Abstract
Most bacteria form biofilms, which are thick multicellular communities covered in extracellular matrix. Biofilms can become thick enough to be even observed by the naked eye, and biofilm formation is a tightly regulated process. Paracoccus denitrificans is a non-motile, Gram-negative bacterium that forms a very thin, unique biofilm. A key factor in the biofilm formed by this bacterium is a large surface protein named biofilm-associated protein A (BapA), which was recently reported to be regulated by cyclic diguanosine monophosphate (cyclic-di-GMP or c-di-GMP). Cyclic-di-GMP is a major second messenger involved in biofilm formation in many bacteria. Though cyclic-di-GMP is generally reported as a positive regulatory factor in biofilm formation, it represses biofilm formation in P. denitrificans. Furthermore, quorum sensing (QS) represses biofilm formation in this bacterium, which is also reported as a positive regulator of biofilm formation in most bacteria. The QS signal used in P. denitrificans is hydrophobic and is delivered through membrane vesicles. Studies on QS show that P. denitrificans can potentially form a thick biofilm but maintains a thin biofilm under normal growth conditions. In this review, we discuss the peculiarities of biofilm formation by P. denitrificans with the aim of deepening the overall understanding of bacterial biofilm formation and functions.
Keywords: Biofilm, BapA, Cyclic-di GMP, Quorum-sensing, Membrane vesicle, Paracoccus denitrificans
Introduction
Most bacteria in the environment form bacterial communities embedded in extracellular polymeric substance matrix known as biofilms (Chan et al. 2016). Biofilms are related to human health as well as infections and have a potential role in colonization of the gut by commensal bacteria which are still largely unexplored (Donlan 2001; Sekirov et al. 2010). Biofilms are also actively used in applications such as biocatalysts (Halan et al. 2012; Rosche et al. 2009), and in wastewater treatment, where bacteria are present in aggregated forms of cells, also known as activated sludges and granules. The cells inside biofilms show a different physiology from that of the planktonic counterparts, leading to increased resistance to environmental stresses such as pH shift, osmotic shock, and UV radiation (Davey and O'toole 2000; Flemming 1993). Hence, studying biofilm would lead to our better understanding of the bacterial ecology and optimization of biofilm applications.
Recent studies highlight the importance of flagella and pili driven motility in biofilm formation. Biofilm formation is first initiated by attachment of planktonic cells to the surface; this step is called initial attachment (Stoodley et al. 2002). In many bacteria, motility accelerates surface attachment of cells. Initial attachment is followed by irreversible attachment, where cells initiate formation of highly structured microcolonies and embed themselves in EPS. In Vibrio cholerae, type IV pili, together with flagella, are utilized for solid surface motility and to accelerate surface attachment (Utada et al. 2014), whereas in Pseudomonas aeruginosa, type IV pili are required for microcolony formation (O'Toole and Kolter 1998). Eventually, the subpopulations of cells in the biofilm disperse through motility and leave the biofilm (Sauer et al. 2002). While flagella and pili driven motility has been shown to play an important role in each step of biofilm formation, little is known about how non-motile bacteria form biofilms, especially Gram-negative bacteria.
Paracoccus species are Gram-negative Alphaproteobacteria often present in soil, activated sludges, and aerobic granular sludges (Cydzik-Kwiatkowska 2015; Pelissari et al. 2017; Xia et al. 2018). One of the most studied Paracoccus species is Paracoccus denitrificans that is a non-motile, denitrifying bacterium and is used as a model organism for studies of denitrification, respiration chains, and polyhydroxyalkanoate production (Kelly et al. 2006; Kojima et al. 2004; Lycus et al. 2018). In contrast to what has been shown in most bacteria that form highly structured architectures, P. denitrificans forms a peculiarly thin biofilm that consists of almost a monolayer of cells (Yoshida et al. 2017). Recent studies show a critical role of an adhesion protein BapA in initiating biofilm formation in P. denitrificans (Yoshida et al. 2017). Further studies showed that the biofilm structure is regulated by an intracellular second messenger cyclic diguanosine monophosphate (Cyclic-di-GMP) and by bacterial communication (Kumar and Spiro 2017; Morinaga et al. 2018; Toyofuku et al. 2017) (Fig. 1). Furthermore, P. denitrificans produces a hydrophobic signal that is carried by membrane vesicles (MVs) (Toyofuku et al. 2017). Here, we review current knowledge on biofilm formation by P. denitrificans and discuss its associated factors and functions.
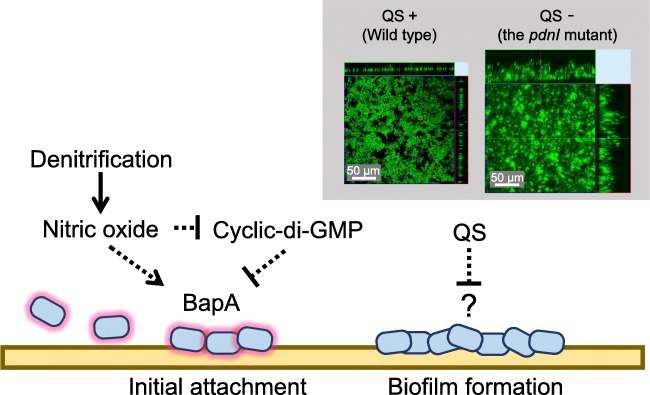
Fig. 1.
Schematic image of biofilm formation in P. denitrificans. BapA protein is necessary for the attachment to initiate biofilm formation (Kumar and Spiro 2017; Yoshida et al. 2017). Cyclic-di GMP inhibits bapA gene expression or BapA protein secretion (Kumar and Spiro 2017). QS inhibits biofilm formation (Morinaga et al. 2018). Confocal laser scanning microscopy images show biofilm structure of wildtype P. denitrificans and the pdnI mutant. Reproduced with permission from (Morinaga et al. 2018) with modifications
Paracoccus biofilms in applications
Paracoccus species are shown to form biofilms that have important applications, especially in bioreactors, while the detailed mechanism of biofilm formation in this genus is largely unknown (Nisha et al. 2015; Singh et al. 2015). Due to their versatile metabolisms, Paracoccus species are considered as important organisms for bioremediation and are suggested to be involved in degrading and removing refractory pollutants such as 4-chlorophenol from wastewater (Gomez-Acata et al. 2018; Zhao et al. 2015). Several Paracoccus species can carry out denitrification, and are frequently isolated from denitrifying reactor (Neef et al. 1996; Shi et al. 2013). In addition, P. denitrificans is isolated from microbial fuel cell anode biofilm communities and can generate electricity using formic acid as an electron donor (Kiely et al. 2010). Furthermore, P. denitrificans can be immobilized in gels with Nitrosomonas europaea for efficient nitrogen removal from wastewater and have great potentials in their applications (Kokufuta et al. 1988; Uemoto and Saiki 1996). Hence, understanding biofilm formation in Paracoccus species may optimize the functions of biofilm-based bioreactors.
Biofilm structure of P. denitrificans
Biofilms are generally a thick layer of cells where microcolonies with mushroom-like structures or mound-like structures could often be observed inside. The thick layer and metabolic activity of cells form microenvironments in biofilms, with micro-scale gradients or patches of chemicals, such as electron acceptors and donors, nutrients, pH, and cell-to-cell communication signals (Flemming et al. 2016). The biofilm architecture could be highly organized, even including water channels to facilitate the transport of liquids (Wilking et al. 2013). One of the remarkable features of P. denitrificans biofilm is that it forms a very thin and flat biofilm with less than ∼4 μm thickness in a liquid-air interface (Yoshida et al. 2017), making it an interesting model for biofilm formation as compared to other model organisms such as P. aeruginosa that form multilayered biofilms (Heydorn et al. 2002; Sauer et al. 2002) Confocal laser scanning microscopy (CLSM) analysis showed that the cells in the biofilm are often densely packed and frequently arranged in chains (Yoshida et al. 2017). Since P. denitrificans is non-motile, it is suggested that the cells spread on the surface by cell division, resulting in dense biofilms with chains of cells. Similarly, a monolayer biofilm is also observed in another Paracoccus species which biofilm morphology changes depending on the culture conditions (Srinandan et al. 2010).
BapA protein is critical in biofilm formation by P. denitrificans
Given the thin layer of cells and lack of motility, initial attachment would have more direct impact on the biofilm structure in P. denitrificans compared to other bacteria that undergo several stages to finally form a mature biofilm. Bacteria secrete various types of EPS to adhere to surfaces and form biofilms. The main EPS components are polysaccharides, proteins, lectins, and nucleic acids (Flemming and Wingender 2010; Schooling and Beveridge 2006). Bacteria often produce several types of EPS that interact with each other and play different roles in forming biofilms. For example, V. cholerae produces an extracellular Vibrio polysaccharide (VPS), biofilm-associated protein 1 (Bap1), and the matrix proteins, rugosity and biofilm structure modulator A (RbmA) and rugosity and biofilm structure modulator C (RbmC) (Berk et al. 2012; Yan et al. 2016). VPS in the biofilms maintain the 3-dimensional biofilm structure by accumulation between cells, whereas Bap1 acts as cell-to-surface adhesion and RbmA functions as cell-to-cell linkage. Bap is a group of proteins that are monomeric repetitive adhesins. Baps in Gram-negative bacteria are repeats-in-toxin (RTX) family of proteins and composed of three domains: N-termini, core large repeats and C-termini (Satchell 2011). Molecular sizes of Baps vary, ranging from 1873 to 8682 amino acids (aa), depending on the core repeat sequences (Lasa and Penades 2006). Baps are localized at cell surfaces and mediate cell-to-surface and/or cell-to-cell adhesion. In Gram-negative bacteria, Baps are secreted directly from the cytoplasm to extracellular space by their ABC transporters known as Type I secretion system. RTX adhesins contain glycine-aspartate-rich peptide repeats at the C-termini and each peptide binds to a calcium ion (Satchell 2011), which is important at least for some Baps to function (Martinez-Gil et al. 2012). One of the best studied Bap proteins in Gram-negative bacteria is LapA, which is involved in cell adhesion of Pseudomonas fluorescens (Hinsa et al. 2003).
An analysis of EPS components in P. denitrificans using EPS-degrading enzymes suggested that an extracellular proteinaceous component influences biofilm formation (Yoshida et al. 2017). In line with this observation, the biofilm formation of P. denitrificans requires BapA for its initial cell attachment (Kumar and Spiro 2017; Yoshida et al. 2017). BapA protein of P. denitrificans contains a large repeat region consisting of 891 tandem repeats of aspartate and alanine although the precise roles of this extremely acidic sequence are still unknown. This repeat sequence is unique to this protein, while other Baps typically contain 80 to 300 aa repeat unit sequences (Satchell 2011). BapA protein of P. denitrificans is secreted extracellularly by Type I secretion system BapBCD. BapA is localized at the cell surface and makes the cell surface more hydrophobic, which presumably drives cells to attach to the substratum (Yoshida et al. 2017). It was further shown that BapA is a calcium-binding protein and it is necessary for cell attachment (Kumar and Spiro 2017). Consistent with these results, divalent cations are shown to enhance biofilm formation in a Paracoccus species and EDTA treatment can detach biofilm formation (Srinandan et al. 2010).
Cyclic-di-GMP regulation of biofilm formation in P. denitrificans
BapA protein regulation in P. denitrificans is not well studied, but nitric oxide (NO) has been reported to control the BapA level that partially involves cyclic diguanosine monophosphate (cyclic-di-GMP). Cyclic-di-GMP is an intracellular second messenger involved in biofilm formation of many bacteria by controlling EPS production and motility. For example, in P. aeruginosa, accumulation of cyclic-di-GMP downregulates cell motility and upregulates EPS production (Valentini and Filloux 2016). Low concentration of cyclic-di GMP due to degradation by phosphodiesterases (PDEs) induces cell dispersal from biofilms. The intracellular cyclic di-GMP level is controlled by its synthesis by diguanylate cyclase (DGC), and degradation by PDE. Cyclic-di-GMP is synthesized from GTP by DGC which has the GGDEF domains and degraded to 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) and/or GMP by PDEs which have EAL or HD-GYP domains (Valentini and Filloux 2016).
P. denitrificans possesses two proteins (Pden_3720; DgcA and Pden_3982; DgcB) with GGDEF domains and two proteins (Pden_0876; PdeA and Pden_2025; PdeB) with EAL domains (Kumar and Spiro 2017). DgcB has an N-terminal response regulator domain, implying a signal cascade from a sensor partner. dgcA overlaps with a gene coding a protein that has heme-nitric oxide/oxygen binding (H-NOX) domain. H-NOX protein commonly functions as a sensor for the gaseous signaling agent nitric oxide (NO) (Plate and Marletta 2013), suggesting that H-NOX protein in P. denitrificans regulates the biosynthesis of cyclic-di GMP. P. denitrificans produces NO as an intermediate of denitrification. Kumar and Spiro showed that NO stimulates biofilm formation in P. denitrificans (Kumar and Spiro 2017). Using a series of mutants, they showed that altering the NO and cyclic-di GMP levels controls the abundance of BapA protein on the cell envelope and biofilm formation. It is suggested that NO controls biofilm formation via both H-NOX/cyclic-di-GMP dependent and independent pathways. In other bacteria, NO often leads to biofilm dispersal (Arora et al. 2015; Barraud et al. 2009), and the opposite reaction or tolerance against NO could be one of the reasons why Paracoccus species become abundant in denitrifying reactors (Singh et al. 2015).
Quorum-sensing down-regulates biofilm formation in P. denitrificans
Another major regulatory system in biofilm formation of P. denitrificans is quorum sensing (QS). Bacteria can communicate with each other using signaling molecules, and the density-dependent bacterial communication is called quorum-sensing, where gene expression is regulated when a signal reaches a threshold concentration.
In many bacteria, QS influences biofilm formation such as shown in P. aeruginosa, where mutants defective in QS form thin and dense biofilm, while the wildtype forms thick biofilms (Davies et al. 1998). One of the signals that is produced in Gram-negative bacteria is N-acyl-homoserine lactones (AHLs). AHL consists of a lactone ring and an N-acyl side chain whose length ranges from 4 to 20 carbons, which may have additional modifications (Arashida et al. 2018). P. denitrificans produces N-hexadecanoyl-L-homoserine lactone (C16-HSL) as its QS signal (Schaefer et al. 2002), which is synthesized by a luxI homolog, pdnI (Toyofuku et al. 2017). The pdnI mutant strongly aggregates and forms a thick biofilm in comparison to the wildtype, indicating that QS down-regulates aggregation in P. denitrificans (Morinaga et al. 2018). It suggests that C16-HSL regulates EPS production, and this relation, once verified, would provide a direct link between QS and biofilm formation.
Interestingly, P. denitrificans can respond to other AHL signals in different manners and control biofilm formation and cell aggregation (Morinaga et al. 2018). Long-chain AHLs (C10 to C18-HSL) inhibit aggregation or biofilm formation similarly to C16-HSL. While P. denitrificans does not respond to short-chain AHLs (C4 to C8-HSL) alone, when C4- and C6-HSL are present together with C16-HSL, it can modulate the threshold concentration and less C16-HSL is required to inhibit cell aggregation of the pdnI mutant. On the other hand, more C16-HSL is required to inhibit aggregation in the presence of C8 and C10-HSL. Such interaction with other signals suggests how this bacterium forms biofilm in a polymicrobial community.
Membrane vesicle mediated bacterial communication
The classical QS model is based on the assumption that diffusible QS signals synchronously change target gene expression when the signal concentration reaches a certain threshold (Fuqua et al. 1994). However, hydrophobic signals such as the long-chain AHLs, including C16-HSL, cannot freely diffuse in an aquatic environment, and how hydrophobic signals are released and transmitted in the environment had always been a question. In P. denitrificans, it was shown that this bacterium uses membrane vesicles (MVs) to release these signal molecules from the cell and to deliver them to other cells. MVs are produced by most bacteria including Gram-negative and Gram-positive bacteria and are abundant in the EPS matrix of biofilms (Schooling and Beveridge 2006; Toyofuku et al. 2012, 2015, 2018).
The size of MVs ranges from 20 to 400 nm and they carry cellular components such as DNA, RNA, proteins, and metabolites as their cargo. MVs are involved in various bacterial and host-bacterial interactions and are also used as a platform for therapeutics such as vaccine development (Toyofuku et al. 2015). MVs isolated from P. denitrificans can induce QS targeted gene expression and can also regulate cell aggregation by delivering C16-HSL (Toyofuku et al. 2017). Furthermore, one MV contains a much higher amount of C16-HSL than the threshold concentration that would induce gene expression in a P. denitrificans cell. Signals packed in MVs would lead to binary signaling, where QS is triggered only in cells that receive MVs, out of the whole cell population. In addition, the calling distance of this digital signaling system would be longer than the free diffusion of signals, as packaging in MVs would prevent the signals from being diluted below the threshold concentration. MVs released from P. denitrificans also show a propensity of cell targeting and have a high affinity to kin species than other species such as P. aeruginosa and P. putida (Toyofuku et al. 2017) (Fig. 2). MVs can also absorb free long-chain AHLs from the environment that can be utilized by P. denitrificans (Morinaga et al. 2018). Hence, MVs are involved in trafficking signals that lead to different socioecological consequences. The delivery of hydrophobic signals by MVs is also reported in other bacteria (Brameyer et al. 2018; Feitosa-Junior et al. 2019; Mashburn and Whiteley 2005) and could be a general way to disperse the signals.
Fig. 2.

MVs isolated from Paracoccus denitrificans. MVs carry AHLs utilized for QS in P. denitrificans. The Transmission Electron Microscopy (TEM) image shows MVs of P. denitrificans. Reproduced with permission from (Toyofuku et al. 2017)
Conclusion
P. denitrificans forms a remarkably thin biofilm, which is unique when compared to other bacteria. Furthermore, in many bacteria, cyclic-di-GMP and QS positively regulate biofilm formation, while these factors inhibit biofilm formation in P. denitrificans. Disruption of QS system leads to thick biofilm formation, indicating that this bacterium possesses the ability to form thick biofilms while repressing it at normal growth conditions (Morinaga et al. 2018). Further understanding of environmental factors that control cyclic-di-GMP levels and QS, together with identification of QS-regulated biofilm formation factors, could be key in understanding biofilm formation. In addition, BapA protein, the key factor in the surface attachment of this bacterium, alters the hydrophobicity of the cells, and it would be of interest to examine the influence of BapA expression on MV delivery and C16-HSL signaling in biofilms. One of the advantages of keeping the biofilm thin is that each cell in the biofilm has more access to nutrients than in thick biofilms. However, this aspect is yet to be explored in detail. Biofilm thickness is a key property of its function (Suarez et al. 2019), and optimizing the uptake of nutrients by biofilms could be applied to industries including wastewater treatment process using biofilms. P. denitrificans has great potentials in its application for biotechnology, and understanding its biofilm formation may ultimately lead to optimize its functionality.
Acknowledgments
MT was supported by a Grant-in-Aid for Scientific Research (19H02866 and 19H05682) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). NN was supported by the Japan Science and Technology Agency, ERATO (JPMJER1502) and Grant-in-Aid for Scientific Research (19H05682 and 16H06382) from MEXT.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arashida N, Shimbo K, Terada T, Okimi T, Kikuchi Y, Hashiro S, Umekage S, Yasueda H. Identification of novel long chain N-acylhomoserine lactones of chain length C20 from the marine phototrophic bacterium Rhodovulum sulfidophilum. Biosci Biotechnol Biochem. 2018;82(10):1683–1693. doi: 10.1080/09168451.2018.1490168. [DOI] [PubMed] [Google Scholar]
- Arora DP, Hossain S, Xu Y, Boon EM. Nitric oxide regulation of bacterial biofilms. Biochemistry. 2015;54(24):3717–3728. doi: 10.1021/bi501476n. [DOI] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191(23):7333–7342. doi: 10.1128/jb.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337(6091):236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameyer S, Plener L, Muller A, Klingl A, Wanner G, Jung K (2018) Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI-1 between Vibrio harveyi cells. J Bacteriol. 10.1128/JB.00740-17 [DOI] [PMC free article] [PubMed]
- Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. The architecture of Iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol. 2016;7:796. doi: 10.3389/fmicb.2016.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cydzik-Kwiatkowska A. Bacterial structure of aerobic granules is determined by aeration mode and nitrogen load in the reactor cycle. Bioresour Technol. 2015;181:312–320. doi: 10.1016/j.biortech.2015.01.101. [DOI] [PubMed] [Google Scholar]
- Davey ME, O'toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847. doi: 10.1128/Mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;7(2):277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitosa-Junior OR, Stefanello E, Zaini PA, Nascimento R, Pierry PM, Dandekar AM, Lindow SE, da Silva AM. Proteomic and metabolomic analyses of Xylella fastidiosa OMV-enriched fractions reveal association with virulence factors and signaling molecules of the DSF family. Phytopathology. 2019;109(8):1344–1353. doi: 10.1094/phyto-03-19-0083-r. [DOI] [PubMed] [Google Scholar]
- Flemming HC. Biofilms and environmental-protection. Water Sci Technol. 1993;27(7–8):1–10. doi: 10.2166/wst.1993.0528. [DOI] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176(2):269–275. doi: 10.1128/JB.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Acata S, Vital-Jacome M, Perez-Sandoval MV, Navarro-Noya YE, Thalasso F, Luna-Guido M, Conde-Barajas E, Dendooven L. Microbial community structure in aerobic and fluffy granules formed in a sequencing batch reactor supplied with 4-chlorophenol at different settling times. J Hazard Mater. 2018;342:606–616. doi: 10.1016/j.jhazmat.2017.08.073. [DOI] [PubMed] [Google Scholar]
- Halan B, Buehler K, Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012;30(9):453–465. doi: 10.1016/j.tibtech.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Heydorn A, Ersboll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol. 2002;68(4):2008–2017. doi: 10.1128/aem.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49(4):905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Rainey FA, Wood AP (2006) The genus Paracoccus. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes: Volume 5: Proteobacteria: alpha to Beta subclasses. Springer New York, New York, pp 232–249
- Kiely PD, Call DF, Yates MD, Regan JM, Logan BE. Anodic biofilms in microbial fuel cells harbor low numbers of higher-power-producing bacteria than abundant genera. Appl Microbiol Biotechnol. 2010;88(1):371–380. doi: 10.1007/s00253-010-2757-2. [DOI] [PubMed] [Google Scholar]
- Kojima T, Nishiyama T, Maehara A, Ueda S, Nakano H, Yamane T. Expression profiles of polyhydroxyalkanoate synthesis-related genes in Paracoccus denitrificans. J Biosci Bioeng. 2004;97(1):45–53. doi: 10.1016/s1389-1723(04)70164-4. [DOI] [PubMed] [Google Scholar]
- Kokufuta E, Shimohashi M, Nakamura I. Simultaneously occurring nitrification and denitrification under oxygen gradient by polyelectrolyte complex-coimmobilized Nitrosomonas europaea and Paracoccus denitrificans cells. Biotechnol Bioeng. 1988;31(4):382–384. doi: 10.1002/bit.260310415. [DOI] [PubMed] [Google Scholar]
- Kumar S, Spiro S (2017) Environmental and Genetic Determinants of Biofilm Formation in Paracoccus denitrificans. mSphere 2(5). 10.1128/mSphereDirect.00350-17 [DOI] [PMC free article] [PubMed]
- Lasa I, Penades JR. Bap: A family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157(2):99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Lycus P, Soriano-Laguna MJ, Kjos M, Richardson DJ, Gates AJ, Milligan DA, Frostegard A, Bergaust L, Bakken LR. A bet-hedging strategy for denitrifying bacteria curtails their release of N2O. Proc Natl Acad Sci U S A. 2018;115(46):11820–11825. doi: 10.1073/pnas.1805000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gil M, Romero D, Kolter R, Espinosa-Urgel M. Calcium causes multimerization of the large adhesin LapF and modulates biofilm formation by Pseudomonas putida. J Bacteriol. 2012;194(24):6782–6789. doi: 10.1128/JB.01094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- Morinaga K, Yamamoto T, Nomura N, Toyofuku M. Paracoccus denitrificans can utilize various long-chain N-acyl homoserine lactones and sequester them in membrane vesicles. Environ Microbiol Rep. 2018;10(6):651–654. doi: 10.1111/1758-2229.12674. [DOI] [PubMed] [Google Scholar]
- Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer KH. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62(12):4329–4339. doi: 10.1128/AEM.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisha KN, Devi V, Varalakshmi P, Ashokkumar B. Biodegradation and utilization of dimethylformamide by biofilm forming Paracoccus sp. strains MKU1 and MKU2. Bioresour Technol. 2015;188:9–13. doi: 10.1016/j.biortech.2015.02.042. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Pelissari C, Ávila C, Trein CM, García J, de Armas RD, Sezerino PH. Nitrogen transforming bacteria within a full-scale partially saturated vertical subsurface flow constructed wetland treating urban wastewater. Sci Total Environ. 2017;574:390–399. doi: 10.1016/j.scitotenv.2016.08.207. [DOI] [PubMed] [Google Scholar]
- Plate L, Marletta MA. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci. 2013;38(11):566–575. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche B, Li XZ, Hauer B, Schmid A, Buehler K. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 2009;27(11):636–643. doi: 10.1016/j.tibtech.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Satchell KJ. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184(4):1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Taylor TA, Beatty JT, Greenberg EP. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J Bacteriol. 2002;184(23):6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188(16):5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shi Z, Zhang Y, Zhou J, Chen M, Wang X. Biological removal of nitrate and ammonium under aerobic atmosphere by Paracoccus versutus LYM. Bioresour Technol. 2013;148:144–148. doi: 10.1016/j.biortech.2013.08.052. [DOI] [PubMed] [Google Scholar]
- Singh S, Nerurkar AS, Srinandan CS. Nitrate levels modulate the abundance of Paracoccus sp. in a biofilm community. World J Microbiol Biotechnol. 2015;31(6):951–958. doi: 10.1007/s11274-015-1849-7. [DOI] [PubMed] [Google Scholar]
- Srinandan CS, Jadav V, Cecilia D, Nerurkar AS. Nutrients determine the spatial architecture of Paracoccus sp. biofilm. Biofouling. 2010;26(4):449–459. doi: 10.1080/08927011003739760. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Suarez C, Piculell M, Modin O, Langenheder S, Persson F, Hermansson M. Thickness determines microbial community structure and function in nitrifying biofilms via deterministic assembly. Sci Rep. 2019;9(1):5110. doi: 10.1038/s41598-019-41542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Roschitzki B, Riedel K, Eberl L. Identification of proteins associated with the Pseudomonas aeruginosa biofilm extracellular matrix. J Proteome Res. 2012;11(10):4906–4915. doi: 10.1021/pr300395j. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Tashiro Y, Hasegawa Y, Kurosawa M, Nomura N. Bacterial membrane vesicles, an overlooked environmental colloid: biology, environmental perspectives and applications. Adv Colloid Interf Sci. 2015;226(Pt a):65–77. doi: 10.1016/j.cis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H, Schmitt-Kopplin P, Eberl L, Nomura N. Membrane vesicle-mediated bacterial communication. ISME J. 2017;11(6):1504–1509. doi: 10.1038/ismej.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2018;17(1):13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- Uemoto H, Saiki H. Nitrogen removal by tubular gel containing Nitrosomonas europaea and Paracoccus denitrificans. Appl Environ Microbiol. 1996;62(11):4224–4228. doi: 10.1128/AEM.62.11.4224-4228.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utada AS, Bennett RR, Fong JC, Gibiansky ML, Yildiz FH, Golestanian R, Wong GC. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun. 2014;5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini Martina, Filloux Alain. Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons fromPseudomonas aeruginosaand Other Bacteria. Journal of Biological Chemistry. 2016;291(24):12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking J. N., Zaburdaev V., De Volder M., Losick R., Brenner M. P., Weitz D. A. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proceedings of the National Academy of Sciences. 2012;110(3):848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Ye L, Ren H, Zhang XX. Microbial community structure and function in aerobic granular sludge. Appl Microbiol Biotechnol. 2018;102(9):3967–3979. doi: 10.1007/s00253-018-8905-9. [DOI] [PubMed] [Google Scholar]
- Yan J, Sharo AG, Stone HA, Wingreen NS, Bassler BL. Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc Natl Acad Sci U S A. 2016;113(36):E5337–E5343. doi: 10.1073/pnas.1611494113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Toyofuku M, Obana N, Nomura N (2017) Biofilm formation by Paracoccus denitrificans requires a type I secretion system-dependent adhesin BapA. FEMS Microbiol Lett 364(4). 10.1093/femsle/fnx029 [DOI] [PubMed]
- Zhao X, Chen Z, Wang X, Li J, Shen J, Xu H. Remediation of pharmaceuticals and personal care products using an aerobic granular sludge sequencing bioreactor and microbial community profiling using Solexa sequencing technology analysis. Bioresour Technol. 2015;179:104–112. doi: 10.1016/j.biortech.2014.12.002. [DOI] [PubMed] [Google Scholar]