Abstract
Teschovirus A belongs to the family Picornaviridae and is a causal agent of the disease Teschovirus encephalomyelitis and other infections that remain asymptomatic. The present study was performed to design epitope-based peptide vaccine against Teschovirus A by identifying the potential T cell and B-cell epitopes from capsid proteins (VP1, VP3 and VP2) of the virus using reverse vaccinology and immunoinformatics approaches. In the current study, hexapeptide T-cell and octapeptide B-cell epitopes were analyzed for immunogenicity, antigenicity and hydrophilicity scores of each epitope. Each potential epitope was further characterized using ExPASy-ProtParam and Antimicrobial Peptide Database (APD3) tools for determining various physical and chemical parameters of the epitope. One linear hexapeptide T-cell epitope, i.e., RPVNDE (epitope position 77–82) and one linear octapeptide B-cell epitope, i.e., AYSRSHPQ (236–243) were identified from the viral capsid protein as they possess the capability to raise effective immunogenic reaction in the host organism against the virus. Pharmaceutical industries could harness the results of this investigation to develop epitope-based peptide vaccines by loading the identified epitopes in combination with targeting signal peptides of T-cells and B-cells and then inserting the combination into virus like particle (vlp) or constructing subunit vaccines for further trial.
Keywords: Teschovirus A, Peptide vaccine, Epitopes, Antigen, Immunogenicity
Introduction
Teschovirus A was formerly known as Porcine Teschovirus (PTV) (Cano-Gómez et al. 2017) and it causes infectious diseases in wild boars and swine. The diseases include enchephalomyelitis, diarrhoea, myocarditis, pneumonia and reproductive loses (Long et al. 1966; Knowles 2008; Chiu et al. 2012). Teschovirus A has been assigned to the genus Teschovirus, family Picornaviridae and order Picornavirales (Deng et al. 2012). A set of 13 serotypes of Teschovirus A (PTV-1 to PTV-13) have been identified (Cano-Gómez and Jiménez-Clavero 2016). Majority of PTV serotypes may cause milder infection, myocarditis, pericarditis or reproductive disorders or may not produce any signs of infection at all in host organism. However, the serotype, i.e., PTV-1 has been known to cause infection of central nervous system (CNS) giving rise to neurological disease which may either show severe infection or mild infection (Edington et al. 1972). Severe infection is known as Teschen disease while mild infection is called Talfan disease (Manuelidis et al. 1954; Knowles 2008). Two different strains of PTV-1 are responsible for these two diseases (Doherty et al. 1999; Prodělalová 2012). Any neurological disorder caused by Teschovirus A is termed as Teschovirus encephalomyelitis. The first outbreak of Teschovirus encephalomyelitis took place in 1929 at Teschen in Czechoslovakia (Trefny 1930). Europe and its neighbouring continents were under the influence of this disease in 1940–1950. This havoc caused high mortality and morbidity of swine resulting in significant loss in swine industries (Kouba 2009). The disease spread widely across Ukraine, Russia, Madagascar, Japan, Maldavis, Romania, China and Uganda (Kouba 2009; Wang et al. 2010; Salles et al. 2011).
Teschovirus A is a non-enveloped virus. It consists of a positive sense single-stranded RNA of size 7.1 kb in its genome and codes for only one polyprotein (Cano-Gómez and Jiménez-Clavero 2016; Cano-Gómez et al. 2017). The virus is round in shape with a diameter of 30 nm (Zell et al. 2005; Cano-Gómez et al. 2017). In Teschovirus A, three precursor viral polypeptides arise when the polyprotein is nicked and further nicking of each results into several components, namely a leader (L) protein, an RNA-dependent RNA polymerase (3D), a protease (3C), a virus-encoded peptide (3B), four structural proteins (VP1, VP2, VP3 and VP4) and four non-structural proteins (3A, 2A, 2B and 2C) (Manuelidis et al. 1954; Zell et al. 2001; Zell et al. 2005). The genome of Teschovirus A also comprises of 5′-UTR and 3′-UTR sequences (Doherty et al. 1999; Zell et al. 2001). The virus has a capsid comprising of four structural proteins viz. VP1, VP2, VP3 and VP4, wherein VP4 protein is located in the interior region of the capsid (Smith and Baker 1999).
The recent outbreak of Teschovirus A was found to occur in Europe, America, Asia, Africa and Oceania (Bangari et al. 2010; Wang et al. 2010; Zhang et al. 2010; Salles et al. 2011). Moreover, an outbreak was reported recently where Teschovirus A affected farrows and caused diseases such as gastroenteritis and respiratory disease (Ventura et al. 2012). Infection by this virus also resulted into abortion and stillbirth (Liu et al. 2011). In recent years, severe outbreak was caused by Teschovirus A in Haiti, Canada and Dominican Republic and this had largely affected the morbidity and mortality rate of swines (Deng et al. 2012; Ventura et al. 2012). It is very difficult to entirely eradicate the virus. In order to control the disease caused by this virus, attenuated or inactivated vaccines were used (Alexandersen et al. 2019).
Traditional vaccines that are prepared on the basis of live attenuated or inactivated whole viruses require specific conditions for their utility, maintenance and preservation. Traditional vaccines are sometimes considered as unsafe since the live attenuated or inactivated viruses may turn to live viruses and cause diseases (Purcell et al. 2007). These vaccines can lead to skin irritation and other side-effects (Harrison et al. 1999). The development of peptide vaccine based on genomics and proteomics has been suggested as a potential intervention against disease causing organism (Jackwood et al. 2008). Various preclinical studies stated that epitope based peptide vaccines formulated by reverse vaccinology showed promising outcomes (Davies and Flower 2007). Bioinformatics tools provide a means for identifying the potential T-cell and B-cell epitopes for designing peptide vaccines against pathogen and moreover, the in vitro experiments for formulating vaccines have significantly decreased in quantity as a consequence of utilizing the in silico approach due to low cost and time requirements (Florea and Halldorsson 2003; Backert and Kohlbacher 2015). Compared to traditional vaccines, epitope-based vaccines are considered to be safer and cheaper (Ellis 1999).
Two lymphocytes, T-cells and B-cells are essentially important for providing immunogenic responses in host organism against any pathogenic agent (Crotty 2015). Development of T-cells takes place in thymus whereas development of B-cells occurs in bone marrow (Zúñiga-Pflücker 2004). Certain components of the antigen, called epitopes of the invading agents are identified by these cells with the help of receptors present on their exterior surface (Weiss and Littman 1994). On recognition of an antigen, the production of antibodies is mediated by B-cells which can destroy the antigenic particle. Moreover, B-cells also develop into memory cells which provide future protection against the foreign invaders (Chaplin 2010). The destruction of foreign antigen by T-cells occurs in an organized fashion where a group of proteins called the major histocompatibility complex (MHC) molecules identify the antigen and exhibit them on APCs’ surfaces and subsequently, T-cell receptor identifies the antigen presented by APCs and initiates its destruction (Valitutti et al. 1995; Jensen 2007; Lafuente and Reche 2009).
Multiple-epitope loaded peptide vaccines based on reverse vaccinology approach for treatment of Teschovirus encephalomyelitis affecting swine and boars has not been developed so far. The present study was performed to identify the T-cell and B-cell epitopes from the viral capsid proteins having potential to develop peptide vaccine against Teschovirus A. The epitopes possessing high immunogenicity could be considered promising as vaccine candidates and could be administered in swine and wild boars, thereby, providing immunity against the disease.
Materials and Methods
Retrieval of Sequences
The capsid proteins of Teschovirus A were selected for the current study as they exist on the external surface of the virion and therefore, the epitopes determined from these proteins would have the capacity to generate immune responses against Teschovirus encephalomyelitis. However, the VP4 protein, one of the components of the capsid, was excluded from the study as the protein lies in the internal region and may have less potential for designing vaccine. The entire set of protein sequences of the viral capsids, namely VP1, VP3 and VP2 were extracted from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) having accession number NP_740352.1, NP_740351.1 and NP_740350.1 respectively.
Prediction of T-cell Epitopes
T-cell epitopes (linear hexapeptides) from each protein of Teschovirus A were predicted using the method developed by Hopp and Woods (Hopp and Woods 1981).
Prediction of B-cell Epitopes
B-cell epitopes (linear octapeptides) from each protein were identified on the basis of a semi-empirical method developed by Kolaskar and Tongaonkar (Kolaskar and Tongaonkar 1990).
Hydropathy Index and Hydrophilicity Score
Hydropathy index is characterized by a numerical value that describes the hydrophobicity or hydrophilicity of side-chains of amino acids (Kyte and Doolittle 1982). Hydrophilicity score is the average hydrophilicity value of the amino acids in a protein or peptide and is useful in predicting protein structure (Hopp and Woods 1981).
Immunogenicity Score
T-cell and B-cell epitopes from the proteins having excessive immunogenic response for the production of high quantity of antibodies were identified using AbDesigner algorithm (Pisitkun et al. 2011) and the hydropathy index of each epitope was evaluated. Further, Chou-Fasman prediction tool for predicting beta-turn of secondary protein structure (Chou and Fasman 1978) was used to determine the immunogenicity of the epitopes as the beta-turn region of proteins are responsible for providing immunogenicity (Rini et al. 1992).
The parameters of each T-cell and B-cell epitope, namely hydrophilicity, antigenicity, immunogenicity and the number of aliphatic amino acids were determined using a PERL program developed by SC (corresponding author).
Physicochemical Characterization of T-cell and B-cell Epitopes
Physicochemical properties of T-cell and B-cell epitopes were determined using ExPASy-ProtParam tool (https://web.expasy.org/protparam/). Amino acid composition of each epitope was determined. Moreover, net charge and potential protein–protein binding interaction, i.e., Boman index of each epitope were estimated using the tool in APD3 (Antimicrobial Peptide Database) (http://aps.unmc.edu/AP/prediction/prediction_main.php). When a protein possesses Boman index > 2.48 kcal/mol, it indicates that the protein has great potential for interaction with other protein.
Results
Identification of T-cell and B-cell Epitopes and Their Hydrophilicity Analysis
Generally, peptides of 6–10 amino acids in linear continuous stretches are the constituents of antigenic determinants or epitopes of any protein. To combat any pathogen, hexapeptide epitopes possess immense potential to generate significant immunogenic reaction in the host organism (Hopp and Woods 1981). For this study, five hexapeptide T-cell epitopes from each capsid protein i.e. VP1, VP3 and VP2 of Teschovirus A were identified using bioinformatic tools.
Using the method of Hopp and Woods, T-cell epitopes for each protein were identified on the basis of maximal hydrophilicity scores of the epitopes (Hopp and Woods 1981). The hexapeptide T-cell epitopes were analyzed for determining their location within the proteins, their hydrophilicity scores and net charges (Table 1). Among the five epitopes of the VP1 protein, three epitopes viz. SREHDE (epitope position 245–250), ESREHD (244–249) and EKAKDS (135–140) were found to carry a net charge of − 2, − 2 and 0 respectively and to have the highest hydrophilicity score of 11.8. Likewise, two hexapeptide epitopes, namely KNREIR (56–61) of VP3 protein and DDTDKH (177–182) of VP2 protein possessed the highest hydrophilicity score of 10.4 and 11.1 respectively while the total net charge of KNREIR was + 2 and that of DDTDKH was − 2. Epitopes with high hydrophilicity scores usually remain exposed on the protein surface and interact easily with antigen and thereby, produce immunogenic response against the antigens.
Table 1.
T-cell epitopes of Teschovirus A showing their hydrophilicity score and total net charge (Hopp and Woods 1981)
| Name of virus | Name of proteins | Accession number | Hexapeptide epitope | Epitope position | Hydrophilicity score | Net charge |
|---|---|---|---|---|---|---|
| Teschovirus A | VP1 | NP_740352.1 | SREHDE | 245–250 | 11.8 | − 2 |
| ESREHD | 244–249 | 11.8 | − 2 | |||
| EKAKDS | 135–140 | 11.8 | 0 | |||
| REHDEG | 246–251 | 11.5 | − 2 | |||
| IEKAKD | 134–139 | 9.7 | 0 | |||
| VP3 | NP_740351.1 | KNREIR | 56–61 | 10.4 | + 2 | |
| STERTK | 64–69 | 8.5 | + 1 | |||
| TERTKP | 65–70 | 8.2 | + 1 | |||
| RPVNDE | 77–82 | 7.7 | − 1 | |||
| DIDASR | 7–12 | 7 | − 1 | |||
| VP2 | NP_740350.1 | DDTDKH | 177–182 | 11.1 | − 2 | |
| EPKFEE | 1–6 | 9.5 | − 2 | |||
| KPEEQP | 185–190 | 9.2 | − 1 | |||
| FDDTDK | 176–181 | 9.1 | − 2 | |||
| KFEEYK | 3–8 | 7.2 | 0 |
The selection of promising B-cell epitopes for each protein was based on maximum hydrophilicity score of the epitopes (Kolaskar and Tongaonkar 1990). The position of each epitope in the protein sequence, along with their hydrophilicity score and net charge were determined (Table 2). Among five epitopes of VP1 protein, two epitopes, namely KDSVDNYA (138–145) and AKDSVDNY (137–144) were found to possess the maximum hydrophilicity score of 5.2 and each possessed a net charge of − 1. For VP3 protein, the epitope, PYTKTDFP (172–179) had the highest hydrophilicity score of 0.4 without any net charge. In case of VP2 protein, the epitope, YKPEEQPY (184–191) possessed the highest hydrophilicity score of 4.6 with a net charge of − 1.
Table 2.
B-cell epitopes of Teschovirus A showing their hydrophilicity score and total net charge (Kolaskar and Tongaonkar 1990)
| Name of virus | Name of proteins | Accession number | Octapeptide epitope | Epitope position | Hydrophilicity score | Net charge |
|---|---|---|---|---|---|---|
| Teschovirus A | VP1 | NP_740352.1 | YSRSHPQE | 237–244 | 4 | 0 |
| PRPGIHDP | 227–234 | 3.7 | 0 | |||
| AYSRSHPQ | 236–243 | 0.5 | + 1 | |||
| KDSVDNYA | 138–145 | 5.2 | − 1 | |||
| AKDSVDNY | 137–144 | 5.2 | − 1 | |||
| VP3 | NP_740351.1 | TPYWSKGY | 191–198 | − 5.1 | + 1 | |
| SSPSYMPG | 33–40 | − 2.7 | 0 | |||
| ASPPNSPA | 208–215 | − 0.2 | 0 | |||
| NLASPPNS | 206–213 | − 1.3 | 0 | |||
| PYTKTDFP | 172–179 | 0.4 | 0 | |||
| VP2 | NP_740350.1 | YKPEEQPY | 184–191 | 4.6 | − 1 | |
| HYHHTFDD | 171–178 | − 0.7 | − 2 | |||
| RPDGQLTY | 135–142 | 1.7 | 0 | |||
| YSTWHYHH | 167–174 | − 9.6 | 0 | |||
| YHGLRQSS | 269–276 | − 0.8 | + 1 |
Antigenicity Analysis of T-cell and B-cell Epitopes
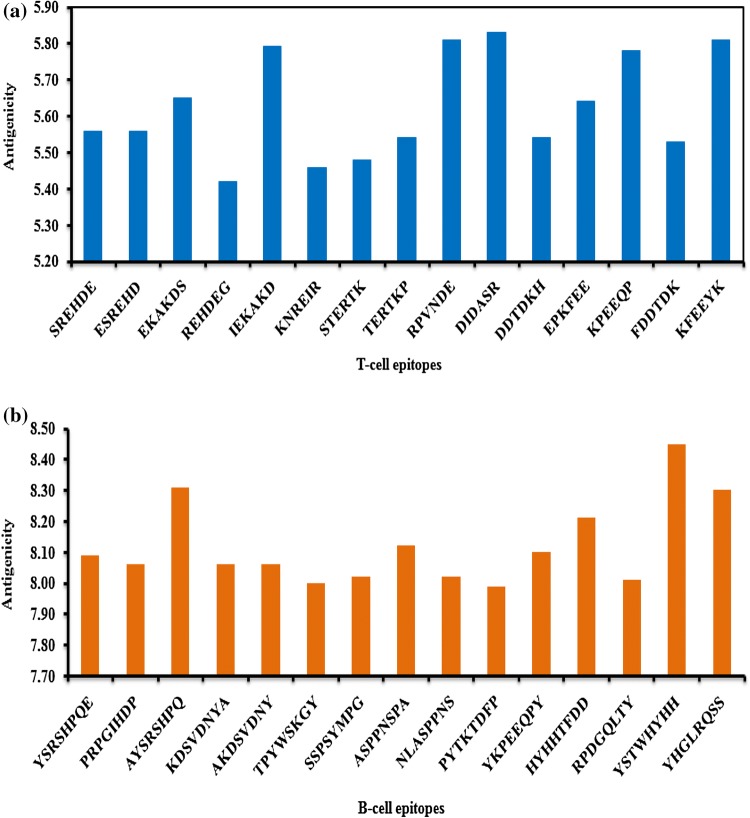
Antigenicity score of each hexapeptide T-cell epitope was estimated (Fig. 1a). All the T-cell epitopes were plotted on the x-axis of the graph while antigenicity scores of all epitopes were plotted on the y-axis. The range of antigenicity score for T-cell epitopes varied from 5.42 (REHDEG of VP1 protein, epitope position 246–251) to 5.83 (DIDASR of VP3 protein, 7–12). High antigenicity score of an epitope indicates that the epitope possesses high antigenic property, an essential property for the development of epitope-based peptide vaccines against pathogen.
Fig. 1.
a Antigenicity of T-cell epitopes of Teschovirus A.b Antigenicity of B-cell epitopes of Teschovirus A
For each octapeptide B-cell epitope, the antigenicity scores were estimated (Fig. 1b). Antigenicity score of the epitopes varied from 7.99 (PYTKTDFP of VP3 protein, 172–179) to 8.45 (YSTWHYHH of VP2 protein, 167–174).
Immunogenicity Analysis of T-cell and B-cell Epitopes
Based on Chou–Fasman beta turn conformation and hydropathy index of each T-cell epitope, the immunogenicity score of individual hexapeptide epitope was estimated and also the number of aliphatic amino acids within the epitopes was determined (Table 3) (Pisitkun et al. 2011). Among the five epitopes of VP1 protein, the epitope, REHDEG (246–251), was found to possess the highest immunogenicity score of 8.13. For VP3 protein, the epitope, RPVNDE (77–82) had the highest immunogenicity score of 7.36 and in case of VP2 protein, the epitope DDTDKH (177–182) was identified with the highest immunogenicity score of 9.21. High immunogenicity score of an epitope from a viral protein signifies that it has the capability to activate significant immune response against the antigen. Epitopes rich in aliphatic amino acids, namely A, G, I, K, L, M and V usually interact effectively with antigens.
Table 3.
T-cell epitopes of Teschovirus A showing number of aliphatic amino acids and immunogenicity (Ig) analysis (Hopp and Woods 1981)
| Name of virus | Name of proteins | Accession number | Hexapeptide epitope | Aliphatic amino acids | Ig analysis of hexapeptide epitopes | ||
|---|---|---|---|---|---|---|---|
| Hydropathy index | Chou-Fasman conformation | Ig value | |||||
| Teschovirus A | VP1 | NP_740352.1 | SREHDE | 0 | 7.67 | 1.05 | 8.05 |
| ESREHD | 0 | 7.67 | 1.05 | 8.05 | |||
| EKAKDS | 3 | 6.8 | 1.05 | 7.14 | |||
| REHDEG | 1 | 7.6 | 1.07 | 8.13 | |||
| IEKAKD | 4 | 5.92 | 0.89 | 5.27 | |||
| VP3 | NP_740351.1 | KNREIR | 2 | 7.07 | 0.95 | 6.72 | |
| STERTK | 1 | 6.85 | 1.01 | 6.92 | |||
| TERTKP | 1 | 6.98 | 1.02 | 7.12 | |||
| RPVNDE | 1 | 6.57 | 1.12 | 7.36 | |||
| DIDASR | 2 | 5.5 | 1.07 | 5.89 | |||
| VP2 | NP_740350.1 | DDTDKH | 1 | 7.55 | 1.22 | 9.21 | |
| EPKFEE | 1 | 6.7 | 0.89 | 5.96 | |||
| KPEEQP | 1 | 7.43 | 1.09 | 8.1 | |||
| FDDTDK | 1 | 6.55 | 1.16 | 7.6 | |||
| KFEEYK | 2 | 6.72 | 0.87 | 5.85 | |||
The immunogenicity score and the number of aliphatic amino acids present in individual octapeptide B-cell epitope were estimated (Table 4) (Pisitkun et al. 2011). The range of immunogenicity scores of B-cell epitopes varied from 6.6 (YHGLRQSS of VP2 protein, 269–276) to 7.87 (YSRSHPQE of VP1 protein, 237–244).
Table 4.
B-cell epitopes of Teschovirus A showing number of aliphatic amino acids and immunogenicity (Ig) analysis (Kolaskar and Tongaonkar 1990)
| Name of virus | Name of proteins | Accession number | Octapeptide epitope | Aliphatic amino acids | Ig analysis of octapeptide epitopes | ||
|---|---|---|---|---|---|---|---|
| Hydropathy index | Chou-Fasman conformation | Ig value | |||||
| Teschovirus A | VP1 | NP_740352.1 | YSRSHPQE | 0 | 6.9 | 1.14 | 7.87 |
| PRPGIHDP | 2 | 5.99 | 1.24 | 7.43 | |||
| AYSRSHPQ | 1 | 6.24 | 1.13 | 7.05 | |||
| KDSVDNYA | 3 | 5.81 | 1.15 | 6.68 | |||
| AKDSVDNY | 3 | 5.81 | 1.15 | 6.68 | |||
| VP3 | NP_740351.1 | TPYWSKGY | 2 | 5.86 | 1.22 | 7.15 | |
| SSPSYMPG | 2 | 5.18 | 1.33 | 6.89 | |||
| ASPPNSPA | 2 | 5.29 | 1.29 | 6.82 | |||
| NLASPPNS | 2 | 5.28 | 1.28 | 6.76 | |||
| PYTKTDFP | 1 | 5.81 | 1.15 | 6.68 | |||
| VP2 | NP_740350.1 | YKPEEQPY | 1 | 7.03 | 1.1 | 7.73 | |
| HYHHTFDD | 0 | 6.48 | 1.06 | 6.87 | |||
| RPDGQLTY | 2 | 5.96 | 1.15 | 6.85 | |||
| YSTWHYHH | 0 | 6.33 | 1.06 | 6.71 | |||
| YHGLRQSS | 2 | 5.84 | 1.13 | 6.6 | |||
Boman Index of T-cell and B-cell Epitopes
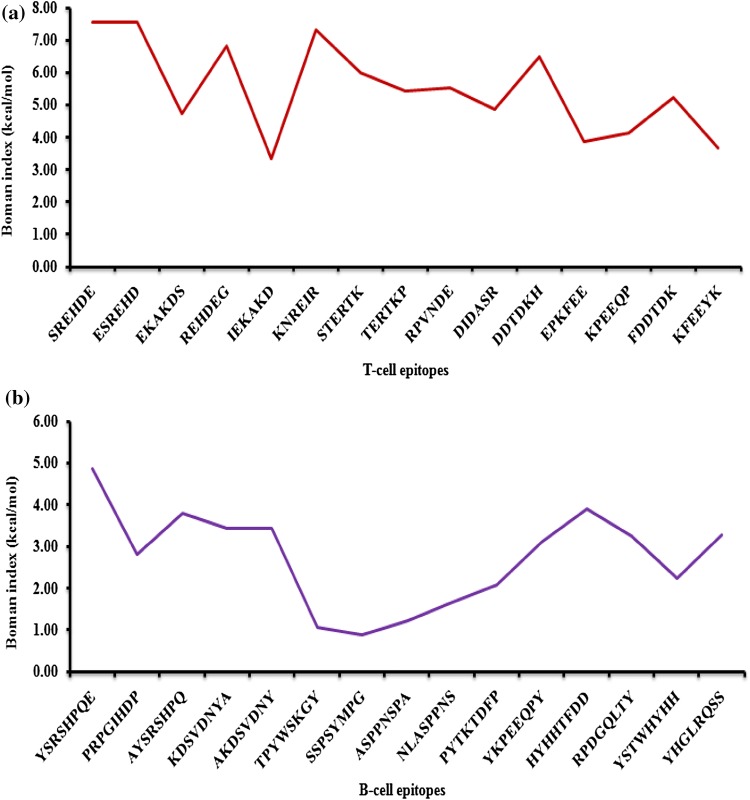
For each hexapeptide T-cell epitope, the Boman index value was determined (Fig. 2a). All the hexapeptide T-cell epitopes were found to possess Boman index > 2.48 kcal/mol which indicated that T-cell epitopes in the present study possessed the ability to interact effectively with other proteins. The values of Boman index of T-cell epitopes ranged from 3.31 (IEKAKD of VP1 protein, 134–139) to 7.55 (SREHDE, 245–250 and ESREHD, 244–249 of VP1 protein).
Fig. 2.
a Boman index of T-cell epitopes of Teschovirus A.b Boman index of B-cell epitopes of Teschovirus A
For each octapeptide B-cell epitopes, the Boman index was determined (Fig. 2b). All the B-cell epitopes of VP1 and VP2 protein had Boman index value greater than 2.48 kcal/mol and the values ranged from 3.12 (YKPEEQPY of VP2 protein, 184–191) to 4.85 (YSRSHPQE of VP1 protein, 237–244). In contrast, all the epitopes of VP3 possessed Boman index value < 2.48 kcal/mol, indicating that these epitopes would not effectively bind with the antibodies.
Amino Acid Composition of T-cell and B-cell Epitopes
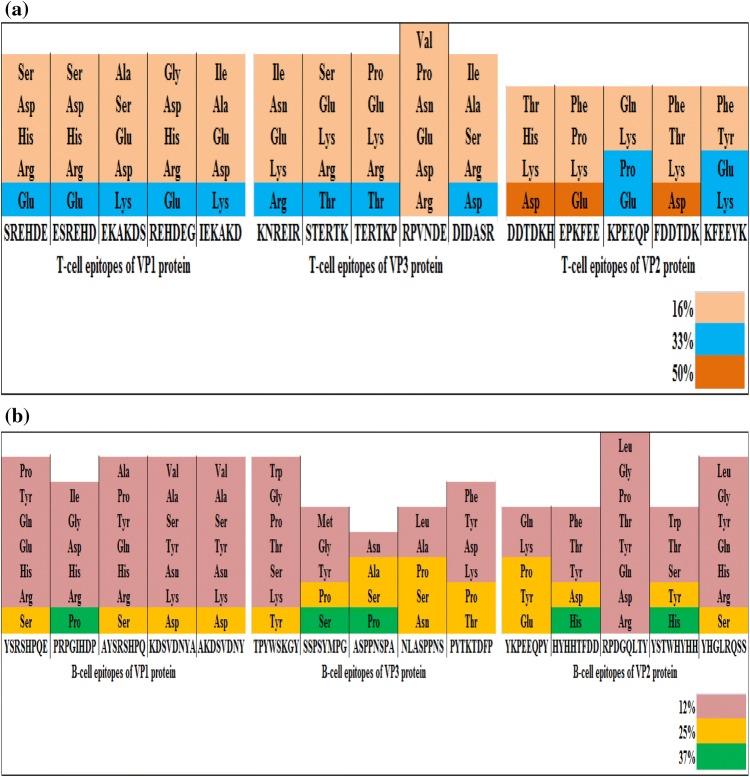
Amino acid composition of each hexapeptide T-cell epitope was analyzed (Fig. 3a). Most of the T-cell epitopes were composed of different amino acids and notably for every epitope, majority of amino acids constituted 16% of the total composition, while a few amino acids constituted 33% or 50% of the total composition. All the five epitopes of VP1 protein contained amino acids at 16% and 33% level respectively, while in case of VP3 protein, the only epitope RPVNDE comprised of six different amino acids (16%). However, in case of VP2 protein, the epitopes i.e. DDTDKH, EPKFEE and FDDTDK were found to contain some amino acids up to 50% level.
Fig. 3.
a Composition of amino acids of each T-cell epitopes of Teschovirus A.b Composition of amino acids of each B-cell epitopes of Teschovirus A
Amino acid composition of each octapeptide B-cell epitope was determined (Fig. 3b). All the epitopes were found to have different composition of amino acids. For individual epitope, majority of amino acids consisted 12% of the total composition. Four epitopes viz, SSPSYMPG and ASPPNSPA of VP3 protein and HYHHTFDD and YSTWHYHH of VP2 protein contained amino acids that occurred at 12%, 25% or 37% level. The epitope, PRPGIHDP of VP1 protein comprised of amino acids that appeared at 12% and 25% level, while the only epitope, RPDGQLTY of VP2 protein was made up of amino acids occurring at 12% level. The remaining epitopes were found to contain amino acids occurring at 12% and 25% level respectively.
Discussion
No peptide vaccine has yet been designed on the basis of immunoinformatics approach for curing Teschovirus encephalomyelitis caused by Teschovirus A in swine and wild boars. In the present study, both potential T-cell and B-cell epitopes from the capsid proteins of Teschovirus A were predicted in order to design peptide vaccines. Individual epitope was analyzed for different parameters using various bioinformatics tools. As various criteria are required to be satisfied by the epitopes for vaccine formulation, only a few promising epitopes of each protein were identified and analyzed for developing multiple-epitope loaded peptide vaccines.
Kaku et al. (2007) reported that in the strain of PTV-1 serotype of Teschovirus A responsible for Talfan disease in swine and wild boars, antigenic sites of the capsid proteins (VP1, VP3 and VP2) were determined by a process based on epitope mapping. From their study, two sites, i.e., GH loop and C-terminal end of VP1 protein were estimated to be antigenic in nature while the antigenic site of VP2 protein was identified to be EF loop (Kaku et al. 2007).
Schmallenberg virus (SBV) has been known to cause infectious diseases in cattle, buffaloes, goats, sheep, etc. Infected animals show clinical symptoms such as diarrhoea, fever as well as low milk production. A study reported that potential T-cell and B-cell epitopes identified from surface glycoproteins (Gn and Gc) of M polyprotein of SBV could aid in the development of peptide vaccine for generating immunogenic response against the virus. The potential T-cell epitopes identified were YMYNKYFKL, SECCVKDDI and IVYVFIPIF and the potential B-cell epitope was QQQACSS (Osman et al. 2016).
Crimean-Congo haemorrhagic fever (CCHF) virus affects livestock animals such as cattle, goats and sheep and causes severe haemorrhagic fever, photophobia and dizziness. A study suggested the prediction of probable T-cell and B-cell epitopes from CCHF viral proteins could be used for formulating epitope-based peptide vaccine for preventing the infection. The outcome of the prediction was a potential T-cell epitope, SVMDLSQMY and a B-cell epitope, LGVEDASESKL from the envelope polyprotein of the virus (Sahrawat and Checkervarty 2019).
Newcastle disease caused by Newcastle disease virus (NDV) occurs in birds. The disease is spread by ingestion or inhalation. In order to design peptide vaccine for preventing this viral infection, a study was carried out to determine the potential T-cell and B-cell epitopes from fusion protein of NDV using bioinformatics tools. From the study, four T-cell epitopes, namely NTSACMYSK, NYGEAVSLI, VAVGKMQQF and YLTELTTVF were identified to be potential use for raising immunity against the virus (Badawi et al. 2016).
Foot and mouth disease (FMD), caused by FMD virus, has long been known to affect the survival rate in livestock. This virus, assigned to the genus Aphthovirus, had severely affected the economy of farmers. In one study, the potential T-cell and B-cell epitopes were identified using immunoinformatics approach in order to design peptide vaccine for developing immunity against the virus. From the study, 18 T-cell and 5 B-cell epitopes were predicted to possess the capability to raise immunity in the host against the virus (Raza et al. 2019).
Conclusion
The current study suggests the possibility of developing peptide vaccines for treatment of Teschovirus encephalomyelitis caused by Teschovirus A. Among the 15 T-cell epitopes of the viral capsid proteins, five epitopes viz, KPEEQP, IEKAKD, KFEEYK, RPVNDE and DIDASR were determined based on high antigenicity score. Further, RPVNDE and KPEEQP with high immunogenicity score, hydrophilicity score and Boman index were identified. Further we could infer that the negatively charged epitope, KPEEQP may not bind accurately with the negatively charged virion and cannot be suggested for formulating vaccines. The epitope RPVNDE having positive charge holds the potential as a T-cell epitope that can be used for epitope-based vaccine development. All the six different amino acids of the epitope occurred equally (16%) and valine is the only aliphatic amino acid.
Out of 15 B-cell epitopes of capsid proteins in the present study, five epitopes i.e. ASPPNSPA, HYHHTFDD, YHGLRQSS, AYSRSHPQ and YSTWHYHH were identified with high antigenicity scores. Further, the epitopes AYSRSHPQ and HYHHTFDD had high immunogenicity and hydrophilicity scores with high Boman index. The epitope AYSRSHPQ had a positive total net charge and hence, this epitope could be used for vaccine formulation, whereas the epitope HYHHTFDD with net negative charge could be excluded. Among the eight amino acids of AYSRSHPQ, only serine occurred with 25% of total composition while the remaining amino acids occurred at 12% level and the epitope also comprised of only one aliphatic amino acid i.e. alanine.
Pharmaceutical industries could develop peptide vaccines against Teschovirus A by combining the identified epitopes viz. RPVNDE and AYSRSHPQ together with signal peptides of both T-cells and B-cells or these epitopes could be incorporated into virus like particle (vlp) for use as subunit vaccine to combat Teschovirus encephalomyelitis in swine and wild boars. However, the designed vaccine must undergo extensive trials to ensure its efficacy and safety.
Acknowledgements
The authors are grateful to Assam University, Silchar, Assam for providing necessary facilities to carry out this research work.
Compliance with Ethical Standards
Conflicts of interest
Authors declare no conflict of interest in the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alexandersen S, Knowles NJ, et al. Picornaviruses. Dis Swine. 2019 doi: 10.1002/9781119350927.ch40. [DOI] [Google Scholar]
- Backert L, Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015;7(1):119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi MM, Fadl Alla A, et al. Immunoinformatics predication and in silico modeling of epitope-based peptide vaccine against virulent Newcastle disease viruses. Am J Infect Dis Microbiol. 2016;4(3):61–71. [Google Scholar]
- Bangari DS, Pogranichniy RM, et al. Genotyping of Porcine teschovirus from nervous tissue of pigs with and without polioencephalomyelitis in Indiana. J Vet Diagn Invest. 2010;22(4):594–597. doi: 10.1177/104063871002200415. [DOI] [PubMed] [Google Scholar]
- Cano-Gómez C, Jiménez-Clavero M. Complete coding genome sequence of a putative novel Teschovirus serotype 12 strain. Genome Announc. 2016;4(2):e00107–e00116. doi: 10.1128/genomeA.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gómez C, Fernández-Pinero J, et al. Characterization of PTV-12, a newly described porcine teschovirus serotype: in vivo infection and cross-protection studies. J Gen Virol. 2017;98(7):1636–1645. doi: 10.1099/jgv.0.000822. [DOI] [PubMed] [Google Scholar]
- Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2):S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S-C, Hu S-C, et al. The role of porcine teschovirus in causing diseases in endemically infected pigs. Vet Microbiol. 2012;161(1–2):88–95. doi: 10.1016/j.vetmic.2012.07.031. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47(1):251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Flower DR. Harnessing bioinformatics to discover new vaccines. Drug Discov Today. 2007;12(9–10):389–395. doi: 10.1016/j.drudis.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Deng MY, Millien M, et al. Diagnosis of Porcine teschovirus encephalomyelitis in the Republic of Haiti. J Vet Diagn Invest. 2012;24(4):671–678. doi: 10.1177/1040638712445769. [DOI] [PubMed] [Google Scholar]
- Doherty M, Todd D, et al. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J Gen Virol. 1999;80(8):1929–1941. doi: 10.1099/0022-1317-80-8-1929. [DOI] [PubMed] [Google Scholar]
- Edington N, Christofinis G, et al. Pathogenicity of Talfan and Konratice strains of Teschen virus in gnotobiotic pigs. J Comp Pathol. 1972;82(4):393–399. doi: 10.1016/0021-9975(72)90038-2. [DOI] [PubMed] [Google Scholar]
- Ellis RW. New technologies for making vaccines. Vaccine. 1999;17(13–14):1596–1604. doi: 10.1016/s0264-410x(98)00416-2. [DOI] [PubMed] [Google Scholar]
- Florea L, Halldorsson B et al (2003). Epitope prediction algorithms for peptide-based vaccine design. Computational Systems Bioinformatics. CSB2003. Proceedings of the 2003 IEEE Bioinformatics Conference. CSB2003, IEEE [PubMed]
- Harrison G, Shakes T, et al. Duration of immunity, efficacy and safety in sheep of a recombinant Taenia ovis vaccine formulated with saponin or selected adjuvants. Vet Immunol Immunopathol. 1999;70(3–4):161–172. doi: 10.1016/s0165-2427(99)00039-2. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci. 1981;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M, Hickle L, et al. Vaccine development using recombinant DNA technology. Counc Agric Sci Technol. 2008;38:1–11. [Google Scholar]
- Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8(10):1041. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- Kaku Y, Murakami Y, et al. Antigenic properties of porcine teschovirus 1 (PTV-1) Talfan strain and molecular strategy for serotyping of PTVs. Adv Virol. 2007;152(5):929–940. doi: 10.1007/s00705-006-0908-7. [DOI] [PubMed] [Google Scholar]
- Knowles N. Manual of standards for diagnostic tests and vaccines. Paris: Office International des Epizoties; 2008. Teschovirus encephalomyelitis (previously enterovirus encephalomyelitis or Teschen/Talfan disease) pp. 1146–1152. [Google Scholar]
- Kolaskar A, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276(1–2):172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- Kouba V. Teschen disease (Teschovirus encephalomyelitis) eradication in Czechoslovakia: a historical report. Vet Med. 2009;54(11):550–560. [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, Reche PA. Prediction of MHC-peptide binding: a systematic and comprehensive overview. Curr Pharm Des. 2009;15(28):3209–3220. doi: 10.2174/138161209789105162. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhao Y, et al. A multiplex RT-PCR for rapid and simultaneous detection of porcine teschovirus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. J Virol Methods. 2011;172(1–2):88–92. doi: 10.1016/j.jviromet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Long J, Koestner A, et al. Infectivity of three porcine polioencephalomyelitis viruses for germfree and pathogen-free pigs. Am J Vet Res. 1966;27(116):274. [PubMed] [Google Scholar]
- Manuelidis EE, Sprinz H, et al. Pathology of Teschen disease (virus encephalomyelitis of swine) Am J Pathol. 1954;30(3):567. [PMC free article] [PubMed] [Google Scholar]
- Osman M, Abdelrazig N, et al. Silico modeling and identification of novel epitopes-based vaccine of M polyprotein (Gn/Gc) against Schmallenberg virus for ruminants. Munich: Grin Publishing; 2016. [Google Scholar]
- Pisitkun T, Hoffert JD, et al. NHLBI-AbDesigner: an online tool for design of peptide-directed antibodies. Am J Physiol-Cell Physiol. 2011;302:C154–C164. doi: 10.1152/ajpcell.00325.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodělalová J. The survey of porcine teschoviruses, sapeloviruses and enteroviruses B infecting domestic pigs and wild boars in the Czech Republic between 2005 and 2011. Infect Genet Evol. 2012;12(7):1447–1451. doi: 10.1016/j.meegid.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Purcell AW, McCluskey J, et al. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6(5):404. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Raza S, Siddique K, et al. In silico analysis of four structural proteins of aphthovirus serotypes revealed significant B and T cell epitopes. Microb Pathog. 2019;128:254–262. doi: 10.1016/j.micpath.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Rini JM, Schulze-Gahmen U, et al. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255(5047):959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- Sahrawat TR, Checkervarty AK. Identification of epitopes for vaccine design against Crimean-Congo Haemorrhagic fever virus (CCHFV): an immuno-informatics approach. JRAR-Int J Res Anal Rev. 2019;6:807–811. [Google Scholar]
- Salles MW, Scholes SF, et al. Porcine teschovirus polioencephalomyelitis in western Canada. J Vet Diagn Invest. 2011;23(2):367–373. doi: 10.1177/104063871102300231. [DOI] [PubMed] [Google Scholar]
- Smith TJ, Baker T. Picornaviruses: epitopes, canyons, and pockets. Adv Virus Res. 1999;52:1–23. doi: 10.1016/s0065-3527(08)60297-3. [DOI] [PubMed] [Google Scholar]
- Trefny L. Massive illness of swine in Teschen area. Zveroleki Obzori. 1930;23:235–236. [Google Scholar]
- Valitutti S, Müller S, et al. Serial triggering of many T-cell receptors by a few peptide–MHC complexes. Nature. 1995;375(6527):148. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- Ventura A, Gonzalez W, et al. Virus and antibody diagnostics for swine samples of the Dominican Republic collected in regions near the border to Haiti. ISRN Virol. 2012;2013:7. [Google Scholar]
- Wang B, Tian Z-J, et al. Isolation of serotype 2 porcine teschovirus in China: evidence of natural recombination. Vet Microbiol. 2010;146(1–2):138–143. doi: 10.1016/j.vetmic.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76(2):263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Zell R, Dauber M, et al. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J Virol. 2001;75(4):1620–1631. doi: 10.1128/JVI.75.4.1620-1631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R, Seitz S, et al. Linkage map of protein–protein interactions of porcine teschovirus. J Gen Virol. 2005;86(10):2763–2768. doi: 10.1099/vir.0.81144-0. [DOI] [PubMed] [Google Scholar]
- Zhang C-F, Cui S-J, et al. Isolation and characterization of the first Chinese strain of porcine Teschovirus-8. J Virol Methods. 2010;167(2):208–213. doi: 10.1016/j.jviromet.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Zúñiga-Pflücker JC. T-cell development made simple. Nat Rev Immunol. 2004;4(1):67. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]