Abstract
Advent of quantitative polymerase chain reaction and its variants have enabled identification and quantification of seven known Eimeria species of poultry in biological samples. Attempts were made in the present study to identify and quantify three important pathogenic Eimeria species responsible for intestinal coccidiosis in domestic farmed chicken, E. necatrix, E. acervulina and E. maxima in droppings collected from thirty one poultry farms of North Indian states of Haryana, Punjab, Uttar Pradesh and Uttarakhand. The study included broiler, layer and backyard rearing units. Overall occurrence of E. necatrix, E. maxima and E. acervulina was 64.5%. E. necatrix was detected in 55% (11/20) broiler farms, 66.7% (4/6) layer farms and 100% (5/5) backyard rearing units studied. Thus, occurrence of E. necatrix was detected in 64.5% (20/31) farms studied. E. maxima and E. acervulina were detected in droppings of 65% (13/20) broiler farms, 66.7% (4/6) layer farms and 60% (3/5) back yard rearing units. Genome counts of each Eimeria species revealed maximum parasite load of E. necatrix followed by E. acervulina in broiler farms and least in layer farms. The mean parasite load (genome) copies for these parasite species were intermediate for backyard units while E. maxima had the lowest number of genome copies in droppings. Mean E. maxima counts were highest in boiler farms, while it was similar for layer and back yard units. However, statistically no significant differences were observed for parasite load existing either between the broiler, layer or back yard units or between the genome counts of E. necatrix, E. acervulina or E. maxima.
Keywords: Eimeria, Species, North India, Quantitative PCR, Detection
Introduction
Among the seven species of Eimeria infecting domestic chicken five species are of great economic importance due to their relatively high pathogenicity (Shirley et al. 1983). Among the five species, E. tenella is the most pathogenic species and is localized within the caecum and causes fatal caecal coccidiosis (Soulsby 2012). E. necatrix, E. acervulina and E. maxima cause intestinal coccidiosis. E. necatrix is the second most pathogenic species after E. tenella and produces a more chronic form of the disease (Soulsby 2012). E. acervulina and E. maxima are moderately pathogenic species and hence cause high morbidity but low mortality. E. acervulina, E. necatrix and E. maxima are most common species of Eimeria responsible for intestinal coccidiosis prevalent in North India (Prakshbabu et al. 2017). E. brunetti, the causative of rectal coccidiosis is highly pathogenic but has a low prevalence in India (Prakshbabu et al. 2017). In farms of South India, highly pathogenic E. necatrix was more prevalent as compared to moderately pathogenic E. acervulina (Prakshbabu et al. 2017). The variation in population dynamics of high, moderate or low pathogenic species in farms of a geographical area will region do impact the farm economics in several ways. This epidemiological factor may determine the possibility of future coccidiosis outbreaks and prophylactic measures to be undertaken. Moreover, higher incidence of high or moderately pathogenic Eimeria species in different farming systems may be indicative development of resistance against anti-coccidial chemotherapy. Hence, quantification of each individual Eimeria species present in chicken faecal droppings is necessary for understanding disease outbreaks and risk factors associated with quantitative presence of virulent species.
Identification of various Eimeria species of chicken in faecal droppings had been carried out based on traditional techniques like morphometry (based on shape and determination of size) and sporulation time based on compound microscope. An online morphometry database software based on size, morphology and texture/granularity of chicken Eimeria species named COCCIMORPH has been developed. The microphotographs of oocysts can be uploaded onto this software and the software predicts the species of oocyst based on characteristics like size and granularity. The software has been used for identification of Eimeria species in droppings of chicken collected from various farms of North India, but its efficiency in correctly identifying the species is low when compared to PCR based diagnostic methods (Kumar et al. 2014). Quantification of various Eimeria species from faecal droppings based on traditional morphometry or sporulation time would require more time and would be tedious for the technical person involved in diagnosis. Moreover the traditional techniques would be less accurate in correct identification and quantification of species.
As of today, quantitative PCR is the most useful technique since it can both identify and quantify Eimeria species in biological samples like faecal droppings or tissues (Blake et al. 2006; Kawahara et al. 2008; Morgan et al. 2009; Vrba et al. 2010; Kundu et al. 2017).
This study was performed to identify and enumerate the genome copies of E. necatrix, E. acervulina and E. maxima with quantitative realtime PCR in faecal droppings from different farming systems (broiler, layer and back yard rearing units) from four North India states of Uttarpradesh, Uttarakhand, Punjab and Haryana.
Materials and methods
Collection of poultry droppings and purification of oocysts
Poultry droppings were collected from 31 commercial farms and backyard units from Uttar Pradesh, Uttarakhand, Punjab and Haryana. There were twenty one broiler farms, six layer farms and five back yard rearing units. Birds in each of these farms under study were in apparently good health and none of the farms had experienced coccidiosis outbreaks during collection. However, as per history collected from the owners, most of the broiler farms had suffered outbreaks of coccidiosis. None of the backyard units or layer farms had experienced outbreaks of coccidiosis in the past. Poultry droppings were collected, oocyst count per gram of droppings were estimated and further processed for separation of oocyts as described by Kumar et al. (2014).
Extraction of DNA from purified oocysts
Oocysts harvested from faecal droppings were allowed to sporulate in 2% potassium dichromate. Sporulated oocysts were thoroughly washed, centrifuged in a 1.5 ml microcentrifuge tube at 5000 g for 5 min. Supernatant water was discarded and equal volume of autoclaved glass beads of approximately 0.5 mm diameter (Sigma-Aldrich, USA) were added to the oocyst pellet. Lysis buffer (ASL buffer provided with QIAmp DNA stool mini kit, Qiagen, USA), 50 μl was added to the oocyst pellet and glass bead mixture and the mixure was vortexed on a Spinix vortexer (Tarsons, India) for 3 min to disrupt the oocysts.. The vortexing time was standardised by checking the presence of disrupted oocyts, sporocysts and sporozoites under 400X magnification of microscope. This method achieved in disrupting more than 90% of the oocysts. ASL buffer was further added to the disrupted oocyst pellet, vortexed thoroughly and incubated at 70 °C for 5 min. Further steps for extraction of DNA were carried out as per the instructions provided in the manual provided with the Qiagen Stool DNA mini kit. DNA extracted from oocysts was stored at − 20 °C for further use.
Quantitative real time PCR (qPCR) for identification and quantification of E. necatrix, E. acervulina and E. maxima
Primers used for identification and quantification of E. necatrix, E. acervulina and E. maxima were same as described earlier by Vrba et al. (2010). The details of primers and annealing temperatures are provided in Table 1. Pure DNA of each Eimeria species provided by Prof. Damer P. Blake of Royal Veterinary College, London were used as template for initial PCR amplification and standardization of PCR.
Table 1.
Primer pairs used for nested and quantitative PCR of Eimeria necatrix, E. acervulina and E. maxima along with estimated linear equation, correlation co-efficient (R2) and efficiency of quantitative PCRs targeting three different Eimeria species
| Primer name | Species | Primer name and sequence (Vrba et al. 2010) | Product size (bp) | Ta (°C) | Linear equation | R2 value | PCR efficiency (%) |
|---|---|---|---|---|---|---|---|
| NECF | E. necatrix | AACGCCGGTATGCCTCGTCG | 134 | 60 | Ct = (42.245–3.210) log copy No | 0.997 | 104.89 |
| NECR | GTACTGGTGCCAACGGAGA | ||||||
| ACEF | E. acervulina | GCAGTCCGATGAAAGGTATTTG | 103 | 60 | Ct = (42.596–3.586) log copy No | 0.997 | 105.58 |
| ACER | GAAGCGAAATGTTAGGCCATCT | ||||||
| MAXF | E. maxima | TCGTTGCATTCGACAGATTC | 138 | 60 | Ct = (37.635–3.193 log copy No | 0.997 | 90.05 |
| MAXR | TAGCGACTGCTCAAGGGTTT |
Ta refers to annealing temperature used for the quantitative PCR; R2 value refers to the correlation co-efficient; No. refers to number
In brief, PCR reactions were performed in 25 μl reaction with 12.5 μl master mix (Takara GC Amp PCR master mix), 02 μl DNA, 01 μl each of forward and reverse primers (10 pmol μl−1) and 8.5 μl nuclease free water. Thermal cycling condition were carried out with initial denaturation at 98 °C for 10 s, 35 cycles of denaturation 98 °C for 10 s, annealing along with extension at 60 °C for 30 s. PCR products were used for ligation into TA vector. Details of primer sequences have been provided in Table 1. The recombinant vector were coned into Escherichia coli and cultured. Recombinant plamids from the positive E. coli clones were extracted and used as standards.
Recombinant plasmids were quantified using Nanodrop spectrophotometer (ND-1000, Thermo Scientific). Copy numbers of recombinant plasmids were calculated from the concentration and size of the plasmids. Ten fold serial dilutions of recombinant plasmids in nuclease free water for all three species were prepared so as to obtain a range containing 106 plasmids per µl to 101 plasmids per µl. These were used as standards in qPCR for preparing the standard curves for absolute quantification during species specific qPCR studies.
For amplification of DNA extracted from faecal samples each qPCR reaction mixture comprised of 10 µl of qPCR master mix with Eva Green dye (BioRad, USA), 03 µl of genomic DNA, 01 µl each of forward and reverse primers as per standardized concentration and nuclease free water to make the volume to 20 µl. Each concentration of plasmids in volume of 01 μl was used as template to prepare standard curve. Forward and reverse primers at concentration of 400 nM, 300 nM and 150 nM each were used for qPCR amplification of E. necatrix, E. maxima and E. acervulina respectively. Amplification for qPCR were initial denaturation at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 05 s and annealing with extension at 60 °C for 10 s, followed by melt curve analysis step at temperature range of 65–95 °C (0.5 °C increment), 5 s per step.
Calculation of genome copy numbers from standard curve of Ct values
The Ct values obtained from each plasmid concentration (logarithm of plasmid copy nymbers) in the qPCR system were plotted. Values of slope (m) of the linear regression curve and square of correlation co-efficient (r2) was determined. The efficiency of the q-PCR was determined from the formula − 1 + 10(−1/slope). The plot of Ct values against the logarithm (log) of plasmid copy numbers provided a standard curve for each Eimeria species. The linear regression equation for each Eimeria species was obtained as Y = mX + c; where Y = Ct values (obtained from the real time data generated by the PCR machine), m = slope of the curve, c = y-intercept and X = log copy numbers (unknown). This equation was used to interpolate the Ct values obtained from DNA of field samples with unknown concentration of genomes of E. necatrix, E. maxima or E. acervulina.
Statistics
Analysis of variance (ANOVA) test supplemented with Tukey’s post hoc was used for using evaluation of statistically significant difference between number of parasite genomes of each of the three species, E. tenella, E. necatrix, E. maxima and E. acervulina estimated by quantitative PCR between each farm types (broiler, layer and backyard units) (IBM SPSS Statistics 20.0, IBM Corp. 2011).
Results
Estimation of efficiency of quantitative real time PCR
The Ct values obtained during standardization of qPCR were plotted against the logarithm of plasmid copy numbers for each species. These analyses provided the standard linear equation of Ct values and log copy number which was used for the quantification of genome copy numbers. Efficiency and co-efficient of correlation (R2) values of qPCR estimated for E. necatrix, E. maxima and E. acervulina were 104.8%/0.997, 105.58%/0.997 and 90.05%/0.997 respectively (Table 1, Fig. 1). Linear equations derived from the standard curve were, Ct = (42.245–3.210) log copy number for E. necatrix, Ct = (42.245–3.210) log copy number for E. acervulina and Ct = (37.635–3.193 log copy number for E. maxima (Table 1).
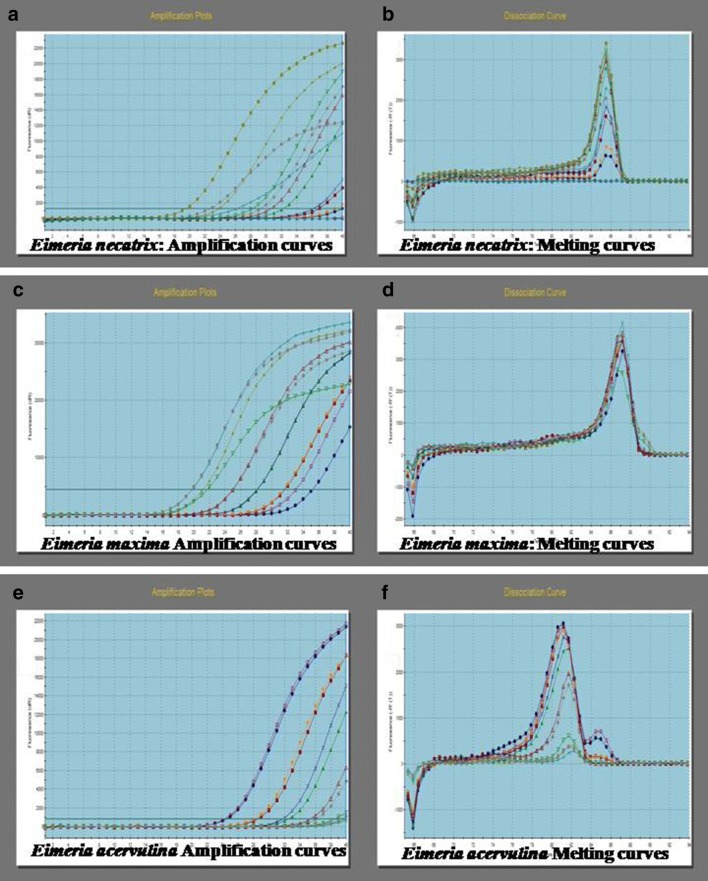
Fig. 1.
Real time amplification of E. necatrix, E.maxima and E. acervulina using different concentrations of cloned plamids (106/μl to 101/μl) as template. Amplification and melting curves have been depicted in the figure: a, b Amplification and melt curves of E. necatrix; c, d Amplification and melt curves of E. maxima; e, f Amplification and melt curves of E. acervulina
Detection and quantification of E. necatrix, E. maxima and E. acervulina with quantitative PCR
The detection and quantification of three species of Eimeria (E. necatrix, E. maxima and E. acervulina) by quantitative PCR are provided in Table 2. E. necatrix was detected in 55% (11/20) broiler farms, 66.7% (4/6) layer farms and 100% (5/5) backyard rearing units studied. Thus, occurrence of E. necatrix was detected in 64.5% (20/31) farms studied. E. maxima and E. acervulina were detected in facial droppings of 65% (13/20) broiler farms, 66.7% (4/6) layer farms and 60% (3/5) back yard rearing units. Thus, overall occurrence of E. maxima and E. acervulina was similar to E. necatrix (64.5%).
Table 2.
Detection and quantification of genomes of Eimerianecatrix, E. maxima and E. acervulinain oocyst DNA extracted from faecal droppings
| State | Sample number | Farm type | OPG | Genome (number of copies) | ||
|---|---|---|---|---|---|---|
| E. necatrix | E. maxima | E. acervulina | ||||
| Uttar Pradesh | 1 | Backyard | 53,000 | 233,707 | x | x |
| 2 | Broiler | 37,330 | x | x | 1219 | |
| 3 | Broiler | 40,929 | 468,655 | 149 | 44,159 | |
| 4 | Broiler | 1324 | x | 28 | 2551 | |
| 5 | Broiler | 1,08,491 | 174,159 | 112 | 70,565 | |
| 6 | Broiler | 1662 | x | x | x | |
| 7 | Broiler | 37,298 | 23,539 | x | 55,286 | |
| 8 | Broiler | 82,386 | x | 2742 | 1863 | |
| 9 | Layer | 1170 | 5409 | 32 | x | |
| 10 | Layer | 950 | x | 9 | 999 | |
| Uttarakhand | 1 | Backyard | 15,818 | 22,547 | 50 | x |
| 2 | Backyard | 7982 | 25,471 | 16 | 8983 | |
| 3 | Backyard | 41,600 | 1046 | 14 | 4376 | |
| 4 | Broiler | 8437 | 486 | 13 | 11,466 | |
| 5 | Broiler | 3,34,082 | x | 75 | x | |
| 6 | Broiler | 2310 | 624 | 11 | 1100 | |
| 7 | Broiler | 19,796 | x | 522 | 19,663 | |
| 8 | Broiler | 23,485 | 4118 | 97 | x | |
| 9 | Broiler | 4700 | 8685 | x | x | |
| Haryana | 1 | Broiler | 4,04,473 | 56,880 | 27 | 84,139 |
| 2 | Broiler breeder | 48,110 | 11,137 | 31 | 1196 | |
| 3 | Layer | 10,710 | 6854 | 18 | 1702 | |
| 4 | Layer | 4924 | x | x | x | |
| 5 | Layer | 5974 | x | x | x | |
| 6 | Layer | 1024 | 3227 | 49 | 793 | |
| Punjab | 1 | Backyard | 76,817 | 577 | x | 248 |
| 2 | Layer | 7355 | 58,536 | x | 1032 | |
| 3 | Broiler | 32,060 | 726 | 41 | 12,448 | |
| 4 | Broiler | 21,050 | 1374 | 33 | 2303 | |
| 5 | Broiler | 52,000 | x | x | x | |
| 6 | Broiler | 1441 | x | x | x | |
OPG refers to oocysts per gram of dropping; Symbol “x” was used to denote cases where no genome copies either of E. necatrix or E. tenella or E. maxima were detected by quantitative PCR
Mean genome copies (mean ± standard error) of E. necatrix in broiler farms was calculated as 71,577.818 ± 42,619.99, in layer farms was 18,506.5 ± 13,369.9 and in back yard rearing units was 56,669.6 ± 44,564 (Table 3). Mean genome copies (mean ± standard error) of E. acervulina in broiler farms was calculated as 23,689.076 ± 8189.49, in layer farms was 1131.5 ± 197.38 and in back yard rearing units was 4535.66 ± 2522. Mean genome copies (mean ± standard error) of E. maxima in broiler farms was calculated as 298.54 ± 207.05, in layer farms was 27 ± 8.72 and in back yard rearing units was 26.66 ± 11.68.
Table 3.
Mean genome copies of E. necatrix, E. maxima and E. acervulina in different farm types (broiler, layer and backyard rearing units)
| Species | Genome copy numbers in different farm types (mean ± SE) | ||
|---|---|---|---|
| Broiler farm | Layer farm | Backyard unit | |
| E. necatrix | 71,577.818 ± 42,619.99aA | 18,506.5 ± 13,369.9aB | 56,669.6 ± 44,564aC |
| E. maxima | 298.54 ± 207.05bA | 27 ± 8.72bB | 26.66 ± 11.68bC |
| E. acervulina | 23,689.076 ± 8189.49cA | 1131.5 ± 197.38cB | 4535.66 ± 2522cC |
No significant difference of genome copy numbers between species within different farm types was observed. Similarly no significant difference in genome copy numbers for same Eimeria species between three farm types was observed
Mean ± SE values, with similar superscripts a or b or c indicate no significant difference of the parasite load (genome count) of one species between three different farm types, using one way ANOVA (at p < 0.05)
Mean ± SE values, with similar superscripts A or B or C indicate no significant difference of the parasite load (genome count) between three different species within the same farm type, using one way ANOVA (at p < 0.05)
No significant difference between the genome copy numbers of E. necatrix, E. acervulina and E. maxima in broiler farms, layer farms and back yard units was observed (Table 3). No significant difference in genome copy numbers of each Eimeria species between the different farm types was observed (Table 3).
Discussion
Eimeria species of domestic cannot be differentiated on the basis of morphology alone, with certainty. PCR based techniques targeting various genes or DNA fragments have been adopted for identification of different species existing in the poultry farms. Correct identification of Eimeria species in a mixed population of oocysts and their quantification in poultry litter or faecal droppings is important. Thus, use of quantitative real time PCR for quantification of the parasite genome in the DNA from mixed oocyst population is the method of choice. An attempt was made to quantify genomes of E. acervulina, E. maxima and E. necatrix in DNA sample extracted from oocysts. Quantitative PCR has been used in chicken coccidiosis primarily to monitor the development of parasitic stages in the intestine of the host (Blake et al. 2008; Kundu et al. 2017; Nolan et al. 2015). Kawahara et al. (2008) and Morgan et al. (2009) developed qPCR technique targeting ITS-1 and ITS-2 region of parasite DNA, as a tool to study molecular epidemiology. However ITS-1 and ITS-2 fragments are multiple nucleic acid regions and hence are not suitable for exact quantification of Eimeria genomes (Morgan et al. 2009). Vrba et al. (2010) used primers targeting sequence characterized amplified region (SCAR) and are single copy nucleic acid fragments. Hence, primers targeting SCARs are most suitable for quantification of species. The same primers were used in this study for quantification of genomes of E. necatrix, E. maxima and E. necatrix in faecal dropping from chicken farms.
Kumar et al. (2014) reported PCR based identification of various species of Eimeria is improved when DNA is extracted from concentrated oocysts rather than the faecal samples directly. Same protocol was used for extraction of DNA from purified oocysts and used for quantification of genomes of three Eimeria species.
Parasite burden of E. necatrix was highest in broiler farms (71,577.818 ± 42,619.99) followed by back yard units (56,669.6 ± 44,564) and layer farms (18,506.5 ± 13,369.9). Genome copies of E. acervulina in highest in broiler farms (23,689.076 ± 8189.49) followed by backyard units (4535.66 ± 2522) and layer units (1131.5 ± 197.38). Similarly, quantity of E. maxima oocysts were more in broiler farms (298.54 ± 207.05) as compared to layer farms (27 ± 8.72) and back yard units (26.66 ± 11.68). The oocyst or parasite load of E. maxima in faecal droppings of layer and back yard units were similar. Though no statistically significant differences were observed between these quantities estimated in faecal droppings of three Eimeria species in the three different farm types (Table 3), still it can be said that oocyst count of E. necatrix was highest in all the three types of farms compared to E. acervulina followed by E. maxima. Parasitic load of either E. necatrix or E. acervulina or E. maxima was highest in broiler farms as compared to backyard and layer farms. In the Indian scenario, particularly in North India layer chickens are reared in cages and have less exposure to the oocyst infection. So it is not surprising that the oocyst burden is the least for any Eimeria species recorded in layer farms in this study. Broiler chickens are reared on deep litter system with more exposure to oocysts while feeding or drinking and hence the chance of picking up infection is more. More birds are infected by the circulating oocysts which sporulate in litter after being excreted by previously infected birds. Backyard chickens are reared in a semi-intensive system, where they are on a partial free ranging system for a considerable part of the day and are housed only in the evening hours. These birds pick up oocysts shed from the infected counterparts in the flock. It may be assumed that the ground/ soil may not provide the suitable moisture and temperature conditions for oocyst sporulation compared to the litter bedding. Moreover, free ranging habits of birds area helps spread of oocysts over a large area, hence minimizing concentrations of infective oocysts within a limited area as observed in the broiler house litter. The lesser number of birds in the free range system of backyard farms results in lower stocking density and hence lower environmental oocyst burden resulting in low oocyst shedding than intensively reared broiler chicken.
Advantages of Q-PCR over the conventional and nPCR include single step amplification and quantification, no requirement of post amplification electrophoresis and visulisation of PCR product, thus saving on time and minimizing the chances of contamination during PCR (Kawahara et al. 2008). The only disadvantage being higher cost of reagents and the real time thermal cycler equipment. Quantitative PCR in the recent past are limited only to species identification (Kawahara et al. 2008; Morgan et al. 2009; Luu et al. 2013). However, application of quantitative PCR would have been more meaningful if the relative abundance of each species could be estimated (Morgan et al. 2009).This may be of immense importance for designing vaccination strategy, identification and proportionate evaluation of circulating vaccine and wild strains in farms or help identify the species more prone to development of anti-coccidial resistance.
Acknowledgements
Authors’ acknowledge the funding provided by BBSRC and DFID, UK for the CIDLID project titled “Anticoccidial vaccine development: the importance of genetic diversity and delivery strategy”under which the work was carried out. The facilities and support provided by ICAR and Director, IVRI are also acknowledged.
Funding
The work was carried out with the funding from BBSRC and DFID, UK under CIDLID project.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest among the authors of this manuscript.
Ethical standard
Experiments carried out in the manuscript are within the ethical standards. The work involved collection of faecal droppings from litter material from poultry sheds and no invasive procedure was involved. Hence no ethical permission was required in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Blake DP, Hesketh P, Archer A, Shirley MW, Smith AL. Eimeria maxima: the influence of host genotype on parasite reproduction as revealed by quantitative real-time PCR. Int J Parasitol. 2006;36:97–105. doi: 10.1016/j.ijpara.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Blake DP, Qin Z, Cai J, Smith AL. Development and validation of real-time polymerase chain reaction assays specific to four species of Eimeria. Avian Pathol. 2008;37:89–94. doi: 10.1080/03079450701802248. [DOI] [PubMed] [Google Scholar]
- IBM Corp. IBM SPSS statistics for Windows, Version 20.0. Armonk: IBM Corp; 2011. [Google Scholar]
- Kawahara F, Taira K, Nagai S, Onaga H, Onuma M, Nunoya T. Detection of five avian Eimeria species by species-specific real-time polymerase chain reaction assay. Avian Dis. 2008;52:652–656. doi: 10.1637/8351-050908-Reg.1. [DOI] [PubMed] [Google Scholar]
- Kumar S, Garg R, MoftahA CE, Macdonald SE, Chaudhary AS, Sparagano O, Banerjee PS, Kundu K, Tomley FM, Blake DP. An optimised protocol for molecular identification of Eimeria from chickens. Vet Parasitol. 2014;199:24–31. doi: 10.1016/j.vetpar.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu K, Garg R, Kumar S, Mandal M, Tomley FM, Blake DP, Banerjee PS. Humoral and cytokine response elicited during immunisation with recombinant Immune Mapped protein-1 (EtIMP-1) and oocysts of Eimeria tenella. Vet Parasitol. 2017;244C:44–52. doi: 10.1016/j.vetpar.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Luu L, Bettridge J, Christley RM, Melese K, Dessie T, Wigley P, Desta TT, Hanotte O, Kaiser P, Terfa ZG, Collins M, Lynch SE. Prevalence and molecular characterisation of Eimeria species in Ethiopian village chickens. BMC Vet Res. 2013;9:208. doi: 10.1186/1746-6148-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Morris GM, Wlodek BM, Byrnes R, Jenner M, Constantinoiu CC, Anderson GR, Lew-Tabor AE, Molloy JB, Gasser RB, Jorgensen WK. Real-time polymerase chain reaction (PCR) assays for the specific detection and quantification of seven Eimeria species that cause coccidiosis in chickens. Mol Cell Probes. 2009;23:83–89. doi: 10.1016/j.mcp.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Nolan MJ, Tomley FM, Kaiser P, Blake DP. Quantitative real-time PCR (qPCR) for Eimeria tenella replication—implications for experimental refinement and animal welfare. Parasitol Int. 2015;64:464–470. doi: 10.1016/j.parint.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakshbabu B, Thenmozhi V, Limon G, Kundu K, Kumar S, Garg R, Clark EL, Srinivasa Rao ASR, Raj DG, Raman M, Banerjee PS, Tomley FM, Guitian DP. Eimeria species occurrence varies between geographicregions and poultry production systems and may influence parasite genetic diversity. Vet Parasitol. 2017;233:62–72. doi: 10.1016/j.vetpar.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley MW, Jeffers TK, Long PL. Studies to determine the taxonomic status of Eimeria mitis, Tyzzer 1929 and E. mivati, Edgar and Seibold 1964. Parasitology. 1983;87:185–198. doi: 10.1017/S0031182000052550. [DOI] [PubMed] [Google Scholar]
- Soulsby EJL. Helminths, arthropods and protozoa of domesticated animals. 7. New Delhi: East-West press Pvt. Ltd.; 2012. pp. 729–737. [Google Scholar]
- Vrba V, Blake DP, Poplstein M. Quantitative real-time PCR assays for detection and quantification of all seven Eimeria species that infect the chicken. Vet Parasitol. 2010;174:183–190. doi: 10.1016/j.vetpar.2010.09.006. [DOI] [PubMed] [Google Scholar]