods 2012; 9: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Zhong J, Chen G, et al. ClusterViz: a Cytoscape APP for cluster analysis of biological network. IEEE/ACM Trans Comput Biol Bioinform 2015; 12: 815–822. [DOI] [PubMed] [Google Scholar]
- 22.Prystowsky EN, Padanilam BJ, Fogel RI. Treatment of atrial fibrillation. JAMA 2015; 314: 278. [DOI] [PubMed] [Google Scholar]
- 23.Lip GYH, Lane DA. Stroke prevention in atrial fibrillation. JAMA 2015; 313: 1950. [DOI] [PubMed] [Google Scholar]
- 24.Wallentin L, Hijazi Z, Andersson U, et al. Growth differentiation factor 15, a marker of oxidative stress and inflammation, for risk assessment in patients with atrial fibrillation: insights from the Apixaban for Reduction in Stroke and Other Thromboembolic Events in Atrial Fibrillation (ARISTOTLE) trial. Circulation 2014; 130: 1847–1858. [DOI] [PubMed] [Google Scholar]
- 25.Hu YF, Yu-Feng H, Yi-Jen C, et al. Inflammation and the pathogenesis of atrial fibrillation. Nat Rev Cardiol 2015; 12: 230–243. [DOI] [PubMed] [Google Scholar]
- 26.Harada M, Van Wagoner DR, Nattel S. Role of inflammation in atrial fibrillation pathophysiology and management. Circ J 2015; 79: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Lip GYH, Apostolakis S. Inflammatory biomarkers and atrial fibrillation: potential role of inflammatory pathways in the pathogenesis of atrial fibrillation-induced thromboembolism. Curr Vasc Pharmacol 2015; 13: 192–201. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Liu L, Hu J, et al. MicroRNA regulatory network revealing the mechanism of inflammation in atrial fibrillation. Med Sci Monit 2015; 21: 3505–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McManus DD, Freedman JE. MicroRNAs in platelet function and cardiovascular disease. Nat Rev Cardiol 2015; 12: 711–717. [DOI] [PubMed] [Google Scholar]
- 30.Llombart V, Garcia-Berrocoso T, Bustamente A, et al. Cardioembolic stroke diagnosis using blood biomarkers. Curr Cardiol Rev 2014; 9: 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhagavatula MRK. Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet 2004; 13: 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y, Yang Y, Zhang X, et al. Relationship of the CAG repeat polymorphism of the MEF2A gene and coronary artery disease in a Chinese population. Clin Chem Lab Med 2007; 45: 987–992. [DOI] [PubMed] [Google Scholar]
- 33.Wang L. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 2003; 302: 1578–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Niu W, Wu Z, et al. Variants in exon 11 of MEF2A gene and coronary artery disease: evidence from a case-control study, systematic review, and meta-analysis. PLoS One 2012; 7: e31406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q. Advances in the genetic basis of coronary artery disease. Curr Atheroscler Rep 2005; 7: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki E, Satonaka H, Nishimatsu H, et al. Myocyte enhancer factor 2 mediates vascular inflammation via the p38-dependent pathway. Circ Res 2004; 95: 42–49. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher FR, Siew K, Zhang J, et al. Characterisation of the Cullin-3 mutation that causes a severe form of familial hypertension and hyperkalaemia. EMBO Mol Med 2015; 7: 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvestri P, Di Russo C, Rigattieri S, et al. MicroRNAs and ischemic heart disease: towards a better comprehension of pathogenesis, new diagnostic tools and new therapeutic targets. Recent Pat Cardiovasc Drug Discov 2009; 4: 109–118. [DOI] [PubMed] [Google Scholar]
- 39.Zorio E, Medina P, Rueda J, et al. Insights into the role of microRNAs in cardiac diseases: from biological signalling to therapeutic targets. Cardiovasc Hematol Agents Med Chem 2009; 7: 82–90. [DOI] [PubMed] [Google Scholar]
- 40.Cai B, Pan Z, Lu Y. The roles of microRNAs in heart diseases: a novel important regulator. Curr Med Chem 2010; 17: 407–411. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda S, He A, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol 2009; 29: 2193–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008; 456: 980–984. [DOI] [PubMed] [Google Scholar]
- 43.Adam O, Löhfelm B, Thum T, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol 2012; 107: 278. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto S, Sakata Y, Suna S, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 2013; 113: 322–326. [DOI] [PubMed] [Google Scholar]
- 45.Fang L, Ellims AH, Moore XL, et al. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med 2015; 13: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol 2009; 10: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y, Jin J, Chang M, et al. Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat Med 2013; 19: 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Zhao M, Guo C, et al. PDCD4 deficiency aggravated colitis and colitis-associated colorectal cancer via promoting IL-6/STAT3 pathway in mice. Inflamm Bowel Dis 2016; 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 49.Peacock O, Lee AC, Cameron F, et al. Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS One 2014; 9: e110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su Q, Li L, Zhou Y, et al. Induction of myocardial PDCD4 in coronary microembolization-related cardiac dysfunction: evidence from a large-animal study. Cell Physiol Biochem 2014; 34: 533–542. [DOI] [PubMed] [Google Scholar]
- 51.Su Q, Li L, Wang J, et al. Mechanism of programmed cell death factor 4/nuclear factor-κB signaling pathway in porcine coronary micro-embolization-induced cardiac dysfunction. Exp Biol Med 2015; 240: 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Q, Li L, Liu Y, et al. Ultrasound-targeted microbubble destruction-mediated microRNA-21 transfection regulated PDCD4/NF-κB/TNF-α pathway to prevent coronary microembolization-induced cardiac dysfunction. Gene Ther 2015; 22: 1000–1006. [DOI] [PubMed] [Google Scholar]
- 53.O’Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A 2007; 104: 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ceppi M, Pereira PM, Dunand-Sauthier I, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A 2009; 106: 2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cremer TJ, Ravneberg DH, Clay CD, et al. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS One 2009; 4: e8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang P, Hou J, Lin L, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol 2010; 185: 6226–6233. [DOI] [PubMed] [Google Scholar]
- 57.McCoy CE, Sheedy FJ, Qualls JE, et al. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem 2010; 285: 20492–20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res 2013; 41: 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu WH, Kang SG, Huang Z, et al. A miR-155-Peli1-c-Rel pathway controls the generation and function of T follicular helper cells. J Exp Med 2016; 213: 1901–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomsen KG, Terp MG, Lund RR, et al. miR-155, identified as anti-metastatic by global miRNA profiling of a metastasis model, inhibits cancer cell extravasation and colonization in vivo and causes significant signaling alterations. Oncotarget 2015; 6: 29224–29239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zadeh MM, Motamed N, Ranji N, et al. Silibinin-induced apoptosis and downregulation of microRNA-21 and microRNA-155 in MCF-7 human breast cancer cells. J Breast Cancer 2016; 19: 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hafez MM, Hassan ZK, Zekri ARN, et al. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac J Cancer Prev 2012; 13: 591–598. [DOI] [PubMed] [Google Scholar]
- 63.Wang JG, Meng X, Han J, et al. [Differential expressions of miRNAs in patients with nonvalvular atrial fibrillation]. Zhonghua Yi Xue Za Zhi 2012; 92: 1816–1819. [PubMed] [Google Scholar]
- 64.Choi GH, Ko KH, Kim JO, et al. Association of miR-34a, miR-130a, miR-150 and miR-155 polymorphisms with the risk of ischemic stroke. Int J Mol Med 2016; 38: 345–356. [DOI] [PubMed] [Google Scholar]
- 65.Lopez-Ramirez MA, Wu D, Pryce G, et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 2014; 28: 2551–2565. [DOI] [PubMed] [Google Scholar]
- 66.Caballero-Garrido E, Pena-Philippides JC, Lordkipanidze T, et al. In vivo inhibition of miR-155 promotes recovery after experimental mouse stroke. J Neurosci 2015; 35: 12446–12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen Y, Zhang X, Dong L, et al. Acetylbritannilactone modulates microRNA-155-mediated inflammatory response in ischemic cerebral tissues. Mol Med 2015; 21: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]