Abstract
Coronavirus disease 2019 (COVID-19) is a global health concern as it continues to spread within China and beyond. The causative agent of this disease, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), belongs to the genus Betacoronavirus, which also includes severe acute respiratory syndrome-related coronavirus (SARSr-CoV) and Middle East respiratory syndrome-related coronavirus (MERSr-CoV). Codon usage of viral genes are believed to be subjected to different selection pressures in different host environments. Previous studies on codon usage of influenza A viruses helped identify viral host origins and evolution trends, however, similar studies on coronaviruses are lacking. In this study, we compared the codon usage bias using global correspondence analysis (CA), within-group CA and between-group CA. We found that the bat RaTG13 virus best matched the overall codon usage pattern of SARS-CoV-2 in orf1ab, spike and nucleocapsid genes, while the pangolin P1E virus had a more similar codon usage in membrane gene. The amino acid usage pattern of SARS-CoV-2 was generally found similar to bat and human SARSr-CoVs. However, we found greater synonymous codon usage differences between SARS-CoV-2 and its phylogenetic relatives on spike and membrane genes, suggesting these two genes of SARS-CoV-2 are subjected to different evolutionary pressures.
Keywords: SARS-CoV-2, coronavirus, codon usage analysis, WCA
1. Introduction
A novel coronavirus outbreak took place in Wuhan, Hubei province, China in December 2019 (Wang et al. 2020). This novel coronavirus (SARS-CoV-2) causes pneumonia in patients (Zhu et al. 2020) and it has rapidly spread to other provinces in China and other countries (WHO 2020). This novel coronavirus outbreak had raised global concern but current knowledge on the origin and transmission route of the pathogen is still limited. The SARS-CoV-2 belongs to the genus Betacoronavirus, which also includes two highly virulent human coronaviruses, SARS-CoV and MERS-CoV. Apart from human, many animal species, such as bat, rat, camel, swine, and hedgehog, can be infected by different types of coronaviruses. Further sequence analyses of this novel and other betacoronaviruses might provide additional information to better understand the evolution of SARS-CoV-2.
Preferential codon usage is commonly seen in different organisms, and it has been evident that the uneven codon usage is not neutral but related to gene expression or other selection pressures (Akashi and Eyre-Walker 1998; Percudani and Ottonello 1999; Pepin, Domsic, and McKenna 2008). There are two levels of codon usage biases, one is at amino acid level and the other is at synonymous codon level. The first one mainly originates from preferential usage of certain amino acids, for example integral membrane proteins maybe enriched in hydrophobic amino acids and respective codons (Perriere 2002). The synonymous codon usage bias in viruses can be driven by selective pressures from the host cells (Wong et al. 2010; Fan et al. 2015;Gu et al. 2019), and studies on different viruses have shown that selection pressures can be dominant in the evolution of codon usage patterns of the virus (Liu et al. 2011; Fan et al. 2015; Kumar et al. 2016; Wang et al. 2016). Correspondence analysis (CA) is a tool for visualization high dimension discrete-state data, which has been applied in many codon usage studies (Perriere 2002; Charif et al. 2005; Wong et al. 2010; Lobry 2018;). The recently developed within-group CA (WCA) and between-group CA (BCA) are derivatives to the conventional CA (Lobry 2018), and the application of block structure in these methods allows separating the codon usage bias at amino acid and synonymous codon levels. As the natural history of the SARS-CoV-2 remains largely unknown, an in-depth codon usage analysis of this newly emerging virus might provide some novel insights.
In this study, we used both CA, BCA, and WCA to analyses codon usage patterns of 3,076 betacoronavirus sequences. We found SARS-CoV-2 and bat SARSr-CoV have similar amino acid usage. However, our analyses suggested that the spike and membrane genes of SARS-CoV-2 have relatively distinct synonymous codon usage patterns to the orf1ab or nucleocapsid genes.
2. Methods
2.1 Sequence data
To construct a reference sequence dataset, available full-length complete genome sequences of coronavirus were collected through Virus Pathogen Resource database (https://www.viprbrc.org/brc/home.spg?decorator=corona, accessed 13 July 2019, ticket 958868915368). The sequences were filtered by the following steps: 1, remove sequences without protein annotation; 2, keep only sequences with complete set of desired replicase and structural proteins (sequences coding for orf1ab, spike, membrane, and nucleocapsid); 3, filter out sequences that are unusually long and short (>130% or <70% of the median length for each group of gene sequences); 4, limit our analysis to genus Betacoronavirus; and 5, concatenate orf1a and orf1b sequences to form orf1ab if necessary.
The final dataset comprised 769 individual strains (3,076 individual gene sequences) that contain complete sets of coding regions for orf1ab, spike, membrane, and nucleocapsid genes. The sequences for envelope gene were not included in the analysis because of the short length and potential bias in codon usage. Corresponding metadata for the sequences were extracted by the sequence name field. Twenty-four complete genome sequences of the newly identified SARS-CoV-2 and its phylogenetically close relatives were retrieved from Genbank and GISAID (accessed 22 Jan 2020). Six genomes in this study were used as references (BetaCoV/bat/Yunnan/RaTG13/2013|EPI_ISL_402131; BetaCoV/pangolin/Guangxi/P1E/2017|EPI_ISL_410539; MG772934.1_Bat_SARS-like_coronavirus_isolate_bat-SL-CoVZXC21; MG772933.1_Bat_SARS-like_coronavirus_isolate_bat-SL-CoVZC45; KY352407.1_Severe_acute_respiratory_syndrome-related_coronavirus_strain_BtKY72 and GU190215.1_Bat_coronavirus_BM48-31/BGR/2008), as they have previously been reported to have close phylogenetic relationship with SARS-CoV-2 (Lam et al. 2020; Lu et al. 2020; Zhou et al. 2020). Detailed accession ID for the above data are provided in Supplementary Table S1.
The codon count for every gene sequence input for the CA was calculated by the SynMut (Gu and Poon 2019) package. The implementation of the different correspondence analyses in this study was performed by functions in the package ade4 (Dray and Dufour 2007). Three stop codons (TAA, TAG, and TGA) were excluded in the CA.
2.2 Global CA on codon usage
Correspondence analysis (CA) is a dimension reduction method applied to a contingency table that is well suited for amino acid and codon usage analysis. The concept in CA is similar to Pearson’s test (i.e., the expected counts are calculated under the hypothesis of independence, based on the observed contingency table). With the deduced expected count table, the distance (Supplementary Method) can be used to evaluate the difference between two observations. The total inertia calculated from the distance is proportional to the statistic used in Pearson’s test (Lobry 2018). The Pearson residuals were applied in the CA as input for singular value decomposition (Suzuki et al. 2008), and resulted eigenvalues were visualized and interpreted in the study. K-means clustering (k = 7) was performed based on the results from CA analysis to provide quantitative measurement on the proximity between data points.
All the correspondence analyses in this study were performed individually for each gene, to achieve better resolution on gene specific codon usage pattern.
2.3 Within-group CA and between-group CA
In contrast to the previous global CA, the within-block CA (Benzécri 1983) (WCA) can segregate the effects of different codon compositions in different amino acids, by introducing a block structure into the analysis. WCA becomes ‘model of choice’ for analysing synonymous codon usage in recent years, as it is more robust than other traditional methods (e.g. CA with relative codon frequency or CA with relative synonymous codon usage values) (Perriere 2002; Suzuki et al. 2008). WCA focuses on the within-amino acid variability, and it technically excludes the variation of amino acid usage differences. The implementation of WCA was based on the existing global CA, with additional information for factoring (details included in Supplementary Method).
Between-group CA (BCA) is complementary to WCA and it focuses on the between-group variability. BCA can be interpreted as the CA on amino acid usage. We used BCA in this study to investigate the amino acid usage pattern in different coronaviruses.
2.4 Grand average of hydropathy score
The grand average of hydropathy (GRAVY) score provides an easy way to estimate the hydropathy character of a protein (Kyte and Doolittle 1982). It was used in this study as a proxy to identify proteins that are likely to be membrane-bound proteins. The GRAVY score was calculated in a linear form on codon frequencies as:
where is the coefficient for a particular amino acid (provided by data EXP in Seqinr package (Charif and Lobry 2007)) encoded by codon , correspond to the relative frequency of codon .
3. Results
3.1 General sequence features in Betacoronavirus
A total of 3,076 individual gene sequences passed the filtering criteria and were included in this study. Viral sequences from three different species (Middle East respiratory syndrome-related coronavirus (MERSr-CoV), Betacoronavirus 1, SARS-related coronavirus (SARSr-Cov)) were the three most dominant species (see Supplementary Fig. S1) in the filtered dataset.
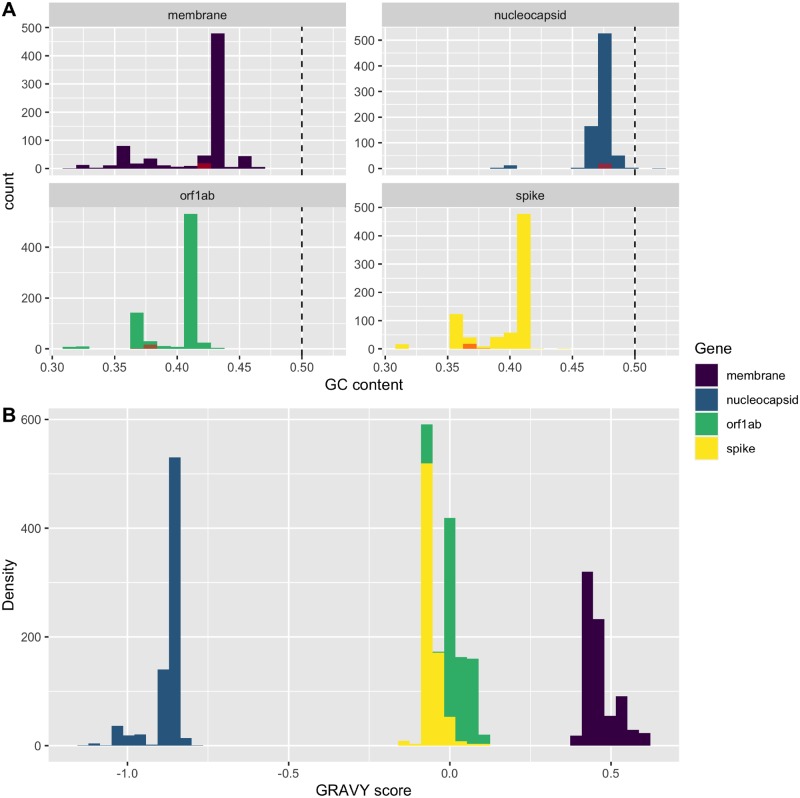
Four conserved protein sequence encoding regions of Betacoronavirus were analysed separately. The median lengths of the studied sequence regions were 21,237 nt for orf1ab gene, 4,062 nt for spike gene, 660 nt for membrane gene, and 1,242 nt for nucleocapsid gene. Spike gene has the lowest average and median G + C contents among these four genes (median: 37.45%, 37.31%, 42.60%, and 47.22% for orf1ab, spike, membrane, and nucleocapsid, respectively). The G+C contents of the orf1ab and spike genes were found distributed in bi-modal patterns, and the G + C contents of SARS-CoV-2 were found located at the lesser half of the data of these two genes. The G + C contents for membrane and nucleocapsid genes of studied viral sequences were distributed in unimodal pattern (Fig. 1A).
Figure 1.
Histogram of (A) G + C content and (B) GRAVY score by different genes in Betacoronavirus. The G + C content values of SARS-CoV-2 were plotted separately in red. The dashed line showed the G + C content of 50 per cent.
The overall amino acid and codon usage of the dataset are plotted in an ascending order (Fig. 2). We observed that leucine and valine were the two most frequently used amino acids in the four studied genes, while tryptophan, histidine, and methionine were the three least used ones. We also found that codons ending with cytosine or guanine were generally less frequent than the codons ending with adenine or thymine. This pattern of uneven usage in synonymous codons is in accordance with the G + C content distribution results (codons ending with guanine or cytosine were less frequently observed).
Figure 2.
Codon usage in Betacoronavirus (Cleveland’s dot plot). Points in green showed the count of codons in a sample SARS-CoV-2 genome (MN908947).
We found a substantial bias in amino acid usage among these four genes, and this bias is well explained by the hydropathy of the encoded proteins (collectively results from global CA on all the four genes, data not shown). We discovered that the nucleocapsid protein sequences had significantly lower GRAVY scores when compared with those from other genes, while the membrane protein sequences had highest GRAVY scores (Fig. 1B).
3.2 The overall codon usage of SARS-CoV-2 in orf1ab, spike, and membrane genes are similar to those of bat and pangolin CoVs
Of all the four global correspondence analyses for the four genes, the extracted first factors explained more than 50 per cent of the total variance (see Supplementary Fig. S2). The first two factors in orf1ab global CA represented 67.7 and 16.8 per cent of total inertia. Similarly, the first two factors of the spike, membrane, and nucleocapsid global CA represented 51.0 and 18.5 per cent, 52.6 and 20.2 per cent, and 54.8 and 14.2 per cent, respectively, of total inertia. With only these two factors, we could extract ∼70 per cent of the variability of the overall codon usage for each studied gene. These levels of representations were higher than or similar to those deduced from other codon usage analyses (Zhou et al. 2005; Suzuki et al. 2008;Lobry 2018).
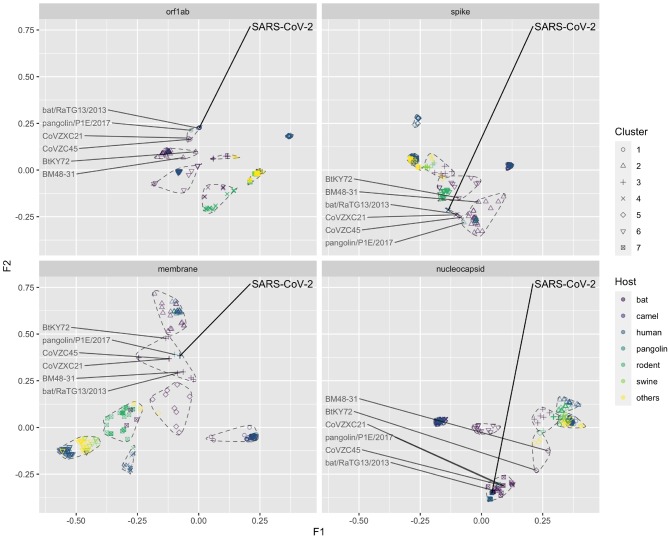
The data points in global CA analysis are shown in different colours that represent different features of the sequences (e.g. viral host or viral species). There were no neighbouring human viruses in the same k-means cluster around SARS-CoV-2 in CA results of orf1ab, spike, and membrane (Fig. 3; Supplementary Fig. 3A), suggesting that the overall codon usage of SARS-CoV-2 in the orf1ab, spike, or membrane gene was significantly different from those of human betacoronaviruses. In contrast, the nucleocapsid genes of SARS coronavirus and SARS-CoV-2 are found to be relatively similar (Supplementary Fig. S3A). Except for the nucleocapsid gene, virus sequences adjacent to the SARS-CoV-2 were all from bat coronaviruses (coloured in purple in Fig. 3). The five groups of viral sequences (SARS-CoV-2, Betacoronavirus 1, human coronavirus HKU 1, MERS-CoV, and SARS-CoV) were well separated from each other in three genes, except that in nucleocapsid, SARS-CoV-2, and SARS-CoV have similar overall codon usage. We also found that SARS-CoV codon usage processed more similarity to SARS-CoV-2 compared with the other three types of human coronaviruses (i.e. yellow point always closest to SARS-CoV-2 in Supplementary Fig. S3A).
Figure 3.
Factorial map of the first and second factors for global CA by different genes, coloured by different viral host. The SARS-CoV-2 and related reference data points were labelled. The seven clusters identified by k-means clustering were circled by dashed lines.
Compared with human coronavirus sequences, the bat coronavirus sequences have more scattered codon usage, even within the same viral species (Supplementary Fig. S3B). Some viral species in bats formed their own clusters in all four genes (e.g. SARSr-CoV). SARSr-CoV is a group of coronavirus that can be found in both humans and bats. The codon usage of SARS-CoV-2 in orf1ab, spike, and membrane were slightly different from the SARS-CoV clusters and these data points are located in between SARSr-CoV and other coronavirus species (e.g. MERSr-CoV and bat coronavirus HKU9, etc.)
The global codon usages of bat RatG13 virus were found most similar to SARS-CoV-2 in orf1ab, spike, and nucleocapsid genes, but not in membrane gene (Fig. 3). In membrane protein, pangolin P1E virus had a more similar codon usage to SARS-CoV-2 than all the other viruses. We found the similarity in codon usage between pangolin P1E and SARS-CoV-2 were also high in orf1ab, where P1E was the second closest data point to SARS-CoV-2. But this is not the case for spike and nucleocapsid genes.
We also observed that the codon usage pattern in spike gene was more complex than in other genes. For example, we found that the P1E virus was in different k-means cluster with SARS-CoV-2, which was not observed in the other three genes. Moreover, data points adjacent to the spike gene of SARS-CoV-2 were coronaviruses not only from bat and human but also from rodent hosts, which is unseen in the other three genes (Fig. 3; Supplementary Fig. S4B). Although the CA results suggested relatively novel codon usage pattern, it captured a large proportion of the variance between hosts or virus species. This was supported by significant differences between the CA distances in both host (ANOSIM statistic R: 0.085, P = 0.001) and virus species groups (ANOSIM statistic R: 0.506, P = 0.001).
The codon usage from camel, swine, and other coronaviruses were found to be well clustered and relatively distant to SARS-CoV-2 (see Supplementary Fig. S4A, C, and D).
3.3 The codon usage at synonymous level suggested novel patterns of SARS-CoV-2 in spike and membrane genes
WCA and BCA were used to further differentiate codon usage of these betacoronaviruses at synonymous codon usage and amino acid usage levels, respectively. We found that most of the variability in codon usage can be explained at synonymous codon usage level (90.36% for orf1ab gene, 85.29% for spike gene, 83.71% for member gene, and 84.07% for nucleocapsid gene) (Table 1).
Table 1.
Variability explained by the synonymous codon usage level and the amino acid level.
| Orf1ab (%) | Spike (%) | Membrane (%) | Nucleocapsid (%) | |
|---|---|---|---|---|
| WCA (synonymous codon level) | 90.36 | 85.29 | 83.71 | 84.07 |
| BCA (amino acid level) | 9.64 | 14.71 | 16.29 | 15.93 |
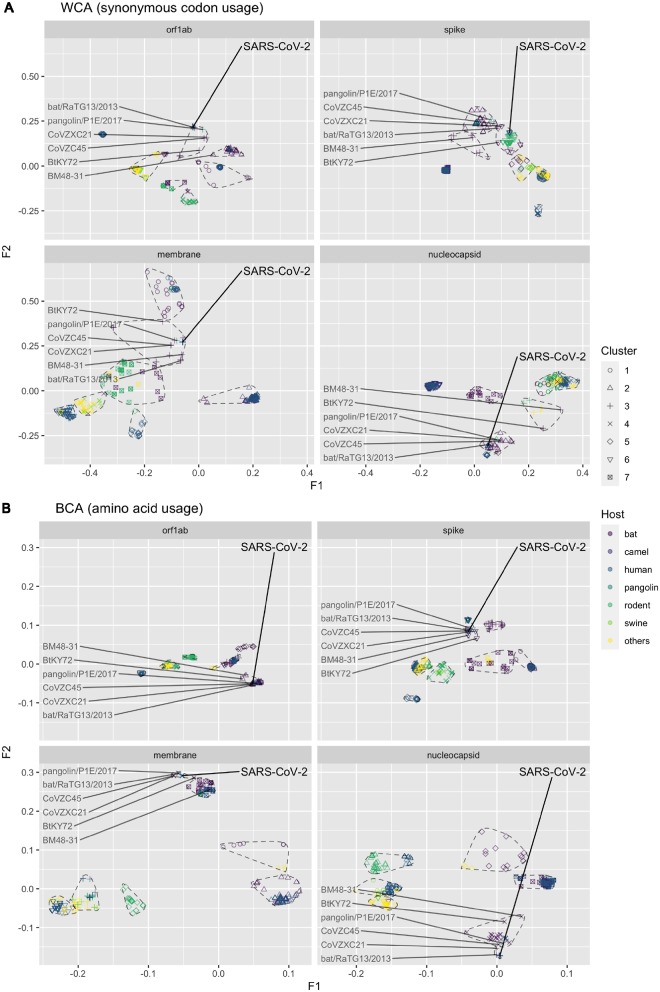
Results from the BCA suggested that the amino acid usage of SARS-CoV-2 is closely related to bat and human SARSr-CoVs in all four genes (Figs 4B and 5B). Specifically, we discovered that the SARS-CoV-2 had amino acid usage pattern most similar to bat RaTG13 virus, followed by pangolin P1E, bat CovVZC45 and bat CoVZXC21. The sequences of BtKY72 and BM48-31 were from a more phylogenetically distant clade, and, accordingly, they had relatively distinct amino acid usage to SARS-CoV-2 as expected in all four studied genes. This result agrees with the result in the full-genome phylogenetic analysis (Supplementary Fig. S5).
Figure 4.
Factorial map of the first and second factors for WCA and BCA by different genes, coloured by different viral host. The SARS-CoV-2 and related reference data points were labelled. The seven clusters identified by k-means algorithm were circled by dashed lines.
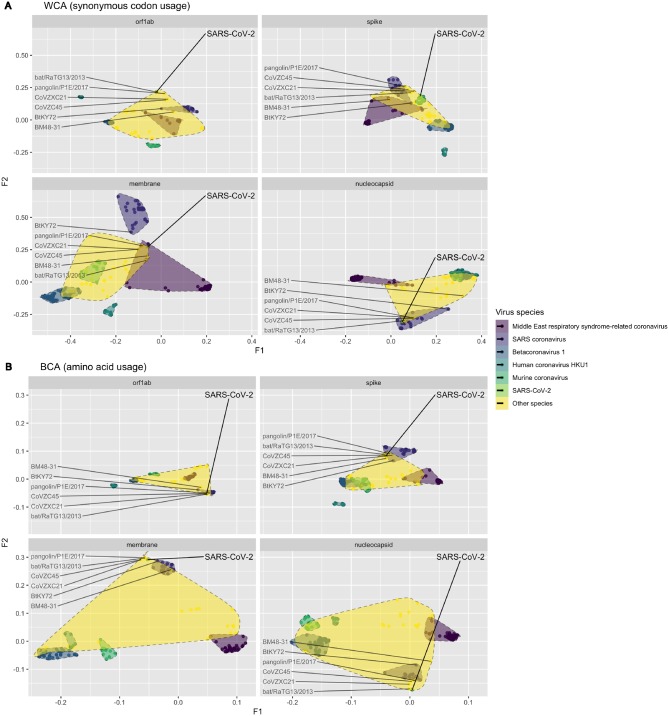
Figure 5.
Factorial map of the first and second factors for WCA and BCA by different genes, coloured by different viral species. The SARS-CoV-2 and related reference data points were labelled.
The difference between SARS-CoV-2 and RaTG13 at synonymous codon usage level was marginal in orf1ab and nucleocapsid sequences. However, our results suggest the synonymous codon usage patterns in the spike and membrane gene of SARS-CoV-2 are different from those of its genetically related viruses (i.e. RaTG13 and other reference relatives). For example, the pangolin P1E virus was not grouped in the same k-means cluster with SARS-CoV-2 in spike, and the synonymous codon usage pattern of SARS-CoV-2 was found to be closer to a cluster of rodent murine coronaviruses at the first two factorial levels (Figs 4A and 5A).
Further analysis on spike gene, however, suggested that the codon usage of SARS-CoV-2 and rodent murine coronaviruses were distinct at the third factorial level (Supplementary Fig. S6A). The results show that although RaTG13 became the most adjacent to SARS-CoV-2 when adding the third dimension WCA values. Our results suggest a complex genomic background in the spike gene of SARS-CoV-2, which made its synonymous codon usage harder to differentiate from other genomic sequences in our WCA analysis (the distances between k-means clusters are smaller). Despite the relative proximity between RaTG13 and SARS-CoV-2 at three-dimensional level, they were still at significantly different positions (Supplementary Fig. S6A). It is evident that the synonymous codon usage pattern of SARS-CoV-2 is distinct from other bat origin coronaviruses. The difference in synonymous codon usage is largely explained by the first factor (more than 50%), and our analysis on codon usages suggest that the first factor maybe highly related to the preferential usage of codons ending with cytosine (Supplementary Fig. S7). We also had similar observation for the membrane gene. Our three-dimensional analysis revealed that the synonymous codon usage of SARS-CoV-2 in membrane was most similar to P1E and CoVZXC21 (Supplementary Fig. S6B). It is worth noting that comparing to RaTG13, P1E, and CoVZXC21 had lower synonymous codon usage similarity to SARS-CoV-2 in the other three genes.
Overall, our WCA results support a more complex synonymous codon usage background on spike and membrane genes, though we identified unique codon usage patterns of SARS-CoV-2 on these two genes.
4. Discussion
Codon usage can be affected by many sequence features, including nucleotide composition, dinucleotide composition, amino acid preference, host adaption, etc. (Hershberg and Petrov 2008; Suzuki et al. 2008; Gu et al. 2019). The codon usages of viral sequences can vary by genes and host origins (Jenkins and Holmes 2003; Wong et al. 2010; Cristina et al. 2015). The bias in codon usage is a unique and distinctive characteristic that can reflect the ‘signature’ of a genomic sequence. Codon usage analyses are often complementary to ordinary sequence alignment-based analyses that focus on the genetic distance at nucleotide level, whereas codon usage analyses enable capturing signals at different sequence parameters. Therefore, codon usage bias can be another good proxy for identifying unique traits (e.g. virus origin, host origin, or some functions of proteins) of a genome. The goal of this study was to investigate the codon usage bias of betacoronaviruses. By studying the codon usages of these viruses in a systematic manner, we identified viral sequences carrying traits similar to those of SARS-CoV-2, which provided useful information for studying the host origin and evolutionary history of SARS-CoV-2.
The codon usage of different genes in betacoronaviruses is very different. The G + C content, especially the GC3 content is known to be influential to the codon usage of some bacteria and viruses (Perriere 2002; Gu et al. 2004; Woo et al. 2010). The GC3 content has pronounced effects on our WCA analysis of the orf1ab and spike genes. The GC3 content was found correlated with high WCA values on the first factor of orf1ab. In contrast, codons ending with cytosine had lower factorial values in the spike gene analysis (Supplementary Fig. S7). The G + C contents in membrane and nucleocapsid genes were less suppressed (Fig. 1A). This can be partly explained by the fact that membrane and nucleocapsid are two genes with shorter lengths which may limit the flexibilities for mutation or codon usage adaptation. In addition to global CA analysis, the application of WCA and BCA can eliminate the effects caused by amino acid compositions and synonymous codon usage, respectively. These alternative analytical tools were important because the amino acid sequences are expected to be more conserved such that they can preserve biological functions of the translated genes. In contrast, mutations at synonymous level tend to be more frequent, as most of these codon alternatives do not affect the biological function of a protein.
Of all the existing genomes in the dataset, RaTG13 best matched the overall codon usage pattern of the SARS-CoV-2. Although the SARS-CoV-2 had amino acid usage similar to bat and human SARSr-CoVs, the synonymous codon usages between them were relatively different, which indicates similar protein characteristics but maybe different evolutionary histories. The codon usage of bat coronaviruses is more scattered than coronaviruses of other hosts. This result agrees with the fact that bat is a major host reservoir of coronavirus (Calisher et al. 2006), thus it harbours coronaviruses with more complex genomic backgrounds.
SARS-CoV-2 was first identified in human, but its codon usage pattern is very different from those of other human betacoroanviruses (Supplementary Fig. S3A). In fact, the codon usage at both the amino acid level and synonymous level denote that the orf1ab gene in SARS-CoV-2 had closest relationship to SARSr-CoV, especially RaTG13. The CoVZX45 and CoVZXC21 had similar amino acid usage but relatively different synonymous codon usage to SARS-CoV-2 (Fig. 4). Besides bat-origin SARSr-CoV, the pangolin P1E also had similar codon usage to SARS-CoV-2 both at amino acid and synonymous codon levels. The result in orf1ab is in accordance with the full-genome phylogenetic analysis (Supplementary Fig. S5), showing a close relationship between SARS-CoV-2 and RaTG13 by the overall backbone of the genome.
The S protein is responsible for receptor binding which is important for viral entry. The genetic variability is extreme in spike gene (Gallagher and Buchmeier 2001), and this highly mutable gene may possess more information about recent evolution history. In our results, the synonymous codon usage of SARS-CoV-2 in spike gene was distinct from those of P1E and other phylogenetic relatives (Fig. 4A), which was not observed in orf1ab or nucleocapsid gene. Although the codon usage in spike of SARS-CoV-2, RaTG13, and P1E were similar at amino acid level, the difference at synonymous codon usage level indicates that they are unlikely to share a very recent common ancestor. It is more likely that SARS-CoV-2, RaTG13, and P1E might have undergone different evolution pathways for a certain period of time. The amino acid usage of SARS-CoV-2 in membrane was clustered with bat SARSr-CoV, however, the synonymous codon usage of SARS-CoV-2 was still distinct to these bat coronaviruses. Notably, in membrane gene, pangolin P1E had a more similar synonymous codon usage to SARS-CoV-2 than RaTG13. These findings suggest that there may be different selection forces between genes. Our result supports different evolutionary background or currently unknown host adaption history in SARS-CoV-2. The codon usage of SARS-CoV-2 in nucleocapsid gene was similar to bat SARSr-CoV both at amino acid level and synonymous level, suggesting that no highly significant mutation happened in this gene.
Codon usage can be shaped by many different selection forces, including the influence from host factors. Some researchers have hypothesized that the codon usage in SARS-CoV-2 maybe directly correlated to the codon usage of its host (Ji et al. 2020). However, our recent study on influenza A viruses implied that these may not be the most influential factors shaping the codon usage of a viral genome (Gu et al. 2019). Our analysis took advantage of the existing genomes of Betacoronavirus to study the complex host effect on codon usage, which warrants more accurate but relatively conserved estimation. However, we also understand that the results from codon usage analysis cannot be deterministic or direct evidence revealing the origin of the virus. As the number of available genomic data is currently limited, and virus with high (>99%) genetic similarity to SARS-CoV-2 was yet to be identified, we cannot draw a conclusion on the origin of SARS-CoV-2 at this stage. Another potential limitation of the study maybe from the CA method itself. Although CA have been widely applied in codon usage studies in many different organisms, the codon usage data are usually not independent observations. The phylogenetic relationship among observations make it hard to differentiate source of the codon usage bias, as the codon usage bias may both affected by selective pressures and descent relationships. Unfortunately, there is currently no established method for adjusting phylogenetic relationships in CA analysis in codon usage data, an alternative method addressing the above concern is needed.
Supplementary Material
Acknowledgements
We thank researchers for making viral sequences available for public access. We gratefully acknowledge the Originating and Submitting Laboratories for sharing genetic sequences and other associated data through the GISAID Initiative, on which this research is based. A list of the authors can be found in Supplementary Table S2.
Funding
This work was supported by RGC (GRF17112117) and National Institutes of Allergy and Infectious Diseases, National Institutes of Health (USA) (contract HHSN272201400006C). LLMP is supported by Croucher Foundation.
Conflict of interest: None declared.
References
- Akashi H., Eyre-Walker A. (1998) ‘Translational Selection and Molecular Evolution’, Current Opinion in Genetics and Development, 8: 688–93. [DOI] [PubMed] [Google Scholar]
- Benzécri J. P. (1983) ‘Analyse de L’inertie Intraclasse Par L’analyse D’un Tableau de Correspondance’, Cahiers de l’Analyse des données, 8: 351–8. [Google Scholar]
- Calisher C. H. et al. (2006) ‘Bats: Important Reservoir Hosts of Emerging Viruses’, Clinical Microbiology Reviews, 19: 531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D. et al. (2005) ‘Online Synonymous Codon Usage Analyses with the ade4 and seqinR Packages’, Bioinformatics, 21: 545–7. [DOI] [PubMed] [Google Scholar]
- Charif D., Lobry J. R. (2007) ‘SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis’, in Bastolla U.et al. (eds.) Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, Biological and Medical Physics, Biomedical Engineering, pp. 207–32. Springer: New York. [Google Scholar]
- Cristina J. et al. (2015) ‘Genome-Wide Analysis of Codon Usage Bias in Ebolavirus’, Virus Research, 196: 87–93. [DOI] [PubMed] [Google Scholar]
- Dray S., Dufour A. B. (2007) ‘The ade4 Package: Implementing the Duality Diagram for Ecologists’, Journal of Statistical Software, 22: 1–20. [Google Scholar]
- Fan R. L. Y. et al. (2015) ‘Generation of Live Attenuated Influenza Virus by Using Codon Usage Bias’, Journal of Virology, 89: 10762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T. M., Buchmeier M. J. (2001) ‘Coronavirus Spike Proteins in Viral Entry and Pathogenesis’, Virology, 279: 371–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H. et al. (2019) ‘Dinucleotide Evolutionary Dynamics in Influenza a Virus’, Virus Evolution, 5: vez038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Poon L. L. (2019) ‘Bioconductor - SynMut’. < 10.18129/B9.bioc.SynMut> accessed 24 Jan 2020. [DOI]
- Gu W. et al. (2004) ‘Analysis of Synonymous Codon Usage in SARS Coronavirus and Other Viruses in the Nidovirales’, Virus Research, 101: 155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A. (2008) ‘Selection on Codon Bias’, Annual Review of Genetics, 42: 287–99. [DOI] [PubMed] [Google Scholar]
- Jenkins G. M., Holmes E. C. (2003) ‘The Extent of Codon Usage Bias in Human RNA Viruses and Its Evolutionary Origin’, Virus Research, 92: 1–7. [DOI] [PubMed] [Google Scholar]
- Ji W. et al. (2020) ‘Homologous Recombination within the Spike Glycoprotein of the Newly Identified Coronavirus May Boost Cross‐Species Transmission from Snake to Human’, Journal of Medical Virology, 2020; 92: 433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N. et al. (2016) ‘Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses’, PLoS One, 11: e0154376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. (1982) ‘A Simple Method for Displaying the Hydropathic Character of a Protein’, Journal of Molecular Biology, 157: 105–32. [DOI] [PubMed] [Google Scholar]
- Lam T. T.-Y. et al. (2020) ‘Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins’, Nature, doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed]
- Liu Y. S. et al. (2011) ‘The Characteristics of the Synonymous Codon Usage in Enterovirus 71 Virus and the Effects of Host on the Virus in Codon Usage Pattern’, Infection, Genetics and Evolution, 11: 1168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry J. R. (2018) Multivariate Analyses of Codon Usage Biases, 1st edn, pp. 26–57. London, UK: Elsevier. [Google Scholar]
- Lu R. et al. (2020) ‘Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding’,The Lancet, 395: 565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K. M., Domsic J., McKenna R. (2008) ‘Genomic Evolution in a Virus under Specific Selection for Host Recognition’, Infection, Genetics and Evolution, 8: 825–34. [DOI] [PubMed] [Google Scholar]
- Percudani R., Ottonello S. (1999) ‘Selection at the Wobble Position of Codons Read by the Same tRNA in Saccharomyces cerevisiae’, Molecular Biology and Evolution, 16: 1752–62. [DOI] [PubMed] [Google Scholar]
- Perriere G. (2002) ‘Use and Misuse of Correspondence Analysis in Codon Usage Studies’, Nucleic Acids Research, 30: 4548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H. et al. (2008) ‘Comparison of Correspondence Analysis Methods for Synonymous Codon Usage in Bacteria’, DNA Research, 15: 357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. (2020) ‘A Novel Coronavirus Outbreak of Global Health Concern’, The Lancet, 395: 470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. (2016) ‘Analysis of Synonymous Codon Usage Bias of Zika Virus and Its Adaption to the Hosts’, PLoS One, 11: e0166260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) Novel Coronavirus – Republic of Korea (ex-China). Geneva: World Health Organization. [Google Scholar]
- Wong E. H. et al. (2010) ‘Codon Usage Bias and the Evolution of Influenza a Viruses. Codon Usage Biases of Influenza Virus’, BMC Evolutionary Biology, 10: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. Y. et al. (2010) ‘Coronavirus Genomics and Bioinformatics Analysis’, Viruses, 2: 1804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P. et al. (2020) ‘A pneumonia outbreak associated with a new coronavirus of probable bat origin’, Nature 579: 270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. et al. (2005) ‘Analysis of Synonymous Codon Usage in H5N1 Virus and Other Influenza a Viruses’, Biosystems, 81: 77–86. [DOI] [PubMed] [Google Scholar]
- Zhu N. et al. (2020) ‘A Novel Coronavirus from Patients with Pneumonia in China, 2019’, New England Journal of Medicine, 382: 727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.