Abstract
The present study reports the molecular and antigenic characterization of 13 bovine herpesvirus type 1 (BoHV-1) field viruses obtained from cattle with different clinical cases in Turkey between 1992 and 2017. We selected glycoprotein C (gC) of BoHV-1 as a target to detect and/or verify presence of the virus in suspect materials followed by virus isolation (VI) in MDBK cells. In seven out of 13 BoHV-1 positive samples, cytophatic effects (CPEs) were observed in MDBK cell cultures, although only four virus samples reached a sufficient titer to use in phylogenetic assay, restriction endonuclease analysis (REA), and virus neutralization test (VNT). According to the results of sequence analysis of the 13 BoHV-1 positive samples, nine BoHV-1 field viruses were determined as BoHV-1.1 and four as BoHV-1.2. Using REA, we demonstrated that two of our isolated viruses could be categorized as BoHV-1.1 while the other two isolates were BoHV-1.2 subtypes. Differences between the BoHV-1.1 and BoHV-1.2 isolates were also detected in the VNT results by assaying 125 suspected serum samples after testing with isolated (KY748023, KY748022, KY748020, and KY748021) and reference viruses (BoHV-1 Cooper and BoHV-5 Texas 89). These results are indicating the need to correctly identify BoHV-1 field isolates to better understand the epidemiology and pathogenesis of infection. In addition, it would be useful to identify the subtypes circulating in the specific geographical area while determining vaccination preferences.
Keywords: BoHV-1, Phylogenetic analysis, Molecular and antigenic characterization, Cattle, Turkey
Introduction
Bovine herpesvirus type 1 (BoHV-1), of the Herpesviridae family and Alphaherpesvirinae subfamily, is distributed worldwide, causing significant economic losses to the cattle industry (Ackermann and Engels 2006).
This virus is the causative agent of various severe conditions in cattle respiratory and genital systems, including infectious bovine rhinotracheitis (IBR) and infectious pustular vulvovaginitis/balanoposthitis (IPV/IBP) (Straub 1990). BoHV-1 genome consists of one linear DNA molecule of about 140 kb, which replicates in the upper respiratory or genital tracts and gains access to neurons to establish latency in the corresponding ganglia (Ackermann et al. 1990). According to its antigenic and genomic characteristics, BoHV-1 is further subdivided into two distinct yet closely related subtypes: 1 (BoHV-1.1) and 2 (BoHV-1.2) (Metzler et al. 1986). Such subtypes may also be associated with distinct manifestations of the disease in cattle (Engels et al.1992). However, all subtypes are antigenically similar. BoHV-1.1 and BoHV-1.2a are mostly related to the respiratory syndrome and abortions. BoHV-1.2b subtypes, which are considered to be moderately pathogenic (Metzler et al. 1985), may cause vulvovaginitis/balanopostitis, but have not yet been linked with abortions (Miller et al. 1991). All subtypes are able to infect cattle respiratory and genital tracts, although it has been suggested that each genotype is more adapted to either one (Rijsewijk et al. 1999; Spilki et al. 2004). Previously, because BoHV-5 was reported as BoHV-1.3 and considered to be associated with neurological disorders, including cattle encephalitis, it was named bovine encephalitic herpesvirus (Muylkens et al. 2007). BoHV-5 has also been subdivided into a, b, and non-a/b or c (Pidone et al. 1999).
BoHV-1 genome encodes for 33 structural proteins, with 13 probably associated with the envelope (Liang et al. 1996), of which 10 have the potential to encode glycoproteins (Schwyzer and Ackermann 1996). BoHV-1.1 and BoHV-1.2 subtypes differ in glycoprotein C (gC) epitopes, which may alter viral attachment and virulence (Rijsewijk et al. 1999). Despite close similarities, BoHV-1.1, 1.2, and BoHV-5 can be differentiated by antigenic or molecular analysis of gC (Claus et al. 2005; Silva et al. 2007; Esteves et al. 2008). BoHV-1 has been eradicated in several European countries, and in addition, control programs based on the use of marker vaccines have been initiated in other European countries, especially those experiencing high seroprevalence. These vaccines allow serological differentiation between naturally infected and vaccinated cattle (Van Oirschot et al. 1997).
In Turkey, serological and virological investigations have indicated the prevalence of BoHV-1 in cattle with different clinical signs (Alkan et al. 2005; Yildirim et al. 2011; Bilge Dağalp 1996; Ozkul et al. 1995). However, there have been few studies on molecular detection of BoHV-1 (Aslan et al. 2015, Bilge Dağalp et al. 2011, Tuncer-Göktuna et al. 2016) and no study to genetically or antigeneitically characterize BoHV-1 strains in Turkey. Thus, to our knowledge, the present study is the first to report the antigenic and molecular characterization of BoHV-1 in different isolated strains (1.1 and 1.2) and map their associations with different clinical and necropsy samples derived from naturally infected cattle in Turkey.
Material and methods
Samples
We analyzed thirteen BoHV-1-positive samples in a phylogenetic assay based on virus isolation and/or PCR obtained from cattle with different clinical signs, including respiratory disease, mastitis, and abortus. The samples were provided to our laboratory between 1992 and 2017 from six different herds in 3 provinces of the Central Anatolia region of Turkey.
Table 1 shows the field viruses and diagnostic details of BoHV-1 used in this study. In addition, 125 blood serum samples were taken from 3 herds suffering from abortion for VNT testing of anti-BoHV-1 cross-neutralizing antibodies among our field isolates, BoHV-1 Cooper, and BoHV-5 Texas 89 strains.
Table 1.
Demographic presentation of clinical and diagnostic details of the BoHV-1 viruses involved in the study
| Herd no. and virus code | Clinical signs | Isolation (± ) |
Year | Clinical samples | GenBank Ac. no./type | Vaccinated/ unvaccinated |
|---|---|---|---|---|---|---|
|
ANK-SKR-Lng (Ankara 1) |
Respiratory tract disease | + | 1992 | Lung | KY748020 (BoHV-1.1) | Unvaccinated |
|
TGM-IZ41-Milk (Ankara 1) |
Mastitis | + | 1993 | Milk | KY748022 (BoHV-1.2) | Vaccinated (inactive conventional) |
|
Halk-THYM-Thm (Ankara 2) |
Abortus | + | 1993 | Thymus | KY748021 (BoHV-1.2) | Unvaccinated |
|
KYS-73765-Milk (Kayseri) |
Mastitis | + | 2001 | Milk | KY748023 (BoHV-1.1) | Unvaccinated |
|
ANK-Ery-Nose (Ankara 3) |
Respiratory tract disease | – | 2006 | Nasal swab | KY748027 (BoHV-1.2) | Unvaccinated |
|
ANK-CGOZ-Eye (Ankara 4) |
Respiratory tract disease | – | 2014 | Ocular swab |
(BoHV-1.2) |
Unvaccinated |
|
ANK-SW1-Nose (Ankara 5) |
Respiratory tract disease | – | 2014 | Nasal swab | KY748025 (BoHV-1.1) | Unvaccinated |
|
ANK-SW3-Nose (Ankara 5) |
Respiratory tract disease | – | 2014 | Nasal swab |
(BoHV-1.1) |
Unvaccinated |
|
KON-133-NS-TR2017 (Konya) |
Respiratory tract disease | + | 2017 | Nasal swab | MF458855 (BoHV-1.1) | Unvaccinated |
|
KON-793-NS-TR2017 (Konya) |
Respiratory tract disease | + | 2017 | Nasal swab | MF458856 (BoHV-1.1) | Unvaccinated |
|
KON-794-NS-TR2017 (Konya) |
Respiratory tract disease | – | 2017 | Nasal swab | MF458857 (BoHV-1.1) | Unvaccinated |
|
KON-795-NS-TR2017 (Konya) |
Respiratory tract disease | + | 2017 | Nasal swab | MF458858 (BoHV-1.1) | Unvaccinated |
|
KON-IBR2-LOK-TR2017 (Konya) |
Respiratory tract disease | – | 2017 | Leucocyte | MF458859 (BoHV-1.1) | Unvaccinated |
PCR assay for virus detection in the samples
Viral DNA extraction was performed according to Sambrook et al. (1989). For PCR detection of the virus, primer sets targeting BoHV-1 gC were selected according to Esteves et al. (2008). Forward primer (5′-CGGCCACGACGCTGACGA-3′) and reverse primer (5′-CGCCGCCGAGTACTACCC-3′) were used to amplify the 575 and 572 bp fragments of BoHV-1 and -5 (873–1447 and 813–1384 nucleotides of the sequences by accession numbers of Z49223 and Z49224), respectively. PCR reactions were performed by Phusion High Fidelity DNA polymerase (ThermoScientific, USA) under the following conditions: initial denaturation at 98 °C for 30 s, 30 cycles of first denaturation at 98 °C for 10 s, annealing at 56 °C for 30 s, extension at 72 °C for 20 s, and final extension at 72 °C for 10 min. To visualize the PCR products under UV light, 1% agarose gel was stained with ethidium bromide (0.5 μg/mL) and run at 80 mV for 1 h in 1X TAE buffer.
Virus isolation
MDBK cells were used to isolate the viruses from the positive samples. The cells were maintained in complete DMEM (Sigma, USA) supplemented with 2% fetal bovine serum (FBS), 0.34 g/L of L-glutamine, 500,000 IU/L of penicillin, and 0.5 g/L of streptomycin sulfate. Samples were homogenized in DMEM and centrifuged at 12,000×g for 10 min at 4 °C. The supernatants were then collected, passed through 0.22 filters and inoculated into MDBK cells for 2 h, followed by the addition of virus growth medium containing 2% FBS and incubation at 37 °C, 5% CO2. The cells were observed daily for the appearence of cytopathic effects (CPEs). In the case of no CPEs, monolayers were blind-passaged at weekly intervals for a total of three passages. The supernatants of all cell cultures were tested using PCR for the presence of BoHV-1 gC.
Sequence analysis
Sequence analysis was performed based on the sequences of 575 bp PCR products, which targeted BoHV-1 gC in our positive samples. Because some positive samples did not propagate in the cell culture, we used their PCR products directly obtained from the samples. The nucleotide sequences of the PCR products were determined using a CEQ8000 genetic analysis system (Beckman Coulter, USA). Sequence assembly and editing were performed using Bioedit (Version 7.0.5.3) and Clustral W (Hall 1999), before comparison with the GenBank nucleotide sequence database for sequence similarities using the Basic Length Alignment Search Tool (BLAST) software of the National Center for Biotechnology Information (NCBI) (Altschul et al. 1997). Phylogenetic analysis based on BoHV-1 gC sequences was performed using MEGA6 software’s neighbor-joining method (Tamura et al. 2011) (Fig. 1), based on the evolutionary distances between different sequences calculated by the Kimura two-parameter model. The confidence level of the NJ tree was assessed by bootstrapping using 1000 replicates.
Fig. 1.
Phylogenetic analysis of BoHV-1 based on gC gene
Restriction endonuclease analysis
The restriction endonuclease analysis (REA) was performed with 4 field isolates (KY748023, KY748022, KY748020, and KY748021). After virus inoculation in MDBK cells for 7–10 days and the CPEs had reached 90%, our viruses were collected followed by 3 freeze-thaw cycles and centrifugation at 5000×g for 15 min at 4 °C. The collected viruses were purified by centrifugation at 25,000×g for 5 h at 4 °C through 40% sucrose in 1× PBS before the pellets were resuspended in 550 μl of molecular grade water. To release the viral DNA, 35 μl of 20% SDS plus 40 μl of 20 mg/mL proteinase K were added to the virus suspension and incubated for 1 h at 37 °C. Then two rounds of phenol-chloroform extraction followed by ethanol precipitation were performed before the pellet was resuspended in TE buffer (pH 8.5) and an additional 0.2 μl of 10 μg/mL RNaseA and incubated for 30 min at 37 °C. After measurement by NanoDrop One/OnecUV-Vis Spectrophotometer (ThermoScientific, USA), the viral DNA was stored at + 4 °C. For REA, fastdigest Hind III, BamHI, and EcoRI (ThermoScientific, USA) restriction enzymes were used according to the manufacturer’s instructions. To visualize viruses, the cut viral DNAs were run in 0.6% agarose gel at 40 mV for 5–8 h with 1 μl of 10 mg/mL Ethidium Bromide per 20 mL of agarose gel.
Virus neutralization test
The cross-neutralization test was conducted using BoHV-1 Cooper, BoHV-5 Texas 89 strains, and 4 field isolates (KY748023, KY748022, KY748020, and KY748021) to analyze 125 blood serum samples with a history of abortion at different pregnancy stages from 3 separate dairy herds that were mostly unvaccinated against BoHV-1. The tests were carried out in 96-well cell culture plates as previously described (Van Oirschot 2000). Serum samples were twofold diluted, and the serum-virus mixtures (equal volumes of 50 μL) were incubated at 37 °C for 2 h before inoculation into MDBK cells. Tests were separately performed against a 100-TCID50 dose of each of the six different virus strains. Plates were incubated at 5% CO2 at 37 °C for up to 5 days until the final reading, based on the presence or absence of cytopathic effects in the virus control wells. Results were calculated by the method of Reed and Muench (Lorenz and Bögel 1973) and expressed as positive when the neutralizing antibody titer was ≥ 2.
Results
VI and/or PCR
Of the suspect BoHV-1 samples, 13 ones were determined as positive in gC PCR. All the positive samples were inoculated in MDBK cells and seven propagated. However, after several passages, only four samples reached sufficient to be considered as our field-isolated strains and used in REA and VNT. The sequences obtained from the gC PCR products of the 6 positive samples along with the seven virus isolates were sequenced and submitted to the NCBI Genbank. Table 1 lists the positive samples and isolates and their relation to clinical symptoms, year of collection, organ from which samples were collected, propagation in the cell culture, Genbank accession number, and vaccination program of the cattle herd from which samples were collected. No positive samples of BoHV-5 were detected from the PCR assays.
Phylogenetic analysis
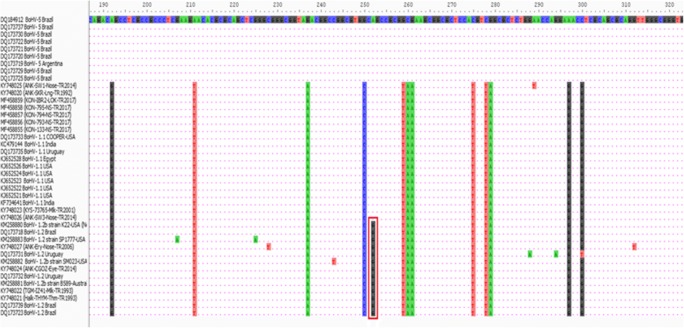
We used the 13 BoHV-1-positive samples from the infected cell culture or the positive samples that had been unable to grow in MDBK cells based on PCR amplification of the gC gene. The phylogenetic analysis results are shown in Fig. 1, with the sequences of the glycoprotein C of different BoHV-1 and BoHV-5 strains included for comparison. The phylogenetic tree inferred by the neighbor-joining method allowed the viruses to be grouped according to their different types or subtypes, as previously reported (Metzler et al. 1986; Miller et al. 1991; Souza et al. 2002). The tree based on nucleotide sequences showed similar topologies in the presence of two main groups: one formed by BoHV-1.1, 1.2, and the other formed by BoHV-5 viruses. The nucleotide (nt) sequence alignment of the gC region of the our field viruses and the published sequences showed variable percentages of homology (98.14 to 99.69%) (Fig. 2). The nucleotide sequence alignments revealed levels of genomic similarity ranging from 99.07 to 100% among our BoHV-1 field viruses (n = 13) and 89.71 to 90.65% between BoHV-1 and BoHV-5 (Table 2).
Fig. 2.
According to partial gC gene sequences nucleotide differences of reference viruses and field isolates of BoHV-1
Table 2.
Nucleotide similarity between our field viruses and BoHV-1.1, BoHV-1.2, and BoHV-5 strains
| Virus code | BoHV-5 DQ173730 |
BoHV-1.1 DQ173733 (Cooper/USA) |
BoHV-1.1 KJ652528 |
BoHV-1.2 DQ1737718 |
BoHV-1.2 DQ173723 |
BoHV-1.2 KM258880 |
|---|---|---|---|---|---|---|
| ANK-SKR-Lng KY748020 (BoHV-1.1) | 90.34% | 99.69% | 99.69% | 99.07% | 99.38% | 98.76% |
| TGM-IZ41-Milk KY748022 (BoHV-1.2) | 90.34% | 99.69% | 99.69% | 99.38% | 100% | 99.38% |
| Halk-THYM-Thm KY748021 (BoHV-1.2) | 90.34% | 99.69% | 99.69% | 99.69% | 99.69% | 99.38% |
|
KYS-73765-Milk KY748023 (BoHV-1.1) |
90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
|
ANK-Ery-Nose KY748027 (BoHV-1.2) |
89.71% | 99.07% | 99.07% | 99.07% | 99.38% | 98.76% |
|
ANK-CGOZ-Eye KY748024 (BoHV-1.2) |
90.03% | 99.38% | 99.38% | 99.38% | 99.69% | 99.07% |
| ANK-SW1-Nose KY748025 (BoHV-1.1) | 90.03% | 99.38% | 99.38% | 98.76% | 99.07% | 98.45% |
|
ANK-SW3-Nose KY748026 (BoHV-1.1) |
90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
| KON-133-NS-TR2017 MF458855 (BoHV-1.1) | 90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
| KON-793-NS-TR2017 MF458856 (BoHV-1.1) | 90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
| KON-794-NS-TR2017 MF458857 (BoHV-1.1) | 90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
|
KON-795-NS-TR2017 MF458858 (BoHV-1.1) |
90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
| KON-IBR2-LOK-TR2017 MF458859 (BoHV-1.1) | 90.65% | 100% | 100% | 99.38% | 99.69% | 99.07% |
Restriction endonuclease analysis
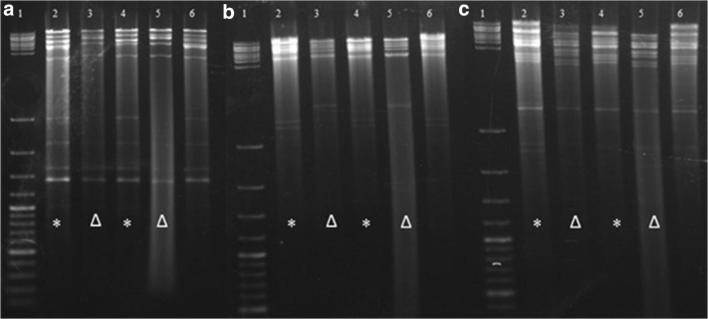
To confirm the results of the phylogenetic analysis based on the gC gene for the four isolated viruses (two BoHV-1.1 and two BoHV-1.2), we performed REA. Based on this assay, two isolates (KY748023 and KY748020) were confirmed as BoHV-1.1 while the other two (KY748022 and KY748021) were found to be members of BoHV-1.2a subtype (Fig. 3).
Fig. 3.
REA of BoHV-1 isolates and reference strain by BamHI (A), HindIII (B), and EcoRI (C). Lane (1):DNA ladder (100 bp plus and 1kbp plus, ThermoScientific), Lane (2): KY748023 (KYS-73675-Milk), Lane (3): KY748022 (TGM-IZ41-Milk), Lane (4): KY748020 (ANK-SKR-Lng), Lane (5): KY748021 (Halk-THYM-Thm), Lane (6): Cooper strain. REA pattern differences indicated by *; KY748022 (Lane3) and KY748021 (Lane 5) were BoHV-1.2a while the other viruses in the test were BoHV-1.1 strains
VNT
A total of 39 serum samples were found positive for neutralizing antibodies for at least one of the viruses tested (our 4 field isolates and two reference viruses) whereas 86 serum samples were negative for all viruses. Comparing the VNT results for our BoHV-1.1 (n = 2) and BoHV-1.2 strains (n = 2), BoHV-1.1 and BoHV-1.2 detected 35/125 (28.0%) and 36/125 (28.8%) of the total 39/125 (31.2%) positive serum samples, respectively (Table 3). Besides, VNT did not allow type-specific differentiation between the neutralizing antibody responses to BoHV-1 and BoHV-5 in most samples, as previously reported (Varela et al. 2010). Out of the 39 antibody-positive serum samples, 33 (33/39; 84.6%) reacted with all the viruses (1.1, 1.2 subtypes, and 5). However, 2 (2/39; 5.1%) specimens were positive for BoHV-1.1 strains but negative for BoHV-1.2 strains while 3 samples (3/39; 7.69%) were positive for BoHV-1.2 strains but negative for BoHV-1.1 strains. In addition, one sample (1/39; 2.5%) was positive only for BoHV-5 (Table 3). Thus, when the test was performed with BoHV-1.1 strains, 4 samples (4/39; 10.2%) were negative, which were documented as false negatives (Table 2 and 4).
Table 3.
Total positivity values and distribution of positive serum samples for BoHV-1.1, BoHV-1.2, and BoHV-5 viruses
| Viruses | TGM-IZ41-Milk | Halk-THYM-Thm | ANK-SKR-Lng | KYS-73675-Milk | BoHV-1 Cooper | BoHV-5 Texas | Common (+) | Total (+) |
|---|---|---|---|---|---|---|---|---|
| Total positivity |
36/125 28.8% |
36/125 28.8% |
35/125 28.0% |
35/125 28.0% |
35/125 28.0% |
37/125 29.6% |
33/125 26.4% |
39/125 31.2% |
| Positive samples |
36/39 (92.3%) |
36/39 (92.3%) |
35/39 (89.74%) |
35/39 (89.74%) |
35/39 (89.74%) |
37/39 (94.87%) |
33/39 (84.61%) |
|
| Negative samples |
3/39 (7. 69%) |
3/39 (7.69%) |
4/39 (10.25%) |
4/39 (10.25%) |
4/39 (10.25%) |
2/39 (5.12%) |
6/39 (15.38%) |
Table 4.
Neutralization results and SN50 values of BoHV-1.1, BoHV-1.2, and BoHV-5 viruses
| Sample no. | TGM-IZ41-Milk | Halk-THYM-Thm | ANK-SKR-Lng | KYS-73675-Milk | BoHV-1 Cooper | BoHV-5 Texas |
|---|---|---|---|---|---|---|
| SN50 | SN50 | SN50 | SN50 | SN50 | SN50 | |
| 1 | 1/3 | 1/3 | 1/3 | 1/3 | 1/2 | 1/2 |
| 2 | 1/4 | 1/6 | 1/12 | 1/6 | 1/12 | 1/8 |
| 3 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 |
| 4 | – | – | 1/3 | 1/3 | 1/3 | – |
| 5 | 1/6 | 1/6 | – | – | – | 1/8 |
| 6 | 1/8 | 1/4 | > 1/64 | 1/16 | 1/24 | 1/32 |
| 7 | 1/48 | 1/12 | > 1/64 | 1/48 | 1/48 | 1/32 |
| 8 | 1/3 | 1/4 | 1/6 | 1/3 | 1/1 | 1/6 |
| 9 | 1/8 | 1/12 | > 1/64 | 1/32 | 1/24 | 1/16 |
| 10 | 1/4 | 1/3 | 1/16 | 1/16 | 1/8 | 1/8 |
| 11 | 1/3 | 1/3 | – | – | – | 1/3 |
| 12 | 1/16 | 1/12 | – | – | – | 1/6 |
| 13 | 1/16 | 1/12 | 1/12 | 1/8 | 1/48 | 1/32 |
| 14 | 1/48 | 1/24 | > 1/64 | 1/24 | 1/64 | 1/64 |
| 15 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 |
| 16 | 1/6 | 1/3 | 1/24 | 1/8 | 1/12 | 1/6 |
| 17 | 1/32 | 1/32 | > 1/64 | 1/48 | 1/64 | 1/64 |
| 18 | 1/8 | 1/6 | 1/24 | 1/12 | 1/12 | 1/6 |
| 19 | 1/8 | 1/4 | 1/24 | 1/8 | 1/12 | 1/16 |
| 20 | 1/12 | 1/4 | > 1/64 | 1/32 | 1/48 | 1/64 |
| 21 | 1/12 | 1/8 | 1/48 | 1/16 | 1/24 | 1/16 |
| 22 | 1/12 | 1/16 | > 1/64 | 1/24 | > 1/64 | 1/32 |
| 23 | 1/24 | 1/16 | 1/64 | 1/48 | 1/32 | 1/48 |
| 24 | – | – | – | – | – | 1/3 |
| 25 | 1/3 | 1/3 | 1/3 | 1/3 | 1/1 | 1/2 |
| 26 | 1/3 | 1/16 | 1/3 | 1/3 | 1/16 | 1/16 |
| 27 | 1/3 | 1/3 | 1/4 | 1/3 | 1/4 | 1/4 |
| 28 | 1/4 | 1/6 | 1/16 | 1/6 | 1/12 | 1/16 |
| 29 | 1/3 | 1/3 | 1/12 | 1/3 | 1/3 | 1/12 |
| 30 | 1/3 | 1/3 | 1/3 | 1/3 | 1/4 | 1/2 |
| 31 | 1/12 | 1/6 | 1/64 | 1/24 | 1/24 | 1/16 |
| 32 | 1/6 | 1/12 | 1/32 | 1/6 | 1/24 | 1/32 |
| 33 | 1/3 | 1/3 | 1/6 | 1/3 | 1/12 | 1/16 |
| 34 | 1/32 | 1/32 | > 1/64 | 1/48 | > 1/64 | 1/24 |
| 35 | 1/12 | 1/3 | 1/24 | 1/16 | 1/16 | 1/16 |
| 36 | 1/24 | 1/3 | 1/64 | 1/48 | 1/48 | 1/32 |
| 37 | – | – | 1/3 | 1/3 | 1/1 | – |
| 38 | 1/3 | 1/3 | 1/3 | 1/3 | 1/1 | 1/3 |
| 39 | 1/24 | 1/8 | 1/48 | 1/16 | 1/48 | 1/48 |
| Total (+) | 36 | 36 | 35 | 35 | 35 | 37 |
Discussion
The present study reported the genomic and antigenic characterization of thirteen BoHV-1-positive samples obtained from different clinical cattle cases in Turkey. Seven field strains of BoHV-1 were isolated from three different clinical syndromes (abortion, respiratory disease, and mastitis) between 1992 and 2017 in Ankara, Konya, and Kayseri provinces of Turkey. However, after several rounds of propagation in MDBK cells, only 4 viruses adapted to the cell culture system. The presence and characteristics of the viruses were firstly detected by a PCR and/or VI before confirmation by VNT, PCR/sequence analysis, and REA (Engels et al. 1986). The REA results from this study indicate that four isolates of BoHV-1 were very closely related with some minor differences between them (Fig. 3). Besides, the restriction patterns in the native isolates were also similar to other BoHV-1 strains described in previous reports (D’Arce et al. 2002; Whetstone et al. 1989). Some researchers have reported that there is no direct relationship between clinical patterns and RE profiles (Ros and Belak 1999). Since we only had a few isolated strains in our study, we made no such comparison. The positive gC PCR products of the 7 isolated viruses and 6 positive samples were subjected to sequencing. The consistent alignment revealed high degrees of identity in all herpesviruses analyzed in the present study. The tree constructed based on nucleotide sequences showed a similar topology and the presence of two main groups: one formed by BoHV-1.1 and 1.2, and the other formed by BoHV-5 viruses. Our positive samples could be clearly separated into BoHV-1.1 (n = 9) and BoHV-1.2 (n = 4) by phylogenetic analysis. The present study established the prevalence of BoHV-1.1 as the predominant circulating subtype of BoHV-1 in Turkey. The close genetic and antigenic relationship that we observed between our BoHV-1.1- and 1.2-positive samples was not unexpected. The nucleotide sequence alignments revealed levels of genomic similarity ranging from 99.07 to 100% among our BoHV-1 field viruses (n = 13), and 89.71 to 90.65% between BoHV-1 and BoHV-5 (Table 2). Our BoHV-1.2 strains showed more than 98% sequence identity with BoHV-1.1 strains, indicating the possibility of very little genomic differences between these two BoHV-1subtypes. Based on gC sequencing, the nucleotide sequence differences between these subtypes lie at position of 252, with the A nucleotide of BoHV-1.1 substituted with G in BoHV-1.2 strains.
The present study detected BoHV-1.1 and BoHV-1.2 from similar clinical cases to those found by previous researchers, such as upper respiratory disease, mastitis, and abortion in cattle. We also found no relationship between BoHV-1 subtypes and clinical presentations (d’Offay et al. 2016; Esteves et al. 2008; Fulton et al. 2015). Thus, our findings indicate the need to correctly identify BoHV-1 field isolates to better understand their epidemiology and pathogenesis, and to develop a better strategy for vaccine development.
BoHV-1.2b strains were not detected in the suspect samples of this study. In addition, none of the detected viruses were closely related to BoHV-5, which has not yet been reported in Turkey.
In one of the herds (Ankara I), BoHV-1.2a (KY748022) was isolated from the milk of one animal that had been immunized with an inactivated conventional IBR vaccine. This is consistent with many previous findings that BoHV-1 can be detected in different secretions and excretions despite the presence of antibodies (Dennett et al. 1976; Sheffy and Rodman 1973).
We also analyzed the presence of anti-BoHV-1 cross-neutralizing antibodies in 125 serum samples of cattle with a history of abortion in different pregnancy stages from 3 dairy herds in the Aegean region. As most laboratories use the BoHV-1.1 subtype in VN tests, we included two isolated strains (KY748023 and KY748020) of this subtype to determine whether sensitivity might also be affected by strain variation within a subtype. Two BoHV-1.2 isolated strains (KY748022 and KY748021) and BoHV-5 (Texas/1989 strain) were also included in the test. Metzler et al. (1985) reported that cross-neutralization studies using hyperimmune serum failed to differentiate BoHV-1.1 from BoHV-1.2, reflecting the two strains’ high degree of structural and antigenic similarity.
In our study, like several previous ones (Varela et al. 2010; Holz et al. 2009), differences between BoHV-1.1 and BoHV-1.2 strains were detected by VNT. This may be related to the predominant strain that causes infection in cattle populations. Accordingly, methods involving two subtypes in the diagnostic assays may eliminate false negative results. On the other hand, this test did not allow type-specific differentiation between the neutralizing antibody responses against BoHV-1 and BoHV-5 in most samples, as reported before (Varela et al. 2010), except one sample (sample no. 24) which was detected positive only for BoHV-5 (Table 3). Regarding VNT, out of 39 antibody-positive serum samples, 33 (33/39; 84.61%) reacted with all viruses (BoHV-1.1, 1.2 subtypes and 5) (Table 3). In addition, among the positive samples, only 2 (2/39; 5.1%) specimens were positive for BoHV-1.1 but negative for BoHV-1.2, while three samples (3/39; 7.69%) were positive for BoHV-1.2 strains but negative for BoHV-1.1 strains (Tables 3, 4, and 5). According to these results, when the test was performed with BoHV-1.1 strains, four samples (4/39; 10.2%) were detected as negative (Tables 3 and 4). Comparing the results obtained with the same subtype showed similar results. We found that if only one of the six viruses (four field isolates, BoHV-1 Cooper, and BoHV-5 Texas strains) were included in the assay, the results could give a 10.2% miss rate of the neutralizing antibody of the positive serum samples (Table 3). Similarly, Holz et al. (2009) reported that it is possible to miss at least 23.6% of all neutralizing antibody-positive serum samples. Cross-neutralization studies using serum samples from naturally infected animals failed to differentiate BoHV-1.1 and BoHV-1.2, reflecting these subtypes’ high degree of structural/antigenic similarity. However, we believe that serological results may vary depending on the subtypes. Varela et al. (2010) reported that the sensitivity of SN50 to a single virus, regardless of their types/subtypes, varied between 77 and 91% giving rise to significantly low sensitivity. Even with viruses belonging to a same subtype, for example, the two BoHV-1.1 strains, sensitivity varied from 86 to 91%. Thus, low sensitivity may comprise the use of VNT to support control or eradication strategies. These results also suggest that type or subtype may have little meaning in terms of antigenic relatedness among such viruses (Varela et al. 2010). Previous studies have been unable to achieve type-differentiation using VNT because antibody titers did not differ significantly (equal to or greater than fourfold) in about 92% of serum samples (Teixeira et al. 2001). Therefore, type-specific tests still wait for further development to overcome this obstacle. In addition, cattle may also be infected with more than one virus types/subtypes, as recently demonstrated (Campos et al. 2009). However, using many distinct viruses in VNT is not practical and difficult to perform in diagnostic laboratories. We also found that both BoHV-1.1 (KY748020) and 1.2a (KY748022) subtypes were circulating simultaneously in the same herd (Ankara 1). As both virus types and most subtypes may circulate in herds, the choice of a challenge virus may significantly influence the test’s sensitivity.
Table 5.
Differences between BoHV-1.1, BoHV-1.2, and BoHV-5 viruses according to neutralization test
| The sample no. | TGM-IZ41-Milk | Halk-THYM-Thm | ANK-SKR-Lng | KYS-73675-Milk | BoHV-1 Cooper | BoHV-5 Texas |
|---|---|---|---|---|---|---|
| SN50 | SN50 | SN50 | SN50 | SN50 | SN50 | |
| 4 | – | – | 1/3 | 1/3 | 1/3 | – |
| 37 | – | – | 1/3 | 1/3 | 1/1 | – |
| 5 | 1/6 | 1/6 | – | – | – | 1/8 |
| 11 | 1/3 | 1/3 | – | – | – | 1/3 |
| 12 | 1/16 | 1/12 | – | – | – | 1/6 |
| 24 | – | – | – | – | – | 1/3 |
BoHV-1 infection is seen widely in Turkey, and control of the disease depends mainly on vaccination, with either gE(−)e inactivated marker vaccine or combined conventional vaccine, which contains a combination of killed BVDV (BVDV1, 2), BRSV, BoHV-1, and PIV-3. Besides, there have been no studies on the molecular characterization of Turkish BoHV-1 strains. Thus, the present study reported the isolation, antigenic/molecular characterization, and phylogenetic analysis of different bovine herpesvirus type 1 (BoHV-1) strains from naturally infected cattle with distinct clinical signs. These findings are helpful for comparing old and new virus strains to determine which strains can be used in future vaccination programs. Further studies on molecular and antigenic characterization using field isolates from different clinical signs will help achieve a better understanding of the virus’s epidemiology in order to develop more effective vaccines and diagnostic assays.
Author contributions
S.B.D designed the study; T.A.F and F.D. conducted the experiments; S.B.D and F.D analyzed the data and prepared the figures; S.B.D, T.A.F., A.O., and F.A wrote the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Ethical approval
All of the materials were sent to us by field veterinarians.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Seval Bilge Dagalp, Email: dagalp@ankara.edu.tr, Email: sevalbilge@hotmail.com.
Touraj Aligholipour Farzani, Email: touraj.farzani@gmail.com.
Firat Dogan, Email: firat9837@gmail.com.
Feray Alkan, Email: falkan@ankara.edu.tr.
Aykut Ozkul, Email: ozkulaykut@gmail.com.
References
- Ackermann M, Engels M. Pro and contra IBR-eradication. Veterinary Microbiology. 2006;113:293–302. doi: 10.1016/j.vetmic.2005.11.043. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Belak S, Bitsch V, Edwards S, Moussa A, Rockborn G, Thiry E. Round table on infectious bovine rhinotracheitis infectious pustular vulvovaginitis: Virus infection, diagnosis and control. Vet. Microbiol. 1990;23:361–363. doi: 10.1016/0378-1135(90)90167-T. [DOI] [PubMed] [Google Scholar]
- Alkan F, Burgu I, Bilge-Dagalp S, Yildirim Y, Gencay A, Güngör B, Ataseven VS, Akça Y. The seroprevalence of BHV-1 infection on selected dairy cattle herds in Turkey. Rev. Med. Vet. 2005;156(3):166–169. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;17:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan M E, Azkur A K, and Gazyagci S. (2015). Epidemiology and genetic characterization of BVDV, BHV-1, BHV-4, BHV-5 and Brucella spp. infections in cattle in Turkey J. Vet. Med. Sci. 77(11): 1371–1377. [DOI] [PMC free article] [PubMed]
- Bilge Dağalp. (1996). Kan ve süt serumu örneklerinde IBR-IPV antikorlarının nötralizasyon testi ile saptanması ve süt örneklerinden virus izolasyonu. Ankara Üniv Sağlık Bilimleri Enstitüsü Doktora Tezi.
- Bilge Dağalp, S., Oğuzoğlu, T.C., Faraji, A., Timurkan, M.Ö., and Alkan, F. (2011). The Investigation of BVDV, BoHV-4 and BoHV-1 as a possible aetiological agents in abortion and neonatal death cases in cattle. The 8th ESVV Pestivirus Symposium in Hannover, Germany.
- Campos FS, Franco AC, Hübner SO, Oliveira MT, Silva AD, Esteves PA, Rijsewijk FAM. High prevalence of co-infections with bovine herpesvirus 1 and 5 found in cattle in southern Brazil. Veterinary microbiology. 2009;139(1):67–73. doi: 10.1016/j.vetmic.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Claus MP, Alfieri AF, Folgueras-Flatschart AV, Wosiacki SR, Médici KC, Alfieri AA. Rapid detection and differentiation of bovine herpesvirus 1 and 5 glycoprotein C gene in clinical specimens by multiplex-PCR. Journal of virological methods. 2005;128(1):183–188. doi: 10.1016/j.jviromet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- D’Arce RCF, Almeida RS, Silva TC, Franco AC, Spilki F, Roehe PM, Arns CW. Restriction endonuclease and monoclonal antibody analysis of Brazilian isolates of bovine herpesviruses types 1 and 5. Vet. Mic. 2002;88(4):315–324. doi: 10.1016/S0378-1135(02)00126-8. [DOI] [PubMed] [Google Scholar]
- d’Offay JM, Eberle R, Fulton RW, Kirkland PD. Complete genomic sequence and comparative analysis of four genital and respiratory isolates of bovine herpesvirus subtype 1.2 b (BoHV-1.2 b), including the prototype virus strain K22. Archives of virology. 2016;161(11):3269–3274. doi: 10.1007/s00705-016-3026-1. [DOI] [PubMed] [Google Scholar]
- Dennett DP, Barasa JQ, Johnson JH. Infectious bovine rhinotracheitis virus: studies on the venereal carrier status range of cattle. Res. Vet. Sci. 1976;20:77–83. doi: 10.1016/S0034-5288(18)33482-9. [DOI] [PubMed] [Google Scholar]
- Engels Monika, Giuliani Carlo, Wild Peter, Beck Thomas M., Loepfe Eva, Wyler Robert. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Research. 1986;6(1):57–73. doi: 10.1016/0168-1702(86)90057-2. [DOI] [PubMed] [Google Scholar]
- Engels M, Palatini M, Metzler AE, Probst U, Kihm U, Ackermann M. Interactions of bovine and caprine herpesviruses with the natural and the foreign hosts. Vet. Microbiol. 1992;33(1–4):69–78. doi: 10.1016/0378-1135(92)90036-S. [DOI] [PubMed] [Google Scholar]
- Esteves PAO, Dellagostin A, Pinto LS, Silva AD, Spilki FR, Ciacci-Zanella JR, Hübner SO, Puentes R, Maisonnave J, Franco AC, Rijsewijk FA, Batista HBCR, Teixeira TF, Dezen D, Oliveira AP, David C, Arns CW, Roehe PM. Phylogenetic comparison of the carboxy terminal region of glycoprotein C (gC) of bovine herpesviruses (BoHV) 1.1., 1.2 and 5 from South America (SA) Virus Res. 2008;131:16–22. doi: 10.1016/j.virusres.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Fulton RW, d’Offay JM, Eberle R, Moeller RB, Van Campen H, O’Toole D, Chase C, Miller MM, Sprowls R, Nydam DV. Bovine herpesvirus-1: evaluation of genetic diversity of subtypes derived from field strains of varied clinical syndromes and their relationship to vaccine strains. Vaccine. 2015;33:549–558. doi: 10.1016/j.vaccine.2014.11.033. [DOI] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids symposium series (Vol. 41, No. 41, pp. 95-98). [London]: Information Retrieval Ltd., c1979-c2000.
- Holz SP, Cibulski TF, Teixeira HBCR, Batista FS, Campos JR, Silva APM, Varela A, Cenci ACF, Roehe PM. Seroprevalence of bovine herpesvirus types 1 and/or 5 in the state of Rio Grande do Sul Pesq. Vet. Bras. 2009;29:767–773. doi: 10.1590/S0100-736X2009000900014. [DOI] [Google Scholar]
- Liang X, Chow B, Raggo C, Babiuk LA. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. Journal of Virology. 1996;70:1448–1454. doi: 10.1128/JVI.70.3.1448-1454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, R. J. and Bögel, K. (1973). Laboratory techniques in rabies: methods of calculation. Monograph series. World Health Organization, (23), 321. [PubMed]
- Metzler AE, Matile H, Gassman U, Engels M, Wyler R. European isolates of bovine herpesvirus 1: A comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol. 1985;85:57–69. doi: 10.1007/BF01317006. [DOI] [PubMed] [Google Scholar]
- Metzler AE, Schudel AA, Engels M. Bovine herpesvirus 1: molecular and antigenic characteristics of variant viruses isolated from calves with neurological disease. Arch. Virol. 1986;87:205–217. doi: 10.1007/BF01315300. [DOI] [PubMed] [Google Scholar]
- Miller JM, Whetstone CA, Van Der Maaten MJ. Abortifacient property of bovine herpesvirus type 1 isolates that represent three subtypes determined by restriction endonuclease analysis of viral DNA. Am. J. Vet. Res. 1991;52:458–461. [PubMed] [Google Scholar]
- Muylkens B, Thiry J, Kirten P, Schynts F, Thiry E. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Veterinary Research. 2007;38:181–209. doi: 10.1051/vetres:2006059. [DOI] [PubMed] [Google Scholar]
- Ozkul A, Cabalar M, Bilge S, Akca Y, Burgu I. Süt sığırcılığı işletmelerinde rastlanan IBR-IPV ve BVD virus enfeksiyonlarının infertilite olgularındaki rolü. Ankara Üniv. Vet. Fak. Derg. 1995;42(3):381–387. [Google Scholar]
- Pidone CL, Galosi CM, Etcheverrigaray ME, Nosetto EO, Etcheverrigaray ME. Restriction endonuclease analysis of BHV1 and BHV-5 strains isolated in Argentina. J. Vet. Med. B. 1999;46(7):453–456. doi: 10.1046/j.1439-0450.1999.00257.x. [DOI] [PubMed] [Google Scholar]
- Rijsewijk, F. A. M J Kaashoek. J P Langeveld. R Meloen. J, Judek. K, Bienkowska-Szewczyk. M, A, Maris-Veldhuis. and J, T, Van Oırschot. (1999). Epitopes on glycoprotein C of bovine herpesvirus-1 (BHV-1) that allow differentiation between BHV-1.1 and BHV-1.2 strains. J. Gen. Virol. 80, 1477–1483. [DOI] [PubMed]
- Ros C, Belak S. Studies of genetic relationships between bovine, caprine, cervine and rangiferine alphaherpes viruses and improved molecular methods for virus detection and identification. J Clin Microbiol. 1999;37:1247–1253. doi: 10.1128/JCM.37.5.1247-1253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, EF., Maniatis, T. (1989). Molecular cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, 914–923.
- Schwyzer M, Ackermann M. Molecular virology of ruminant herpesviruses. Vet. Microbiol. (1996) 1996;53:17–29. doi: 10.1016/S0378-1135(96)01231-X. [DOI] [PubMed] [Google Scholar]
- Sheffy, B. E., Rodman, S. (1973). Activation of latent infectious bovine rhinotracheitis infection. Journal of the American Veterinary Medical Association. 163 pp. 850-851.
- Silva M.S., Brum M.C.S., Loreto E.L.S., Weiblen R., Flores E.F. Molecular and antigenic characterization of Brazilian bovine herpesvirus type 1 isolates recovered from the brain of cattle with neurological disease. Virus Research. 2007;129(2):191–199. doi: 10.1016/j.virusres.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Souza V.F., Melo S.V., Esteves P.A., Schmidt C.S., Gonçalves D.A., Schaefer R., Silva T.C., Almeida R.S., Vicentini F., Franco A.C., Oliveira E.A., Spilki F.R., Weiblen R., Flores E.F., Lemos R.A., Alfieri A.A., Pituco E.M., Roehe P.M. Caracterização de herpesvírus bovinos tipos 1 (BHV-1) e 5 (BHV-5) com anticorpos monoclonais. Pesquisa Veterinária Brasileira. 2002;22(1):13–18. doi: 10.1590/S0100-736X2002000100004. [DOI] [Google Scholar]
- Spilki Fernando Rosado, Esteves Paulo Augusto, Lima Marcelo de, Franco Ana Cláudia, Chiminazzo Cláudio, Flores Eduardo Furtado, Weiblen Rudi, Driemeier David, Roehe Paulo Michel. Comparative pathogenicity of bovine herpesvirus 1 (BHV-1) subtypes 1 (BHV-1.1) and 2a (BHV-1.2a) Pesquisa Veterinária Brasileira. 2004;24(1):43–49. doi: 10.1590/S0100-736X2004000100010. [DOI] [Google Scholar]
- STRAUB O.C. Virus Infections of Ruminants. 1990. Infectious Bovine Rhinotracheitis Virus; pp. 71–108. [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MFB, Esteves PA, Schmidt CS, Spilki FR, Silva TC, Dotta MA, Roehe PM. A monoclonal blocking ELISA for the serological diagnosis of bovine herpesvirus type 1 (BHV-1) infections. Pesq. Vet. Bras. 2001;21(1):33–37. doi: 10.1590/S0100-736X2001000100007. [DOI] [Google Scholar]
- Tuncer-Göktuna P, Alpay G, Öner EB, Yeşilbağ K. The role of herpesviruses (BoHV-1 and BoHV-4) and pestiviruses (BVDV and BDV) in ruminant abortion cases in western Turkey. Trop Anim Health Prod. 2016;48:1021–1027. doi: 10.1007/s11250-016-1050-5. [DOI] [PubMed] [Google Scholar]
- Van Oirschot, J.T. (2000). Infectious bovine rhinotracheitis/infectious pustular vulvovaginitis. In: Manual of Standards for Diagnostic Tests and Vaccines. 4th ed. Paris: Office International des Epizooties 381–391.
- Van Oirschot JT, Kaashoek MJ, Marisveldhuis MA, Weerdmeester K, Rijsewijk FAM. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J. Virol. Methods. 1997;67:23–34. doi: 10.1016/S0166-0934(97)00073-6. [DOI] [PubMed] [Google Scholar]
- Varela A.P.M., Holz C.L., Cibulski S.P., Teixeira T.F., Antunes D.A., Franco A.C., Roehe L.R., Oliveira M.T., Campos F.S., Dezen D., Cenci A., Brito W.D., Roehe P.M. Neutralizing antibodies to bovine herpesvirus types 1 (BoHV-1) and 5 (BoHV-5) and its subtypes. Veterinary Microbiology. 2010;142(3-4):254–260. doi: 10.1016/j.vetmic.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Whetstone CA, Miller JM, Bortner DM, Van der Maaten MJ. Changes in the bovine herpesvirus 1 genome during acute infection, after reactivation from latency and after superinfection in the host animal. Arch Virol. 1989;106:261–279. doi: 10.1007/BF01313957. [DOI] [PubMed] [Google Scholar]
- Yildirim, Y., Yilmaz, V., Kalaycıoglu A.T., Bilge Dagalp, S., Faraji, A.R., Celebi, Ö., Akca D. (2011). An Investigation of A Possible Involvement of BVDV, BHV-1 and BHV-4 Infections in Abortion of Dairy Cattle in Kars District of Turkey Kafkas Univ Vet Fak Derg 17 (6): 879–883,