Abstract
Bacteria are prime cell factories that can efficiently convert carbon and nitrogen sources into a large diversity of intracellular and extracellular biopolymers, such as polysaccharides, polyamides, polyesters, polyphosphates, extracellular DNA and proteinaceous components. Bacterial polymers have important roles in pathogenicity, and their varied chemical and material properties make them suitable for medical and industrial applications. The same biopolymers when produced by pathogenic bacteria function as major virulence factors, whereas when they are produced by non-pathogenic bacteria, they become food ingredients or biomaterials. Interdisciplinary research has shed light on the molecular mechanisms of bacterial polymer synthesis, identified new targets for antibacterial drugs and informed synthetic biology approaches to design and manufacture innovative materials. This Review summarizes the role of bacterial polymers in pathogenesis, their synthesis and their material properties as well as approaches to design cell factories for production of tailor-made bio-based materials suitable for high-value applications.
Subject terms: Bacterial pathogenesis, Bacterial synthetic biology, Metabolic engineering, Applied microbiology, Polymers
Bacteria produce diverse polymers, such as polysaccharides, polyesters, polyphosphates and extracellular DNA. In this Review, Moradali and Rehm discuss the types of bacterial polymers and their role in bacterial physiology and pathogenesis as well as their production and use as novel biomaterials.
Introduction
Biopolymers are produced by living organisms and are synthesized by processive enzymes that link building blocks such as sugars, amino acids or hydroxy fatty acids to yield high molecular weight molecules. Bacteria can synthesize various classes of these biopolymers, such as polysaccharides (composed of sugars and/or sugar acids connected by glycosidic linkages), polyamides (composed of amino acids connected by peptide bonds), polyesters (composed of hydroxy fatty acids linked by ester bonds) and polyphosphates (polyPs; composed of inorganic phosphates linked by anhydride bonds). For decades, scientific efforts have been focusing on understanding biosynthesis pathways of bacterial polymers because of their involvement in bacterial pathogenicity and persistence. These polymeric substances can function as storage molecules, as protective capsular layers surrounding cells and as major matrix components of biofilms, which are involved in 60–80% of all human bacterial infections1–3. However, research on the physicochemical properties of biopolymers also sheds light on their utility for medical and industrial applications. Recent advances in synthetic biology and bioengineering methods allowed the production of innovative biopolymers with uses or potential applications in medicine (for example, hyaluronate as a biomaterial), as additives in cosmetic products, as additives in food (for example, xanthan and dextran) and as biopolyesters in packaging4–6. The rational design of biopolymer-producing cell factories has increasingly attracted research and commercial interest.
Although biopolymer synthesis consumes chemical energy and nutrients, it is maintained by bacteria as biopolymers enable them to persist and grow under a wide range of often unfavourable conditions, including exposure to immune responses of the host during infection. These polymers have diverse biological functions, such as adhesion, energy storage or protection, and their synthesis is regulated in response to environmental stimuli7. Their physicochemical properties are important for bacterial behaviours, such as translocation, attachment onto biotic or abiotic surfaces, invasion, protection and persistence. For example, bacteria produce extracellular polymeric substances, which is a general term referring to various bacterial polymeric substances that entangle themselves into a matrix that encases bacterial cells. Production of extracellular polymeric substances is essential for the formation of biofilms, which are highly structured microbial communities2,8 and one of the most persistent forms of life on Earth. As biofilm formation is the hallmark of many chronic infections2,9, a large body of research has been conducted to understand the role of bacterial biopolymers in biofilm formation and in pathogenesis. Such bacterial biopolymers and their biosynthesis and biological functions provide targets for developing novel antibacterial drugs.
On the other hand, extensive research has been focused on harnessing the unique material properties of bacterial polymers, such as cellulose10, dextran11, xanthan12 and polyesters13, in industrial production for medical and technical uses. Over the past few decades, genome sequencing and advanced molecular techniques have generated a large set of data not only providing insight into the role of bacterial polymers in pathogenesis but also for engineering bacteria as cell factories that produce tailor-made bio-based materials. Such renewable and biodegradable materials could replace oil-based commodity materials and would also advance development of novel high-value biomaterials to provide solutions for unmet medical needs as they are often inherently biocompatible14. This Review highlights recent advances in our understanding of bacterial biopolymers, reflecting on their biological function and their use as bio-based materials.
Main classes of bacterial polymers
Polysaccharides
Polysaccharides are polymers composed of sugars and/or sugar acids. They are classified into homopolymers and heteropolymers and they can be charged or non-charged, non-repeating or repeating, and branched or unbranched. Diverse bacteria produce polysaccharides and store them inside cells (for example, glycogen) or secrete them either as capsular polysaccharides that are linked to the cell surface or as free exopolysaccharides that contribute to the biofilm matrix (for example, alginate and cellulose)4. When motile, pathogens produce virulence factors and toxic molecules (for example, flagella and exotoxins). However, when they switch to a sessile lifestyle, they produce different types of exopolysaccharides as matrix components, such as alginate, cellulose and hyaluronate. This switch to the sessile biofilm lifestyle underlies the progression of many chronic infections as embedded or encapsulated cells are protected from immune cells and antibacterial drugs2 (Fig. 1). For example, alginate within the biofilm matrix of Pseudomonas aeruginosa provides a survival advantage by protecting cells from phagocytosis1. Alginates (Table 1) interact with divalent cations to form dense hydrogels with high water-holding capacity15,16. Production of cellulose (Table 1) provides similar advantages to enterobacterial pathogens17,18. Escherichia coli produces phosphoethanolamine cellulose, which forms mortar-like structures to stabilize proteinaceous curli fibres. These fibres mediate strong connections between cells in complex biofilms and provide resistance in high-shear conditions19,20. Some pathogens, such as Streptococcus pyogenes and Bacillus cereus G9241, produce a capsule of hyaluronate, a linear negatively charged heteropolysaccharide (Table 1) that mimics the structure of hyaluronate found in human connective tissues. Thereby, these pathogens can hide their antigenic surface from opsonization and phagocytosis21,22. Serogroup B Neisseria meningitidis causes invasive meningococcal disease and produces a capsular polysaccharide composed of homopolymers of sialic acid (N-acetylneuraminic acid) with (α2→8)-sialic acid linkages. This structure resembles polysialic acid moieties of human tissue antigens, and such molecular mimicry imparts poor immunogenic properties on these capsular polysaccharides, making the pathogen invisible to the host23. Streptococcus pneumoniae causes severe lung infections and comprises more than 100 serotypes that produce different capsular polysaccharides to evade adaptive immune responses24. Secreted and capsular polysaccharides are used as antigens in conjugate vaccines (Table 2). As newly emerging serotypes of pathogens such as S. pneumoniae and N. meningitidis reduce the efficacy of existing vaccines, the development of serotype-independent vaccines is becoming increasingly attractive25.
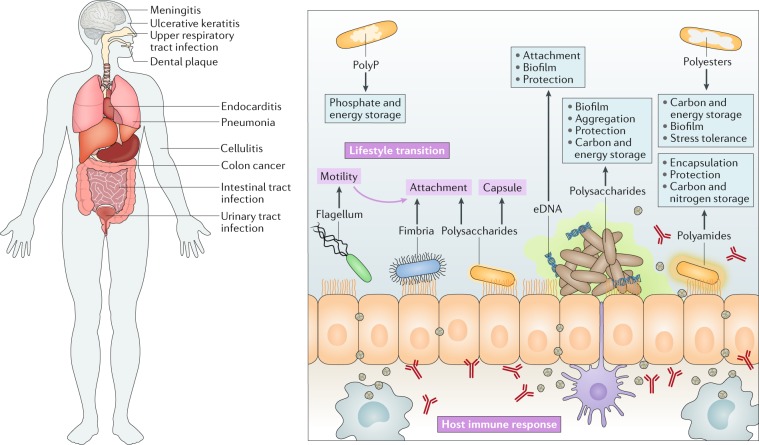
Fig. 1. Bacterial biopolymers and their functions.
Bacteria can survive in diverse ecosystems and infect a variety of living organisms. When produced by bacterial pathogens, secreted biopolymers can function as virulence factors, whereas intracellular polymers are mainly reserve materials that increase survival during starvation. The switch from motility to sessility of bacterial pathogens is a strategic decision that is often connected with the production of exopolysaccharides. Pathogens benefit from the production of high molecular weight polysaccharides as they are an integral part of the biofilm matrix and interact with counterions and other polymers to form a hydrogel-like niche2,16. Furthermore, they protect embedded bacterial cells from environmental stresses, the immune systems and antimicrobial treatment. This lifestyle transition underlies the establishment of many chronic and hard to eradicate infections. Capsular polysaccharides are attached to the cell surface and protect the pathogen from phagocytosis and antimicrobial drugs. Glycogen is an intracellular storage polysaccharide that promotes the survival of some pathogens during the intracellular phase of infection. Polyhydroxyalkanoates (PHAs) are highly reduced biopolyesters that function as storage compounds that increase bacterial fitness and potentially function as an electron sink in anaerobic zones of biofilms54,56. PHA-metabolizing enzymes are produced under specific nutritional and environmental stresses to enhance bacterial survival. Polyamides function as bacterial capsules or slimes to protect cells45 or as intracellular storage material. Bacillus anthracis, which can cause lethal infections, produces such a capsule. Polyphosphates (polyPs) are chains of condensed phosphates that function as a storage material with high energy-rich bonds. The metabolism of polyP is positively correlated with the production of virulence factors64,67. Extracellular DNA (eDNA) mediates the surface adhesion of cells and stabilizes the biofilm matrix through interaction with other secreted polymers and cations1. Proteinaceous components such as fimbriae, pili and flagella are extracellular self-assembling nanostructures that contribute to surface attachment, the formation of the biofilm matrix and/or bacterial motility.
Table 1.
Key bacterial biopolymers and their applications as bio-based materials
| Polymer | Structure | Bacterial producersa and yield | Unique characteristics | Potential applications | Commercial applications |
|---|---|---|---|---|---|
| Alginate | Non-repeating negatively charged heteropolymer, β-(1,4)-linked β-d-mannuronic acid and α-l-guluronic acid | Pseudomonas aeruginosa (P), other pseudomonads and Azotobacter spp. (NP); ~26 g l−1 (P. aeruginosa PGN5 (ref.152)) | Acetylated, HMW (molecular mass >1 MDa), polydispersity index close to 1.0, tailor-made production | Hydrogels, fibres, films and nanoparticles for various purposes, such as drug delivery, cell encapsulation and tissue engineering | Bacterial alginates do not have GRAS status; algal alginates are widely used as biomaterials for food, cosmetic, pharmaceutical and biomedical purposes (for example, wound dressings and antacids) |
| Cellulose | Homopolymer, unbranched β-(1,4)-linked glucose units | Escherichia coli (P), Salmonella enterica (P), Sarcina spp. (P and NP), Agrobacterium spp. (NP), Rhizobium spp. (NP), Pseudomonas fluorescens (NP), Komagataeibacter hansenii (also known as Gluconacetobacter hansenii or Gluconacetobacter xylinus) (NP) and Komagataeibacter rhaeticus iGEM (NP); E. coli (NP) as recombinant host; ~18 g l−1 (Acetobacter xylinum KJ1 (ref.88)) | Acetylated and/or pEtN-ated, lignin-free or hemicellulose-free, HMW (molecular mass ~1 MDa), high tensile strength, high water-holding capacity, high crystallinity index, thin fibrils, high porous structure, forming ribbon cellulose, high mouldability, tailor-made production | Hydrogels, fibres, films and nanoparticles for various purposes, such as drug delivery and cell encapsulation | Bacterial cellulose produced by certain bacteria (for example, K. hansenii) has GRAS status; widely used in food, biomedical and packaging products (for example, wound dressings, surgical and dental implants and textile fibres) |
| Hyaluronate | Unbranched negatively charged heteropolymer, alternating β-d-glucuronic acid and β-d-N-acetylglucosamine residues linked via β-(1→4)-glycosidic bonds and β-(1→3)-glycosidic bonds | Group A Streptococcus (P), Pasteurella multocida (P), Bacillus cereus G9241 (P) and Streptococcus equi (NP); Lactococcus lactis, Bacillus subtilis and E. coli (NP) and Corynebacterium glutamicum (NP) as recombinant hosts; ~12 g l−1 (S. equi157) | HMW (>1 ×106), highly viscous, gelling properties, high stability, high water-holding capacity, tailor-made and cell-free production | Hydrogels, surface-modified liposomes, nanoparticles and microparticles for many medical, pharmaceutical, food and cosmetic applications | Bacterial hyaluronate produced by certain bacteria (for example, S. equi) has GRAS status; widely used in cosmetic, topical, ophthalmologic and viscosupplementation formulations (for example to treat osteoarthritis of the knee) |
| γ-PGA | Homopolyamide composed of d-glutamic acid and/or l-glutamic acid units connected by amide linkages between α-amino and γ-carboxylic acid groups | Mostly Gram-positive bacteria, Bacillus anthracis (P), Streptococcus epidermidis (P) and Fusobacterium nucleatum (P); B. subtilis, Bacillus licheniformis strains and E. coli (NP) as recombinant hosts; ~44 g l−1 (B. licheniformis134) | Highly hygroscopic, negatively charged, chelating cations | Hydrogels and blends for biomedical, industrial and bioremediation applications | Antiscalant, dispersing agent, thickener, food additives, humectant and drug carriers |
| PHAs | Inclusions, homopolymer or heteropolymer of (R)-3-hydroxy fatty acids | Gram-positive and Gram-negative bacteria and archaea; Pseudomonas putida, Aeromonas hydrophila, Ralstonia eutropha and Alcaligenes latus (NP) as industrial hosts; E. coli and Ralstonia eutropha (NP) as recombinant hosts; ~168 g l−1 (E. coli CGSC 4401 (ref.158)) | Various thermoplastics with melting temperature of 60–180 °C, glass transition temperature of ~4–40 °C, 10−80% crystallinity, elongation to break 3–450% | Nanoparticles, fibres, films, blends and composites for various purposes, including vaccine development, regenerative medicine, implants and tissue engineering | Some PHA-based products are FDA approved (for example, sutures); applied in medical products, cosmetics, food packaging, coatings, agricultural films and bioplastic based materials |
| PolyP | Homopolymer of orthophosphate residues linked by anhydride bonds | Most bacteria; 127 mg per gram of cell dry weight (Citrobacter freundii159) | Negatively charged, rich in high-energy bonds, PolyPs (~1,000–1,300 phosphate units) are potent modulators of the human blood clotting system | Hydrogels or nanoparticles for biomedical applications (for example, regenerative medicine and drug delivery); for delivering high-energy phosphate for synthesis reactions | Generally, polyP has GRAS status as a direct food additive; widely used for industrial purposes such as in liquid phosphate fertilizers, water filter cartridges and wastewater treatment |
GRAS, generally recognized as safe; HMW, high molecular weight; γ-PGA, poly(γ-d-glutamic acid); PHAs, polyhydroxyalkanoates; pEtN, phosphoethanolamine; polyP, polyphosphate. aPathogenic (P) or non-pathogenic (NP) in humans.
Table 2.
Bacterial exopolysaccharides as virulence factors and drug and vaccine targets
| Exopolysaccharidea | Structure | Pathogen | Pathogenic mechanism | Potential drugs and targets | Vaccines on the market or in developmentb |
|---|---|---|---|---|---|
| Alginate | β-(1,4)-linked non-repeating heteropolymer of partially acetylated mannuronic acid and guluronic acid | Pseudomonas spp., Pseudomonas aeruginosa | Biofilm matrix component; antiphagocytic factor, protection from free radicals and antibiotics | Alginate oligomers induce a greater negative charge on the cell surface, reduce bacterial adherence and biofilm formation, bind to bacterial flagella and inhibit motility, and disrupt the extracellular polymeric matrix114,160c; alginate lyase degrades alginate and reduces the viscoelasticity of mucoid biofilms161; a class of mercaptobenzotriazoloquinazolinones inhibits the Alg44–c-di-GMP interaction and alginate production112 | Polymannuronic acid; alginate; conjugated with various carriers; AR-105c,d (IgG1 mAb) targets alginate |
| Pel | Partially acetylated (1→4)-glycosidic linkages of N-acetylgalactosamine and N-acetylglucosamine | P. aeruginosa | Biofilm matrix component; protection from certain aminoglycoside antibiotics | The glycoside hydrolase PelAh targets Pel and inhibits biofilm formation, reduces biofilm thickness and increases neutrophil killing162 | NA |
| Psl | Repeating pentasaccharide of d-mannose, d-glucose and l-rhamnose | P. aeruginosa | Adhesion of cells to surface for biofilm formation; protection from phagocytosis, oxidative stress and antibiotics | The glycoside hydrolase PslGh inhibits biofilm formation and reduces biofilm thickness162 | MEDI3902d (IgG1 mAb) targets Psl |
| Cellulose | β-(1,4)-linked homopolymer of d-glucose | Many enterobacterial species (for example, Escherichia coli, Salmonella enterica and Citrobacter freundii) | Biofilm matrix component; phosphoethanolamine cellulose has a mortar-like function and stabilizes curli attachment to bladder cells; protects Salmonella enterica subsp. enterica serovar Typhimurium inside macrophages; antiphagocytic factor | NA | NA |
| Hyaluronate | β-(1,4)-linked repeating heteropolymer of glucoronate and N-acetylglucosamine | Group A and group C Streptococcus spp.; Bacillus cereus G9241; Pasteurella multocida Carter type A | Biofilm matrix component; antiphagocytic factor; supports pneumococcal growth on digestion | NA | Group A Streptococcus: Lancefield group A carbohydrate conjugated with TT carrier; N-acetylglucosamine -deficient polysaccharide conjugated with recombinant pneumococcal protein SP0435 |
| K antigens (CPS) | >80 serotypes; differ from each other by their sugar composition, linkage between the sugars and their stereoisomeric formse | E. coli (groups 1, 2, 3 and 4) | Capsular; antiphagocytic factor; some are structurally similar to host cell polysaccharides; adhesion to the host cells; anionic CPS binds cationic antimicrobial peptides and prevents cell lysis | Unnatural cyclic octasaccharide (6-deoxy-6-amino) cyclomaltooctaose inhibits K30 polysaccharide transport in E. coli E69 by binding to the transporter Wza163 | NA |
| Colanic acid | β-(1,4)-linked repeating heteropolymer of fucose, glucose, glucuronate and galactose, decorated with O-acetyl and pyruvate side chains | E. coli, Shigella spp., Salmonella spp. and Enterobacter spp. | Biofilm matrix component; stress tolerance; protection from antibiotic treatment | NA | NA |
| GBS polysaccharides | 9 serotypes, heteropolymer of glucose, galactose and N-acetylneuraminic acid; N-acetylglucosamine or rhamnose are variable | Group B Streptococcus (also known as Streptococcus agalactiae) | Capsular, antiphagocytic factor; interfering with complement-mediated killing | NA | Various capsular polysaccharides conjugated to carrier proteins are currently under investigation as vaccinesc |
| Polysialic acid capsular polysaccharides | Polymer of N-acetylneuraminic acid with (α2→8)-sialic acid linkages | Neisseria meningitidis and E. coli K1 | Capsule component; molecular mimicry with polysialic acid moieties of human tissue antigens; antiphagocytic factor | γ-Cyclodextrin blocks the Wza transporter163 | N. meningitidis serogroup A, C, Y and W-135 capsular polysaccharide or oligosaccharide antigens individually conjugated to DT, TT or CRM197; synthetic glycoconjugate of Hep2Kdo2 (the core tetrasaccharide in LPS) and DT164f |
| Pneumococcal polysaccharides | >91 serotypes; differ from each other by their sugar composition, their linkage between the sugars and their stereoisomeric forms | Streptococcus pneumonia (8 genetic groups due to variation in cps loci) | Capsule component; required for pathogen transition from the lung to the bloodstream; antiphagocytic factor | The glycoside hydrolase Pn3Pase degrades the type 3 capsular polysaccharide of S. pneumonia165 | Mixture of purified CPSs from multiple types of S. pneumonia; conjugate vaccines containing CPSs |
| Staphylococcus CPS | >10 serotypes; CP5 and CP8 serotypes have repeating units of N-acetyl mannosaminuronic acid, N-acetyl l-fucosamine and N-acetyl d-fucosamine; variable based on glycosidic linkages between the sugars and the sites of O-acetylation | Staphylococcus aureus | Capsule component; antiphagocytic factor | NA | CPS (that is, CP5 and CP8) conjugated to P. aeruginosa exotoxin Ac |
CPS, capsular polysaccharide; DT, diphtheria toxoid; LPS, lipopolysaccharide; mAb, monoclonal antibody; NA, not available; TT, tetanus toxoid. aLipopolysaccharides (for example, O antigen) are not included. bA complete list of glycoconjugate vaccines is provided in ref.166. cIn clinical trials. dPassive immunization. eSee EK3D database devoted to E. coli K antigens. fAntibacterial vaccination strategy.
Glycogen, a water-soluble polymer of α-1,4-linked and α-1,6-linked glucose, is a widespread form of carbon and energy storage that promotes survival during starvation26. During the intracellular phase of infection, glycogen can have an important role in the survival of pathogens, such as Mycobacterium tuberculosis, Salmonella enterica subsp. enterica serovar Typhimurium and Chlamydia trachomatis27,28.
Besides their role as virulence factors, bacterial polysaccharides exhibit unique material properties (Fig. 2). Chemical synthesis of polysaccharides is very laborious, costly and limited to low molecular weights and has been achieved for only a few types of polysaccharides. Hence, microbial cell factories are required for the manufacture of these polymers. The presence of hydrophilic groups (for example, hydroxy and carboxyl groups) on polysaccharides confers high water-binding capacity and allows intermolecular interactions and crosslinking (for example, polymer–drug, polymer–polymer and polymer–host tissue and cell interactions). Polysaccharides can form porous hydrogels that can be used for drug delivery and controlled release of anticancer drugs29,30, immobilization of enzymes31, tissue engineering30, therapeutic cell entrapment and protection of transplanted cells from the host immune system32,33 (Fig. 2; Table 1). Hydrogels made of bacterial cellulose form efficient matrices, hydrogel nanofibrillar network scaffolds or fibre composites for biomedical applications; for example, in wound dressings that deliver human epidermal keratinocytes and dermal fibroblasts24,34–36. Production and application of cellulose produced by Komagataeibacter xylinus have been extensively studied37,38, and a process for large-scale production of bacterial cellulose-based ‘rayon fibres’ for use as wearable textiles has been developed. A successful example of a bacterial polysaccharide used in biomedical applications is hyaluronate produced by non-pyogenic Streptococcus zooepidemicus39. Commercial formulations of a gel-like fluid of hyaluronate were used for injection into the knee joint to mitigate arthritis pain.
Fig. 2. Bacterial polysaccharides as biomaterials and their properties.
High molecular weight exopolysaccharides, such as alginate, cellulose and hyaluronate, are well-known virulence factors constituting the biofilm matrix. The interaction of polysaccharides and other polymeric substances can determine the properties of the biofilm matrix. Bacterial polysaccharides are very diverse, and their diversity and material properties are determined by the constituent sugars or sugar acids, the type of glycosidic linkages and whether they are unbranched or branched, the length of the polymer (and thus the molecular weight), the type of side group (for example, acetyl, pyruvate or succinate) and the degree of substitution7,16. Bacterial polysaccharides are important biomaterials due to their unique material properties, including solubility, rheological characteristics, viscoelastic properties, interaction with cations, ionic strength, crosslinking, gelation, water retention, extendibility and stability under different conditions. Hence, polysaccharides have been applied as natural viscosifiers, thickeners, stabilizers, gel and film formers, and additives or have been processed into nanostructures (for example, nanoparticles and nanotubes), microspheres, microcapsules, sponges, hydrogels, foams, elastomers and fibres43,44,156. Besides the desired material properties, high purity and the purification process are crucial for the use of bacterial polysaccharides as high-value biomaterials32,43. d-Glc, d-glucose; d-GlcA, d-glucuronic acid; GlcNAc, N-acetylglucosamine; l-GulA, l-guluronic acid; d-ManA, d-mannuronic acid.
Specific enzymes naturally modify bacterial polysaccharides to change their material properties and support their biological functions (Table 1). For example, the presence of acetyl groups on polysaccharide chains notably alters the structural conformation and affects chain–chain interactions, solubility, water-holding capacity, viscoelasticity and molecular weight16,40–42. Genetic engineering of polysaccharide-modifying enzymes in bacterial cell factories or the use of such enzymes for in vitro modification of polysaccharides allows production of tailor-made polysaccharides. The design of materials becomes even more versatile through blending with other polymeric and non-polymeric components (for example, citric acid (crosslinking), stearic acid (esterifying) and plasticizers). Blending allows tailoring of properties such as viscoelasticity, gelation degree, porosity and material strength. Such materials have gained much attention as feedstock materials or bioinks for 3D bioprinting with numerous biomedical and engineering applications, including tissue engineering, drug delivery and drug testing43. Cell-loaded 3D scaffolds of alginate or hyaluronate have been used successfully as an artificial extracellular matrix that provides a temporary environment to support infiltration, adhesion, proliferation and differentiation of various cell types, including mesenchymal stem cells, fibroblasts, chondrocytes, osteoblasts and embryonic stem cells44. Overall, bacteria are a major natural resource for the production of a vast variety of polysaccharides with many potential industrial and medical uses (Table 1).
Polyamides
Bacteria can produce polyamides or poly(amino acid) chains (Fig. 3a), such as secreted poly(γ-d-glutamic acid) (γ-PGA) and poly(ε-l-lysine) (ε-PL) or the intracellular cyanophycin (a copolymer of l-aspartic acid and l-arginine), which can function as capsules or biofilm matrix45,46, or as storage materials47,48, respectively (Fig. 1). Similarly to the role of polysaccharides in the biofilm matrix, a polyamide capsule or biofilm is poorly immunogenic and conceals pathogens such as Bacillus anthracis from surveillance by the immune system45. Many non-pathogenic polyamide-producing bacteria, such as Bacillus licheniformis, Bacillus megaterium and most cyanobacteria, have been considered as hosts for the production of polyamide-based materials49. Polyamides are highly charged and can be polyanionic (for example, γ-PGA) or polycationic (for example, ε-PL). They are biodegradable, non-toxic and renewable. Metabolic engineering allowed enhanced biotechnological production of polyamides. Polyamides have been considered as substitutes for chemically synthesized polymers in industrial applications. For example, γ-PGA can be used as a flocculant to replace synthetic flocculants (for example, polyaluminium chloride and polyacrylamide) in wastewater treatment50,51 (Table 1). ε-PL has antibacterial properties as it disrupts membrane integrity, and its crosslinked form was used in antimicrobial coatings52,53.
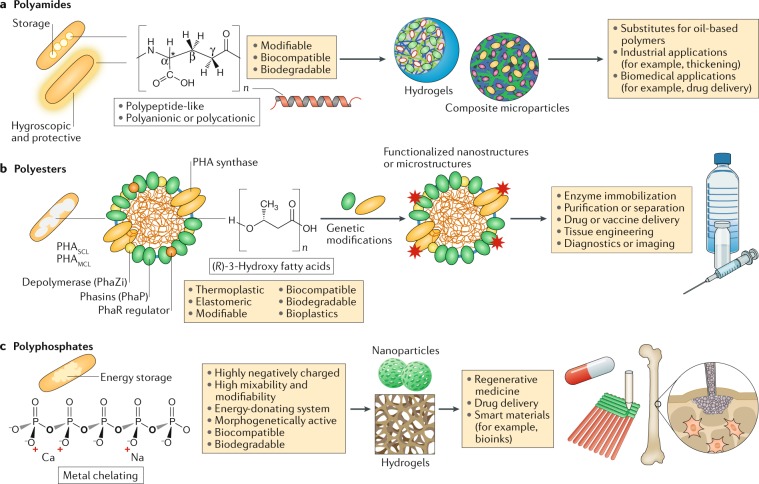
Fig. 3. Bacterial polymer granules as biomaterials.
a | Polyamides are composed of amino acids and are non-ribosomally synthesized by specific synthetases. They are found as intracellular granules without confining membranes and decorating proteins or as secreted extracellular capsules and slimes45,47. Due to their biodegradability, non-toxicity and modifiability, bacterial polyamides have been considered as substitutes for chemically synthesized polymers that can be processed into formulations for industrial, biomedical, pharmaceutical and cosmetic applications50. b | Polyhydroxyalkanoates (PHAs), such as polyhydroxybutyrate, are natural polyesters that are synthesized into hydrophobic spherical inclusions from (R)-3-hydroxybutyric acid. PHAs have been classified into short-chain-length PHAs (PHASCL; containing constituents with 3–5 carbon atoms) and medium-chain-length PHAs (PHAMCL; containing constituents with 6–14 carbon atoms), which are primarily produced by pseudomonads58. Synthases and other PHA-binding proteins decorate the surface of PHA inclusions. PHAs are unique bio-based materials processed as bioplastics or bioengineered functionalized nanoparticles for uses in medicine and industry. PHA nanobeads can function as effective platforms for enzyme immobilization, protein purification, bioseparation, drug or vaccine delivery, tissue engineering, diagnostics and imaging58. c | Polyphosphates are composed of orthophosphates (inorganic phosphates, three to several hundred phosphates) linked by phosphoanhydride (P–O–P) bonds. They contribute to energy storage and can be processed into hydrogels or nanoparticles for various applications (Table 1). The phosphate and counterions such as Ca2+ and Sr2+ are released on hydrolysis and can be used for bone biomineralization, as smart bioinks for generating 3D scaffolds and for cell bioprinting of regeneratively active patient-specific osteoarticular implants69–71,73. Polyphosphate or collagen hydrogels were formulated for improving tissue integration of meshes to improve the outcome of surgical hernia repair72.
Polyesters
Polyhydroxyalkanoates (PHAs) such as poly((R)-3-hydroxybutyrate) are bacterially synthesized bioplastics. They are linear polyesters that are synthesized and assembled into hydrophobic spherical inclusions and they function in carbon and energy storage7 (Fig. 3b; Table 1). Although a wide range of Gram-positive and Gram-negative bacteria produce PHAs, the roles of PHAs and their metabolizing genes in the context of persistence and pathogenesis remain largely unknown. PHA– mutants of P. aeruginosa showed reduced attachment to glass surfaces and reduced stress tolerance in biofilms, suggesting a possible contribution of PHA to persistence during infection54. In the plant-pathogen Xanthomonas oryzae pv. oryzae, which causes major losses in rice production, the regulatory protein PhaR not only represses PHA synthesis but also affects production of extracellular polymeric substances, the bacterial lifestyle, phenotypic changes and virulence55. PHAs are proposed as a sink for electrons under anaerobic conditions; that is, in the absence of terminal electron acceptors such as oxygen, they enhance survival56,57 (Fig. 1). PHA synthesis and mobilization are regulated in response to environmental stimuli, such as nutritional and environmental stresses, providing a survival advantage55.
PHAs have been considered as unique bio-based plastics that can be bioengineered, chemically modified and processed into high-value medical materials (for example, sutures, tissue engineering scaffolds, drug carriers and particulate vaccines) or low-value commodity bioplastics (Table 1). Production of tailor-made PHAs via bioengineering, physical blending and chemical modification resulted in improved material properties, which met specifications for industrial and medical applications58,59. An exciting innovative approach engineered bacterial cell factories to assemble PHA inclusions that are densely coated with functional proteins of interest. These functionalized PHA beads were stable after separation from the bacterial cell mass and showed promising performance as vaccines, immunodiagnostics, bioseparation resins, enzyme carriers and tools for recombinant protein production60,61. The functionality of these non-porous protein-coated PHA beads was further tunable by controlled encapsulation into porous alginate microspheres, which allowed flow-through applications. This study is an example of the tremendous materials design space provided by bioengineering of polymers and by blending of polymers to generate functional composite materials62.
Polyphosphates
PolyP is a polymer of condensed phosphates (three to several hundred inorganic phosphates) that is highly negatively charged and rich in ‘high-energy’ anhydride bonds. It functions as an energy-storage polymer63,64 (Fig. 3c; Table 1). PolyP synthesis is an evolutionarily ancient ability of bacteria, and polyPs, besides functioning in phosphate storage, also provide chemical energy for biosynthesis pathways, function as a buffer against alkalis and as a metal-chelating agent and contribute to channel complexes for the uptake of DNA7,64,65. PolyPs also regulate cell signalling and thereby affect bacterial lifestyle, persistence, viability, growth, stress tolerance and virulence66–68 (Fig. 1).
Due to their eminent energy-storage feature, industry has increasingly considered polyPs to drive energy-consuming enzyme-catalysed reactions (Table 1). They are also considered as morphogenetically active biomaterials in regenerative medicine, such as in cartilage repair and bone regeneration69,70, and as drug delivery vehicles71. Owing to their capacity to interact with positively charged polymers (for example, alginate and hyaluronate), inorganic cations (for example, Ca2+, Mg2+, Zn2+, Fe2+, Na+ and K+), or basic organic components (for example, amino acids, polyamines and peptides), polyPs can be processed into hydrogels or nanoparticles for bone biomineralization69,70 and other biomedical applications72. Importantly, the physical properties, such as mechanical strength, stability and functionality, of polyP-based complex hydrogels or nanoparticles vary with the type of interacting counterions or blended polymers. This variation provides substantial design space to generate a range of desired material properties for ‘smart biomaterials’ and bioinks in regenerative medicine71,73 (Fig. 3c). As no abiotic polyP minerals have been found on Earth, living organisms, in particular bacteria, are unique sources of polyP. Bacteria belonging to the genera Mycobacterium and Corynebacterium produce polyP granules with a high yield and therefore are potential production strains for the manufacture of polyPs74.
Other polymers produced by bacteria
Bacteria also produce other types of biopolymers, including extracellular DNA and proteinaceous components (Fig. 1). They are not only important in bacterial pathogenesis but are also considered for development of bio-based materials. Extracellular DNA arises when a cell lyses and releases intracellular DNA. In biofilms, lysis of a subpopulation of cells contributes extracellular DNA to the biofilm architecture, for example in the stalks of mushroom-shaped microcolonies of P. aeruginosa75. Due to its high negative charge, extracellular DNA has multifaceted roles, including in the adhesion and stability of the biofilm matrix through interaction with positively charged polysaccharides (for example, Pel) and cations, it is a nutrient source during starvation and it confers antibiotic resistance1.
Secreted polypeptides, for example composed of alternating hydrophilic and hydrophobic amino acid residues, and proteins, such as fimbrillin, pilin and flagellin, can be molecular building blocks of extracellular self-assembling structures, such as functional amyloids (for example, curli fibres), fimbriae, pili and flagella. These self-assembling structures can form nanofibres or nanotubes and mediate cell adhesion to biotic and abiotic surfaces, development of the biofilm matrix or motility during pathogenesis. Several features of these structures make them attractive for applications, including the precise arrangement of protein building blocks in self-assembling structures, their high surface area to volume ratio and polymorphic transformation in response to physical and chemical stimulations. These features render them valuable bio-based materials and biotemplates for fabrication of novel nanostructures, nanodevices and multilayer lattices applicable in bioengineering and nanomedicine (for example, drug delivery)76–78.
The genetic programmability and ease of engineering of extracellular DNA, polypeptides and proteins make them fascinating programmable biomaterial platforms that are hardly achievable for other biopolymer types, including polysaccharides and polyesters79. Straightforward genetic programmability has generated much recent interest in developing engineered living materials; that is, living cells are engineered to autonomously self-assemble entire materials with novel and tunable properties for a variety of purposes, such as microbial electrosynthesis, biosensors, electronic monitoring devices and bioremediation79,80. We only scratch the surface of the vast scope of self-assembling structures produced by bacteria and their applications, but they have been reviewed extensively elsewhere80–83.
Bacterial polymer synthesis
Synthesis pathways and their regulation
Genome sequencing, functional genomics, advanced molecular tools and techniques, and new biochemical and biophysical approaches enhance our understanding of the biosynthesis of bacterial biopolymers. Vast DNA and protein databases combined with in silico approaches provide insights into biosynthesis pathways and the structure and function of key biosynthesis proteins. All these advancements have created a solid foundation for the design of cell factories for enhanced production of polymers or to produce tailor-made polymers. Current synthetic biology approaches that use DNA foundries are the next-generation tools for the design of cell factories and allow precision engineering of production strains84. Biosynthesis pathways for representative bacterial polymers are illustrated in Fig. 4. In addition to a better understanding of biosynthesis pathways and enzymes, knowledge of the molecular mechanisms of synthesis, modification and, if required, secretion informs production of novel tailor-made polymers. For example, many bacterial polysaccharides are enzymatically modified at the polymer level, such as acetylated, deacetylated, epimerized or phosphoethanolaminated, and these modifications affect material properties, such as viscoelasticity and gel-forming capacity40. Genes encoding enzymes involved in polymerization and modifications of polysaccharides are usually co-clustered in one main operon (Fig. 4). Potent specific promoters often control these operons and the transcription of the entire biosynthesis gene cluster4.
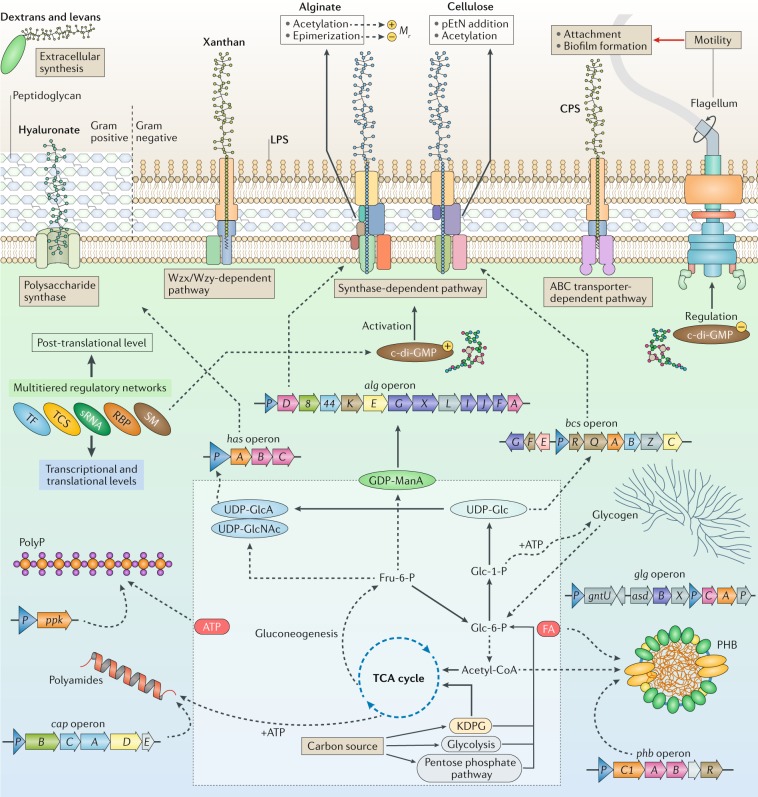
Fig. 4. The main metabolic routes for the synthesis of bacterial biopolymers.
Intermediates of central metabolism are diverted towards the provision of precursors for polymer synthesis. Four general mechanisms for the production of polysaccharides in bacteria are shown. Synthesis of some secreted non-repeating polysaccharides, such as alginate and cellulose, is mediated by multiprotein complexes, usually consisting of a polymerase, a copolymerase, carbohydrate-modifying enzymes and secretion subunits. The genes encoding such functionally related protein subunits are co-clustered in large operons, such as the alg and bcs operons. Some polysaccharides, such as xanthan, are produced through the Wzy-dependent polysaccharide synthesis mechanism. In this pathway the repeating sugar units and their linked lipid carriers are assembled by several glycosyltransferases at the cytoplasmic membrane, followed by flipping across the cytoplasmic membrane, the final polymerization step in the periplasm and secretion. However, the synthesis of some polysaccharides, such as hyaluronate, dextrans and levans, is less complex and is mediated by a single enzyme. Dextrans and levans are synthesized outside the cell by sugar transferases that convert disaccharides into polysaccharides and use the energy that is released by hydrolysis of the glycosidic bond of the disaccharides. Modification of secreted polysaccharides (for example, acetylation, deacetylation, epimerization and phosphoethanolamine (pEtN) addition) can occur during translocation of nascent polymers across the cell envelope. Polyhydroxyalkanoates (for example, polyhydroxybutyrate (PHB)) are synthesized by a polyhydroxyalkanoate synthase that coverts hydroxyacyl-CoA derivatives of central metabolism into intracellular polyesters. Enzymatic processes independent of ribosomal protein biosynthesis synthesize polyamides. Dashed lines indicate multiple enzymatic steps, a circled plus sign indicates positive correlation and a circled minus sign indicates negative correlation. ABC, ATP-binding cassette; CPS, capsular polysaccharide; FA, fatty acid de novo biosynthesis; Fru-6-P, fructose 6-phosphate; Glc, glucose; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; Glc-1-P, glucose 1-phosphate; Glc-6-P, glucose 6-phosphate; KDPG, 2-keto-3-deoxy-6-phosphogluconate pathway; LPS, lipopolysaccharide; ManA, mannuronic acid; polyP, polyphosphate; RBP, RNA-binding protein; SM, second messenger; sRNA small non-coding RNA; TCA, tricarboxylic acid; TCS, two-component system; TF, transcription factor.
Biosynthesis of polymers in bacteria is controlled by regulatory networks that process environmental signals and mediate responses through transcriptional and post-translational regulation (Fig. 4). At the transcriptional level, transcription factors activate promoters that control the expression of functionally related genes. Such transcription factors include sigma factors, which are subunits of RNA polymerase, and regulatory proteins that bind to DNA regions upstream of biosynthesis genes. Some sigma factors are sequestered by anti-sigma factors and are released on exposure to external stimuli. For example, AlgU is a sigma factor that binds to the core RNA polymerase and thereby mediates binding to a specific promoter region upstream of the alginate biosynthesis gene cluster. AlgU is sequestered by the membrane-bound anti-sigma factor MucA in P. aeruginosa85 and likely other pseudomonads under conditions that are not permissive to alginate production. On environmentally induced destabilization of this complex, such as in response to cell envelope stress or on mutation of the mucA gene (for example, adaptive mutation during chronic infection), AlgU is released and activates transcription of the alginate biosynthesis gene cluster. An engineered MucA-inactivated strain86 constitutively produced high yields of alginate, which might be an avenue for enhanced production of bacterial alginate. Furthermore, small non-coding RNAs, two-component systems, regulatory RNA-binding proteins and second messengers (such as c-di-GMP and c-di-AMP) are involved in signal processing and complex regulatory networks that control polymer synthesis in bacteria1,87. Improved understanding of these regulatory complexities via systems biology will inform synthetic biology approaches for efficient production of polymeric materials.
Processive synthesis of high molecular weight biopolymers
Bacteria have highly processive enzymes for the production of biopolymers with high molecular weights (molecular mass >100 kDa) that cannot be achieved by chemical synthesis. Many bacterial exopolysaccharides have high molecular weights, for example molecular mass ~3.9 MDa for alginate from P. aeruginosa40, ~1,000 KDa for cellulose from Komagataeibacter sucrofermentans DSM88 and ~2.1 MDa for hyaluronate from S. zooepidemicus89, and this affects polymer properties, bacterial pathogenesis and evasion of host immunity and antimicrobial treatment. In the context of bio-based material development, bacterial polysaccharides with high molecular weights have gained much attention as materials that are biocompatible and have high water retention capacity, excellent gelling properties and a long half-life under physiological conditions. Bacterial biopolymers can also be a source of biologically active oligomers (molecular mass usually <10 kDa). Such oligomers can be used as therapeutic drugs for applications such as promotion of angiogenesis, inhibition of tumour progression or induction of the production of proinflammatory mediators, anti-inflammatory substances and antibiofilm agents90–96. They can also be a source of valuable monomers, such as rare sugar monomers (for example, fucose)97, which are in high demand as precursors for pharmaceuticals and nutraceuticals. Another example is hydroxyalkanoic acid monomers that can be obtained from the hydrolysis of PHAs, which are precursors for several antibiotics98. There are various mechanisms for controlling the degree of polymerization of polysaccharides. They include substrate tethering, as described for mycobacterial galactan. In this example, the acceptor of a lipid-linked initiator oligosaccharide binds to a specific site on the glycosyltransferase and facilitates processive polymerization resulting in longer polymer chains99. Another mechanism is the coupling of polymerization with modifications, for example the processivity of alginate polymerization in P. aeruginosa, which is linked to in situ enzymatic modifications (that is, epimerization and acetylation)40,100,101. The chain length determinant protein Wzz also controls the degree of polymerization of, for example, lipopolysaccharide O antigens. In the absence of Wzz, O antigens are randomly distributed and of shorter chain length than in the presence of Wzz102,103. Substrate concentration is another regulatory mechanism, as it affects the rate of polymerization, the yield of the final product and the molecular weight. For example, high concentrations of the UDP-N-acetylglucosamine substrate increased the molecular weight of hyalorunate104. Finally, the degree of polymerization also depends on the copy numbers of polymerase and synthases. For example, the presence of a number of different synthases competing for substrates reduced polymer chain lengths of alginate40 and polyhydroxybutyrate (PHB)105.
Biopolymer synthesis as a target for new antibacterial drugs
In pathogenic bacteria, biopolymers are often major virulence factors and contribute to the persistence of infections in different ways, including antigenic mimicry (for example, polysialic acid and hyaluronate)23, hiding of the antigenic cell surface to evade opsonization and phagocytosis (for example, hyaluronate, alginate and γ-PGA)1,106,107 and as a barrier against antimicrobial drugs and toxic molecules108. Therefore, biopolymer synthesis and function might be new targets for antimicrobial drugs to overcome persistent infection, antibiotic resistance and antibiotic persistence109–111. Although this is an emerging field, there is evidence for the success of such approaches. For example, inhibition of the interaction between Alg44 and c-di-GMP, which is required for alginate polymerization, by a class of thiol-benzo-triazolo-quinazolinone compounds reduced alginate secretion by P. aeruginosa by up to 30% (ref.112). OligoG CF-5/20 (an alginate oligomer) decreased the thickness of mucus in cystic fibrosis lungs and destabilized the biofilm matrix and the extracellular polymeric network113,114. Furthermore, biopolymer-degrading enzymes (such as alginate lyases115,116 and amylases117) degraded the biofilm matrix. Finally, natural products (such as quercetin and curcumin) reduced the production of alginate and other polysaccharides by reducing the expression of the quorum sensing-regulated genes and concomitantly virulence factors such as pyocyanin, protease and elastase in P. aeruginosa, Klebsiella pneumoniae and Yersinia enterocolitica118–120 (Table 2).
Producing novel bio-based materials
Design of cell factories for the production of novel biopolymers
Over the past decade, knowledge of the biosynthesis of bacterial polymers together with systems biology and synthetic biology has revolutionized the rational engineering of cell factories, which has increased production yields and/or led to production of innovative bio-based materials (Fig. 5). As mentioned earlier, biosynthesis of bacterial polymers requires the engagement of complex cellular processes from gene expression to provision of enzymes and proteins, central metabolism, and regulatory and signalling systems leading to intracellular assembly or secretion across the cell envelope. Hence, design of cell factories for production of novel biopolymers requires integration of the complexity of cellular and metabolic process and extensive experimentation to combine the relevant genetic information. DNA foundries use advanced software, robotic and analytical approaches to allow automated ‘design–build–test’ engineering cycles for the high-throughput development of desired cell factories through synthetic assembly of genetic elements. DNA foundries have higher experimental consistency and lower costs than manual operations. Furthermore, modular genetic elements, such as promoters, terminators, ribosome-binding sites, orthogonal polymerases, untranslated regions, signal peptides, putative stabilization modules, genetic effectors and protein folding enhancers, provide a dynamic platform to tune gene expression and protein production. Striking advancements include the introduction of inducible and/or controllable genetic switches, such as T7 polymerase-based expression systems121, programmable T7-based synthetic transcription factors122, the RiboTite system123, vector engineering124 and CRISPR–Cas125 tools, to allowed fine-tuned expression of endogenous or heterologous genes. Furthermore, CRISPR–Cas9 has been successfully used to simultaneously manipulate several genes. CRISPR interference has been successfully used to redirect metabolic flux towards PHA biosynthesis6. Other examples are the rational reprogramming of Komagataeibacter rhaeticus iGEM for the production of cellulose-based materials126 and the engineering of broad-host-range vector systems to use cyanobacteria for the production of renewable bioproducts127. Recombinant production of hyaluronate, PHAs, γ-PGA and cyanophycin has been successfully achieved as these required reconstructions of relatively simple pathways. E. coli, Ralstonia eutropha, Pseudomonas putida and Alcaligenes latus are industrial workhorses for commercial production of PHA, and Bacillus subtilis has been used for commercial production of hyaluronate128. Novel inducible systems such as light-sensing or temperature-sensing systems can act as logic gates, timers, switches and oscillators to precisely control the expression or production of desired products in response to specific inputs or inducers129,130. These inducers are alternatives to chemical induction, which suffers from loss of directionality and poor control over the induction period. Light-sensing systems from cyanobacteria have been adapted for the photoinducible expression of specific genes in E. coli131 and P. aeruginosa132.
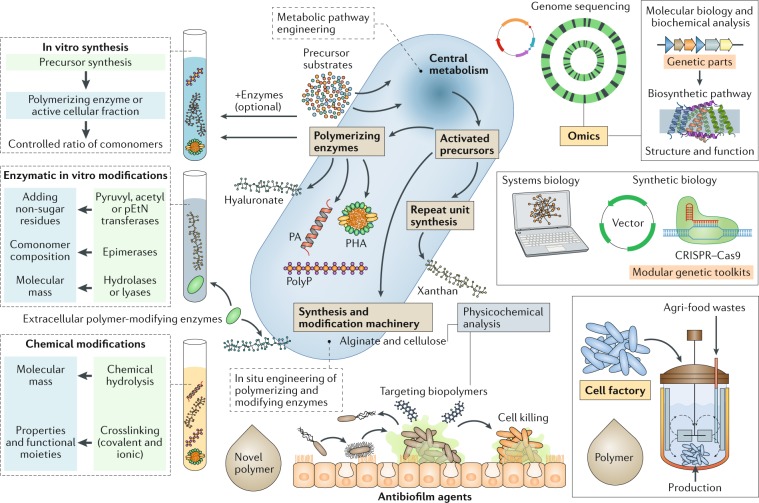
Fig. 5. Production of novel enhanced biopolymers and biopolymer synthesis as a target for drug discovery.
Production of novel biopolymers can be achieved by synthetic biology for the development of cell factories. In vitro enzymatic synthesis or modification of biopolymers as well as chemical modifications can achieve novel biopolymers with altered material properties and functions. Molecular biology and biochemical or biophysical approaches have provided insight into biosynthesis pathways of bacterial biopolymers. Selective inhibition of biopolymers that function as virulence factors offers targets for antimicrobial drug discovery. Systems biology, synthetic biology and metabolic engineering tools have accelerated the construction of novel cell factories for the production of novel bio-based materials. PA, polyamide; pEtN, phosphoethanolamine; PHA, polyhydroxyalkanoate; polyP, polyphosphate.
As illustrated in Fig. 4, the biosynthesis of biopolymers is linked to central metabolism, which means that the engineering of highly productive cell factories requires integration of carbon, nitrogen and energy fluxes133. Initially, several cell factories need to be generated to then efficiently convert precursor substrates into polymers. For example, a genetically engineered mutant of P. aeruginosa produced ~125 g of alginate from 1 g of dry cells40, which suggested a predominant flux of precursor substrates into the polymer. Indeed, understanding the major points that control the flux in the biosynthesis of biopolymers and the energetic state and relevant metabolites in the cells is a key step for increasing productivity. Thus, the metabolic engineering of synthesis pathways aims to enhance substrate and energy flux towards biosynthesis of the desired polymer. Biosynthesis of active precursors is an energy-consuming process and is commonly based on diversion of metabolites from central metabolism and primary cellular functions (Fig. 4). Therefore, determining the redox state of the cells is important when one is amending metabolic pathways and redirecting metabolites towards the desired biosynthesis pathway. The redox state is determined by factors such as electron carriers (for example, NADH and NAD+), oxygen availability, the carbon and nitrogen uptake rates and the kinetics of enzymes involved in metabolic flux. For example, the balance between the concentration of hyaluronate precursors and ATP levels, which are linked to the recycling of electron carriers, was crucial for optimal production of hyaluronate39. In B. licheniformis, increasing the ATP content of cells increased the production of γ-PGA. This was achieved by improving the respiratory electron transport chain (through the Vitreoscilla sp. haemoglobin), ATP synthesis and nitrate metabolism134. In another example, weakening or abolishment of competing pathways (for example, β-oxidation of fatty acids) and boosting of NADH (or NADPH) levels increase carbon flux towards PHA biosynthesis125. In E. coli, a combination of multiple gene deletions and additions coupled lactate utilization and conversion with the formation of GDP-fucose and, in combination with blocking of the competing colonic acid biosynthesis pathway, this strategy led to high yields of fucosylated N-acetyllactosamine oligosaccharides135. Such control elements should enhance carbon and energy flux towards the synthesis of the biopolymer, but not towards cell biomass and/or metabolic by-products40,136.
Systems biology combined with metabolic engineering using computational methods linked with high-throughput measurements of cellular processes (including metabolic pathways and gene regulatory and signalling networks) and omics data (that is, transcriptomics, proteomics, metabolomics and fluxomics data) has greatly advanced the development and improvement of cell factories and their products5,137 (Fig. 5). The number of in silico tools and computational frameworks to support synthetic biology approaches is growing and these include the iGEM Registry of Standard Biological Parts (a collection of genetic parts), COBRA and Cameo (for gene target identification, gene knockout and gene overexpression)138,139 and macromolecular expression models (for computing the optimal proteome composition of a growing cell)140. In particular for biopolymers with a complex biosynthesis pathway such as polysaccharides, computational modelling of the interplay between central metabolism and biosynthesis pathways can strongly improve bioengineering strategies. In silico genome-scale metabolic flux analysis identified metabolic engineering targets in E. coli to enhance the yields of polylactic acid and poly(3-hydroxybutyrate-co-lactate), contributing to 11% and 56% of cellular dry weight, respectively141. Accordingly, metabolic engineering of E. coli achieved the production of non-natural tailor-made polymers such as poly(lactate-co-glycolate) with a broad range of material properties142.
Rational engineering increased the range of production hosts; for example, developing the halophilic bacterium Halomonas smyrnensis AAD6T as a biotechnological production platform that does not require costly sterilization steps (high-salt media prevent growth of other living organisms) for the production of levan, Pel exopolysaccharide, PHAs and osmoprotectants143.
Challenges in bacterial production of bio-based materials
The application of bacterial biopolymers as bio-based materials is expanding (Table 1). Despite inherent properties such as biocompatibility and biodegradability, some bacterial bio-based materials have shortcomings; for example, they do not meet specifications (such as consistency and purity) that are required for medical applications. In addition, bacterial fermentation is inherently expensive and associated high production costs often prohibit commercial use.
The basic chemical structure has a major role in determining the biophysical properties of a biopolymer and its applications. For example, some PHAs have high crystallinity that causes stiffness, brittleness, poor thermomechanical properties (high melting temperature and low glass transition temperature), high hydrophobicity and stickiness and therefore restricts their application58,144. For some polysaccharides, poor mechanical stability, a lack of elastomeric properties and reduced solubility due to neutral charge or a high molecular weight restrict their utility. However, these biopolymers are naturally diverse in structure and can be enzymatically or chemically modified, which provides a wide range of physicochemical properties suitable for various applications. Improvements of biopolymers have been successfully achieved by genetic manipulation of cell factories, improving fermentation conditions and enzymatic modifications142,145–147 as well as blending with other biopolymers and/or chemical modifications such as crosslinking, chlorination, epoxidation, hydroxylation, carboxylation, etherification and esterification144,148,149. These approaches have extensively improved biopolymer properties such as stability, solubility, crystallinity, glass transition temperature, elasticity and permeability and have expanded the utility for biomedical applications, such as drug delivery and regenerative medicine43,58.
For medical applications, the cell factory and the biopolymer must be certified as ‘generally recognized as safe’ (GRAS), a designation determined by the FDA that applies to substances accepted as safe. Despite advantages (for example, biocompatibility) over synthetic materials, biomedical and biotechnological applications of bacterial biopolymers are constrained by the GRAS status of the production strain. For example, despite the extensive study of bacterial alginate biosynthesis, these alginates cannot be regarded as GRAS so far. This is also true for many products derived from Gram-negative bacteria, for which host cell-derived impurities such as endotoxins might reduce product quality150,151.
The GRAS standard of biopolymers requires the establishment of standard assays to demonstrate that polymers derived from bacteria meet purity criteria and are safe to be used as a medical device. Furthermore, the safety profiles should include that long-term use will not induce undesirable immune responses and potentially autoimmune diseases. Currently, the FDA and contract research organizations lack such standard assays for quality control. Development of these standard safety assays and their validation in relevant animal models will be important. Compositional analysis of bacterial biopolymers using next-generation, high-end analytical instruments such as advanced chromatographs and mass spectrometers will help further improve their quality control.
Also, it is clear that development of safe bacterial cell factories (for example, novel endotoxin-free and non-pathogenic strains) through synthetic biology and bioengineering as well as efficient purification methods can lead to a plethora of new polymers and high-value biomaterials. Successful examples include generation of endotoxin-free E. coli ClearColi150 and the commercial use of highly attenuated P. aeruginosa PGN5 for production of alginate152 and non-pyogenic S. zooepidemicus for production of hyaluronate, which was purified through extensive filtration and diverse adsorbents to eliminate impurities153 (Table 1).
Successful industrial-scale production of biopolymers depends on various factors, including the cost of precursor substrates, yield over substrate rate, volumetric productivity and the cost of downstream processing (purification). Whereas bioengineering aims at improving the upstream process (use of low-cost substrates and increased productivity), bioprocess optimization of the upstream and downstream processes is required for scalable and cost-effective manufacture154,155. Production of extracellular biopolymers is challenging because of associated high viscosity of the culture liquids, which reduces the diffusion of dissolved oxygen and ATP formation. Therefore, strategies that could enhance tolerance of anaerobic conditions or boost energy-generating systems may enhance productivity.
Future perspectives
Extracellular polymers that are produced by bacterial pathogens are major virulence factors. Thus, inhibition of their biosynthesis pathways represents a strategy for the treatment of bacterial infections. Owing to rising rates of antimicrobial resistance, the development of novel strategies to fight bacterial infections is in high demand. Insights into the synthesis, secretion and regulation of biopolymers will disclose new and specific targets suitable for drug discovery; for example, for targets that weaken bacterial defences against the host immune defences or antimicrobial treatment (Fig. 5).
Polymers that are produced by non-pathogenic bacteria are considered safe materials for a range of applications. Despite great advances in the design of cell factories for enhanced biopolymer production as well as production of tailored biopolymers, challenges remain. Because of a plethora of interacting components and multiple feedback loops in complex biological systems, rational engineering of novel GRAS-certified cell factories and biopolymers remains challenging. It is important to reduce this complexity through systems biology to better inform genome-scale metabolic models, metabolic network modelling and computational simulations of large data sets that feed into synthetic biology approaches. This work will provide the foundation for efficient bioengineering strategies and accurate predictions for cell factory and bioprocess development.
In this Review, we have highlighted the advances in understanding the roles of bacterial biopolymers in pathogenesis and their current and potential applications as bio-based materials. We hope that this Review will guide both drug discovery programmes and the development of new bio-based materials by outlining strategies to overcome pitfalls and challenges associated with biopolymers as virulence factors and as innovative bio-based materials.
Acknowledgements
B.H.A.R. thanks past and present members of his research group at the Centre for Cell Factories and Biopolymers at Griffith University for contributions to the work on biopolymer synthesis.
Glossary
- Processive
Continuous catalytic reactions by an enzyme without releasing its substrate.
- Synthetic biology
An interdisciplinary research field that involves the application of engineering principles to biology aiming at (re)designing and fabricating biological components and systems.
- Cell factories
Engineered cells that have been reprogrammed for enhanced production of desired compounds.
- Biocompatible
A characteristic of biomaterials defined by their non-toxicity and lack of activating the immune responses.
- Homopolymers
Polymers composed of one building block.
- Heteropolymers
Polymers composed of at least two building blocks.
- Sessile lifestyle
A bacterial growth mode associated with biofilm formation and loss of motility.
- Opsonization
An immune response by which antigens are marked by specific proteins and antibodies to facilitate their recognition and engulfment by phagocytes.
- Serotypes
An epidemiological classification in which groups belonging to a single species of microorganism share distinctive immunogenic surface structures.
- Regenerative medicine
A branch of medicine aiming at regrowing, repairing or replacing damaged or diseased cells, organs or tissues.
- Smart biomaterials
Biomaterials with the ability to respond to stimuli or physiological conditions at the micrometre to nanometre scale.
- DNA foundries
Facilities using advanced software, automation or robotics and analytical approaches for faster, easier and scalable assembly of DNA to develop advanced cell factories.
- Second messengers
Small molecules that relay signals received by cell-surface receptors to effector proteins and regulate cellular processes.
- Systems biology
A research field focusing on understanding relationships between networks of biological processes through computational and mathematical approaches.
- Nutraceuticals
Food additives that not only supplement the diet but also provide health and medical benefits.
- RiboTite system
A multilayered modular genetic control circuit using standard inducible promoters and orthogonal riboswitches for tunable control of T7 RNA polymerase activity and recombinant expression of genes of interest.
- CRISPR–Cas
A genetic system for gene editing based on naturally occurring genome editing in bacteria and archaea that confers adaptive immunity against invading phages.
- CRISPR interference
A method that uses a catalytically inactive Cas9 protein and a customizable single guide RNA that binds to DNA and blocks transcription of a gene of interest.
- Glass transition
A reversible physical transition (for example, from a hard, glassy state to a soft, rubbery state) that an amorphous material undergoes in a particular temperature range.
Author contributions
Both authors researched data for the article, discussed the content, wrote the article and reviewed and edited the manuscript before submission.
Competing interests
B.H.A.R. is a co-founder and shareholder of PolyBatics Ltd, which commercializes veterinary tuberculosis diagnostic products related to protein-coated polyester spheres assembled in engineered E. coli. M.F.M. declares no competing interests.
Footnotes
Peer review information
Nature Reviews Microbiology thanks Guo-Qiang Chen, Hongwei Yu and the other, anonymous, reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
EK3D database: http://www.iith.ac.in/EK3D/
References
- 1.Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC. EPS-then and now. Microorganisms. 2016;4:41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid J, Sieber V, Rehm BHA. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi KR, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019 doi: 10.1016/j.tibtech.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Lv L, Ren Y-L, Chen J-C, Wu Q, Chen G-Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P(3HB-co-4HB) biosynthesis. Metab. Eng. 2015;29:160–168. doi: 10.1016/j.ymben.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Rehm BHA. Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010;8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 8.Flemming HC, Wuertz S. Bacteria and archaea on earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 9.Bartell JA, et al. Evolutionary highways to persistent bacterial infection. Nat. Commun. 2019;10:629. doi: 10.1038/s41467-019-08504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao M, et al. A natural in situ fabrication method of functional bacterial cellulose using a microorganism. Nat. Commun. 2019;10:437. doi: 10.1038/s41467-018-07879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Eyckmans J, Chen CS. Designer biomaterials for mechanobiology. Nat. Mater. 2017;16:1164–1168. doi: 10.1038/nmat5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondal S, et al. A minimal length rigid helical peptide motif allows rational design of modular surfactants. Nat. Commun. 2017;8:14018. doi: 10.1038/ncomms14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Romain C, Williams CK. Sustainable polymers from renewable resources. Nature. 2016;540:354–362. doi: 10.1038/nature21001. [DOI] [PubMed] [Google Scholar]
- 14.Boom time for biomaterials. Nat. Mater.8, 439 (2009). [DOI] [PubMed]
- 15.Bruchet M, Melman A. Fabrication of patterned calcium cross-linked alginate hydrogel films and coatings through reductive cation exchange. Carbohydr. Polym. 2015;131:57–64. doi: 10.1016/j.carbpol.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Moradali, M. F., Ghods, S. & Rehm, B. H. A. in Alginates and Their Biomedical Applications (eds Rehm, B. H. A. & Moradali M. F.) (Springer, 2018).
- 17.Pontes MH, Lee EJ, Choi J, Groisman EA. Salmonella promotes virulence by repressing cellulose production. Proc. Natl Acad. Sci. USA. 2015;112:5183–5188. doi: 10.1073/pnas.1500989112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015;23:545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thongsomboon W, et al. Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science. 2018;359:334–338. doi: 10.1126/science.aao4096. [DOI] [PubMed] [Google Scholar]
- 20.Hollenbeck EC, et al. Phosphoethanolamine cellulose enhances curli-mediated adhesion of uropathogenic. Proc. Natl Acad. Sci. USA. 2018;115:10106–10111. doi: 10.1073/pnas.1801564115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh SY, Budzik JM, Garufi G, Schneewind O. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol. Microbiol. 2011;80:455–470. doi: 10.1111/j.1365-2958.2011.07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkening RV, Federle MJ. Evolutionary constraints shaping Streptococcus pyogenes-host interactions. Trends Microbiol. 2017;25:562–572. doi: 10.1016/j.tim.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzeng Y-L, Thomas J, Stephens DS. Regulation of capsule in Neisseria meningitidis. Crit. Rev. Microbiol. 2016;42:759–772. doi: 10.3109/1040841X.2015.1022507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geno KA, et al. Pneumococcal capsules and their types: past, present, and future. Clin. Microbiol. Rev. 2015;28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehm BHA. Bioengineering towards self-assembly of particulate vaccines. Curr. Opin. Biotechnol. 2017;48:42–53. doi: 10.1016/j.copbio.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Liu Z, Dai S, Yan J, Wise MJ. The sit-and-wait hypothesis in bacterial pathogens: a theoretical study of durability and virulence. Front. Microbiol. 2017;8:2167. doi: 10.3389/fmicb.2017.02167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambou T, et al. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Mol. Microbiol. 2008;70:762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Best A, Abu Kwaik Y. Nutrition and bipartite metabolism of intracellular pathogens. Trends Microbiol. 2019;27:550–561. doi: 10.1016/j.tim.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Mooney DJ. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016;1:16071. doi: 10.1038/natrevmats.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao T, Wang J, Zeng Y, Liu G, Chen X. Polysaccharide-based controlled release systems for therapeutics delivery and tissue engineering: from bench to bedside. Adv. Sci. 2018;5:1700513. doi: 10.1002/advs.201700513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan T, et al. Designing hydrophobically modified polysaccharide derivatives for highly efficient enzyme immobilization. Biomacromolecules. 2015;16:2403–2411. doi: 10.1021/acs.biomac.5b00638. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, et al. Hyaluronate and its derivatives for customized biomedical applications. Biomaterials. 2017;123:155–171. doi: 10.1016/j.biomaterials.2017.01.029. [DOI] [PubMed] [Google Scholar]
- 33.Mitrousis N, Fokina A, Shoichet MS. Biomaterials for cell transplantation. Nat. Rev. Mater. 2018;3:441–456. [Google Scholar]
- 34.Kang D-H, Kim D, Wang S, Song D, Yoon M-H. Water-insoluble, nanocrystalline, and hydrogel fibrillar scaffolds for biomedical applications. Polym. J. 2018;50:637–647. [Google Scholar]
- 35.Loh EYX, et al. Development of a bacterial cellulose-based hydrogel cell carrier containing keratinocytes and fibroblasts for full-thickness wound healing. Sci. Rep. 2018;8:2875. doi: 10.1038/s41598-018-21174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee KY, Buldum G, Mantalaris A, Bismarck A. More than meets the eye in bacterial cellulose: biosynthesis, bioprocessing, and applications in advanced fiber composites. Macromol. Biosci. 2014;14:10–32. doi: 10.1002/mabi.201300298. [DOI] [PubMed] [Google Scholar]
- 37.Bottan S, et al. Surface-structured bacterial cellulose with guided assembly-based biolithography (GAB) ACS Nano. 2015;9:206–219. doi: 10.1021/nn5036125. [DOI] [PubMed] [Google Scholar]
- 38.Valera MJ, Torija MJ, Mas A, Mateo E. Cellulose production and cellulose synthase gene detection in acetic acid bacteria. Appl. Microbiol. Biotechnol. 2015;99:1349–1361. doi: 10.1007/s00253-014-6198-1. [DOI] [PubMed] [Google Scholar]
- 39.de Oliveira JD, et al. Genetic basis for hyper production of hyaluronic acid in natural and engineered microorganisms. Microb. Cell Fact. 2016;15:119–119. doi: 10.1186/s12934-016-0517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moradali MF, Donati I, Sims IM, Ghods S, Rehm BHA. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. MBio. 2015;6:e00453–e00515. doi: 10.1128/mBio.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pauly M, Ramírez V. New insights into wall polysaccharide O-acetylation. Front. Plant. Sci. 2018;9:1210. doi: 10.3389/fpls.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sychantha D, et al. PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides. Nat. Chem. Biol. 2018;14:79–85. doi: 10.1038/nchembio.2509. [DOI] [PubMed] [Google Scholar]
- 43.Rehm, B. H. A. & Moradali, M. F. (eds) Alginates and Their Biomedical Applications (Springer, 2018).
- 44.Guarino, V. et al. in Alginates and their biomedical applications (eds Rehm, B. H. A. & M. F. Moradali) (Springer, 2018).
- 45.Jang J, et al. Monoclonal antibody against the poly-gamma-D-glutamic acid capsule of Bacillus anthracis protects mice from enhanced lethal toxin activity due to capsule and anthrax spore challenge. Biochim. Biophys. Acta. 2013;1830:2804–2812. doi: 10.1016/j.bbagen.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Yu Y, et al. Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front. Microbiol. 2016;7:1811. doi: 10.3389/fmicb.2016.01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watzer B, Forchhammer K. Cyanophycin synthesis optimizes nitrogen utilization in the unicellular Cyanobacterium synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2018 doi: 10.1128/AEM.01298-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Yang C. Arginine and nitrogen mobilization in cyanobacteria. Mol. Microbiol. 2019;111:863–867. doi: 10.1111/mmi.14204. [DOI] [PubMed] [Google Scholar]
- 49.Du J, Li L, Zhou S. Microbial production of cyanophycin: from enzymes to biopolymers. Biotechnol. Adv. 2019 doi: 10.1016/j.biotechadv.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Lee SY, et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2019;2:18–33. [Google Scholar]
- 51.Luo Z, et al. Microbial synthesis of poly-γ-glutamic acid: current progress, challenges, and future perspectives. Biotechnol. Biofuels. 2016;9:134. doi: 10.1186/s13068-016-0537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyldgaard M, et al. The antimicrobial mechanism of action of epsilon-poly-l-lysine. Appl. Env. Microbiol. 2014;80:7758. doi: 10.1128/AEM.02204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ushimaru K, Hamano Y, Katano H. Antimicrobial activity of ε-poly-l-lysine after forming a water-insoluble complex with an anionic surfactant. Biomacromolecules. 2017;18:1387–1392. doi: 10.1021/acs.biomac.7b00109. [DOI] [PubMed] [Google Scholar]
- 54.Pham TH, Webb JS, Rehm BHA. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology. 2004;150:3405–3413. doi: 10.1099/mic.0.27357-0. [DOI] [PubMed] [Google Scholar]
- 55.Long JY, et al. Mutagenesis of PhaR, a regulator gene of polyhydroxyalkanoate biosynthesis of Xanthomonas oryzae pv. oryzae caused pleiotropic phenotype changes. Front. Microbiol. 2018;9:3046. doi: 10.3389/fmicb.2018.03046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campisano A, Overhage J, Rehm BHA. The polyhydroxyalkanoate biosynthesis genes are differentially regulated in planktonic- and biofilm-grown Pseudomonas aeruginosa. J. Biotechnol. 2008;133:442–452. doi: 10.1016/j.jbiotec.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Coats ER, Watson BS, Brinkman CK. Polyhydroxyalkanoate synthesis by mixed microbial consortia cultured on fermented dairy manure: effect of aeration on process rates/yields and the associated microbial ecology. Water Res. 2016;106:26–40. doi: 10.1016/j.watres.2016.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Draper, J. et al. in Bionanotechnology: Biological Self-assembly and its Applications (ed. Rehm, B. H. A.) (Caister Academic Press, 2013).
- 59.Kai D, Loh XJ. Polyhydroxyalkanoates: chemical modifications toward biomedical applications. ACS Sustain. Chem. Eng. 2014;2:106–119. [Google Scholar]
- 60.Parlane NA, et al. Self-assembled protein-coated polyhydroxyalkanoate beads: properties and biomedical applications. ACS Biomater. Sci. Eng. 2017;3:3043–3057. doi: 10.1021/acsbiomaterials.6b00355. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Miro M, et al. Polyester as antigen carrier toward particulate vaccines. Biomacromolecules. 2019;20:3211–3212. doi: 10.1021/acs.biomac.9b00509. [DOI] [PubMed] [Google Scholar]
- 62.Ogura K, Rehm BHA. Alginate encapsulation of bioengineered protein-coated polyhydroxybutyrate particles: a new platform for multifunctional composite materials. Adv. Funct. Mater. 2019;29:1901893. [Google Scholar]
- 63.Kulaev, I. S., Vagabov, V. M. & Kulakovskaya, T. V. in The Biochemistry of Inorganic Polyphosphates Wiley Online Books (eds Kulaev, I. S., Vagabov, V. M. & Kulakovskaya T. V.) (John Wiley & Sons, Ltd., 2004).
- 64.Moreno SN, Docampo R. Polyphosphate and its diverse functions in host cells and pathogens. PLoS Pathog. 2013;9:e1003230. doi: 10.1371/journal.ppat.1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoo NG, et al. Polyphosphate stabilizes protein unfolding intermediates as soluble amyloid-like oligomers. J. Mol. Biol. 2018;430:4195–4208. doi: 10.1016/j.jmb.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rao NN, Gómez-García MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, et al. Distribution patterns of polyphosphate metabolism pathway and its relationships with bacterial durability and virulence. Front. Microbiol. 2018;9:782. doi: 10.3389/fmicb.2018.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varas MA, et al. Inorganic polyphosphate is essential for Salmonella Typhimurium virulence and survival in Dictyostelium discoideum. Front. Cell Infect. Microbiol. 2018;8:8. doi: 10.3389/fcimb.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, et al. Polyphosphate as a bioactive and biodegradable implant material: induction of bone regeneration in rats. Adv. Eng. Mater. 2016;18:1406–1417. [Google Scholar]
- 70.Müller WEG, et al. Fabrication of amorphous strontium polyphosphate microparticles that induce mineralization of bone cells in vitro and in vivo. Acta Biomater. 2017;50:89–101. doi: 10.1016/j.actbio.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 71.Wang X, Schröder HC, Müller WEG. Amorphous polyphosphate, a smart bioinspired nano-/bio-material for bone and cartilage regeneration: towards a new paradigm in tissue engineering. J. Mater. Chem. B. 2018;6:2385–2412. doi: 10.1039/c8tb00241j. [DOI] [PubMed] [Google Scholar]
- 72.Ackermann M, et al. Collagen-inducing biologization of prosthetic material for hernia repair: polypropylene meshes coated with polyP/collagen. J. Biomed. Mater. Res. B Appl. Biomater. 2018;106:2109–2121. doi: 10.1002/jbm.b.34016. [DOI] [PubMed] [Google Scholar]
- 73.Müller WEG, et al. Transformation of amorphous polyphosphate nanoparticles into coacervate complexes: an approach for the encapsulation of mesenchymal stem cells. Small. 2018;14:1801170. doi: 10.1002/smll.201801170. [DOI] [PubMed] [Google Scholar]
- 74.Lindner SN, Knebel S, Pallerla SR, Schoberth SM, Wendisch VF. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2010;87:703–713. doi: 10.1007/s00253-010-2568-5. [DOI] [PubMed] [Google Scholar]
- 75.Jennings LK, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl Acad. Sci. USA. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imada K. Bacterial flagellar axial structure and its construction. Biophys. Rev. 2018;10:559–570. doi: 10.1007/s12551-017-0378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao B, Xu H, Mao C. Controlled self-assembly of rodlike bacterial pili particles into ordered lattices. Angew. Chem. Int. Ed. 2011;50:6264–6268. doi: 10.1002/anie.201102052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bera S, et al. Rigid helical-like assemblies from a self-aggregating tripeptide. Nat. Mater. 2019;18:503–509. doi: 10.1038/s41563-019-0343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen PQ, Courchesne ND, Duraj-Thatte A, Praveschotinunt P, Joshi NS. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018;30:e1704847. doi: 10.1002/adma.201704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilbert C, Ellis T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 2019;8:1–15. doi: 10.1021/acssynbio.8b00423. [DOI] [PubMed] [Google Scholar]
- 81.Nguyen PQ. Synthetic biology engineering of biofilms as nanomaterials factories. Biochem. Soc. Trans. 2017;45:585–597. doi: 10.1042/BST20160348. [DOI] [PubMed] [Google Scholar]
- 82.Nguyen PQ, Botyanszki Z, Tay PK, Joshi NS. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014;5:4945. doi: 10.1038/ncomms5945. [DOI] [PubMed] [Google Scholar]
- 83.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rehm BHA. Synthetic biology towards the synthesis of custom-made polysaccharides. Microb. Biotechnol. 2015;8:19–20. doi: 10.1111/1751-7915.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BHA. Genetics and regulation of bacterial alginate production. Env. Microbiol. 2014;16:2997–3011. doi: 10.1111/1462-2920.12389. [DOI] [PubMed] [Google Scholar]
- 86.Mathee K, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 87.Martínez LC, Vadyvaloo V. Mechanisms of post-transcriptional gene regulation in bacterial biofilms. Front. Cell Infect. Microbiol. 2014;4:38–38. doi: 10.3389/fcimb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsouko E, et al. Bacterial cellulose production from industrial waste and by-product streams. Int. J. Mol. Sci. 2015;16:14832–14849. doi: 10.3390/ijms160714832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duan XJ, Yang L, Zhang X, Tan WS. Effect of oxygen and shear stress on molecular weight of hyaluronic acid. J. Microbiol. Biotechnol. 2008;18:718–724. [PubMed] [Google Scholar]
- 90.Zeng C, Toole BP, Kinney SD, Kuo J-w, Stamenkovic I. Inhibition of tumor growth in vivo by hyaluronan oligomers. Int. J. Cancer. 1998;77:396–401. doi: 10.1002/(sici)1097-0215(19980729)77:3<396::aid-ijc15>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 91.Stern R, Asari AA, Sugahara KN. Hyaluronan fragments: an information-rich system. Eur. J. Cell Biol. 2006;85:699–715. doi: 10.1016/j.ejcb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Park D, et al. Hyaluronic acid promotes angiogenesis by inducing RHAMM-TGFβ receptor interaction via CD44-PKCδ. Mol. Cell. 2012;33:563–574. doi: 10.1007/s10059-012-2294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sze JH, Brownlie JC, Love CA. Biotechnological production of hyaluronic acid: a mini review. 3 Biotech. 2016;6:67–67. doi: 10.1007/s13205-016-0379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Safrankova B, Gajdova S, Kubala L. The potency of hyaluronan of different molecular weights in the stimulation of blood phagocytes. Mediators Inflamm. 2010;2010:380948. doi: 10.1155/2010/380948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crupi, R. & Cuzzocrea, S. in Alginates and Their Biomedical Applications (eds Rehm B. H. A. & Moradali M. F.) (Springer, 2018).
- 96.Rye, P. D. et al. in Alginates and Their Biomedical Applications (eds Rehm, B. H. A. & Moradali M. F.) (Springer, 2018).
- 97.Ghods S, Sims IM, Moradali MF, Rehm BHA. Bactericidal compounds controlling growth of the plant pathogen Pseudomonas syringae pv. actinidiae, which forms biofilms composed of a novel exopolysaccharide. Appl. Env. Microbiol. 2015;81:4026–4036. doi: 10.1128/AEM.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruth K, et al. Efficient production of (R)-3-hydroxycarboxylic acids by biotechnological conversion of polyhydroxyalkanoates and their purification. Biomacromolecules. 2007;8:279–286. doi: 10.1021/bm060585a. [DOI] [PubMed] [Google Scholar]
- 99.May JF, Splain RA, Brotschi C, Kiessling LL. A tethering mechanism for length control in a processive carbohydrate polymerization. Proc. Natl Acad. Sci. USA. 2009;106:11851–11856. doi: 10.1073/pnas.0901407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BHA. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl. Env. Microbiol. 2013;79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moradali MF, Ghods S, Rehm BHA. Activation mechanism and cellular localization of membrane-anchored alginate polymerase in Pseudomonas aeruginosa. Appl. Env. Microbiol. 2017;83:e03499–e03516. doi: 10.1128/AEM.03499-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murray GL, Attridge SR, Morona R. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 2003;47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 103.Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 104.Chen WY, Marcellin E, Hung J, Nielsen LK. Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J. Biol. Chem. 2009;284:18007–18014. doi: 10.1074/jbc.M109.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sim SJ, et al. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat. Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- 106.Stollerman GH, Dale JB. The importance of the group a Streptococcus capsule in the pathogenesis of human infections: a historical perspective. Clin. Infect. Dis. 2008;46:1038–1045. doi: 10.1086/529194. [DOI] [PubMed] [Google Scholar]
- 107.Tonello F, Zornetta I. Toxins. 2012. Bacillus anthracis factors for phagosomal escape; pp. 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Simpson BW, Trent MS. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019;17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Balaban NQ, et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019;17:479–496. doi: 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 111.Wang TZ, Kodiyanplakkal RPL, Calfee DP. Antimicrobial resistance in nephrology. Nat. Rev. Nephrol. 2019;15:463–481. doi: 10.1038/s41581-019-0150-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou E, et al. Thiol-benzo-triazolo-quinazolinone inhibits Alg44 binding to c-di-GMP and reduces alginate production by Pseudomonas aeruginosa. ACS Chem. Biol. 2017;12:3076–3085. doi: 10.1021/acschembio.7b00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hengzhuang W, et al. OligoG CF-5/20 disruption of mucoid Pseudomonas aeruginosa biofilm in a murine lung infection model. Antimicrob. Agents Chemother. 2016;60:2620. doi: 10.1128/AAC.01721-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Powell LC, et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms. Microbi. 2018;4:13. doi: 10.1038/s41522-018-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang Y, Moradali MF, Goudarztalejerdi A, Sims IM, Rehm BHA. Biological function of a polysaccharide degrading enzyme in the periplasm. Sci. Rep. 2016;6:31249. doi: 10.1038/srep31249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Daboor SM, Raudonis R, Cohen A, Rohde JR, Cheng Z. Marine bacteria, a source for alginolytic enzyme to disrupt. Mar. Drugs. 2019;17:307. doi: 10.3390/md17050307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Watters CM, Burton T, Kirui DK, Millenbaugh NJ. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect. Drug. Resist. 2016;9:71–78. doi: 10.2147/IDR.S103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gopu V, Meena CK, Shetty PH. Quercetin influences quorum sensing in food borne bacteria: in-vitro and in-silico evidence. PLoS One. 2015;10:e0134684. doi: 10.1371/journal.pone.0134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kali A, Bhuvaneshwar D, Charles PMV, Seetha KS. Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates. J. Basic. Clin. Pharm. 2016;7:93–96. doi: 10.4103/0976-0105.183265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Quecan BXV, et al. Effect of quercetin rich onion extracts on bacterial quorum sensing. Front. Microbiol. 2019;10:867. doi: 10.3389/fmicb.2019.00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kar S, Ellington AD. Construction of synthetic T7 RNA polymerase expression systems. Methods. 2018;143:110–120. doi: 10.1016/j.ymeth.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 122.Hussey BJ, McMillen DR. Programmable T7-based synthetic transcription factors. Nucleic Acids Res. 2018;46:9842–9854. doi: 10.1093/nar/gky785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Horga LG, et al. Tuning recombinant protein expression to match secretion capacity. Microb. Cell Fact. 2018;17:199. doi: 10.1186/s12934-018-1047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nora LC, et al. The art of vector engineering: towards the construction of next-generation genetic tools. Microb. Biotechnol. 2019;12:125–147. doi: 10.1111/1751-7915.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen G-Q, Jiang X-R. Engineering microorganisms for improving polyhydroxyalkanoate biosynthesis. Curr. Opin. Biotechnol. 2018;53:20–25. doi: 10.1016/j.copbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 126.Florea M, et al. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl Acad. Sci. USA. 2016;113:E3431–E3440. doi: 10.1073/pnas.1522985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Taton A, et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014;42:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gallo N, et al. Hyaluronic acid for advanced therapies: promises and challenges. Eur. Polym. J. 2019;117:134–147. [Google Scholar]
- 129.Smanski MJ, et al. Synthetic biology to access and expand nature's chemical diversity. Nat. Rev. Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Majerle A, Schmieden DT, Jerala R, Meyer AS. Synthetic biology for multiscale designed biomimetic assemblies: from designed self-assembling biopolymers to bacterial bioprinting. Biochemistry. 2019;58:2095–2104. doi: 10.1021/acs.biochem.8b00922. [DOI] [PubMed] [Google Scholar]
- 131.Jin X, Riedel-Kruse IH. Biofilm Lithography enables high-resolution cell patterning via optogenetic adhesin expression. Proc. Natl Acad. Sci. USA. 2018;115:3698. doi: 10.1073/pnas.1720676115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pu L, Yang S, Xia A, Jin F. Optogenetics manipulation enables prevention of biofilm formation of engineered Pseudomonas aeruginosa on surfaces. ACS Synth. Biol. 2018;7:200–208. doi: 10.1021/acssynbio.7b00273. [DOI] [PubMed] [Google Scholar]
- 133.Dandekar T, Fieselmann A, Majeed S, Ahmed Z. Software applications toward quantitative metabolic flux analysis and modeling. Brief Bioinform. 2012;15:91–107. doi: 10.1093/bib/bbs065. [DOI] [PubMed] [Google Scholar]
- 134.Cai D, et al. Enhanced production of poly-γ-glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis. Biotechnol. Bioeng. 2018;115:2541–2553. doi: 10.1002/bit.26774. [DOI] [PubMed] [Google Scholar]
- 135.Dumon C, et al. In vivo fucosylation of lacto-N-neotetraose and lacto-N-neohexaose by heterologous expression of Helicobacter pylori alpha-1,3 fucosyltransferase in engineered Escherichia coli. Glycoconj. J. 2001;18:465–474. doi: 10.1023/a:1016086118274. [DOI] [PubMed] [Google Scholar]
- 136.Linton JD. Metabolite production and growth efficiency. Antonie Van Leeuwenhoek. 1991;60:293–311. doi: 10.1007/BF00430371. [DOI] [PubMed] [Google Scholar]
- 137.Ates O. Systems biology of microbial exopolysaccharides production. Front. Bioeng. Biotechnol. 2015;3:200–200. doi: 10.3389/fbioe.2015.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cardoso JGR, et al. Cameo: a Python library for computer aided metabolic engineering and optimization of cell factories. ACS Synth. Biol. 2018;7:1163–1166. doi: 10.1021/acssynbio.7b00423. [DOI] [PubMed] [Google Scholar]
- 139.Heirendt L, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lloyd CJ, et al. COBRAme: a computational framework for genome-scale models of metabolism and gene expression. PLoS Comput. Biol. 2018;14:e1006302. doi: 10.1371/journal.pcbi.1006302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jung YK, Kim TY, Park SJ, Lee SY. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol. Bioeng. 2010;105:161–171. doi: 10.1002/bit.22548. [DOI] [PubMed] [Google Scholar]
- 142.Choi SY, et al. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat. Biotechnol. 2016;34:435. doi: 10.1038/nbt.3485. [DOI] [PubMed] [Google Scholar]
- 143.Diken E, et al. Genomic analysis reveals the biotechnological and industrial potential of levan producing halophilic extremophile, Halomonas smyrnensis AAD6T. Springerplus. 2015;4:393–393. doi: 10.1186/s40064-015-1184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li Z, Yang J, Loh XJ. Polyhydroxyalkanoates: opening doors for a sustainable future. NPG Asia Mater. 2016;8:e265. [Google Scholar]
- 145.Chen GQ, Jiang XR. Engineering bacteria for enhanced polyhydroxyalkanoates (PHA) biosynthesis. Synth. Syst. Biotechnol. 2017;2:192–197. doi: 10.1016/j.synbio.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meng DC, Chen GQ. Synthetic biology of polyhydroxyalkanoates (PHA) Adv. Biochem. Eng. Biotechnol. 2018;162:147–174. doi: 10.1007/10_2017_3. [DOI] [PubMed] [Google Scholar]
- 147.Yang JE, et al. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 2018;9:79. doi: 10.1038/s41467-017-02498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cobo I, Li M, Sumerlin BS, Perrier S. Smart hybrid materials by conjugation of responsive polymers to biomacromolecules. Nat. Mater. 2014;14:143. doi: 10.1038/nmat4106. [DOI] [PubMed] [Google Scholar]
- 149.Schreiber CL, Smith BD. Molecular conjugation using non-covalent click chemistry. Nat. Rev. Chem. 2019;3:393–400. doi: 10.1038/s41570-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mamat U, et al. Endotoxin-free protein production — ClearColi™ technology. Nat. Methods. 2013;10:916. [Google Scholar]
- 151.Schwarz H, et al. Biological activity of masked endotoxin. Sci. Rep. 2017;7:44750. doi: 10.1038/srep44750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Valentine ME, et al. Generation of a highly attenuated strain of Pseudomonas aeruginosa for commercial production of alginate. Microb. Biotechnol. 2019 doi: 10.1111/1751-7915.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Choi S, et al. Purification and biocompatibility of fermented hyaluronic acid for its applications to biomaterials. Biomater. Res. 2014;18:6. doi: 10.1186/2055-7124-18-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ko SH, et al. Nanofluidic device for continuous multiparameter quality assurance of biologics. Nat. Nanotechnol. 2017;12:804. doi: 10.1038/nnano.2017.74. [DOI] [PubMed] [Google Scholar]
- 155.Wong BG, Mancuso CP, Kiriakov S, Bashor CJ, Khalil AS. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018;36:614. doi: 10.1038/nbt.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Basu A, Kunduru KR, Abtew E, Domb AJ. Polysaccharide-based conjugates for biomedical applications. Bioconjug. Chem. 2015;26:1396–1412. doi: 10.1021/acs.bioconjchem.5b00242. [DOI] [PubMed] [Google Scholar]
- 157.Güngör G, et al. Bacterial hyaluronic acid production through an alternative extraction method and its characterization. J. Chem. Technol. Biotechnol. 2019;94:1843–1852. [Google Scholar]
- 158.Favaro L, Basaglia M, Casella S. Improving polyhydroxyalkanoate production from inexpensive carbon sources by genetic approaches: a review. Biofuels Bioprod. Biorefin. 2019;13:208–227. [Google Scholar]
- 159.Wang X, et al. Highly effective polyphosphate synthesis, phosphate removal, and concentration using engineered environmental bacteria based on a simple solo medium-copy plasmid strategy. Environ. Sci. Technol. 2018;52:214–222. doi: 10.1021/acs.est.7b04532. [DOI] [PubMed] [Google Scholar]
- 160.Smith WD, et al. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol. Lett. 2017;364:fnx121. doi: 10.1093/femsle/fnx121. [DOI] [PubMed] [Google Scholar]
- 161.Islan GA, Bosio VE, Castro GR. Alginate lyase and ciprofloxacin co-immobilization on biopolymeric microspheres for cystic fibrosis treatment. Macromol. Biosci. 2013;13:1238–1248. doi: 10.1002/mabi.201300134. [DOI] [PubMed] [Google Scholar]
- 162.Baker P, et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2016;2:e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kong L, et al. Single-molecule interrogation of a bacterial sugar transporter allows the discovery of an extracellular inhibitor. Nat. Chem. 2013;5:651–659. doi: 10.1038/nchem.1695. [DOI] [PubMed] [Google Scholar]
- 164.Kong L, et al. An antibacterial vaccination strategy based on a glycoconjugate containing the core lipopolysaccharide tetrasaccharide Hep2Kdo2. Nat. Chem. 2016;8:242. doi: 10.1038/nchem.2432. [DOI] [PubMed] [Google Scholar]
- 165.Middleton DR, et al. Identification and characterization of the Streptococcus pneumoniae type 3 capsule-specific glycoside hydrolase of Paenibacillus species 32352. Glycobiology. 2018;28:90–99. doi: 10.1093/glycob/cwx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Micoli F, Costantino P, Adamo R. Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 2018;42:388–423. doi: 10.1093/femsre/fuy011. [DOI] [PMC free article] [PubMed] [Google Scholar]