Abstract
Pandemics caused by influenza A virus (IAV) are responsible for the deaths of millions of humans around the world. One of these pandemics occurred in Mexico in 2009. Despite the impact of IAV on human health, there is no effective vaccine. Gene mutations and translocation of genome segments of different IAV subtypes infecting a single host cell make the development of a universal vaccine difficult. The design of immunogenic peptides using bioinformatics tools could be an interesting strategy to increase the success of vaccines. In this work, we used the predicted amino acid sequences of the neuraminidase (NA) and hemagglutinin (HA) proteins of different IAV subtypes to perform multiple alignments, epitope predictions, molecular dynamics simulations, and experimental validation. Peptide selection was based on the following criteria: promiscuity, protein surface exposure, and the degree of conservation among different medically relevant IAV strains. These peptides were tested using immunological assays to test their ability to induce production of antibodies against IAV. We immunized rabbits and mice and measured the levels of IgG and IgA antibodies in serum samples and nasal washes. Rabbit antibodies against the peptides P11 and P14 (both of which are hybrids of NA and HA) recognized HA from both group 1 (H1, H2, and H5) and group 2 (H3 and H7) IAV and also recognized the purified NA protein from the viral stock (influenza A Puerto Rico/916/34). IgG antibodies from rabbits immunized with P11 and P14 were capable of recognizing viral particles and inhibited virus hemagglutination. Additionally, intranasal immunization of mice with P11 and P14 induced specific IgG and IgA antibodies in serum and nasal mucosa, respectively. Interestingly, the IgG antibodies were found to have neutralizing capability. In conclusion, the peptides designed through in silico studies were validated in experimental assays.
Electronic supplementary material
The online version of this article (10.1007/s00705-020-04537-2) contains supplementary material, which is available to authorized users.
Introduction
Influenza A virus (IAV) is a lipid-enveloped, single-stranded, negative-sense RNA virus belonging to the family Orthomyxoviridae. The viral envelope contains three transmembrane proteins (NA [neuraminidase], HA [hemagglutinin] and M2 [proton channel]) on the viral surface and one protein (M1 [matrix protein]) below the membrane. The viral core contains the nucleoprotein (NP), viral RNA, and three polymerase proteins (PB1, PB2, and PA) [1]. IAV is classified into subtypes based on two major antigens: the surface spike glycoproteins NA and HA [2]. All IAV subtypes are known to cause infections in birds, which are their natural reservoir [3]. Humans are infected principally by the IAV subtypes H1N1, H2N2, H3N2, H7N9, and H5N1 [4]. Influenza pandemics have become serious socioeconomic and public-health problems worldwide. Moreover, seasonal flu causes approximately 250,000 to 500,000 deaths per year [5, 6]. IAV epidemics and pandemics are attributed to mutations in the viral RNA genome. Mutations involving surface proteins (NA and HA) result in structural protein changes that cause a loss of antibody recognition against the virus. This is one reason why new flu vaccines need to be designed for each seasonal influenza or pandemic influenza strain. The development of vaccines is the major method used to prevent IAV infection and represents one of the most important contributions by the immunology field to public health [7]. An important strategy is to identify conserved epitopes that may be used to design new vaccines that are capable of conferring broad protection.
Currently, the primary goal is to develop vaccines that protect by eliciting antibody responses against multiple subtypes and strains of influenza viruses [8–10]. These broadly neutralizing antibodies (bnAbs) generally target conserved and functional regions or epitopes on the major surface glycoproteins: hemagglutinin (head and stem), neuraminidase, and M2e [10–12]. The hemagglutinin (HA) is the main surface glycoprotein of influenza virus, which mediates the adsorption and penetration of the virus into host cells [13]. Each molecule of HA comprises a membrane distal globular head composed of HA1, which contains the receptor-binding site (RBS), and a stem region, which encompasses the fusion machinery [14]. Most bnAbs are directed against the HA protein. The receptor-binding site is a functionally conserved region on the HA1 globular head domain that is a target for bnAbs that inhibit viral entry by preventing HA binding to its host receptor [15, 16]. Since the stem region contains the most conserved epitopes for antibody recognition, antibodies produce against this region have a higher neutralization breadth than RBS-targeted bnAbs. These stem-binding bnAbs inhibit virus replication by blocking attachment and preventing conformational changes that are essential for membrane fusion [15–17]. NA is the second most abundant glycoprotein on the surface of influenza A and B viruses, and conserved domains or epitopes in NA induce bnAbs that protect against viruses of a single subtype [17]. Thus, NA epitopes may use in universal influenza vaccines [12, 17–20]. Although NA-specific antibodies can control infection by several mechanisms, the main mechanism is the inhibition of enzyme activity [12, 18, 21].
Thus, “universal vaccines consisting of conserved domains or epitopes from HA and NA could be more broadly protective than single proteins.
The rational design of peptide-based vaccines is based on computational procedures that employ knowledge of the antigen recognition of different protein targets, such as the major histocompatibility complex (MHC), T-cell receptor, and/or B-cell receptor. These procedures are performed at the molecular level [22] using information from the Protein Data Bank. This strategy offers many advantages, such as the production of short immunogenic peptides without the risk of infection and safe handling of biological samples under room-temperature conditions. Additionally, peptides can be synthesized in vitro to decrease production costs and can be handled without strict conditions due to their stability [23]. Peptide vaccines are classified according to the immunological cell type involved in the immune response (i.e., B-cell or T-cell epitopes) and are used to induce specific immune responses. T cells recognize peptides coupled to MHC from antigen-presenting cells, and the MHC is then coupled to the T-cell receptor (TCR). Following the formation of the peptide-MHC-TCR complex, T cells receive several biological signals that are capable of activating the immune system to act against microorganisms or cancer cells [24]. In addition, activation of B cells by epitopes can increase the protection spectrum of vaccines, making them more protective than those with limited immune system activation capacity [25].
In this study, we selected a set of peptides (P) from HA and NA (including hybrids) that were capable of generating antibodies against IAV. First, a protein sequence bioinformatics study was performed using the HA and NA proteins to obtain multi-epitope peptides characterized by conserved residues and promiscuous properties. The epitope selection process included criteria related to their protein surface localization. Then, three-dimensional (3D) models of the target epitopes were constructed and subjected to molecular dynamics (MD) simulations to explore their thermodynamic properties. Finally, the epitopes were used in experimental assays to explore their immunogenic properties in animal models.
Materials and methods
Multiple alignments of HA and NA protein sequences
We retrieved the full primary sequences of the HA and NA proteins reported between 1918 and 2014 from the NCBI influenza virus resource [26]. We aligned the HA and NA sequences for each subtype using the Muscle/EBI server [27] and the Clustal X 2.0.11 program [28].
Building a quaternary structure model based on consensus NA and HA sequences
We constructed a three-dimensional (3D) structure model based on the NA consensus sequence (YP_009118627.1) of the H1N1 virus by employing the Swiss Model server [29–32]. The NA monomer A (PDB ID: 3TI4) was used as a template. For the H1N1 HA consensus sequence (Table 1), we used the Modeller 9.10 program to build 3D models of multi-chain HA by employing a multi-trimer template HA (PDB ID: 1RUY). The stereochemical quality of the models was evaluated using Ramachandran plots and PDBSUM [33] (data not show) and ERRAT servers [34] (data not shown).
Table 1.
HAs of different influenza subtypes used to determine the antibody response of rabbits immunized with hybrid peptides P11 and P14 coupled to KLH
| Key | Strain Information (ID: NCBI) | Strain | Hemagglutinin group (G) | *Catalog number |
|---|---|---|---|---|
| H1-1 | A/Puerto Rico/8/34 (ID: AF389118_1) | H1N1 | G1 | 11684-V08H |
| H2-2 | A/Shanghai/2/2013 (ID : YP_009118475.1) | H7N9 | G2 | 40239-V08B |
| H3-3 | A/Cambodia/S1211394/2008 (ID: ADM95445.1) | H5N1 | G1 | 40026-V08H1 |
| H4-1A | A/Brisbane/59/2007 (ID: ACA28844.1) | H1N1 | G1 | 11052-V08H |
| H5-2A | A/Aichi/2/1968 (ID: BAN81712.1) | H3N2 | G2 | 11707-V08H |
| H6-3A | A/Japan/305/1957 (ID: ABI84959.1) | H2N2 | G1 | 11088-V08H |
| H8-8 | A/California/07/2009 (ID: ACP44189.1) | H1N1 | G1 | 11085-V08H |
*Sino Biological Inc. (Beijing, China)
Prediction of epitopes in the consensus sequence
We submitted the NA (YP_009118627.1) and HA consensus sequences (Table 1) to the NetMHC 3.2 the NetMHCII 2.2 and the ABCpred Prediction servers to identify epitopes capable of interacting with MHC I, MHC II, and B cells, respectively. We focused on some of the most important alleles of MHC I (HLA-B*39:01, HLA-B*1501, HLA-A*0201, HLA-A*0301, HLA-A*2601, HLA-B*0702, and HLA-B*5801) and MHC II (HLA-DRB1*0101, HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1*0701, HLA-DRB1*0801, HLA-DRB1*1101, HLA-DRB1*1301 and HLA-DRB1*1501).
Selection of epitopes and design of multi-epitopes
We selected epitopes based on promiscuity criteria among the IAV subtypes, the degree of conservation, and their surface localization on the quaternary structure models of the proteins that were built (Fig. 1S), selecting six peptides (P1, P2, P3, P4, P5 and P6) without conjugation to keyhole limpet hemocyanin (KLH). P1 is located in the middle of HA, whereas P2-P4 and P6 are located in globular heads of HA. P5 is located in in globular head of NA (Fig. 1S). We determined the degree of exposure of the regions by visual inspection, using the VMD program [35]. Once the most promising epitopes were identified, we added linkers of different length and structure (6-9 glycine residues) to control the distance between two selected epitopes and to maintain multi-epitope properties, discarding hybrids in which the added glycines or the neighboring peptides were predicted to be immunogenic. Additionally, epitopes previously identified by our research group were selected as candidates for the construction of hybrid peptides to evaluate their capacity to increase immunological responses [19, 36].
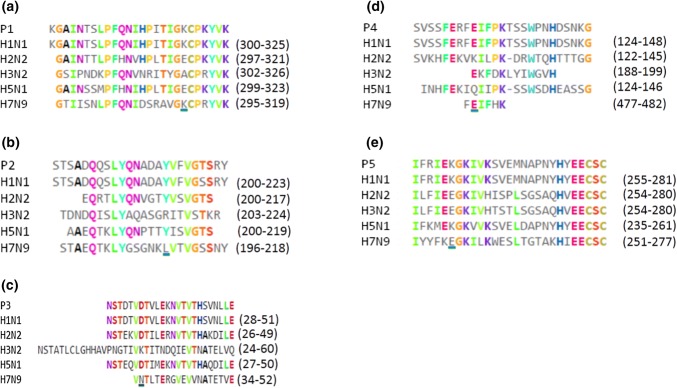
Fig. 1.
Sequence alignment of peptides A) P1, B) P2, C) P3 and D) P4 from hemagglutinin and E) P5 from neuraminidase with the corresponding peptides from different influenza virus subtypes (H1N1, H2N2, H3N2, H5N1 and H7N9). These peptides have structural regions that are important for MHCI and MHC II recognition according to epitope predictions
Proteasome and immunoproteasome cleavage predictions
We predicted proteasome and immunoproteasome cleavage using the Proteasome Cleavage Prediction Server, PCPS (http://imed.med.ucm.es/Tools/pcps/index.html), to verify epitope structural stability. The PCPS analysis identified fragments of peptides ranging from 9 to 21 residues in length. These peptide sequences were submitted to the NetMHC 3.2 [37, 38], NetMHCII 2.2 [37, 39] and PREDBALB/C (http://antigen.i2r.a-star.edu.sg/predBalbc/.) servers to determine whether they maintained their immunogenic properties.
Molecular dynamics simulations of multi-epitopes
We built 3D models of the selected multi-epitopes using the PEPstrMOD server (http://osddlinux.osdd.net/raghava/pepstrmod/, accessed on March 2018). We subjected these multi-epitopes to MD simulations using the NAMD 2.5 program [40] with CHARMM 27 as the force field [41]. The constant number-of-particles, pressure, and temperature (NPT) ensemble and the periodic boundary conditions were used. A constant temperature (310 K) was maintained using a Langevin thermostat set at a constant pressure (1 atm) maintained by an isotropic Langevin barostat [42]. These pressure and temperature values represented standard physiological conditions. Energy minimization was performed with 1000 steps in the conjugate-gradient algorithm with restraints on the peptide backbone, followed by 1000 steps without restraints. The system was heated for 20 ps and equilibrated for 60 ps with restraints to the α-carbon and then finished with no restraints and 310 K. These MD simulations were run for 500 ns.
Analysis of MD simulations
We calculated the root mean square deviation (RMSD), which measures whole-protein motions while generally considering the α-carbon coordinates in relation to MD simulation times, root mean square fluctuations (RMSF), which measure the α-carbon coordinates in relation to MD simulation time per residue, and radius of gyration (Rg), which measures the protein compactness from the center of the protein to the periphery according to the 3D coordinates in the structural analyses of multi-epitope MD simulations from the Carma64 program [43]. The secondary structure was determined using the Visual Molecular Dynamics (VMD) program [35].
Experimental procedures
Peptides
Nine peptides were designed, but eight peptides (including P6 in P14) were selected (Table 1) to be synthesized (Peptide 2.0 Inc.) according to theoretical studies for experimental testing as described in the following experimental procedures (Table 2). The conjugation of peptides with the KLH molecule was performed using succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC), which is an heterobifunctional crosslinker that contains N-hydroxysuccinimide (NHS) ester and maleimide groups that allow covalent conjugation of amine molecules in KLH with the sulfhydryl groups of cysteine residues located in the middle of each hybrid peptide.
Table 2.
Peptides selected for synthesis and experimental analysis
| Peptide name (protein) | Sequence | Solvent |
|---|---|---|
| P1 (HA) | KGAINTSLPFQNIHPITIGKCPKYVK | H2O |
| P2 (HA) | STSADQQSLYQNADAYVFVGTSRY | H2O |
| P3 (HA) | NSTDTVDTVLEKNVTVTHSVNLLE | DMSO |
| P4 (HA) | SSFERFEIFPKTSSWPNHDSNKG | DMSO |
| P5 (NA) | IFRIEKGKIVKSVEMNAPNYHYEECSC | H2O |
| P6 (HA)$ | KTSSWPNHDSNKGVTAASPHAGAKSFYKN | — |
| P9* | KGAINTSLPFQNIHPITIGKCPKYVKGGGGGCGGGGSTSADQQSLYQNADAYVFVGTSRY | DMSO |
| P11* | IFRIEKGKIVKSVEMNAPNYHYEESSSGGGCGGGKGAINTSLPFQNIHPITIGKCPKYVK | H2O |
| P14* | VNSDTVGWSWPDGAELPFTIDKGGGGCGGGGKTSSWPNHDSNKGVTAASPHAGAKSFYKN | H2O |
P9 = hybrid P1 and P2, P11 = hybrid P5 and P1, P14 = hybrid peptide reported by Loyola PK et al. 2013 and P6
*Peptides conjugated with the KLH protein
Underlines indicate polyglycine linkers
$P6 is included in P14
Rabbit immunization
We immunized eight rabbits (from Harlan Mexico) with peptides (P1, P2, P3, P4 and P5) without conjugation to KLH and hybrid peptides (P9, P11 and P14) conjugated to KLH at three time points with during seven-day intervals.
For the primary immunization, 400 µg of peptide plus complete Freund’s adjuvant (Sigma Chemical Co.) was administered by the subcutaneous route. The second immunization (7 days later) included 300 µg of peptide plus incomplete Freund’s adjuvant administered by the subcutaneous route. The third immunization was administered by the intramuscular route 15 days after the primary immunization and contained 300 µg of peptide in 5 mL of saline solution. Seven days after the last immunization, we anesthetized the rabbits with pentobarbital via the intraperitoneal route. Serum samples were obtained from blood extracted by cardiac puncture and stored at -70 °C.
Mouse immunization
Screening was carried out to select the peptides that induced the highest antibody titer in rabbit serum. For this purpose, five groups of peptides were used. Six BALB/c mice for each peptide were injected three times at seven-day intervals via the intranasal route with 100 µg of the peptide (P1, P4, P5, P11 and P14) plus 2 µg of cholera toxin (CT) mucosal adjuvant. Seven days after the last immunization, we anesthetized the mice with pentobarbital. Samples were obtained from blood extracted by cardiac puncture and washes of the nasal cavity with PBS, respectively.
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals, in accordance with Mexican Federal regulations for animal use and care (NOM-062-ZOO-1999) and was authorized by the Comité Interno del Cuidado y Uso de los Animales de Laboratorio (CICUAL) at ESM-IPN. The protocol was approved by the Committee on Ethics of the Escuela Nacional de Medicina y Homeopatia of Instituto Politecnico Naciocal, IPN (protocol number ENMH-CB-0095-2014). Mice were anesthetized with ether for intranasal immunization, and for biological assays, mice and rabbits were killed using pentobarbital. All efforts were made to minimize suffering and to use biological samples immediately.
ELISA for detection of IgG and IgA antibodies against different peptides in the sera from rabbits or sera and nasal washes from mice
We determined anti-peptide antibody levels in serum samples from immunized rabbits and serum samples or nasal washes of mice immunized with peptides, using an indirect enzyme-linked immunosorbent assay (ELISA). For biological assays, the mice or rabbits were killed using pentobarbital, and their biological samples were used immediately. Each sample was tested in duplicate. Briefly, 96-well plates were coated with 100 µL of different peptides (20 µg of peptide per mL in carbonate-bicarbonate buffer [15 mM Na2CO3 and 35 mM NaHCO3, pH 9.6]). The plates were incubated overnight at 4 °C, followed by three washes with phosphate-buffered saline containing 0.05% Tween-20 (PBST). Blocking was performed using PBST plus 6% fat-free milk for 1 h, and the samples were then washed with PBST. Serial dilutions of serum samples from nine immunized rabbits (1:100 to 1:25000), serum from immunized mice (from 1:100 to 1:3200) and nasal washes (from 1:1 to 1:8) were prepared and added to the plates, which were incubated overnight at 4 °C and washed with PBST, after which 50 μL of a 1:3000 dilution of goat anti-rabbit IgG peroxidase (Thermo Scientific) for rabbit serum and rabbit anti-mouse IgG peroxidase (1:3000) or rabbit anti-mouse IgA (1:500) peroxidase (Thermo Scientific) for mouse samples was added to each well. The plates were incubated for 4 h at room temperature and washed with PBST. The enzymatic reactions were initiated by adding 100 µL of substrate solution (0.5 mg of o-phenylenediamine per mL plus 0.01% H2O2 in 0.05 M citrate buffer, pH 5.2). After 10 min, the reaction was stopped by adding 50 μL of 2.5 M H2SO4. The absorbance was measured at 490 nm using a microplate reader (Benchmark, Bio-Rad). The endpoint titer was defined as the reciprocal of the highest analyzed dilution that gave an absorbance value above 0.5.
Influenza viruses
Influenza A virus strain Puerto Rico/916/34 was grown in Madin-Darby canine kidney (MDCK) cells. The cells were cultured in Dulbecco’s modified Eagle medium nutrient mixture F-12 (DMEM/F-12) (Caisson) supplemented with 5% fetal bovine serum (SFB) (Mediatech) at 37 °C and 5% CO2. The viral stock was prepared in serum-free medium and frozen at -20 °C for later use as described previously [36].
ConA envELISA
Viral particles that had been isolated from infected MDCK cells and purified using serum-free medium were trapped in wells coated with concanavalin A (Con A), which immobilizes the detergent-solubilized viral glycoproteins of the viral envelope. Briefly, 96-well plates (Costar) were coated with 100 μL of Con A (Sigma-Aldrich) per well at a concentration of 50 μg/mL in PBS, pH 7.4, for 1 h. The wells were washed three times with PBS containing 0.1% Triton X-100 (PBS-TX) and incubated with solubilized and inactivated influenza A virus strain Puerto Rico/916/34 (serum-free virus stock) in PBS-TX for 1 h. After the wells were washed with PBS-TX, the unreacted ConA binding sites were blocked with RPMI medium 1640 containing 10% FBS (RPMI-10% SFB) for 1 h. Dilutions of the heat-inactivated rabbit serum samples were made in RPMI-10% SFB and incubated for 1 h at room temperature. The positive control was a 1:500 dilution of serum from a patient who was positive for influenza virus [21]. Rabbit pre-immune serum samples were used as negative controls. The wells were washed again and incubated with 100 μL of the appropriate peroxidase-conjugated anti-IgG (H+L) (Santa Cruz) as the secondary antibody. The wells were washed again and then incubated for 1 h with 100 μL of 2, 2’-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) (Sigma-Aldrich), and H2O2 was added as a substrate. Absorbance values were determined at 405 nm [36].
Specific IgG levels in the sera of rabbits immunized with peptides 11 and 14 against hemagglutinin and neuraminidase
Levels of antibody against HA and NA were determined using recombinant HA (rHA) proteins purchased from Sino Biological Inc. (Beijing, China). Group 1 included H1N1, H5N1, H2N2 strains, and group 2 included H7N9, H3N2 strains (Table 1). Neuraminidase was purified from a stock of strain Puerto Rico/916/34 by preparative SDS-polyacrylamide gel electrophoresis. The antibody levels in the serum samples were determined using an ELISA assay. Briefly, 96-well plates were coated with 0.5 µg of protein (hemagglutinin or neuraminidase, 500 μg/mL) in carbonate-bicarbonate buffer (15 mM Na2CO3 and 35 mM NaHCO3, pH 9.6), incubated overnight at 4 °C, and washed three times with 0.05% PBST. Blocking was performed with PBST plus 6% fat-free milk for 1 h, and the plates were then washed with PBST. Serial dilutions of serum samples from immunized rabbits (1:100 to 1:12,800) were added to the plates, which were incubated overnight at 4 °C and washed with PBST. A 1:3000 dilution of the goat anti-rabbit IgG peroxidase (Thermo Scientific) was added to each well, and the plates were incubated for 4 h at room temperature and then washed with PBST. The enzymatic reactions were initiated by adding 100 µL of substrate solution (0.5 mg of o-phenylenediamine per mL plus 0.01% H2O2 in 0.05 M citrate buffer, pH 5.2). After 10 min, we stopped the reactions by adding 50 μL of 2.5 M H2SO4. The absorbance was measured at 490 nm using a microplate reader (Benchmark, Bio-Rad). The endpoint titer was defined as the reciprocal of the highest analyzed dilution that gave an absorbance value above 0.5.
Hemagglutination inhibition (HI) assay
Serum samples from immunized rabbits were inactivated by heating at 56 ± 2 °C in a water bath for 30 min, and the total IgG were purified by affinity chromatography using immobilized protein A Sepharose CL-4B (Sigma-Aldrich) as described by Contis-Montes de Oca et al. [44]. Dilutions were made in PBS and added to a final volume of 25 μL in 96-well round-bottom plates using pre-immune serum as a control. The virus concentration was then adjusted to 4 hemagglutination units (HAU) per 25 μL in PBS and added to samples, except for the negative controls, which consisted of PBS without virus. The plates were incubated for 1 h at 37 °C while human red blood cells (HRBCs) of type “O” were washed twice with 1x PBS, pH 7.2, and centrifuged at 200 × g for 10 min at 4–8 °C. The cells were then resuspended in a final concentration of 0.75% in 1x PBS, and 50 μL of this suspension was added to each well. The plates were incubated again for 1 h at 37 °C, and the effects were observed [45]. The HI titer was the highest dilution that caused total inhibition of agglutination. HRBCs with PBS were used as a negative control, and HRBCs plus virus at 4 HAU were used as a positive control. Sera were tested at dilutions from 1:12 to 1:7680.
Plaque reduction neutralization test (PRNT50)
Neutralizing antibodies were titrated as described previously [46, 47] Briefly, serial dilutions of the heat-inactivated test sera in duplicate starting from 1:100 were mixed with 20 PFU of influenza A virus strain Puerto Rico/916/34 for 1 h at 37 °C and added to MDCK cells at a density of 80,000 cells per well in 24-well plates. Positive and negative control sera and virus back titration to confirm the viral inoculum were included. At 1 h after infection, serum-free DMEM/F-12 medium with 2 µg of TPCK (L-1-tosylamide-2-phenylethyl chloromethyl ketone)-treated trypsin was added to each well, and the plates were incubated for 1 h at 37 °C. Then, 500 µL of overlay medium was added to each well, and the plates were incubated for 3 days under the same conditions. The supernatant was discarded, and plaques were visualized by staining with naphthol blue-black dye. The result is reported as the reciprocal of the dilution that protects 50% of the cells from a cytopathic effect due to infection.
Statistical analysis
Three independent assays were conducted in duplicate, and the results are presented as the mean and standard deviation of one representative assay. The statistical significance of differences among groups was determined by ANOVA, followed by Tukey’s test. Differences with p values less than 0.01 were considered significant. The analysis was performed using the PRISM program (GraphPad, San Diego, CA.). The half-maximal effective concentration (EC50) was determined using the statistical program SigmaPlot for Windows version 11 (Systat Software Inc., San Jose, CA, USA).
Results and discussion
Degree of conservation of peptides
A protein sequence search for NA and HA from IAV in the Influenza Virus Resource at NCBI [26] yielded the following IAV subtypes: H1N1 (8560 HA sequences and 8184 NA sequences), H7N9 (79 HA sequences and 75 NA sequences), H5N1 (216 HA sequences and 259 NA sequences), H2N2 (95 HA sequences and 132 NA sequences) and H3N2 (8124 HA sequences and 8372 NA sequences). These subtypes were related to human influenza. The consensus protein sequences (Fig. 1A-D) of the influenza A H1N1 glycoproteins NA and HA (Fig. 1E) were obtained from multiple alignments. These consensus protein sequences were used to identify the immunogenic regions in HA and NA (Fig. 1). Sequences that were conserved during evolution (data not shown), present in different influenza virus subtypes, or located on the surface (exposed to solvent, see Fig. 1S) were predicted to be easily accessible to antibodies with neutralizing potential [48], as demonstrated for HIV [49]. Six peptides (from H1N1 as reference), including four from HA (P1, P2, P3, P4 and P6), one from NA (P5), and one from NA reported previously by us [19] were selected as described above.
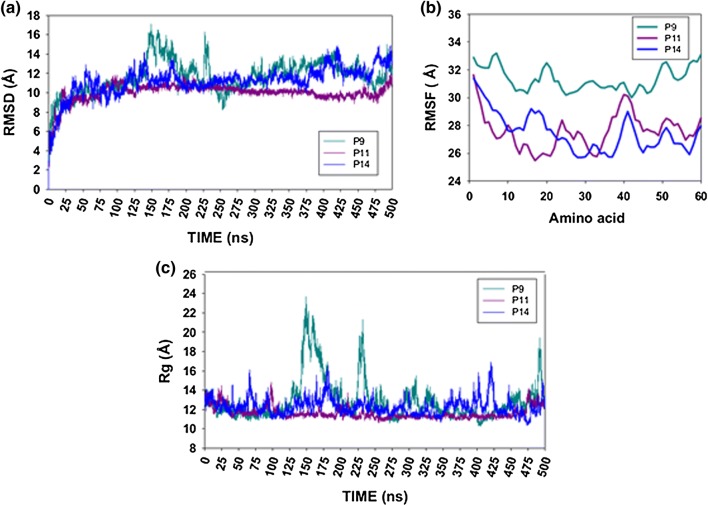
Multiple sequence alignments of HA peptides from different subtypes of influenza A virus (H1N1, H7N9, H5N1, H2N2 and H3N2) showed a degree of conservation for each of the selected peptides when H1N1 (A/California/07/2009) was used as a reference (100% identity, Fig. 1). P1 was conserved to different degrees with respect to subtypes H1N1 (A/Puerto Rico/8/1934), H7N9 and H5N1 (Table 3). The most similarity was observed with H1N1 (A/Puerto Rico/8/1934) and the lowest with H3N2 (see Fig. 1A), suggesting that P1 could induce immunogenic responses against H1N1, H2N2, H3N2, H7N9 and H5N1 as a multi-vaccine. P2 and P4 were most similar to the corresponding sequences of pandemic H1N1 (A/Puerto Rico/8/1934), see Table 3), suggesting that they were potential candidates for specific vaccines and immunodiagnostics, whereas P3 was most similar to the corresponding sequence of H1N1 (A/Puerto Rico/8/1934) and had a low degree of similarity to H3N2 sequences. However, multiple alignment of NA sequences showed that P5 was more similar to the corresponding sequences of the H1N1 (A/Brisbane/59/2007 [H1N1]) and H5N1 subtypes than to those of other influenza virus subtypes (see Table 3). Furthermore, P1, P3 and P5 could work as a multi-vaccine epitope to provide broad protection against unspecified influenza A virus infections. One additional parameter that the epitopes needed to achieve for good recognition by MHC and TCR was low structural mobility [50]. This was investigated using MD simulations [50] measuring structural motions dependent on structural flexibility of hybrid peptides with long sequences (Fig. 2). It is known that lower flexibility of peptides is associated with a better immune response [36]. As shown in Fig. 2, P9 yielded larger RMSD values in MD simulations than compared to P11 and P14 (approximately 125-200 ns and higher fluctuation close to 225 ns) (Fig. 2A). From 275 ns to 500 ns, P11 showed less motion with minimal oscillations compared to P9 and P14, suggesting that it might be more immunogenic. Based on RMSF values, P9 exhibited more mobility than P11 and P14. Additionally, P9 had higher Rg values (approximately 175 ns) compared to P11 and P14. These results agree with some of the biological results described in the following paragraphs.
Table 3.
Degrees of similarity (S), identity (I) and gaps (G) for the region of peptides P1-P4 against each region of HA and P5 against each neuraminidase from influenza virus
| P1 | P2 | P3 | P4 | P5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | G | S | I | G | S | I | G | S | I | G | S | I | G | |
|
A/California/04/2009 [H1N1] |
100 | 100 | 0 | 100 | 95.8 | 0 | 100 | 100 | 0 | 100 | 100 | 0 | 100 | 100 | 0 |
|
A/Brisbane/59/2007 [H1N1] |
100 | 76.9 | 0 | 58.3 | 37.5 | 0 | 100 | 100 | 0 | 64.5 | 61.3 | 25.8 | 96.3 | 77.8 | 0 |
|
A/PuertoRico/8/1934 [H1N1] |
96.2 | 76.9 | 0 | 70.8 | 58.3 | 0 | 100 | 100 | 0 | 87.0 | 78.3 | 4.3 | 92.6 | 77.8 | 0 |
|
A/Japan/305/1957 [H2N2] |
92.3 | 76.9 | 0 | 58.3 | 45.8 | 0 | 87.5 | 66.7 | 0 | 47.8 | 34.8 | 4.3 | 59.3 | 44.4 | 0 |
|
A/Aichi/2/1968 [H3N2] |
69.2 | 57.7 | 0 | 54.2 | 45.8 | 0 | 43.2 | 24.3 | 35.1 | 34.8 | 21.7 | 0 | 59.3 | 48.1 | 0 |
|
A/Cambodia/S211394/2008 [H5N1] |
92.3 | 76.9 | 0 | 62.5 | 45.8 | 0 | 91.7 | 66.7 | 0 | 78.3 | 47.8 | 4.3 | 100 | 77.8 | 0 |
|
A/Shanghai/02/2013 [H7N9] |
73.1 | 57.7 | 0 | 62.5 | 37.5 | 0 | 58.3 | 33.3 | 4.2 | 30.4 | 21.7 | 0 | 59.3 | 40.7 | 0 |
Fig. 2. A.
) Root mean square deviation (RMSD), B) root mean square fluctuations (RMSF), and C) radius of gyrations of hybrid peptides P9, P11 and P14
Prediction of the immunogenicity of peptides from HA and NA
T-cell epitopes
Binding of peptides to human and mouse MHC-II alleles in silico
The peptides that bind to HLA-DR, HLA-DQ, HLA-DP, human MHC class II alleles and mouse I-Ed and I-Ad molecules are shown in Table 4. All of the peptides contain identical or very similar sequences (epitopes) that are predicted to bind to both human and mouse MHC-II alleles. For example, P1 is an epitope that binds to human HLA-DRB1*0101, HLA-DRB1*0701, HLA-DRB1*1302 alleles and mouse I-Ad allele. In summary, the selected peptide contains epitopes that are capable of interacting with human and mouse (BALB/c) MHC-II alleles. These results show that the predicted peptides contain helper T-cell epitopes, which could activate T-cell-dependent responses, such as IgG and IgA antibody responses. The peptides P2, P3 and P4 contain three epitopes, whereas P1 only contains one epitope. The hybrid peptides P9 and P11 contain four epitopes, and P14 contains only one that is likely to induce an immune response against HA and NA proteins [51, 52].
Table 4.
Peptide binding to human and mouse MHC class II molecules (alleles)
| Human MHCII | Mouse MHCII | |||
|---|---|---|---|---|
| Core | Alleles (NetMHCII 2.3 server) | Score | Alleles (PREDBALB/C server) | Score |
| P1[ (KGAINTSLPFQNIHPITIGKCPKYVK) | ||||
| FQNIHPITI |
HLA-DRB1*0701 HLA-DRB1*1302 HLA-DRB1*1001 |
5.50 9.00 9.00 |
I-Ad | 8.72 |
| INTSLPFQN | HLA-DRB1*1001 | 8.50 | ||
| AINTSLPFQ | HLA-DRB4*0101 | 5.50 | ||
| SLPFQNIHP | HLA-DRB4*0101 | 5.50 | ||
| KGAINTSL | I-Ed | 9.20 | ||
| IHPITIGKC | I-Ed | 8.40 | ||
| P2 (STSADQQSLYQNADAYVFVGTSRY) | ||||
| YVFVGTSRY | HLA-DRB3*0101 | 5.00 |
I-Ad I-Ed [AYVFVGTSR] |
8.28 9.60 |
| YQNADAYVF | HLA-DRB3*0101 | 1.40 | I-Ad | 9.08 |
| LYQNADAYV |
HLA-DRB1*1302 HLA-DRB3*0202 |
9.00 5.00 |
I-Ad [SLYQNADAY] I-Ad [QSLYQNADAY] I-Ed [QSLYQNADA] |
8.74 8.30 8.82 |
| TSADQQSLY | I-Ad | 8.16 | ||
| DQQSLYQNA | I-Ad | 8.60 | ||
| DAYVFVGTS | I-Ad | 9.60 | ||
| P3(NSTDTVDTVLEKNVTVTHSVNLLE) | ||||
| TVLEKNVTV | I-Ed..[VLEKNVTVT] | 9.40 | ||
| KNVTVTHSV | I-Ad [NVTVTHSVN] | 8.42 | ||
| VTVTHSVNL | HLA-DRB1*0701 | 5.00 | I-Ad [NVTVTHSVN] | 8.42 |
| VTHSVNLLE | HLA-DRB1*0405 | 5.50 | I-Ed | 8.70 |
| NSTDTVDTV | I-Ad | 8.34 | ||
| STDTVDTVL | I-Ad | 8.20 | ||
| TDTVDTVLE | I-Ad | 8.16 | ||
| DTVDTVLEK |
I-Ad I-Ed. |
8.14 8.70 |
||
| EKNVTVTHS | I-Ad | 10.00 | ||
| P4 (SVSSFERFEIFPKTSSWPNHDSNKG) | ||||
| FERFEIFPK |
HLA-DRB1*1001 HLA-DRB1*0405 HLA-DRB5*0101 |
6.50 9.50 7.50 |
I-Ed | 9.90 |
| FEIFPKTSS |
HLA-DRB1*1101 HLA-DRB1*1602 HLA-DRB1*0802 |
4.00 6.00 5.00 |
||
| IFPKTSSWP | I-Ed | 8.24 | ||
| TSSWPNHDS | I-Ed | 9.16 | ||
| P5 (IFRIEKGKIVKSVEMNAPNYHYEECSC) | ||||
| FRIEKGKIV |
HLA-DRB1*0101 HLA-DRB1*0801 HLA-DRB1*1101 HLA-DRB1*1602 HLA-DRB3*0101 HLA-DRB3*0301 HLA-DRB5*0101 |
5.00 8.50 4.00 6.00 4.00 6.50 3.50 |
I-Ed [RIEKGKIVK] | 8.74 |
| GKIVKSVEM | HLA-DRB1*0701 | 2.50 |
I-Ad I-Ed [KIVKSVEMN] |
8.04 9.72 |
| SVEMNAPNY | HLA-DRB1*1201 | 6.00 | I-Ad | 9.02 |
| VEMNAPNYH |
HLA-DRB1*1302 HLA-DRB3*0202 |
4.00 8.00 |
I-Ed [KSVEMNAPN] | 9.46 |
| IFRIEKGKI |
HLA-DRB1*0103 HLA-DRB4*0103 |
2.50 4.50 |
||
| RIEKGKIVK | HLA-DRB1*0103 | 4.00 | I-Ed | 8.74 |
| VKGKIVKSVE | HLA-DRB1*0103 | 8.50 | ||
| IVKSVEMNA |
HLA-DRB1*0802 HLA-DRB1*0901 HLA-DRB1*1602 HLA-DRB4*0101 |
7.50 7.50 9.50 6.50 |
||
| IEKGKIVKS | HLA-DRB1*1602 | 9.00 | I-Ad [EKGKIVKSV] | 8.30 |
| MNAPNYHYE | I-Ad | 8.50 | ||
| APNYHYEEC | I-Ad | 9.40 | ||
| KIVKSVEMN | I-Ed | 9.72 | ||
| KSVEMNAPN | I-Ed | 9.46 | ||
| P9 (KGAINTSLPFQNIHPITIGKSPKYVKGGGGGCGGGGSTSADQQSLYQNADAYVFVGTSRY) | ||||
| FQNIHPITI |
HLA-DRB1*1001 HLA-DRB3*0101 HLA-DRB1*0701 HLA-DRB1*1302 |
9.00 5.50 5.50 9.00 |
I-Ad | 8.72 |
| YQNADAYVF | HLA-DRB3*0101 | 1.80 | I-Ad | 9.08 |
| YVFVGTSRY | HLA-DRB3*0101 | 5.00 |
I-Ad I-Ad [DAYVFVGTS] I-Ed [AYVFVGTSR] |
8.28 9.60 9.60 |
| LYQNADAYV |
HLA-DRB1*1302 HLA-DRB3*0202 |
9.00 5.00 |
I-Ad [SLYQNADAY] I-Ad [QSLYQNADA] I-Ed [QSLYQNADA] |
8.74 8.30 8.82 |
| AINTSLPFQ | HLA-DRB4*0101 | 5.50 | ||
| INTSLPFQN | HLA-DRB1*1001 | 8.50 | ||
| ITIGKSPKY | HLA-DRB1*1201 | 8.50 | ||
| SLPFQNIHP | HLA-DRB4*0101 | 6.00 | ||
| P11 (IFRIEKGKIVKSVEMNAPNYHHYEESSSGGGCGGGKGAINTSLPFQNIHPITIGKSPKYVK) | ||||
| FRIEKGKIV |
HLA-DRB1*0101 HLA-DRB1*1101 HLA-DRB1*0801 HLA-DRB1*1602 HLA-DRB3*0101 HLA-DRB3*0101 HLA-DRB5*0101 |
5.0 4.00 8.50 6.00 4.00 6.5 3.5 |
||
| FQNIHPITI |
HLA-DRB1*0701 HLA-DRB1*1001 HLA-DRB1*1302 |
5.50 9.00 9.00 |
I-Ad | 8.72 |
| GKIVKSVEM | HLA-DRB1*0701 | 2.50 |
I-Ad I-Ed [RIEKGKIVK] |
8.04 8.74 |
| VEMNAPNYH |
HLA-DRB1*1302 HLA-DRB3*0101 |
8.00 8.00 |
I-Ad [SVEMNAPNY] I-Ed [KSVEMNAPN] |
9.02 9.46 |
| IFRIEKGKI | HLA-DRB1*0103 | 2.50 | ||
| RIEKGKIVK | HLA-DRB1*0103 | 4.00 | ||
| KGKIVKSVE | HLA-DRB1*0103 | 8.50 | ||
| IVKSVEMNA |
HLA-DRB1*0802 HLA-DRB1*0901 HLA-DRB1*1302 HLA-DRB1*1602 HLA-DRB4*0101 |
7.50 7.50 4.50 9.50 6.50 |
||
| INTSLPFQN | HLA-DRB1*1001 | 8.50 | ||
| SVEMNAPNY | HLA-DRB1*1201 | 6.00 | ||
| ITIGKSPKY | HLA-DRB1*1201 | 8.50 | ||
| IEKGKIVKS | HLA-DRB1*1602 | 9.00 | ||
| GAINTSLPF | HLA-DRB3*0202 | 6.5 | ||
| AINTSLPFQ | HLA-DRB4*0101 | 5.50 | ||
| SLPFQNIHP | HLA-DRB4*0101 | 5.50 | ||
| IHPITIGKS |
I-Ad I-Ed |
8.70 8.40 |
||
| P14 (VNSDTVGWSWPDGAELPFTIDKGGGGCGGGGKTSSWPNHDSNKGVTAASPHAGAKSFYKN) | ||||
| VTAASPHAG |
I-Ad [AASPHAGAK] I-Ed [AASPHAGAK] |
8.34 8.20 |
||
| FTIDKGGGG | HLA-DRB3*0101 | 6.50 | ||
| HAGAKSFYK | HLA-DRB5*0101 | 8.00 | I-Ad [SPHAGAKSF] | 8.32 |
| VNSDTVGWS | I-Ad | 8.30 | ||
| SDTVGWSWP | I-Ad | 9.20 | ||
| WSWPDGAEL | I-Ed | 9.18 | ||
| DSNKGVTAA | I-Ad | 9.30 | ||
| SNKGVTAAS | I-Ad | 8.16 | ||
| NKGVTAASP | I-Ad | 9.70 | ||
| KGVTAASPH | I-Ad | 8.06 | ||
| TSSWPNHDS | I-Ed | 9.16 | ||
| DSNKGVTAA | I-Ed | 9.40 | ||
| WSWPDGAEL | I-Ed | 9.18 | ||
Binding of peptides to human and mouse MHC I alleles in silico
The peptides that bind to representative HLA class I (HLA-B*39:01, HLA-B*1501, HLA-A*0201, HLA-A*0301, HLA-A*2601, HLA-B*0702, and HLA-B*5801) and mouse H2-Kd H2-Ld and H2-Dd alleles are shown in Table 5. These peptides are predicted to contain several T-cell epitopes, some of which are predicted to bind to both human and mouse MHC-I alleles. For example, P1 contains the sequence FQNIHPITI, which binds to human HLA-B*3901 and mouse H2-Dd alleles.
Table 5.
Peptide binding to human and mouse MHC class I molecules (alleles)
| Human MHCI | Mouse MHCI | |||
|---|---|---|---|---|
| Core | Alleles (NetMHC 4.0 server) | Score | Alleles (PREDBALB/C server) | Score |
| P1 (KGAINTSLPFQNIHPITIGKCPKYVK) | ||||
| FQNIHPITI |
HLA-B*3901 HLA-B*4001 |
0.12 2.00 |
H2-Dd | 8.88 |
| ITIGKCPKY |
HLA-A*2601 HLA-B*5801 |
0.30 1.90 |
H2-Dd | 8.62 |
| LPFQNIHPI |
HLA-B*3901 HLA-B*0702 HLA-B*0801 |
0.30 0.50 0.60 |
||
| GAINTSLPF |
HLA-B*1501 HLA-A*2601 HLA-B*5801 |
0.06 0.90 0.60 |
H2-Dd | 8.26 |
| NIHPITIGK | HLA-A*0301 | 1.10 | ||
| AINTSLPFQ | HLA-A*2601 | 0.90 | ||
| IGKCPKYVK | H2-Dd | 9.06 | ||
| NTSLPFQN | H2-Dd | 8.84 | ||
| TIGKCPKYV | H2-Kd | 8.50 | ||
| P2 (STSADQQSLYQNADAYVFVGTSRY) | ||||
| TSADQQSLY |
HLA-A*0101 HLA-A*2601 |
0.03 0.20 |
||
| YVFVGTSRY |
HLA-A*0101 HLA-A*2601 HLA-B*1501 HLA-A*0301 |
0.60 0.08 0.80 1.70 |
H2-Dd [DAYVFVGTS] | 8.86 |
| YQNADAYVF |
HLA-B*3901 HLA-B*1501 HLA-A*2402 HLA-B*4001 |
0.08 0.08 0.60 1.60 |
H2-Kd [LYQNADAYV] H-2Dd |
9.10 8.82 |
| SLYQNADAY |
HLA-A*0101 HLA-A*2601 HLA-B*1501 HLA-A*0301 |
1.20 1.00 0.80 1.70 |
H-2Dd H2-Kd |
9.40 8.12 |
| LYQNADAYV | HLA-A*2402 | 2.00 | H2-Kd | 9.10 |
| P3 (NSTDTVDTVLEKNVTVTHSVNLLE) | ||||
| STDTVDTVL |
HLA-B*3901 HLA-A*0101 |
0.90 0.90 |
H2-Dd | 8.06 |
| TVLEKNVTV | HLA-A*0201 | 1.20 | ||
| DTVDTVLEK | HLA-A*2601 | 1.40 | ||
| VTVTHSVNL | H2-Dd | 8.84 | ||
| P4 (SVSSFERFEIFPKTSSWPNHDSNKG) | ||||
| SSFERFEIF |
HLA-A*2402 HLA-A*2601 HLA-B*0801 HLA-B*5801 HLA-B*1501 |
1.10 1.50 2.00 1.60 0.70 |
H2-Dd | 9.20 |
| EIFPKTSSW |
HLA-A*2601 HLA-B*5801 |
0.80 1.30 |
H2-Dd [IFPKTSSWP] | 8.30 |
| KTSSWPNHD | HLA-B*5801 | 1.70 | H2-Dd [SWPNHDSNK] | 8.62 |
| IFPKTSSWP | H2-Dd | 8.30 | ||
| VSSFERFEI | HLA-B*5801 | 0.90 | ||
| P5 (IFRIEKGKIVKSVEMNAPNYHYEECSC) | ||||
| SVEMNAPNY | HLA-A*0101 | 0.90 | ||
| EMNAPNYHY |
HLA-A*0101 HLA-B*1501 |
1.10 0.70 |
H2-Kd | 8.50 |
| FRIEKGKIV |
HLA-B*2705 HLA-B*3901 HLA-A0301 |
1.40 0.70 1.20 |
H2-Kd[IFRIEKGK] H2-Dd [IFRIEKGK] |
9.40 8.30 |
| PNYHYEECS | H2-Dd | 8.30 | ||
| P9 (KGAINTSLPFQNIHPITIGKSPKYVKGGGGGCGGGGSTSADQQSLYQNADAYVFVGTSRY) | ||||
| TSADQQSLY |
HLA-A*0101 HLA-A*2601 HLA-A*0101 |
0.03 0.20 0.03 |
||
| LPFQNIHPI |
HLA-B*0702 HLA-B*3901 |
0.50 0.30 |
||
| FQNIHPITI | HLA-B*3901 | 0.12 | H2-Dd | 8.88 |
| YQNADAYVF |
HLA-B*3901 HLA-B*1501 |
0.08 0.08 |
H2-Kd | 8.82 |
| GAINTSLPF | HLA-B*1501 | 0.06 | H2-Dd | 8.26 |
| SLYQNADAY |
HLA-A*0101 HLA-A*2601 HLA-B*1501 HLA-A*0101 |
1.20 1.00 0.80 1.2 |
H2-Dd H2-Kd [LYQNADAYV] H2-Kd |
9.40 9.10 8.12 |
| YVFVGTSRY |
HLA-A*0101 HLA-A*2601 |
0.60 0.08 |
||
| ITIGKSPKY | HLA-A*2601 | 0.20 | H2-Dd | 9.10 |
| YVKGGGGGC |
HLA-A*2601 HLA-B*1501 HLA-A*0101 |
1.40 0.80 0.60 |
||
| STSADQQSL | HLA-B*3901 | 1.70 | ||
| NTSLPFQNI | H2-Dd | 8.84 | ||
| IHPITIGKS | H2-Dd | 9.20 | ||
| IGKSPKYVK | H2-Dd | 9.14 | ||
| TIGKSPKYV | H2-Kd | 9.40 | ||
| P11 (IFRIEKGKIVKSVEMNAPNYHYEESSSGGGCGGGKGAINTSLPFQNIHPITIGKSPKYVK) | ||||
| ITIGKSPKY |
HLA-A*2601 HLA-B*5801 |
0.20 0.90 |
H2-Kd [TIGKSPKYV] H2-Dd |
9.40 9.10 |
| LPFQNIHPI |
HLA-B*0702 HLA-B*3901 HLA-B*0801 |
0.50 0.30 0.60 |
H2-Kd [NTSLPFQNI] | 8.84 |
| FQNIHPITI |
HLA-B*3901 HLA-B*4001 |
0.12 2.00 |
H2-Dd | 8.88 |
| GAINTSLPF |
HLA-B*1501 HLA-A*2601 HLA-B*5801 |
0.06 0.90 0.60 |
H2-Dd | 8.26 |
| SVEMNAPNY | HLA-A*0101 | 0.90 | ||
| YHYEESSSG | HLA-B*3901 | 0.50 | ||
| FRIEKGKIV |
HLA-B*3901 HLA-B*2705 |
0.70 1.40 |
H2-Dd [IFRIEKGKI] H2-Kd [IFRIEKGKI] |
8.30 9.40 |
| GKGAINTSL | HLA-B*3901 | 1.50 | ||
| EMNAPNYHY | HLA-A*0101 | 1.10 | H2-Kd | 8.50 |
| RIEKGKIVK | HLA-A*0301 | 1.20 | ||
| NIHPITIGK | HLA-A*0301 | 1.10 | ||
| EESSSGGGC | HLA-B*4001 | 1.60 | ||
| TIGKSPKYV | H2-Kd | 9.40 | ||
| P14 (VNSDTVGWSWPDGAELPFTIDKGGGGCGGGGKTSSWPNHDSNKGVTAASPHAGAKSFYKN) | ||||
| WPDGAELPF |
HLA-B*0702 HLA-B*3901 |
0.50 0.70 |
H2-Dd [DGAELPFTI] | 9.60 |
| SPHAGAKSF |
HLA-B*0702 HLA-B*3901 |
0.06 1.90 |
H2-Dd | 8.38 |
| NSDTVGWSW | HLA-B*5801 | 0.04 | ||
| WSWPDGAEL | HLA-B*5801 | 2.00 | H2-Ld | 9.80 |
| KTSSWPNHD | HLA-B*5801 | 1.70 | ||
| NSDTVGWSW | HLA-A*0101 | 0.90 | ||
| AASPHAGAK | HLA-A*0301 | 0.90 | H2-Dd [ASPHAGAKS] | 9.14 |
| HAGAKSFYK | HLA-A*0301 | 0.60 | H2-Dd [AGAKSFYKN] | 8.54 |
| DTVGWSWPD | HLA-A*2601 | 1.40 | ||
| FTIDKGGGG | HLA-A*2601 | 1.00 | ||
NetMHCII 2.3 Server Score: Strong binder <2.0 weak binder >2.0 to 10.0
NetMHC 4.0 Server Score: Strong binder <0.5 weak binder >0.5 to 2.0
PREDBALB/C server Score: Peptides with prediction scores above or equal to 8
The peptides were found to contain different number of epitopes that are recognizable by cytotoxic T cells. P1, P9 and P11 have four epitopes, and P2 and P14 have three. The epitopes that bind to MHC-I molecules could activate cytotoxic T cells that contribute to protection against invasion by influenza virus [51–53].
B-cell epitopes
Using the B-cell epitope prediction program ABCpred, we identified several linear B-cell epitopes (Table 6). P1, P3 and P5 were predicted to contain two B-cell epitopes, whereas the hybrid peptides were predicted to contain 4-5 epitopes. Therefore, hybrid peptides could induce stronger mucosal and systemic humoral immune responses [54].
Table 6.
Epitopes recognized by B cells according to the ABCpred Prediction Server
| Name | Sequence | CORE | Score |
|---|---|---|---|
| P1 | KGAINTSLPFQNIHPITIGKCPKYVK | KGAINTSLPFQNIHPI | 0.93 |
| PFQNIHPITIGKCPKY | 0.55 | ||
| P2 | STSADQQSLYQNADAYVFVGTSR | QSLYQNADAYVFVGTS | 0.82 |
| P3 | NSTDTVDTVLEKNVTVTHSVNLLE | TDTVDTVLEKNVTVTH | 0.76 |
| VLEKNVTVTHSVNLLE | 0.71 | ||
| P4 | SVSSFERFEIFPKTSSWPNHDSNKG | RFEIFPKTSSWPNHDS | 0.86 |
| P5 | IFRIEKGKIVKSVEMNAPNYHYEECSC | KGKIVKSVEMNAPNYH | 0.88 |
| SVEMNAPNYHYEECSC | 0.88 | ||
| P9 | KGAINTSLPFQNIHPITIGKSPKYVKGGGGGCGGGGSTSADQQSLYQNADAYVFVGTSRY | KGAINTSLPFQNIHPI | 0.93 |
| GGGGGCGGGGSTSADQ | 0.86 | ||
| QSLYQNADAYVFVGTS | 0.82 | ||
| GGGSTSADQQSLYQNA | 0.77 | ||
| PITIGKSPKYVKGGGG | 0.76 | ||
| P11 | IFRIEKGKIVKSVEMNAPNYHYEESSSGGGCGGGKGAINTSLPFQNIHPITIGKSPKYVK | KGAINTSLPFQNIHPI | 0.93 |
| KGKIVKSVEMNAPNYH | 0.88 | ||
| SVEMNAPNYHYEESSS | 0.86 | ||
| PFQNIHPITIGKSPKY | 0.43 | ||
| P14 | VNSDTVGWSWPDGAELPFTIDKGGGGCGGGGKTSSWPNHDSNKGVTAASPHAGAKSFYKN | GGGGKTSSWPNHDSNK | 0.93 |
| TIDKGGGGCGGGGKTS | 0.88 | ||
| SDTVGWSWPDGAELPF | 0.80 | ||
| SNKGVTAASPHAGAKS | 0.79 | ||
| SWPNHDSNKGVTAASP | 0.69 |
Note: a higher score indicates a higher probability that the peptide is an epitope
Immunogenicity of peptides in rabbits and mice
To confirm the immunogenicity of these peptides in vivo, we analyzed sera obtained from rabbits, and the peptides that induced the highest antibody titers in rabbits were used to immunize mice intranasally. IgG in sera and IgA in nasal washes, as well as neutralizing antibodies, were then detected.
The unconjugated P1 induced the strongest IgG immune response in rabbits (Fig. 3A). This result means that P1 could be immunogenic because it induced IgG antibodies in the absence of KLH [19]. According to epitope predictions, P1 contains T-cell (MHC II) and B-cell epitopes and is located in the middle of protein with 360º of exposure. P1 was included in peptides P9 and P11. P1 did not induce an immunogenic response when it was hybridized to P2 to yield P9; however, it induced a stronger IgG immune response when it was hybridized to P5 (NA) to yield P14.
Fig. 3.
Peptide-specific IgG antibody titers in the sera of rabbits immunized with unconjugated peptides or peptides conjugated with KLH. Serial dilutions of sera from rabbits inoculated with unconjugated peptides (panel A) or peptides conjugated with KLH (panel B) were added to microplates that had been coated with different peptides. IgG antibodies were detected using a secondary Ab specific for rabbit IgG (1:3000). Sera from rabbits immunized with either the unconjugated peptide 1 or with hybrid peptides P11 and P14 conjugated with KLH showed the highest titer (1:800), whereas sera against the remaining peptides had a lower titer (A490 <0.5; 1:800). Samples of serum from a pre-immune rabbit were used as controls. No peptide-specific IgG responses were found in pre-immune sera. Individual samples were run in duplicate, and the data are shown as the mean ± SD
As expected, the peptides P11 and P14 induced the strongest immune response when conjugated to KLH. However, P9 induced a poor immune response (Fig. 3). P5 hybridized with P1 to yield P11 resulted in an immune response that could be attributed to P1. Although P1 is present in P9, it is located far from KLH. The position of the peptide in the hybrid could therefore explain the experimental results.
P1 in the P9-KLH complex could be located at the KLH protein surface, and this location makes proteasome processing for presentation by MHC more difficult than the P11-KLH complex. The better immunogenic response obtained with P1 alone or P1 hybridized with P5 to yield P11 might have been due to the fact that P1 is very rich in basic residues, which have been shown to be important for immunogenicity [36]. P1 was capable of inducing an immune response despite having a lower molecular weight than P11 and P14. Similar results have been reported previously for another peptide [19].
As reported previously, the immunogenicity of P14 could be due to its NA epitope [19]. As mentioned above, we tested the immunogenicity of peptides conjugated to KLH to investigate the effect of KLH on the response to P9. Surprisingly, P9 still induced a weaker immune response than P11 despite the presence of P1 (Fig. 3, panel B). These results confirmed that the peptide sequence was of key importance for immunogenicity. The carriers could play an important role due to their ability to affect the recognition pattern on MHC regardless of the presence of basic residues that are capable of recognizing important sites, as reported by Carrillo-Vazquez et al. [36].
The IgG immune response is a T-cell-dependent response because the class switch of the IgG isotype requires the interaction of peptides with MHC II molecules and T helper (CD4 T) cells [55, 56]. Thus, our results indirectly demonstrate that immunogenic peptides induce T helper cells restricted to MHC-II.
Using ELISA, the immunogenicity of peptides in the mucosal compartment was examined in mice that were immunized intranasally with peptides mixed with CT (Fig. 4). The IgG and IgA antibody responses to the peptides were similar in the serum samples. Intranasal immunization induced higher titers of IgG and IgA antibodies to P11, lower titers to P14, and very low titers to the unconjugated peptides P1, P4, and P5 (Fig. 4, panels A and B). These results suggest that P1 requires other structural changes for proteasome protection or immunological transport in the P11 hybrid. To investigate the antibody responses in the mucosal compartment, the levels of IgG and IgA antibodies against the peptides were measured in nasal washes (Fig. 4, panels C and D). We observed that P11, P1 and P14 induced higher titers of IgG antibody, whereas the IgA antibody response was highest against P1 and P4, followed by P11, P14 and P5.
Fig. 4.
Peptide-specific IgG or IgA antibody titers in sera and nasal washes of mice immunized with different peptides. Serial dilutions of sera (panels A-B) or nasal washes (panels C-D) from mice inoculated with different peptides were added to microplates that had been coated with specific peptides. Antibodies were detected using a secondary Ab specific for mouse IgG (1:3000) or IgA (1:500). Samples of serum from pre-immune mice were used as controls, and no peptide-specific IgG or IgA responses were found in pre-immune sera or nasal washes, respectively. Individual samples were run in triplicate, and the data are shown as the mean ± SD
Surprisingly, all peptides (both conjugated [P11 and P14] and unconjugated [P1, P4, and P5] peptides) induced high IgA antibody titers in nasal washes, but peptide 1 yielded the highest titers.
The production of IgA antibodies in nasal mucosa requires that dendritic cells process and present peptides on class II MHC molecules (pMHC) to CD4+ T cells. In turn, these helper cells promote IgA class-switching recombination and affinity maturation of IgA-committed B-cells [57]. Thus, our results indicate that the peptides induced CD4+ T helper cells that recognize peptides in the form of an MHC-II–peptide complex [55, 57, 58].
In summary, intranasal immunization of mice with P11 and P14 along with CT induced specific IgG and IgA antibodies in serum and nasal mucosa. These antibodies could protect the host against virus invasion. According to the current concept, IgA is the dominant antibody involved in protection against infection in the nasal compartment (upper respiratory tract), whereas serum IgG, which diffuses into the lower respiratory tract, is predominantly involved in the protection of the lungs [59–61].
The peptide-induced heterosubtypic antibody response
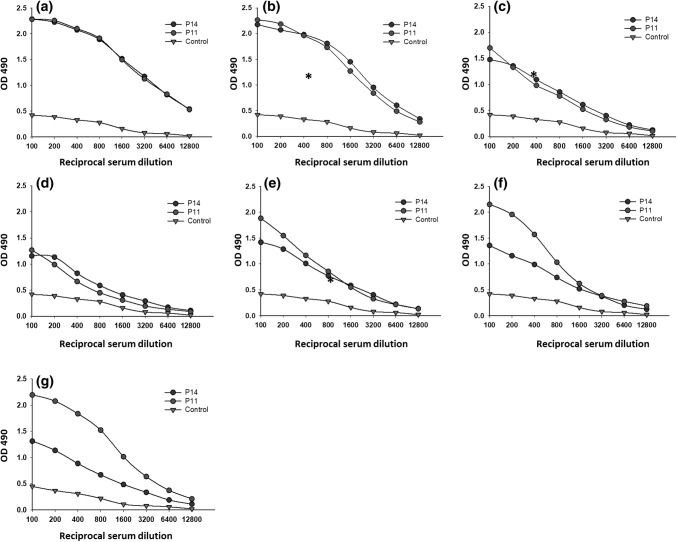
Finally, to test for substantial heterosubtypic binding activity, sera against P11 and P14 were tested for reactivity with purified hemagglutinin proteins from different subtypes and strains. We focused on testing P11 and P14 due to the promising results obtained with these peptides. HAs from a seasonal virus (A/Brisbane/59/2007(H1N1)), three pandemic viruses (A/California/04/2009 (H1N1), A/Aichi/2/1968(H3N2), and A/Japan/305/1957(H2N2)) from the 2009, 1968 and 1957 pandemics, and an avian virus (A/Shanghai/2/2013 (H7N9)) were used in these experiments.
As shown in Fig. 5A-G, the response patterns of the sera anti-P11 and anti-P14 against the hemagglutinins was similar for most hemagglutinins, with the exception of A/Brisbane/59/2007 (H1N1) and A/Puerto Rico/8/34 (H1N1), both of which are H1N1 strains (Fig. 5F and G). Both sera reacted similarly against the H7N9 subtype. The responses of P11 and P14 against the H7N9 subtype were not clear from the bioinformatics studies (see Fig. 1). The linear epitopes could not be evaluated due to their low similarity to the H7N9 protein sequence, suggesting that a conformational epitope was involved. Conformational epitopes are capable of inducing immune responses despite being discontinuous epitopes and are recognized by antibodies as linear epitopes in a tertiary structure [62].
Fig. 5.
Rabbit IgG antibodies against hybrid peptides P11 and peptide P14 recognized recombinant hemagglutinins (rHA) from several influenza A viruses: A) A/California/07/2009, B) A/Shanghai/2/2013, C), A/Aichi/2/1968 D) A/Cambodia/S1211394/2008 E) A/Japan/305/1957, F) A/Puerto Rico/8/34 and G) A/Brisbane/59/2007 . Serial dilutions of sera from rabbits inoculated with peptides conjugated to KLH were added to microplates that had been coated with purified recombinant hemagglutinins (0.5 μg). Antibodies against P11 and P14 had similar binding activity to the majority of the rHAs, except for A/Brisbane/59/2007 and A/Puerto Rico/8/1934, both of which are H1N1 subtypes. As controls, serum samples from pre-immunized rabbits were tested against the specific peptide
Antibodies produced in response to the A/California/04/09 pandemic virus cross-reacted with several influenza virus strains, including A/Brisbane/59/2007 (H1N1), A/Puerto Rico/8/34 (H1N1), A/Aichi/2/1968 (H3N2), and A/Anhui/1/13 (H7N9) [63–66]. The origin of these cross-reactive antibodies may be cross-reactive T and B cells specific for conserved epitopes in the hybrid peptides P11 and P14. Various studies have demonstrated that cross-protection against influenza virus strains may be due to cross-reactive T cells [67–69]. A previous in silico analysis provided a list of potential cross-reactive T-cell epitopes, including the sequences CPKYVKSTK and HAGAKSFYKNL, which were present in P1, P11 and P14 [70]. Also, as shown in Fig. 6, P11 and P14 induced antibodies capable of recognizing purified NA protein from a viral stock in ELISA studies measuring IgG antibodies.
Fig. 6.
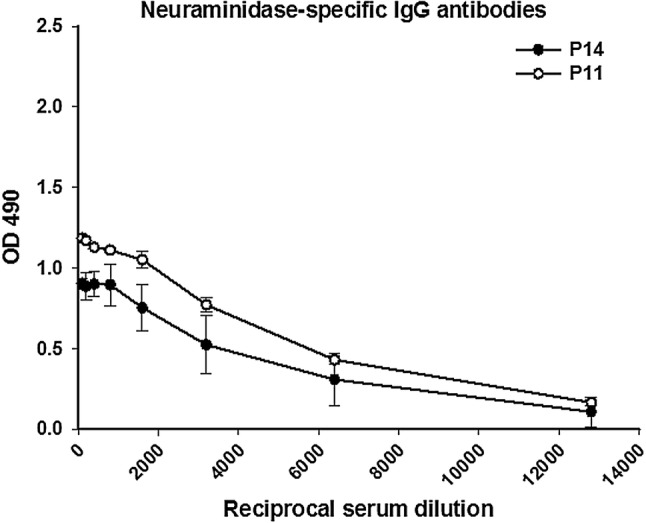
NA-specific IgG antibody titers in sera from rabbits immunized with different peptides. Serial dilutions of sera from rabbits inoculated with peptide 14 (P14), and peptide 11 (P11) were added to microplates previously coated with neuraminidase protein (0.5 µg). IgG antibodies were detected using a secondary Ab specific for rabbit IgG (1:3000). Individual samples were run in duplicate, and the data are shown as the mean ± SD
According to ConA envELISA studies, P11 and P14 are capable of inducing antibodies in rabbits that recognize wild-type viral antigens (immobilized with ConA) from an influenza virus H1N1 strain at dilutions of 1:200 and 1:800 (Fig. 7) with significant statistical differences (one-way ANOVA, p < 0.0001) compared to pre-immune sera at the same dilutions. These absorbance values are equal to values others have reported for patients who were previously infected with influenza virus [36].
Fig. 7.

Trapping ELISA. Antibodies from rabbits immunized with P11 or P14 recognized influenza virus immobilized with ConA. Pre-immune serum samples were used as negative controls, and a human serum was used as a positive control (p < 0.0001, pre-immune vs. immunized rabbit)
In the hemagglutination inhibition assay, it was found that sera from rabbits that had been inoculated with P11 and P14 inhibited hemagglutination at a dilution of 1:384 and 1:960, respectively (Table 7). There are reports in which viral antigens (surface glycoproteins) can be immobilized to be recognized by antibodies in an ELISA, as reported elsewhere for influenza virus and dengue virus [36, 71]. In this study, antibodies from rabbits immunized with P11 and P14 recognized viral particles of strain (A Puerto Rico/916/34) immobilized with ConA (Fig. 7), and they also inhibited hemagglutination with titers higher than 40 (Fig. 7), which decreases the risk of infection to 50% [72]. The above coincides with the neutralizing capacity of antibodies induced in mice immunized with peptides 11 and 14 (reciprocal of PRNT50 titer of 380 and 1600, respectively; Table 7). These results indicate that in silico studies can be used to identify potential protective epitopes.
Table 7.
Hemagglutination inhibition and virus neutralization assays. The hemagglutination inhibition assay was carried out using antibodies from rabbits immunized with P11 and P14. The neutralizing antibody titer was determined using a pool of serum from mice immunized with P11 and P14
| Peptide | HI | Reciprocal of PRNT50 titer |
|---|---|---|
| P11 | 1:384 | 380 |
| P14 | 1:960 | 1600 |
In summary, we compared amino acid sequences (Fig. 1) to provide a plausible explanation for the observed cross-protection between 2009 H1N1 and other influenza viruses [64]. Only P1 alone produced a response in vivo. The observed responses to P11 and P14 could have been due to coupling to KLH. Additionally, P11 contained P1 and P14 contained a recently reported peptide [19]. However, the responses against H7N9 were apparently not due to the presence of linear epitopes, suggesting that the peptides were capable of adopting 3D structures that formed a conformational epitope [62]. Rabbit antibodies to P11 and P14 recognized HA proteins from both group 1 (H1, H2, and H5) and group 2 (H3 and H7) influenza A viruses belonging to three different clades: the H1 clade (H1, H2, and H5), the H3 clade (H3), and the H7 clade (H7). Additionally, the antibodies also recognized purified neuraminidase from the viral stock and viral particles immobilized in the trapping ELISA, and they were able to inhibit hemagglutination and neutralize the virus.
Conclusions
The humoral immune responses against HA and NA are the primary means of resistance to influenza virus infection [68]; therefore, hybrid peptides composed of HA and NA peptides can be used as potent synthetic vaccines. Because antibodies against the hybrid peptides P11 and P14 had broad heterosubtypic activity against the antigenically diverse H1, H2, H3, H5, and H9 influenza subtypes, these peptides might be used as broad-spectrum agents for prophylaxis (to induce neutralizing antibodies) and also for treatment of human or avian influenza infections, since the antibodies recognize the neuraminidase protein and could potentially prevent the release of new viral progeny during infection. The use of bioinformatics tools enabled the identification of potential epitopes for vaccine purposes. Some of these epitopes could act as immunogenic agents alone, offering several advantages, such as low cost and ease of handling without special treatment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors were supported by ICyTDF (Dr. Campos), the Instituto Politécnico Nacional (IPN), BEIFI, COFAA-IPN, and grants CB-254600, PDCPN-782 (CONACYT), 241339 (CONACYT), and SIP-IPN (SIP20171881).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
G. Lizbeth Ramírez-Salinas, Jazmín García-Machorro, Saúl Rojas-Hernández, Rafael Campos-Rodríguez contributed equally to this work.
Deceased; this work is a dedication in memoriam to Dr. Rafael Campos-Rodríguez.
Contributor Information
Jazmín García-Machorro, Email: jazzgama@hotmail.com.
Rafael Campos-Rodríguez, Email: citlicampos@gmail.com.
José Correa-Basurto, Email: corrjose@gmail.com.
References
- 1.Szewczyk B, Bienkowska-Szewczyk K, Krol E. Introduction to molecular biology of influenza a viruses. Acta Biochim Pol. 2014;61(3):397–401. doi: 10.18388/abp.2014_1857. [DOI] [PubMed] [Google Scholar]
- 2.Schild GC. The influenza virus: Antigenic composition and immune response. Postgrad Med J. 1979;55(640):87–97. doi: 10.1136/pgmj.55.640.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrauwen EJ, Fouchier RA. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microb Infect. 2014;3(2):e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sym D, Patel PN, El-Chaar GM. Seasonal, avian, and novel H1N1 influenza: prevention and treatment modalities. Ann Pharmacother. 2009;43(12):2001–2011. doi: 10.1345/aph.1M557. [DOI] [PubMed] [Google Scholar]
- 5.Palache A, Oriol-Mathieu V, Abelin A, Music T. Seasonal influenza vaccine dose distribution in 157 countries (2004–2011) Vaccine. 2014;32(48):6369–6376. doi: 10.1016/j.vaccine.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol. 2014;385:359–375. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 7.Ping J, Lopes TJ, Nidom CA, Ghedin E, Macken CA, Fitch A, et al. Development of high-yield influenza A virus vaccine viruses. Nat Commun. 2015;6:8148. doi: 10.1038/ncomms9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashem AM. Prospects of HA-based universal influenza vaccine. Biomed Res Int. 2015;2015:414637. doi: 10.1155/2015/414637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krammer F, Palese P, Steel J. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol. 2015;386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 10.Laursen NS, Wilson IA. Broadly neutralizing antibodies against influenza viruses. Antiviral Res. 2013;98(3):476–483. doi: 10.1016/j.antiviral.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schepens B, De Vlieger D, Saelens X. Vaccine options for influenza: thinking small. Curr Opin Immunol. 2018;53:22–29. doi: 10.1016/j.coi.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Eichelberger MC, Morens DM, Taubenberger JK. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol. 2018;53:38–44. doi: 10.1016/j.coi.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328(5976):357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 15.Wu NC, Wilson IA. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol. 2017;429(17):2694–2709. doi: 10.1016/j.jmb.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu NC, Wilson IA. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol. 2018;25(2):115–121. doi: 10.1038/s41594-018-0025-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, et al. NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? MBio. 2018 doi: 10.1128/mbio.02332-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 19.Loyola PK, Campos-Rodriguez R, Bello M, Rojas-Hernandez S, Zimic M, Quiliano M, et al. Theoretical analysis of the neuraminidase epitope of the Mexican A H1N1 influenza strain, and experimental studies on its interaction with rabbit and human hosts. Immunol Res. 2013;56(1):44–60. doi: 10.1007/s12026-013-8385-z. [DOI] [PubMed] [Google Scholar]
- 20.Marcelin G, Sandbulte MR, Webby RJ. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol. 2012;22(4):267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López S, Arias C (2010) Influenza A: Biología, vacunas, y origen del virus pandémico A/H1N1. Revista Digital Universitaria
- 22.Oany AR, Emran AA, Jyoti TP. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Des Devel Ther. 2014;8:1139–1149. doi: 10.2147/DDDT.S67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gori A, Longhi R, Peri C, Colombo G. Peptides for immunological purposes: design, strategies and applications. Amino Acids. 2013;45(2):257–268. doi: 10.1007/s00726-013-1526-9. [DOI] [PubMed] [Google Scholar]
- 24.Pellicci DG, Uldrich AP, Le Nours J, Ross F, Chabrol E, Eckle SB, et al. The molecular bases of delta/alphabeta T cell-mediated antigen recognition. J Exp Med. 2014;211(13):2599–2615. doi: 10.1084/jem.20141764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinilla C, Appel JR, Judkowski V, Houghten RA. Identification of B cell and T cell epitopes using synthetic peptide combinatorial libraries. Curr Protoc Immunol. 2012 doi: 10.1002/0471142735.im0905s99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 31.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 33.de Beer TA, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42(Database issue):D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macarthur MW, Laskowski RA, Thornton JM. Knowledge-based validation of protein-structure coordinates derived by X-ray crystallography and nmr-spectroscopy. Curr Opin Struc Biol. 1994;4(5):731–737. doi: 10.1016/S0959-440x(94)90172-4. [DOI] [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Carrillo-Vazquez JP, Correa-Basurto J, Garcia-Machorro J, Campos-Rodriguez R, Moreau V, Rosas-Trigueros JL, et al. A continuous peptide epitope reacting with pandemic influenza AH1N1 predicted by bioinformatic approaches. J Mol Recognit. 2015;28(9):553–564. doi: 10.1002/jmr.2470. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130(3):319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen M, Lund O. NN-align An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 42.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101(5):4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 43.Glykos NM. Software news and updates. Carma: a molecular dynamics analysis program. J Comput Chem. 2006;27(14):1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 44.Contis-Montes de Oca A, Carrasco-Yépez M, Campos-Rodríguez R, Pacheco-Yépez J, Bonilla-Lemus P, Pérez-López J, Rojas-Hernández S. Neutrophils extracellular traps damage Naegleria fowleri trophozoites opsonized with human IgG. Parasite Immunol. 2016;38(8):481–495. doi: 10.1111/pim.12337. [DOI] [PubMed] [Google Scholar]
- 45.Kawaoka Y, Neumann G. Influenza viruses: an introduction. Methods Mol Biol. 2012;865:1–9. doi: 10.1007/978-1-61779-621-0_1. [DOI] [PubMed] [Google Scholar]
- 46.Morens DM, Halstead SB, Repik PM, Putvatana R, Raybourne N. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: comparison of the BHK suspension test with standard plaque reduction neutralization. J Clin Microbiol. 1985;22(2):250–254. doi: 10.1128/JCM.22.2.250-254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KH, To KK, Hung IF, Zhang AJ, Chan JF, Cheng VC, Tse H, Che XY, Chen H, Yuen KY. Differences in antibody responses of individuals with natural infection and those vaccinated against pandemic H1N1 2009 influenza. Clin Vaccine Immunol. 2011;18(5):867–873. doi: 10.1128/CVI.00555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinne M, Haake D. Immuno-fluorescence assay of leptospiral surface-exposed proteins. JoVE. 2011 doi: 10.3791/2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klasse PJ. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv Biol. 2014 doi: 10.1155/2014/157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camacho CJ, Katsumata Y, Ascherman DP. Structural and thermodynamic approach to peptide immunogenicity. PLoS Comput Biol. 2008;4(11):e1000231. doi: 10.1371/journal.pcbi.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altenburg AF, Rimmelzwaan GF, de Vries RD. Virus-specific T cells as correlate of (cross-)protective immunity against influenza. Vaccine. 2015;33(4):500–506. doi: 10.1016/j.vaccine.2014.11.054. [DOI] [PubMed] [Google Scholar]
- 52.Hufford MM, Kim TS, Sun J, Braciale TJ. The effector T cell response to influenza infection. Curr Top Microbiol Immunol. 2015;386:423–455. doi: 10.1007/82_2014_397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hufford MM, Kim TS, Sun J, Braciale TJ. Antiviral CD8+ T cell effector activities in situ are regulated by target cell type. J Exp Med. 2011;208(1):167–180. doi: 10.1084/jem.20101850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu C, Ellebedy AH, Wrammert J, Ahmed R. B cell responses to influenza infection and vaccination. Curr Top Microbiol Immunol. 2015;386:381–398. doi: 10.1007/82_2014_425. [DOI] [PubMed] [Google Scholar]
- 55.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Groot AS. Exploring the immunome: a brave new world for human vaccine development. Hum Vaccin. 2009;5(12):790–793. doi: 10.4161/hv.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bienenstock J, McDermott MR. Bronchus- and nasal-associated lymphoid tissues. Immunol Rev. 2005;206:22–31. doi: 10.1111/j.0105-2896.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 58.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asahi Y, Yoshikawa T, Watanabe I, Iwasaki T, Hasegawa H, Sato Y, et al. Protection against influenza virus infection in polymeric Ig receptor knockout mice immunized intranasally with adjuvant-combined vaccines. J Immunol. 2002;168(6):2930–2938. doi: 10.4049/jimmunol.168.6.2930. [DOI] [PubMed] [Google Scholar]
- 60.Langley JM, Aoki F, Ward BJ, McGeer A, Angel JB, Stiver G, et al. A nasally administered trivalent inactivated influenza vaccine is well tolerated, stimulates both mucosal and systemic immunity, and potentially protects against influenza illness. Vaccine. 2011;29(10):1921–1928. doi: 10.1016/j.vaccine.2010.12.100. [DOI] [PubMed] [Google Scholar]
- 61.van Riet E, Ainai A, Suzuki T, Hasegawa H. Mucosal IgA responses in influenza virus infections; thoughts for vaccine design. Vaccine. 2012;30(40):5893–5900. doi: 10.1016/j.vaccine.2012.04.109. [DOI] [PubMed] [Google Scholar]
- 62.Lucchese G, Sinha AA, Kanduc D. How a single amino acid change may alter the immunological information of a peptide. Front Biosci. 2012;4:1843–1852. doi: 10.2741/e506. [DOI] [PubMed] [Google Scholar]
- 63.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skountzou I, Koutsonanos DG, Kim JH, Powers R, Satyabhama L, Masseoud F, et al. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J Immunol. 2010;185(3):1642–1649. doi: 10.4049/jimmunol.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen JR, Yu YH, Tseng YC, Chiang WL, Chiang MF, Ko YA, et al. Vaccination of monoglycosylated hemagglutinin induces cross-strain protection against influenza virus infections. Proc Natl Acad Sci USA. 2014;111(7):2476–2481. doi: 10.1073/pnas.1323954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. 2015;125(3):1255–1268. doi: 10.1172/JCI74374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenbaum JA, Kotturi MF, Kim Y, Oseroff C, Vaughan K, Salimi N, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci USA. 2009;106(48):20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol. 2010;84(7):3312–3319. doi: 10.1128/jvi.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richards KA, Nayak J, Chaves FA, DiPiazza A, Knowlden ZA, Alam S, et al. Seasonal influenza can poise hosts for CD4 T-cell immunity to H7N9 Avian influenza. J Infect Dis. 2015;212(1):86–94. doi: 10.1093/infdis/jiu662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Groot AS, Ardito M, McClaine EM, Moise L, Martin WD. Immunoinformatic comparison of T-cell epitopes contained in novel swine-origin influenza A (H1N1) virus with epitopes in 2008–2009 conventional influenza vaccine. Vaccine. 2009;27(42):5740–5747. doi: 10.1016/j.vaccine.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Machorro J, Lopez-Gonzalez M, Barrios-Rojas O, Fernandez-Pomares C, Sandoval-Montes C, Santos-Argumedo L, et al. DENV-2 subunit proteins fused to CR2 receptor-binding domain (P28)-induces specific and neutralizing antibodies to the Dengue virus in mice. Hum Vaccines Immunother. 2013;9(11):2326–2335. doi: 10.4161/hv.25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fox A, le Mai Q, le Thanh T, Wolbers M, Le Khanh Hang N, Thai PQ, et al. Hemagglutination inhibiting antibodies and protection against seasonal and pandemic influenza infection. J Infect. 2015;70(2):187–196. doi: 10.1016/j.jinf.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.