Abstract
Objective and methods
This study reviewed the concept of in silico prediction of chemical toxicity for prevention of occupational cancer and future prospects in workers’ health. In this review, a new approach to determine the credibility of in silico predictions with raw data is explored, and the method of determining the confidence level of evaluation based on the credibility of data is discussed. I searched various papers and books related to the in silico prediction of chemical toxicity and carcinogenicity. The intention was to utilize the most recent reports after 2015 regarding in silico prediction.
Results and conclusion
The application of in silico methods is increasing with the prediction of toxic risks to human and the environment. The various toxic effects of industrial chemicals have triggered the recognition of the importance of using a combination of in silico models in the risk assessments. In silico occupational exposure models, industrial accidents, and occupational cancers are effectively managed and chemicals evaluated. It is important to identify and manage hazardous substances proactively through the rigorous evaluation of chemicals.
Keywords: Chemical toxicity, In silico, Prediction, Review, Workers’ health
Introduction
The method of identifying the hazards of chemicals has been almost entirely dependent on animal tests. Obtaining all the toxicological results for a single chemical using laboratory animals requires the sacrifice of at least thousands of animals. The tremendous cost and time required for animal testing are also major barriers to evaluating chemicals known to be commercially available at home and abroad [1].
The EU has established a new chemical assessment paradigm (REACH, enacted in 2007) to evaluate chemicals globally, including the staged registration and evaluation of all existing chemicals that have been manufactured and imported without safety information [2]. In the USA, quantitative structure–activity relationships (QSARs) predictions are used to evaluate two to three thousand chemicals each year and to assess a significant portion of the toxicity information [3]. Due to the uncertainty in the predictions, their use is extremely limited. While the EU and other countries have been very passive in introducing QSARs, they are now actively investing in the development and expansion of QSARs programs with the introduction of the REACH system that declares ‘No Data, No Market’ for all chemicals. In addition, institutional support has been established to require that non-testing methods, such as QSARs, be identified first, before conducting a new toxicity test for REACH registration [2]. In Korea, the enactment of the Law on the Registration and Evaluation of Chemical Substances (the K-REACH Act, enacted in 2015) has strengthened the EU’s obligation to evaluate not only new chemicals, but also existing chemicals that are manufactured or imported of over 1 ton quantity [4]. In order to improve the evaluation of chemicals to the level of developed countries, this law was introduced to strengthen the responsibility for demonstrating the hazards and risks of industries. As a result of this institutional strengthening, the enacted law also allows the submission of data by non-testing methods, such as QSARs as defined in EU REACH. In addition, the K-REACH Act stipulates omission of the toxicity test data for small quantities (less than 1 tonne per year) of new chemicals with a small amount of manufacture or import, which means that it must be absolutely dependent on the results of QSARs for the evaluation of small quantities of new scientific substances [4]. In this respect, many efforts have been made to collect, interpret, and link large amounts of known chemicals to each other, and to link chemicals to evaluations.
QSAR correlates proportionally with the structure of the chemicals and their physiological activity, starting with the premise that similar chemicals have similar physical properties or toxicity. The QSARs program, which can predict the toxicity of other chemicals based on the molecular structure of existing chemicals that are already known to be toxic, can save more time and money than performing the test to determine the hazards of the chemical [1, 2]. This review examines several applications of in silico toxicological approaches that many industries and institutions are taking, reviewing the examples of in silico tools they are using for their predictions. When the in silico toxicology (IST) protocol is fully developed and implemented, the IST assessment will be able to be used to evaluate the chemicals in the workplace and the regulatory body. The development of the IST protocol is an initiative developed in collaboration between international consortia and reflects the latest in silico toxicology for risk identification and characterization. In this review, a new approach to determine the credibility of in silico predictions with raw data is explored, and the method of determining the confidence level of evaluation based on the credibility of data is discussed.
This study examines the concept of in silico prediction of chemical toxicity on workers’ exposure and future prospects in occupational health. This review summarizes articles and books on the concept of in silico and the prediction of chemical toxicity and carcinogenicity. I searched the major Web sites used in most reviews for the necessary data search, specifically Google Scholar (https://scholar.google.com), ScienceDirect (www.sciencedirect.com), Scopus (www.scopus.com), NDSL (https://www.ndsl.kr/index.do), and PubMed (https://www.ncbi.nlm.nih.gov/pubmed). The search was performed using the key terms “in silico,” “prediction,” “chemical toxicity,” and “workers’ health.” Among the literature items searched through this process, the literature concerning in silico prediction was intended to utilize the most recent reports (after 2010), and the data on the research process and basic concepts are presented for ease of explanation date not only from 2010, but also include some data from before 2010.
Alternative tools for chemical toxic tests
Predictive toxicology is used in in silico and in vitro models, and in computational and other tools, in a more cost-effective manner than conventional tools. Here is a brief review of the issues involved in using predictive toxicology in regulatory alternative analysis (AA). It is good practice to use case studies to integrate predictive toxicology with AA, to start with prioritization of chemicals of interest, and to leverage existing resources to integrate predictive toxicology into AA practices using existing resources. Incorporating predictive toxicity into AA will enhance the ability of workplaces and regulators to choose alternatives to hazardous components and further increase the use of toxicity prediction in regulation [5]. Computational toxicology, also known as “in silico toxicology,” is based on knowledge gained from various scientific disciplines, and its activity is known due to the premise that the toxicity of chemicals is predictable in the molecular structure, and can be inferred from the properties. In order to provide a faster, more economic, and animal-free tool for predicting the toxicity of analogous compounds, structure–activity relationships (SARs) play an important role as the application of chemical genotoxicity and carcinogenicity assessments increases. The development of SAR algorithms is an ongoing process, and new models, as well as new versions of applications, are constantly being made available [6].
Research and development (R&D) for new chemicals are long and expensive processes, with efforts going on for decades; but safety issues (especially cardiac and liver toxicity) are a major challenge for chemical industries. To this end, the development of predictive toxicity assays and models has become a strategic problem for these companies, and an integrated approach to in vitro and in vivo experimental data and knowledge-based data sources should be undertaken [7]. As the size and diversity of biological data have increased over the decades, computerized predictions of toxicity have reached new levels. Emerging theories of machine learning for chemistry allow chemical structures, toxicology, simulation, and physical data to be integrated when predicting health risks and other toxicological information [8]. The developed in silico model has been fully validated and released online via the Enalos Cloud platform, which can be accessed at https://enalos.insilicotox.com/MouseTox/. This ready-to-use Web service provides free use of model results through a user-friendly interface, which can serve as a toxicity prediction tool for the risk assessment of new compounds, without special requirements or prior programming skills [9].
In silico prediction for chemical toxicity
The goal of in silico toxicity is to predict chemical toxicity through computational modeling, QSARs, and to predict algorithms with toxicity data. It is considered to be one of the alternatives to animal testing, which, in broad terms, utilizes a variety of computational techniques that correlate the toxicity or efficacy of a chemical with its structure. Computational techniques make it possible to quantitatively and qualitatively predict the toxicity of chemicals, mixtures, nanomaterials, and so on. The methods have strengths and limitations, specifics of scope and interpretation, so it is important to find the most effective method for each problem. Chemicals are toxic by interacting with other substances, and risk assessments can overlook the side effects of the mixture. For example, although adding other metals together increases lead (Pb) toxicity and reports accumulative risk assessment, there is a lack of raw data on the toxicity of the mixture, due to chemical exposure types and complex interactions, and it is not possible to test all combinations of these elements [10].
Prediction of the in silico toxicity of chemicals using machine learning and structural alert is a major issue, including the various toxicity and adverse effects that should be assessed in toxicology. In addition to predictive models made for various toxicological predictions, a database was presented, and Web server that can be used [11]. Efforts have been made to develop in silico models, such as computational tools that are efficient, fast, inexpensive, and accurate, and more than anything, that can be performed before the compound is synthesized [12]. In recent years, chemical toxicity prediction models have been developed in the field of chemical risk assessment, along with an admetSAR Web server with machine learning methods and structural alerts that have been developed for public use at no cost [13]. In order to model chemical carcinogenesis, several papers have shared protocols, and the authors have recently developed CarcinoPred-EL, a Web server that can predict carcinogenicity online [14]. More than 8000 compounds with Ames mutagens are currently known, and in recent years, these toxicity data have been used to extend predictive models and structural alerts [15]. Acute oral toxicity is the most widely studied in computational predictions, and several models were developed to predict acute oral toxicity. Several machine learning methods were developed and applied to construct classification and regression models to predict LD50 or its toxicity categories [16].
Quantitative structure–activity relationship (QSAR)
QSAR is an important tool in bioinformatics and chemistry information and is based primarily on data generated from molecular modeling and computational chemistry to predict the toxicities of chemicals [17]. With the help of QSAR, a correlation is established between the experiment and the biological activity of the molecules obtained by various molecular properties [18, 19]. The fundamental approach to developing, validating, and using QSAR models to predict toxicity is medium consistency. Since mathematical models are used to develop the equations, the quantification of various properties of the molecular set is also easily performed. These models work with new technologies to build models related to structure, activity, and toxicity. QSAR-based research shows application in a variety of fields, including ecotoxicity, antitumor, biotoxicity, and biochemical action, as well as occupational toxicology [20–22].
Structural alert (SA)
Structural alert (SA) is the molecular structure of the molecule that is associated with one of the adverse consequences or toxicological endpoints. SA for mutagenicity is a group of molecular functions or substructures associated with the mutagenic activity of chemical compounds. The SA used in the QSAR model has demonstrated the potential to predict various endpoints, including mutagenicity. Derek and Toxtree are software that use SAs to predict mutagenicity [23]. The Toxicity Forecaster (ToxCast) project uses quantitative high-efficiency screening to predict the chemical toxicity of several biological pathways that follow the NTP guidelines [24].
In silico prediction for nanotoxicology
First of all, in in silico modeling methods in the field of nanotoxicity, quantitative structure–nanotoxicity relationships (also known as quantitative structure–nanotoxicity relationships (QSNR), or nano-QSAR) are specific to nanomaterials, such as the size, shape, surface area, and solubility of nanomaterials. A linear QSNR model has been provided to predict EC50 (effective concentration for 50% enzymatic inhibition) for silver nanoparticles from size and surface charge, and a QSNR model using a quantum chemical descriptor is a fullerene C60 derivative. Integrating high-throughput screening methods with biosafety and in silico modeling emphasizes a system biology approach that ensures quality in nanosafety research, bridges the mechanical gap in fundamental research, and makes recommendations for predictive biological responses in nanotoxicology. In particular, “nano-QSAR” should be systematic and standardized as information collected in consideration of nano-characteristics. When developing toxicity prediction models, system interactions should be considered, and the EU NanoPUZZLES project was one of the global collaborations that improved the data availability and modeling approach to support the evaluation of nanomaterials [25]. Strategic development that combines multiple alternative tests can be useful for NM risk assessment and can assist decision-making [26].
OECD toolbox for the prediction of chemical toxicity
OECD and ECHA provide the OECD Toolbox for predicting or assessing health and environmental hazards for chemicals that have not been identified as toxic, using information from existing chemicals whose health and environmental hazards have been identified [27]. OECD Toolbox predicts (evaluates) methods using read-across, trend analysis, and QSAR, assuming that chemicals with similar mechanisms and structural features have similar toxicity. Health and environmental hazard information that is predictable through the OECD Toolbox includes carcinogenicity, germ cell mutagenicity, developmental toxicity, skin and eye irritation, acute toxicity, and aquatic toxicity. To ensure the credibility of the forecast results, the OECD Toolbox report is prepared by applying five principles to ensure the credibility of the forecast program presented by the OECD. It is expected that this will be useful for collecting hazard and risk information of chemicals, and preparing new safety data sheets for not only researchers, but also companies that manufacture or import chemicals. In addition, it may be used to confirm the results of previous studies, when selecting candidate substances for inhalation toxic test [28].
In the future, it is considered that further developments are needed to transform the toolbox into a more comprehensive chemical management system [29]. This OECD toolbox can be used to calculate not only physicochemical properties, but also endpoints for environmental toxicity and human toxicity.
Other packages
QSARs are estimates of test results based on correlations or relationships between chemical structure, physicochemical properties, and toxic effects in vivo, and predictive values are derived through various algorithms. Programs based on professional rule-of-thumb include Derek, HarzardExpert, Toxtree, and OECD toolbox, while statistics-based programs include MultiCASE, TOPKAT, Lazar, CAESAR, and OASIS TIMES as a mixed model [30]. Many models that predict toxicity through the structure of compounds have been published. Table 1 summarizes the features for each packages.
Table 1.
List of models to predict toxicity through the structure of chemicals
| Models | Features |
|---|---|
| Deductive estimate of risk from existing knowledge (Derek) | Developed by Lhasa Ltd., Derek is a type of QSAR model based on professional experience rules. Derek is a knowledge-based toxicity prediction program that divides a series of categories based on chemical structure, predicts the correlation between structure and biological activity, and can predict various toxicity indicators, including genotoxicity. Toxicophore that causes toxicity in target chemicals is identified to predict toxicity [27]. At this time, the risk assessment is defined on the basis of the relevant literature, which ensures strong confidence in the prediction. Knowledge-based toxicity prediction programs are used to determine the potential toxicity of a substance by obtaining information on the toxic functional groups [4]. In particular, the company is conducting research to improve the predictive power by applying multiple prediction programs, such as Sarah and Toxtree, and multiple applications [28]. Because in silico systems predict more complex phenomena that can use limited data, maximizing data accessibility is becoming increasingly important. In particular, the company is conducting research to improve the predictive power by applying multiple prediction programs, such as Sarah and Toxtree, and multiple applications [29] |
| EPA toxicity estimation software tool (T.E.S.T.) | The Chemistry Development Kit Java open-source and chemical data interworking is characterized by validating results by applying eight characteristic QSAR methods. Input query uses chemical name, CAS No., structure text file, etc., and the range includes acute oral toxicity, gene mutations, and environmental toxicity. Software provided by the US EPA includes human rat LD50 developmental toxicity and genotoxicity models |
| DanishQSAR | This is a repository-based model with data of more than 600,000 chemicals, applicable to 200 QSAR models. Input query can be chemical name, structure, CAS No., SMILES form, Mol file, etc. The range of prediction is physicochemical characteristics, acute toxicity, skin corrosion/irritation, and environmental toxicity |
| VegaHubQSAR | This provides prediction results optimized for REACH requirements and holds 40,000 kinds of chemical data, enabling the simultaneous batch prediction of a large number of substances and supporting the read-across approach. The input query uses the SMILES form, and the prediction range is mutagenic, carcinogenic, skin sensitized, BCF, logP, etc. Vega is a model for predicting human toxicity, which includes models for mutagenicity, carcinogenicity, developmental toxicity, endocrine binding, and skin sensitization, and physicochemical property prediction models. Vega implements and provides models to ensure that the in silico method is used correctly and that professionals use the in silico model |
| Toxtree | Toxtree is the software that implements the decision tree (DT) proposed by Cramer. Cramer DT is classified into three classes according to the metabolism of the compound, information on toxicity data, and information on whether it is used as a component of traditional food. Class 1 substances are known for their metabolic information and are very toxic compounds, and substances such as alcohols, ketones, and aldehydes belong to the first class. Class 2 is intermediate and is more toxic than class 1, but class 2 substances do not exhibit the same toxicity as class 3. Substances belonging to class 2 fall into one of two categories, with functional groups similar to those of class 1, but with higher reactivity, or more complex structure, than class 1. Class 3 is a highly toxic structure that contains compounds with highly reactive functional groups. Cramer’s method consists of 33 questions, and the answer to each question is yes or no. The compounds are classified according to the answers to these questions |
| PreADMET | The PreADMET package provides carcinogenicity prediction models and genotoxicity prediction models. A carcinogenicity prediction model was developed using data from mice administered with a chemical for two years, to determine whether cancer developed. A genotoxicity model was developed using data from the Ames test |
Applications of in silico prediction
Although chemical toxicity is an important reason for late-stage failure in R&D, it is important to establish accurate predictive models for the compound’s toxicity profile. Accurate predictive models can provide guidance for designing low-toxic chemicals [31]. In silico genotoxicity predictions have made significant progress over the past few years and are provided in the ICH M7 guideline for the assessment of reactive impurities in DNA as a major driver of these industries. This has been made possible by an increase in reliable Ames data, attempts to understand the active pathways, and subsequent development of computer-based prediction systems [32]. The new opportunity for “in silico environmental chemistry” goes beyond calculating specific chemical properties, to use statistical models to integrate in silico models, strain paths, product predictions, and environmental factors into model predictions, to make exposure assessments more comprehensive and efficient, and to extend to all applications [33].
Prediction of the acute toxicity of chemicals
To date, great advances have been made using the QSAR/quantitative structure–toxicity relationship (QSTR) model to predict the acute toxicity of chemicals in laboratory animals. Taking into account the reliability of the conditions, the first multitasking (mtk)-QSTR model was developed to simultaneously predict the acute toxicity of a compound. The mtk-QSTR model can classify a compound’s toxicity based on artificial neural networks (ANNs) and make predictions about various chemicals used in life and the workplace [34]. Toxic modeling has been a challenge for developers of QSAR models, due to the complexity of the endpoints; but recently, some new in silico methods have been developed that introduce the possibility of evaluating the integration of existing methods using various modeling perspectives. Model users should be aware of the considerations and assumptions made about these specific models as well as the assumptions made by evaluating the predictions on a case-by-case basis [35].
Prediction of the mutagenicity of chemicals
In the last decade or so, many companies have developed their own internal genotoxicity models built using chemical structure and bacterial mutagenicity data, but the analysis uses internal in-house compounds in the training model, and trains by combining in-house and public data [36]. The genotoxic impurities play a critical role in carcinogenesis, and impurities that affect pharmaceutical, safety, efficacy, and drug quality are attracting increasing interest in regulatory agencies and related industries [37]. Toxicity is defined as a series of events resulting from exposure to, and the distribution and metabolism of chemicals, and ultimately, interactions with other macromolecules in the cell, that is, interactions with DNA or proteins with various toxicity endpoints [38]. Mutagenicity is a major toxic endpoint for chemical risk assessment. The nature of a chemical that alters DNA or RNA sequences is mutagenicity, and these mutations can actually be fixed or transmitted [39]. DNA can be affected by ultraviolet light and by a variety of chemicals, such as industrial chemicals, pesticides, and the combustion by-products of fossil fuels [40]. The accurate identification of potentially toxic chemicals represents an important problem, and chemicals are proven to be mutagens mainly through battery experiments such as in vitro and in vivo. The electrophilic theory of Miller and Miller’s chemical carcinogens suggests that most of the carcinogens are electrophilic or nucleic acids or proteins [41]. Further reacting with nucleophilic groups leads to the fact that electrophilic properties are common between mutations and carcinogens [42].
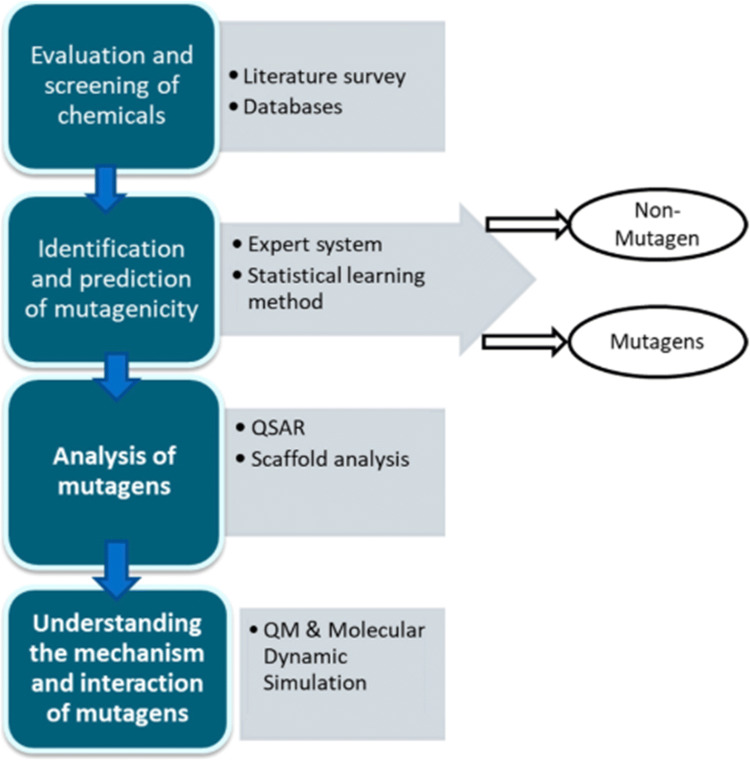
There are various in silico approaches for identifying and predicting chemically induced mutagens [43]. Although the in vivo genotoxicity of a compound does not always correlate with its activity in an in vitro test system, there may be good overlap between two endpoints for certain compounds. However, the difficulty lies in selecting the most appropriate protocol to establish an example of this relationship and to highlight potential in vivo risks. With this in mind, a project has begun to assess the relevance of existing structural alerts for ex vivo chromosomal damage from expert system Derek Nexus to its in vivo activity, by assessing the predictability of in vivo chromosomal damage datasets. Detailed information related to the in vivo activities and protocols added to the alert along with the mechanism information provided is useful for directing further testing of the compound of interest [44]. Various software is currently available for mutagenesis prediction and mutagenesis analysis. Table 2 lists the available resources, and Fig. 1 shows the workflow involved in the search for mutagenicity.
Table 2.
List of software resources available for predicting mutagenicity
| Available resources | Description | Accessibility |
|---|---|---|
| Derek Nexus v 2.0 | Expert rule-based and statistic-based predictions for mutagenicity based on in vitro endpoints | www.lhasalimited.org |
| TEST v 4.0.1/Ames test | Toxicity Estimation Software Tool (TEST) evaluates the toxicity including mutagenicity using QSAR methods | www.epa.gov/chemical-research/toxicity-estimation-software-tool-test |
| TOPKAT (Accelrys discovery studio v 3.1) | TOPKAT predicts various ranges of toxicological endpoints, such as mutagenicity, developmental toxicity, rodent carcinogenicity, rat chronic lowest observed adverse effect level (LOAEL), etc. | www.omictools.com/toxicity-prediction-by-komputer-assisted-technology-tool |
| Toxtree v 2.5.0 | Toxtree estimates various toxic hazards by the structural rules | www.toxtree.sourceforge.net/index.html |
| VEGA Caesar v 2.1.10 | Statistically based models, i.e., CEASAR, which predicts five endpoints: developmental toxicity, skin sensitization, mutagenicity (Ames), carcinogenicity, and the bioconcentration factor | www.caesar-project.eu / |
| VEGA SARpy v 1.0.5-Beta | VEGA SARpy uses a rule-based approach to predict both mutagenicity and non-mutagenicity | www.vegahub.eu |
| Mutagenicity (Ames test) model (ISS) prediction | Predicts mutagenicity | www.vegahub.eu |
| Lazar | Using structural fragment analysis, Lazar predicts toxicological endpoints, such as mutagenicity, human liver toxicity, rodent and hamster carcinogenicity, etc. | www.lazar.in-silico.ch/predict |
| HazardExpert | HazardExpert predicts toxicity based on bioavailability parameters and toxic fragments covering mutagenicity, teratogenicity, carcinogenicity, membrane irritation, immunotoxicity, and neurotoxicity endpoints | https://www.compudrug.com/hazardexpertpro |
| MultiCASE/MC4PC | Based on hierarchical statistics, MC4PC analyzes and compares active (mutagenic) and inactive (non-mutagenic) molecules, and identifies biophores | www.multicase.com/case-ultra-models |
| Leadscope model applier (LSMA) | Pretrained models of Leadscope model applier (LSMA) predict genetic toxicity | www.leadscope.com/model_appliers/ |
| ChemSilico | ChemSilico gives online calculation of various biological and physiochemical parameters for mutagenicity prediction | https://www.labhoo.com/labhoosite.asp?SID=68327280 |
| SciQSAR (formerly MDL-QSAR) software | Using E-state descriptors and nonparametric discriminant, SciQSAR predicts Ames mutagenicity | www.pharmaceuticalonline.com/doc/sciqsar-2d-0001 |
| Sarah nexus | Uses a unique machine-learning approach to build a statistical model for Ames mutagenicity | www.lhasalimited.org |
| Sarah and Derek Nexus | This combination provides toxicological evaluation from an intuitive interface | www.lhasalimited.org |
| AMBIT | Generates a toxicity report based on various precalibrated toxicity models | www.ambit.sourceforge.net/ |
| OpenTox | Predicts mutagenicity, and provides a transparent reasoning behind each prediction | www.opentox.net/ |
| eTOX | Predicts mutagenicity and carcinogenicity of human relevance | www.etoxproject.eu/ |
| ToxBoxes | Based on machine learning, mutagenicity prediction is done using fragment-based Advanced Algorithm Builder (AAB) | www.acdlabs.com/ |
| MDL QSAR | On the basis of new properties, a new compound library is generated, which predicts toxicity endpoints | www.mdl-qsar1.software.informer.com/ |
| Scaffold Hunter | Open-source tool for scaffold analysis of chemicals | www.scaffoldhunter.sourceforge.net/ |
| Bioclipse | Predicts mutagenicity, having modules for structural alerts, similarity searches, and QSARs | www.bioclipse.net/decision-support |
| PreADMET | Calculates descriptors and neuronal network for toxicity prediction system | www.preadmet.bmdrc.kr |
Sourced and modified from Ref. [45]. Adapted with permission
Fig. 1.
Framework for the identification and analysis of mutagenicity/mutagenicity predictions. Sourced and
Predictions for other toxicity
The assessment of eye irritation is an essential component of any risk assessment. This highlights the need for a reliable eye stimulation model that takes into account the mechanism of action and the individual structural classification, and the value of the profiling compound in relation to its chemical reactivity and physicochemical properties, which, together with the existing model, provide better prediction of severe stimulants [46]. Potential endocrine-disrupting chemicals present difficult regulatory challenges, and endocrine-disrupting chemicals can interfere with hormonal receptors associated with various health hazards. By integrating QSAR, docking, and systems biology approaches as virtual screening tools for use in risk assessment, systems biology pathways and network analysis approaches will provide a means to more critically assess the potential impact of endocrine-disrupting chemicals [47]. QSARs can contribute to the assessment of chemicals under REACH, where appropriate, and in some cases can be applied instead of experimental tests to fill in data gaps for information requirements. These results are similar to those of early QSAR and experimental studies of chemicals and show how high-throughput virtual screening can be used to identify potential genotoxic carcinogens, mutagens, and developmental toxins using QSAR predictions [48]. The US EPA Endocrine Disorder Screening Program (EDSP) uses in vitro data sourced from ToxCast/Tox21 high-throughput screening assays to evaluate the endocrine activity of chemicals. The QSAR model estimated more potent estrogen activity for most of the known estrogen metabolites compared to the parent chemical, and the method proposed in the in silico approach is an inexpensive and fast strategy for the detection of chemicals using estrogen metabolites [45]. A rapid test strategy for reproductive toxicity testing, avoiding the use of animals, combines knowledge of important processes affected by reproductive toxicants with the knowledge of the mechanical basis of those effects. As an additional step, examples were generated of how to predict the reproductive toxicity of chemicals using available data, formal validation of panel components was performed, and mechanical approaches used to test and verify, using more innovative approaches [49].
In summary, any approach that provides the information needed to establish an in silico tool for the safety assessment of chemicals can identify and predict a variety of toxicities from chemicals. Early predictions for safety assessments are moving from experiments to in silico knowledge and information-based tools. However, these changes are completely dependent on the stability and success applied to the in silico tools, and despite the benefits provided by the computational approach, extensive corrections and improvements are needed to extract accurate information on various toxicological endpoints in the near future.
Discussion
Efforts to efficiently predict chemical hazards
Since 2008, the US EPA has conducted a mid-to-long-term R&D project called “ToxCast” (Toxicity Forecaster), to use computers to target genomes and proteins in a short time. It has developed a method to evaluate and predict the harmful effects of chemicals, and Japan has announced that it will develop a method for evaluating the safety of artificial intelligence (AI) and big data-based chemicals in stages starting from 2019. This review undertook a comprehensive overview of the properties of in silico models tools for prediction, and the characteristics of the computational tools, and examined the future prospects. The various methods have their different strengths and weaknesses with varied scope of application, and each goal needs to find the most effective method among them. QSNRs (also known as nano QSARs) that use nanomaterial-specific descriptors are related to their structural and physicochemical properties. A linear QSNR model was used to predict the effective concentration for EC50 enzymatic inhibition of silver nanoparticles using size and surface charge. The toxicity of a chemical is related to its interaction with other chemicals. For example, it has been found that the toxicity of lead (Pb) increases with the coadministration of other metals, and a “cumulative risk assessment” has been developed to evaluate the toxicity of mixtures. But due to chemical exposure types and complex interactions, insufficient experimental data on the toxicity of mixtures are available, and it is not possible to test all combinations of these elements [50].
Advances in toxicology prediction for occupational health
Machine learning has been widely applied to classification and regression, and SA can be considered as alternative tools for optimization. These methods and models are areas where further development is expected for future chemical safety assessments. Recently, in vitro and in vivo tests have been performed to evaluate chemical safety and a variety of toxic side effects [11], to develop in vitro models, such as “in vivo organs.” Although efforts are being made, the process is still costly and time-consuming. Comparing the experimental methods, computational techniques show lots of advantages, because of their natural, fast, cheap, and accurate performances before the compound of interest is synthesized [12].
Many software and Web servers can be used to predict chemical toxicity, and OECD Toolbox is a product for predicting and modeling toxicity using QSTR. The Web server is easy to use and favors toxicologists. Lazar can predict many toxic endpoints using a user interface to describe the chemical structure [51]. Toxtree is an open-source tool for using a decision tree (DT) approach to predict toxicity. In silico toxicity predictions continue to evolve through the introduction of new methods, and improvements to current methods. The future of chemical hazard assessment emphasizes the replacement of animal experiments with human-related assays in vitro or in silico, and intelligently combining and using information for toxicity assessment and decision-making through an increasing variety of alternative assays. Certainly, useful components in the process of evaluating in silico toxicology, and in the future, mathematical calculations, will be extended to include special new types of toxicology items and models for chemicals. Toxicity evaluation of chemicals is essential for workers’ health and safety. Regulations for assessing the toxicity of genotoxic impurities lead to the integration of genotoxicity and carcinogenicity in vivo, and in vitro assays are becoming integrated with computational predictions [52]. One of the ICH M7 guidelines focuses on the identification of mutagens using in silico prediction, instead of in vitro studies [53]. Good prediction depends on two main factors: sensitivity and specificity. Sensitivity is the ability of a program to accurately identify true positive mutagens, which are chemicals that are likely to cause mutations, and specificity is the ability to identify true negative mutagens, such as safe chemicals that are not likely to be mutagenic [54]. In silico screening techniques can be applied to the toxicity assessment of chemicals, and the number of chemicals can be filtered by evaluating large libraries of compounds. Virtual screening is a cheminformatics tool applied using two categories of ligand-based and structure-based screening. The approach of virtual screening also utilizes target and ligand information to predict ADMET properties and other molecular characterization data [55].
In silico freeware models (OSIRIS v.2.0, LAZAR and Toxtree) have been presented for predicting the carcinogenicity and mutagenicity of volatile organic compounds (VOCs) related to chemical risk assessment for occupational exposure, and the results found in other software. Increased importance with be attached to using a combination of in silico models in the risk assessment of industrial chemicals. Several occupational exposure models are recommended in accordance with EU REACH legislation, and the availability of high-quality exposure data is limited, and verification is underway. These can also be used in future studies to increase accuracy, along with occupational exposure measurements. In developed countries, such as Japan, the USA, and Europe, industrial accidents and occupational diseases are effectively managed [3]. In recent years, with changes in regulations on industrial chemicals and cosmetics, significant advances have been made in the development, application, and evaluation of non-test approaches, such as QSAR. Much effort has been made to establish guidelines for conducting assessments within this approach, with a view to the current state of non-test approaches taken in these sectors. High-throughput approaches, such as the potential relevance of adverse outcome pathway (AOP) and chemicals R&D stage, suggest a workflow that integrates non-test approaches into test and evaluation strategies in practice [56]. New approaches have been required to assess the health effects of inhaled substances, which are based on toxic mechanisms, in silico modeling, in vitro methods as well as AOP [57].
The introduction of the REACH legislation in the European Union (EU) adds a requirement for toxicity data on chemicals produced in or imported into the EU at levels above 1 tonne/year, which means the increase in in silico forecast for such data [58]. In the future, much effort will still be needed to better understand the biological mechanisms of toxicity and to develop more accurate predictive models for screening compounds [59]. An essential element for the successful use of non-animal and purified safety tests is to improve understanding of chemical exposure and to determine the chemical concentrations measured during cell-based analysis and in vivo studies, and how the predicted human exposure relates to the tested levels. Approaches include understanding this and helping to design toxicology studies using existing information about human exposure [60].
In silico modeling applies to risk assessments for regulatory decisions and has developed due to industry pressure to enable government regulators to conduct risk assessments for chemicals. Understanding the development and appliance of QSAR models is critical to the powerful uses of such tools [61]. Using computer models to accurately predict toxicological outcomes is an important task, but current in silico toxicology is now based on advances in mechanotoxicity and predictive models that have grown remarkably in computing resources over the last decade. The integrated computer-aided discovery and redesign (CADRE) platform is used as a model for key molecular interactions in the toxic pathway [62]. CADRE offers distinct advantages over the primary screening of chemicals and suggests that it could be performed as an alternative in silico tool that is allowed in legislative programs [62]. In silico models are essential for identifying organ-level toxicity and developing integrated alternative methods for the replacement of animal experiments. SA can be directly predictive of toxicity, can support the formation of categories to facilitate readability, and is particularly important in deciphering the myriad mechanisms of action that cause organ-level toxicity [63].
Conclusion
Despite the obvious benefits that new chemicals bring to many industries, there are serious concerns about the potential health effects of these chemicals. As the number of workers or end users who use them for commercial purposes and are exposed to these chemicals every day increases, the need to assess their potential side effects in a cost-effective manner is growing. These data-driven models can be used to accelerate the decision-making process by screening hazards, and making early identification and management of potential toxicity, and physicochemical properties; and maximizing the use of information that these models provide can also support the industrial, regulatory, and public needs for safer chemicals [64]. In addition, further research is needed on how to use the in silico programs to protect workers’ health in industries that manufacture, use, and distribute various chemicals. Since skin sensitization is a complex immune disease that has a significant impact on workers’ health, the computational QSARs provide a screening tool and provide useful information for chemical toxicity assessment [65]. Carcinogenic substances are becoming increasingly problematic in the development of chemicals, because they have a serious impact on workers’ health. Cheminformatics provides a computer method for learning from chemistry data and for the modeling tasks facing chemists. The development of methods for predicting toxicity and assessing risks can meet social concerns about the effects of chemicals on workers’ health. Together with bioinformatics, our understanding of events in living organisms can deepen, so that new strategies can be developed to treat diseases.
Acknowledgements
This study was supported by the Korean Occupational Safety and Health Agency (Ulsan, Republic of Korea), the Ministry of Employment and Labor (Sejong, Republic of Korea), and a Grant-in-Aid for chemical hazard evaluation (2019).
Compliance with ethical standards
Conflict of interest
Kyung-Taek Rim declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.ECETOC (2003) (Q)SARs: evaluation of the commercially available software for human health and environmental endpoints with respect to chemical management applications, Technical report No. 89
- 2.European CHemicals Agency (ECHA) (2008) Guidance on information requirements and chemical safety assessment Chapter R.6: QSARs and grouping of chemicals. https://echa.europa.eu/reach_en.asp
- 3.Ministry of Environment, Republic of Korea (2001) Study on improvement of new chemical substance hazard assessment system
- 4.National Institute of Environmental Research, Republic of Korea (2016) Study on Improving Prediction of Genotoxicity through QSAR Program
- 5.Malloy T, et al. Advancing alternatives analysis: the role of predictive toxicology in selecting safer chemical products and processes. Integr Environ Assess Manag. 2017;13:915–925. doi: 10.1002/ieam.1923. [DOI] [PubMed] [Google Scholar]
- 6.Benigni R. In silico approaches to genetic toxicology: progress and future. Mutagenesis. 2019;34:1–2. doi: 10.1093/mutage/gey018. [DOI] [PubMed] [Google Scholar]
- 7.Loiodice S, Nogueira da Costa A, Atienzar F. Current trends in silico, in vitro toxicology, and safety biomarkers in early drug development. Drug Chem Toxicol. 2019;42:113–121. doi: 10.1080/01480545.2017.1400044. [DOI] [PubMed] [Google Scholar]
- 8.Luechtefeld T, Hartung T. Computational approaches to chemical hazard assessment. Altex. 2017;34:459–478. doi: 10.14573/altex.1710141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varsou DD, Melagraki G, Sarimveis H, Afantitis A. MouseTox: an online toxicity assessment tool for small molecules through Enalos Cloud platform. Food Chem Toxicol. 2017;110:83–93. doi: 10.1016/j.fct.2017.09.058. [DOI] [PubMed] [Google Scholar]
- 10.Kar S, Leszczynski J. Exploration of computational approaches to predict the toxicity of chemical mixtures. Toxics. 2019;7:15. doi: 10.3390/toxics7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segall MD, Barber C. Addressing toxicity risk when designing and selecting compounds in early drug discovery. Drug Discov Today. 2014;19:688–693. doi: 10.1016/j.drudis.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Cheng F, et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, et al. CarcinoPred-EL: novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci Rep. 2017;7:2118. doi: 10.1038/s41598-017-02365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, et al. Evaluation of different methods for identification of structural alerts using chemical ames mutagenicity data set as a benchmark. Chem Res Toxicol. 2017;30:1355–1364. doi: 10.1021/acs.chemrestox.7b00083. [DOI] [PubMed] [Google Scholar]
- 16.Gadaleta D, et al. SAR and QSAR modeling of a large collection of LD50 rat acute oral toxicity data. J Cheminform. 2019;11:58. doi: 10.1186/s13321-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parthasarathi R, et al. Application of quantum chemical descriptors in computational medicinal chemistry and chemoinformatics. Ind J Chem. 2006;45A:111–125. [Google Scholar]
- 18.Hansch C, Hoekman D, Gao HC. Comparative QSAR: toward a deeper understanding of chemicobiological interactions. Chem Rev. 1996;96:1045–1076. doi: 10.1021/cr9400976. [DOI] [PubMed] [Google Scholar]
- 19.Hansch C, et al. Chem-bioinformatics: comparative QSAR at the interface between chemistry and biology. Chem Rev. 2002;102:783–812. doi: 10.1021/cr0102009. [DOI] [PubMed] [Google Scholar]
- 20.Cronin MT, et al. Use of QSARs in international decision-making frameworks to predict health effects of chemical substances. Environ Health Perspect. 2003;111:1391. doi: 10.1289/ehp.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cronin MTD (2012) In silico tools for toxicity prediction. In: New horizons in predictive toxicology: current status and application. R Soc Chem 9–25
- 22.Raies AB, Bajic VB. In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip Rev Computa Mol Sci. 2016;6:147–172. doi: 10.1002/wcms.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridings J, et al. Computer prediction of possible toxic action from chemical structure: an update on the DEREK system. Toxicology. 1996;106:267–279. doi: 10.1016/0300-483X(95)03190-Q. [DOI] [PubMed] [Google Scholar]
- 24.Kavlock R, Dix D. Computational toxicology as implemented by the US EPA: providing high throughput decision support tools for screening and assessing chemical exposure, hazard and risk. J Toxicol Environ Health B. 2010;13:197–217. doi: 10.1080/10937404.2010.483935. [DOI] [PubMed] [Google Scholar]
- 25.Richarz AN, et al. Compilation of data and modelling of nanoparticle interactions and toxicity in the nanoPUZZLES project. Adv Exp Med Biol. 2017;947:303–324. doi: 10.1007/978-3-319-47754-1_10. [DOI] [PubMed] [Google Scholar]
- 26.Shatkin JA, Ong KJ. Alternative testing strategies for nanomaterials: state of the science and considerations for risk analysis. Risk Anal. 2016;36:1564–1580. doi: 10.1111/risa.12642. [DOI] [PubMed] [Google Scholar]
- 27.Hong MK, Jo JH, Choi BK, Kim KW (2018) Study of utilization of OECD toolbox in the field of chemical substance information. Research Project No. 2018-OSHRI-923. OSHRI, KOSHA
- 28.Seed MJ, Agius RM. Progress with structure-activity relationship modelling of occupational chemical respiratory sensitizers. Curr Opin Allergy Clin Immunol. 2017;17:64–71. doi: 10.1097/ACI.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 29.Schultz TW, Diderich R, Kuseva CD, Mekenyan OG. The OECD QSAR toolbox starts its second decade. Methods Mol Biol. 2018;1800:55–77. doi: 10.1007/978-1-4939-7899-1_2. [DOI] [PubMed] [Google Scholar]
- 30.Serafimova R, Fuart Gatnik M, Worth A (2010) Review of QSAR models and software tools for predicting of genotoxicity and carcinogenicity: JRC Scientific and Technical Reports. European Commission Joint Research Centre, Ispra, Italy
- 31.Lu J, et al. In silico prediction of chemical toxicity profile using local lazy learning. Comb Chem High Throughput Screen. 2017;20:346–353. doi: 10.2174/1386207320666170217151826. [DOI] [PubMed] [Google Scholar]
- 32.Wichard JD. In silico prediction of genotoxicity. Food Chem Toxicol. 2017;106(Pt B):595–599. doi: 10.1016/j.fct.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Tratnyek PG, Bylaska EJ, Weber EJ. In silico environmental chemical science: properties and processes from statistical and computational modelling. Environ Sci Process Impacts. 2017;19:188–202. doi: 10.1039/c7em00053g. [DOI] [PubMed] [Google Scholar]
- 34.Kleandrova VV, Luan F, Speck-Planche A, Cordeiro MN. In silico assessment of the acute toxicity of chemicals: recent advances and new model for multitasking prediction of toxic effect. Mini Rev Med Chem. 2015;15:677–686. doi: 10.2174/1389557515666150219143604. [DOI] [PubMed] [Google Scholar]
- 35.Marzo M, et al. In silico model for developmental toxicity: how to use QSAR models and interpret their results. Methods Mol Biol. 2016;1425:139–161. doi: 10.1007/978-1-4939-3609-0_8. [DOI] [PubMed] [Google Scholar]
- 36.Jolly R, et al. An evaluation of in-house and off-the-shelf in silico models: implications on guidance for mutagenicity assessment. Regul Toxicol Pharmacol. 2015;71:388–397. doi: 10.1016/j.yrtph.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Rim KT. In vitro models for chemical toxicity: review of their applications and prospects. Toxicol Environ Health Sci. 2019;11:94–103. doi: 10.1007/s13530-019-0402-8. [DOI] [Google Scholar]
- 38.Cunny H, Hodgson E (2004) Toxicity testing. A textbook of modern toxicology. 353
- 39.Benigni R, et al. Mutagenicity, carcinogenicity, and other end points. Methods Mol Biol. 2013;930:67–98. doi: 10.1007/978-1-62703-059-5_4. [DOI] [PubMed] [Google Scholar]
- 40.Benigni R, Bossa C. Mechanisms of chemical carcinogenicity and mutagenicity: a review with implications for predictive toxicology. Chem Rev. 2011;111:2507–2536. doi: 10.1021/cr100222q. [DOI] [PubMed] [Google Scholar]
- 41.Miller EC, Miller JA. Searches for ultimate chemical carcinogens and their reactions with cellular macromolecules. Cancer. 1981;47:2327–2345. doi: 10.1002/1097-0142(19810515)47:10<2327::AID-CNCR2820471003>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 42.Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci. 1973;70:2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giarratano JC, Riley G (1998) Expert systems. PWS publishing Co
- 44.Canipa SJ, et al. A quantitative in silico model for predicting skin sensitisation using a nearest neighbours approach within expert-derived structure-activity alert spaces. J Appl Toxicol. 2017;37:985–995. doi: 10.1002/jat.3448. [DOI] [PubMed] [Google Scholar]
- 45.Pinto CL, Mansouri K, Judson R, Browne P. Prediction of estrogenic bioactivity of environmental chemical metabolites. Chem Res Toxicol. 2016;29:1410–1427. doi: 10.1021/acs.chemrestox.6b00079. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, et al. In silico prediction of serious eye irritation or corrosion potential of chemicals. RSC Adv. 2017;7:6697–6703. doi: 10.1039/c6ra25267b. [DOI] [Google Scholar]
- 47.Ruiz P, et al. Integration of in silico methods and computational systems biology to explore endocrine-disrupting chemical binding with nuclear hormone receptors. Chemosphere. 2017;178:99–109. doi: 10.1016/j.chemosphere.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wedebye EB, et al. QSAR screening of 70,983 REACH substances for genotoxic carcinogenicity, mutagenicity and developmental toxicity in the ChemScreen project. Reprod Toxicol. 2015;55:64–72. doi: 10.1016/j.reprotox.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 49.van der Burg B, et al. The ChemScreen project to design a pragmatic alternative approach to predict reproductive toxicity of chemicals. Reprod Toxicol. 2015;55:114–123. doi: 10.1016/j.reprotox.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 50.Rider CV, et al. Predicting mixture toxicity with models of additivity. In: Rider C, Simmons J, et al., editors. Chemical mixtures and combined chemical and nonchemical stressors. Cham: Springer; 2018. [Google Scholar]
- 51.Maunz A, et al. lazar: a modular predictive toxicology framework. Front Pharmacol. 2013;4:38. doi: 10.3389/fphar.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.New Computational Method for Predicting Mutagenicity by Chemicals. Ch. 8 in mutagenicity: assays and applications. Ed. By Shraddha Pandit, Alok Dhawan, and Ramakrishnan Parthasarathi. CSIR-Indian Institute of Toxicology Research, Lucknow, India
- 53.Barber C, et al. Establishing best practise in the application of expert review of mutagenicity under ICH M7. Regul Toxicol Pharmacol. 2015;73(1):367–377. doi: 10.1016/j.yrtph.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 54.Lapenna S, Gatnik MF, Worth AP. Review of QSAR models and software tools for predicting acute and chronic systemic toxicity. Luxembourg: Publications Office of the European Union; 2010. [Google Scholar]
- 55.Ekins S, Nikolsky Y, Nikolskaya T. Techniques: application of systems biology to absorption, distribution, metabolism, excretion and toxicity. Trends Pharmacol Sci. 2005;26:202–209. doi: 10.1016/j.tips.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Patlewicz G, Fitzpatrick JM. Current and future perspectives on the development, evaluation, and application of in silico approaches for predicting toxicity. Chem Res Toxicol. 2016;29:438–451. doi: 10.1021/acs.chemrestox.5b00388. [DOI] [PubMed] [Google Scholar]
- 57.Clippinger AJ, et al. Pathway-based predictive approaches for non-animal assessment of acute inhalation toxicity. Toxicol In vitro. 2018;52:131–145. doi: 10.1016/j.tiv.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dearden JC, Rowe PH. Use of artificial neural networks in the QSAR prediction of physicochemical properties and toxicities for REACH legislation. Methods Mol Biol. 2015;1260:65–88. doi: 10.1007/978-1-4939-2239-0_5. [DOI] [PubMed] [Google Scholar]
- 59.Lu J, et al. Machine learning-based modeling of drug toxicity. Methods Mol Biol. 2018;1754:247–264. doi: 10.1007/978-1-4939-7717-8_15. [DOI] [PubMed] [Google Scholar]
- 60.Sewell F, et al. The current status of exposure-driven approaches for chemical safety assessment: a cross-sector perspective. Toxicology. 2017;389:109–117. doi: 10.1016/j.tox.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 61.Steinbach T, et al. (Q)SAR: a tool for the toxicologist. Int J Toxicol. 2015;34:352–354. doi: 10.1177/1091581815584914. [DOI] [PubMed] [Google Scholar]
- 62.Kostal J, Voutchkova-Kostal A. CADRE-SS, an in silico tool for predicting skin sensitization potential based on modeling of molecular interactions. Chem Res Toxicol. 2016;29:58–64. doi: 10.1021/acs.chemrestox.5b00392. [DOI] [PubMed] [Google Scholar]
- 63.Mellor CL, Steinmetz FP, Cronin MT. Using molecular initiating events to develop a structural alert based screening workflow for nuclear receptor ligands associated with hepatic steatosis. Chem Res Toxicol. 2016;29:203–212. doi: 10.1021/acs.chemrestox.5b00480. [DOI] [PubMed] [Google Scholar]
- 64.Oksel C, et al. Literature review of (Q)SAR modelling of nanomaterial toxicity. Adv Exp Med Biol. 2017;947:103–142. doi: 10.1007/978-3-319-47754-1_5. [DOI] [PubMed] [Google Scholar]
- 65.Braga RC, et al. Pred-skin: a fast and reliable web application to assess skin sensitization effect of chemicals. J Chem Inf Model. 2017;57:1013–1017. doi: 10.1021/acs.jcim.7b00194. [DOI] [PubMed] [Google Scholar]