Abstract
Filoviruses such as Ebola virus continue to pose a substantial health risk to humans. Advances in the sequencing and functional characterization of both pathogen and host genomes have provided a wealth of knowledge to clinicians, epidemiologists and public health responders during outbreaks of high-consequence viral disease. Here, we describe how genomics has been historically used to investigate Ebola virus disease outbreaks and how new technologies allow for rapid, large-scale data generation at the point of care. We highlight how genomics extends beyond consensus-level sequencing of the virus to include intra-host viral transcriptomics and the characterization of host responses in acute and persistently infected patients. Similar genomics techniques can also be applied to the characterization of non-human primate animal models and to known natural reservoirs of filoviruses, and metagenomic sequencing can be the key to the discovery of novel filoviruses. Finally, we outline the importance of reverse genetics systems that can swiftly characterize filoviruses as soon as their genome sequences are available.
Subject terms: Ebola virus, Metagenomics, Marburg virus
Filoviruses such as Ebola virus pose a substantial health risk to humans. Advances in genomic technologies have enabled the rapid, large-scale generation of virus sequence data at the location of disease outbreaks and thus the use of reverse functional genomics to swiftly characterize the threat of, and treatment for, filovirus disease.
Introduction
Infections with viruses of the mononegaviral family Filoviridae (in particular, members of the genera Ebolavirus and Marburgvirus) are an increasing threat to mankind. Until recently, the infrequent spillover of these viruses into humans and the fact that spillover often occurred in remote locations, coupled with a limited knowledge of non-human reservoir hosts, the use of low-output genomic sequencing, and biosafety and biosecurity restrictions on filovirus research, contributed to the paucity of publicly available data on filovirus genome sequences1. In December 2013, complete genome sequences for only 29 ebolaviruses and 65 marburgviruses were available2 despite the fact that 35 outbreaks of natural filovirus disease had been recorded3.
Since December 2013, atypically extensive filovirus disease outbreaks, from 2013 to 2016 in Western Africa and from 2018 to present in the Democratic Republic of the Congo, have profoundly impacted public health systems. At least 13,675 fatalities from filovirus disease were reported between December 2013 and April 5, 2020 (refs4,5). By leveraging the continued development and improvement of next-generation sequencing technology, >800 complete filovirus genome sequences and over 2,000 draft genomes (that is, genomes with >80% coverage) across classified and unclassified filovirus family members have become available since 2013 (ref.2). Indeed, among high-consequence, Risk Group 4 viruses, the genomic diversity of filoviruses is arguably becoming the best characterized.
The impact and importance of genomics in pathogen characterization is routinely demonstrated, but the rapid prediction of, response to and mitigation of outbreaks requires more detailed genomic information than virus consensus-genome sequencing. Indeed, as predicted6, metagenomic sequencing has become a powerful tool for identifying novel viruses and, crucially, for predicting pathogen emergence7. Targeted or unbiased sequencing of individual clinical samples aids in the identification of outbreaks, the determination of outbreak aetiology and the definition of virus transmission chains by identifying chain-defining single nucleotide polymorphisms (SNPs). Furthermore, field transcriptomics improves our understanding of host responses to virus infection and will be important in deciphering the differences between asymptomatic and symptomatic disease states and in predicting whether patients with acute and chronic disease will survive8. Functional genomics is becoming the tool of choice for the rapid characterization of patient-specific viruses that have not been isolated in culture or that cannot be equitably shared among laboratories across borders9. Finally, the genomic analysis of patient-specific viruses also enables precision medicine by predicting the efficacy of available medical countermeasures (MCMs) against these individual viruses.
Here, we review how recent advances in genomic technologies have shaped past and current responses to outbreaks of Ebola virus disease (EVD), including insights into filovirus diversity and evolution. We emphasize the importance of accurate and rapid large-scale data generation and its implications for the development of MCMs and outbreak response. We also examine the phenomena of Ebola virus (EBOV) persistence in human hosts and provide an overview of recent genomic advances in threat characterization, vaccine development and immunotherapy. Although we focus primarily on EBOV, these practices can apply to all pathogenic filoviruses and other high-consequence viruses capable of sustaining human-to-human transmission.
Identifying filovirus reservoirs
Although the global distribution and diversity of filoviruses remains largely undefined, metagenomic sequencing is becoming a valuable tool for identifying filovirus reservoirs. Until 1989, disease outbreaks owing to infection by ebolaviruses (including EBOV, Sudan virus (SUDV) and marburgviruses (including Marburg virus (MARV) and Ravn virus (RAVV)) had only been recorded on the African continent (Fig. 1). As the natural reservoir hosts of all of these viruses remained unidentified, despite extensive ecological studies, filoviruses were thought to be African viruses1. This view changed after 1989, when Reston virus (RESTV; of the genus Ebolavirus) was discovered and repeatedly identified as a lethal pathogen of captive crab-eating macaques (Macaca fascicularis) in non-human primate (NHP) breeding facilities in the Philippines10–12 (Fig. 1). However, although RESTV can infect humans, it appears to be apathogenic13. RESTV was subsequently considered to be an Asian anomaly to the African filovirus dogma.
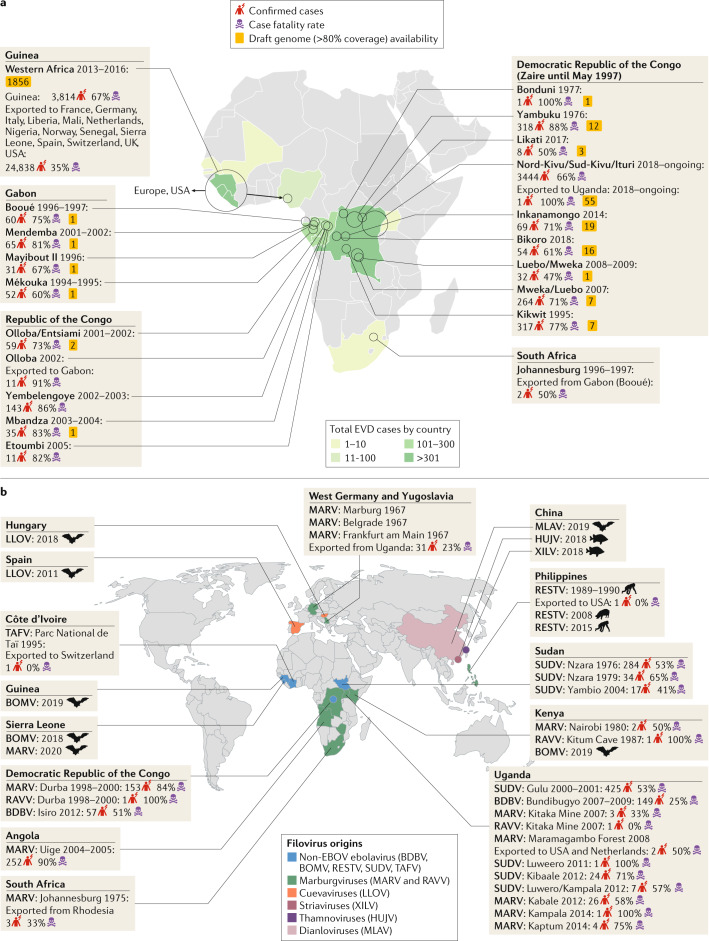
Fig. 1. Geographical overview of filovirus discovery.
a | Outbreaks of Ebola virus disease (EVD) in Africa, including the number of confirmed cases, the case fatality rates and the number of publicly available Ebola virus (EBOV) draft genome sequences per outbreak, are depicted3. Circles represent the relative size (in terms of the number of cases) of the outbreaks. Documented accidental laboratory-acquired infections have been excluded from this figure. b | Overview of global filovirus distribution, excluding EBOV. The place of isolation, known or suspected reservoir host and year of discovery are shown. The description of the distribution of non-EBOV filovirus disease outbreaks includes the total number of confirmed cases and the case fatality rate. BDBV, Bundibugyo virus; BOMV, Bombali virus; HUJV, Huángjiāo virus; LLOV, Lloviu virus; MARV, Marburg virus; MLAV, Měnglà virus; RAVV, Ravn virus; RESTV, Reston virus; SUDV, Sudan virus; TAFV, Taï Forest virus; XILV, Xīlǎng virus.
Classical filovirus-targeted genome sequencing and, later, unbiased broad-scale metagenomic sequencing, shed new light on filovirus ecology. In 2009, the sequencing of samples obtained from Egyptian rousettes (Rousettus aegyptiacus) in Africa revealed that these bats, which are cavernicolous and frugivorous pteropodids, are natural reservoir hosts of both MARV and RAVV. Coding-complete or complete genomic sequences of both viruses were repeatedly obtained from Egyptian rousette populations in Uganda, Sierra Leone and South Africa14–17, and genomic fragments of these viruses were also detected in populations of these bats in the Democratic Republic of the Congo18 and in Zambia19.
Around and after 2008, sequence-based evidence obtained using a range of techniques began to support the hypothesis that RESTV is an Asian virus. RESTV genome sequences were obtained from captive domestic pigs (Sus scrofa domesticus) in the Philippines and China20,21, and RESTV genome fragments were sequenced from samples from molossid, pteropodid and vespertilionid bats in the Philippines22. Next-generation sequencing further enabled the discovery of a highly divergent filovirus, Lloviu virus (LLOV, genus Cuevavirus), in deceased Schreibers’s long-fingered bats (Miniopterus schreibersii) in Hungary23 and in Spain24. The Ebolavirus genus was expanded owing to the discovery (via next-generation sequencing) of Bombali virus (BOMV) in molossid little free-tailed bats (Chaerephon pumilus) and Angolan free-tailed bats (Mops condylurus) in Guinea, Kenya and Sierra Leone25–27. Finally, a new filovirus genus, Dianlovirus, was recently established for a highly divergent filovirus, Měnglà virus (MLAV), which was discovered in unspecified rousettes in China. Preliminary metagenomics studies indicate the existence of other, highly divergent, filoviruses in the same area of China28–30. The most unexpected finding of genomics-based filovirus discovery was the genomic description of Huángjiāo virus (HUJV; genus Thamnovirus) and Xīlǎng virus (XILV; genus Striavirus) in striated frogfish (Antennarius striatus) and greenfin horse-faced filefish (Thamnaconus septentrionalis), respectively, captured in the East China Sea31 (Fig. 1).
In short, genomics has clarified that highly divergent filoviruses, frequently with unknown pathogenic potential, are likely to be distributed widely over the African, Asian and European continents in highly diverse host reservoirs (Fig. 2, Box 1). Furthermore, expanded animal sampling and unbiased host virome sequencing is likely to enable this diversity and distribution to be described in more detail. Of note, the natural host reservoirs of three ebolaviruses that are human pathogens, namely Bundibugyo virus (BDBV), SUDV and Taï Forest virus (TAFV), are still unclear. Furthermore, although bats are suspected to be hosts of EBOV owing to the detection of short EBOV genomic fragments and/or antibodies to EBOV in certain bats32, no complete EBOV genome has yet been sequenced from any bat sample. The genomic investigation of archived or newly acquired samples could also support or refute the often-repeated hypothesis that Middle African central chimpanzee (Pan troglodytes troglodytes), duiker (Cephalophus spp.) and western lowland gorilla (Gorilla gorilla gorilla) populations are frequently decimated by EBOV infection33–35. Thus, genomics may enable the prevention of future filovirus disease outbreaks by identifying filovirus natural hosts and by limiting host–human contacts as well as the initial introduction of filoviruses into the human population.
Fig. 2. Filovirus host reservoirs.
Complete or coding-complete filovirus genome sequences have been obtained from cave-dwelling and house-dwelling bats and highly diverse fish on the African, Asian and European continents (see Fig. 1 for continental distribution). The pathogenic potential of most filoviruses remains unclear, as does the transmission route of pathogenic filoviruses proven to infect humans and pigs or of pathogenic filoviruses suspected to infect chimpanzees, duikers and gorillas. Animals that have been proven to be infected by filoviruses are indicated in black; grey animals are suspected but unproven reservoirs of the indicated viruses. Solid arrows indicate highly likely transmission routes; dashed arrows indicate hypothesized transmission routes. BDBV, Bundibugyo virus; BOMV, Bombali virus; EBOV, Ebola virus; HUJV, Huángjiāo virus; LLOV, Lloviu virus; MARV, Marburg virus; MLAV, Měnglà virus; RAVV, Ravn virus; RESTV, Reston virus; SUDV, Sudan virus; TAFV, Taï Forest virus; XILV, Xīlàng virus.
Box 1 Endogenous filovirus elements.
The bioinformatic analysis of higher animal genome sequences, the number of which is steadily increasing, reveals that a marked percentage of these sequences are derived from ancient retroviruses142. Many animal genomes are mosaics that are likely to have evolved through the accidental integration of retroviral genes or gene fragments into germ cell genomes and the subsequent inheritance of this genetic material by descendants. In some cases, these sequences were positively selected for and were (or are still) expressed and their functions were exapted by hosts for novel functions142. A famous example of exaptation is the use of the human endogenous retrovirus W-derived syncytin, which was once a retrovirion surface glycoprotein that mediated virion host cell entry but is now essential for placental morphogenesis in pregnant women143.
During the past decade, scientists have been increasingly aware that such ‘viral fossils’ or ‘paleoviruses’ can derive from viruses other than retroviruses. Indeed, non-retroviral integrated RNA viruses (NIRVs; also known as endogenous viral elements) were derived from the ancestors of numerous extant virus families144. Prominent examples of negative-sense RNA virus-derived NIRVs are bornavirus sequences, which are found in the genomes of bats, fish, hyraxes, marsupials, primates, rodents, ruminants and elephants145,146, and rhabdovirus sequences, which are found in the genomes of crustaceans, mosquitoes, ticks and plants147,148. Interestingly, filovirus-derived NIRVs also appear to be widespread as they have been located in the genome of afrosoricids, bats, eulipotyphlans, marsupials and rodents145,149,150. The function of these stably inherited filovirus sequences remains to be determined. However, the existence of NIRVs indicates that filoviruses are at least several million years old and that highly divergent mammals were exposed to, and at least occasionally infected by, these viruses and perhaps by the descendants of these viruses that exist today.
Identifying and characterizing outbreaks
Genomics-based techniques have been central in the identification and characterization of filovirus disease outbreaks.
In-country outbreak identification
The largest known filovirus disease outbreak occurred from 2013 to 2016 in Western Africa and was caused by a novel EBOV variant, Makona (EBOV/Mak) (Fig. 1). Genomic sequencing efforts during this EVD outbreak showcased various platforms and strategies to characterize thousands of human clinical samples containing EBOV/Mak36. Early efforts relied on exporting positive samples to high-complexity genomic centres abroad37,38 (Box 2). However, in December 2014, EBOV/Mak genome sequencing using benchtop sequencers, such as the MiSeq System (Illumina) and the Ion Torrent system (Thermo Fisher Scientific) in-country (namely, in Liberia and Sierra Leone), became standard practice39–41. In addition, field laboratories used the iSeq 100 (Illumina), a portable bench-top sequencer with low error rates that can be transported in a suitcase, to obtain complete EBOV genome sequences to determine virus transmission in the Democratic Republic of the Congo42,43. Use of the portable nanopore sequencing technology MinION (Oxford Nanopore Technologies) markedly reduced the time required to obtain the genome sequence from patient samples and enabled the reintroduction of EBOV into Guinea and Sierra Leone to be rapidly confirmed41. Similarly, the EBOV variants causing the 2018 Équateur Province EVD (EBOV/“Tum”) and the ongoing Nord-Kivu/Sud-Kivu/Ituri Province EVD outbreak caused by EBOV/“Itu” in the Democratic Republic of the Congo were quickly identified by the use of MinION42,44.
The timing and establishment of in-country genomic sequencing capabilities determine which information can be captured and disseminated during an EVD outbreak. Early sequencing efforts can provide an informative ‘snapshot’ of the genomic epidemiology of EBOV during the initial phase of the outbreak45. The extent of the genomic diversity of the virus at the beginning of an outbreak can be used to determine whether single or multiple virus spillover events have occurred and to provide a crude estimate of the time at which a virus emerged in a human population37,38,46,47. Highly accurate genomic data have been used to characterize intra-host EBOV populations and genetic drift and even to evaluate, in silico and in real time, the available diagnostic measures and MCMs42,47,48. As an outbreak progresses and the sampling size increases, phylodynamic and spatiotemporal analyses reveal broader trends in the intra-outbreak evolutionary rate of the virus, its geographical migration and factors contributing to virus transmission, disease outcome and virus–host adaptation36,49–54.
Ideally, the viral agent is initially identified using highly portable sequencing platforms on site. After this identification, considerations other than sequencing speed (for example, sequencing accuracy and processivity) become paramount in determining virus transmission networks and in detecting changes in the viral genome (between cases in the current outbreak and between the current and previous outbreaks) that could subvert MCMs. However, whereas unbiased sequencing approaches using high fidelity platforms can lead to the discovery of co-infections and reveal important clinical considerations during the treatment of patients near the point of need, targeted methods of pathogen characterization using the portable sequencing platforms iSeq 100 and MiSeq (which use bait-enrichment techniques) and MinION (which uses amplicon sequencing) can still provide useful genomic data albeit with a lower sequencing output (that is, a lower number of reads) than unbiased sequencing.
Box 2 Recent advances in pathogen genome sequencing.
Over the past two decades, high-throughput benchtop platforms (for example, 454 sequencing (Roche), MiSeq, NextSeq and HiSeq (Illumina), Sequel and RS (PacBio) and Ion Torrent (Thermo Fisher)) have been the backbone of metagenomics and targeted sequencing approaches for pathogen identification151. Genomic centres generally had to allocate a notable portion of space to house and maintain sequencers and transporting sequencers was impractical. For example, MiSeq, which is the smallest of the sequencers, weighs 91 kg and occupies ~ 200,000 cm3. Recently, ‘capacity building’ efforts have established on-site genomics centres in low-resource settings39,40. However, the international shipping of oversized sequencers that rely on precise optical alignments for functionality can be challenging and cost-prohibitive and may cause irreparable damage to the sequencer. The maintenance and repair of internationally shipped sequencers may also be challenging as they do not come with in-country service contracts from the manufacturers. Moreover, in low-resource settings, trained local staff may not be permanent, and they will periodically require guidance and retraining in sample handling and storage and in how to ensure the continued service and activity of the sequencers.
The technological improvements in, and practicality of, portable sequencing technologies have been embraced in recent viral disease outbreaks, including in the 2018–present Ebola virus disease outbreak in the Democratic Republic of the Congo and a 2018 Lassa fever outbreak in Nigeria. The iSeq 100 (Illumina) and MinION (Oxford Nanopore) platforms both conform to on-site field requirements as they are miniature, rapid and easy to use. The ZiBRA Zika virus (ZIKV; a flavivirus of the family Flaviviridae) sequencing project in Brazil also demonstrated the feasibility of a ‘trailer laboratory’, but produced limited complete genome sequences owing to the small amounts of ZIKV RNA in clinical samples152. Several commercialized mobile laboratories incorporated into trucks or trailers shield equipment and temperature-sensitive reagents from austere conditions while providing ample power and laboratory workspace for sequencing. Within such mobile laboratories, smaller PCR thermocyclers, such as the programmable Mini16 thermal cycler with a smartphone (miniPCR bio™), and miniature centrifuges further bolster portability and ease space requirements. Other innovations that limit the need of a ‘cold chain’ for supplies will greatly improve on-site sequencing in hot climates and remote communities that are prone to outbreaks of Ebola virus disease.
Biosurveillance
Sequencing only a single target during an enduring and large outbreak of EVD may result in the detection of co-infections and/or superinfections being missed. The earliest evidence of an EBOV co-infection was obtained in Gabon in 2002, where a patient with EVD also tested positive for human immunodeficiency virus 1 (HIV-1; a lentivirus of the Retroviridae family)55. During the 2013–2016 Western African outbreak, co-infections of patients with EBOV with malaria-causing plasmodia or viruses such as GB virus C (a pegivirus of the family Flaviviridae), hepatitis B virus (an orthohepadnavirus of the family Hepadnaviridae) and Epstein–Barr virus (a lymphocryptovirus of the family Herpesviridae) were reported56–58. Additionally, EBOV infections in patients with a Gram-negative septicaemia or with bacterial translocation have been described59–61. Moreover, an extensive infectious disease due to one particular virus could plausibly conceal a simultaneous outbreak caused by a different pathogen62. Notoriously, there have been outbreaks of cholera, plague, measles and malaria, and sporadic cases of monkeypox and yellow fever, alongside the 2018‒present EVD outbreak in the north-eastern region of the Democratic Republic of the Congo. Detecting and characterizing co-infections during a disease outbreak can provide clinicians with crucial point-of-care information and identify differences in patient outcomes that may result from co-occurring infections.
The characterization of unexpected, co-circulating viruses during large-scale viral outbreaks requires established and reliable sequencing strategies that can be applied without knowledge of which virus is present. The discovery of novel viruses is ideally facilitated by metagenomic sequencing, which often relies on the pre-processing of clinical samples by depleting host-derived genomic material, followed by single or random primer amplification by PCR and deep sequencing63–65. However, metagenomic sequencing is limited by the requirement for computational and bioinformatics resources, which are not always readily available under field conditions. Instead, target-enrichment approaches using a wide breadth of bait probes for known pathogenic viruses, including filoviruses, have led to the successful characterization of known viruses in clinical samples from patients with disease of unknown etiology66–68. Moreover, target-enrichment sequencing is more cost-effective than metagenomic approaches. The percentage of sequencing data matching the target pathogen or pathogens can range from 50% to 99% with target enrichment, whereas with metagenomics approaches often <1% of sequencing data matches the target69,70.
Molecular genomic epidemiology
Punctual and highly accurate sequencing efforts have revealed the molecular genomic epidemiology of disease and thus enabled the characterization of pathogen transmission during disease outbreaks71. The first application of in-country, real-time genomic epidemiology started well into the EVD outbreak of 2013–2016 in Western Africa; portable sequencing was performed and data were analysed in tangent with up-to-date public health data41. The abundance of EBOV genome sequences determined during recent large-scale EVD outbreaks (including the 2013–2016 outbreak and the 2018–present outbreak in the Democratic Republic of the Congo) enabled real-time and retrospective investigations, using median-joining haplotype network establishment and phylodynamic inferences, to reveal cryptic human EBOV transmission chains in humans42,49,72,73. In coordination with classical epidemiological data (for example, that obtained by manual contact tracing), individual virus transmission events identified by genomic analysis such as median-joining haplotype networks can be temporally and spatially linked to determine likely transmission pathways, including the mode of virus diffusion and the identification of ‘superspreaders’ (as reviewed in ref.45)73,74 (Fig. 3). As the quality and quantity of available, complete genome sequences improves, new methods identifying intra-host SNPs may provide more granular analyses of person-to-person transmission than previous techniques39. Such SNPs can distinguish between almost identical consensus sequences of two or more patients75. However, acute infections resulting from direct contact with a recently infected and symptomatic individual, and for which primary infection occurred <21 days before the onset of symptoms, is not the only route for sustained person-to-person transmission during filovirus disease outbreaks.
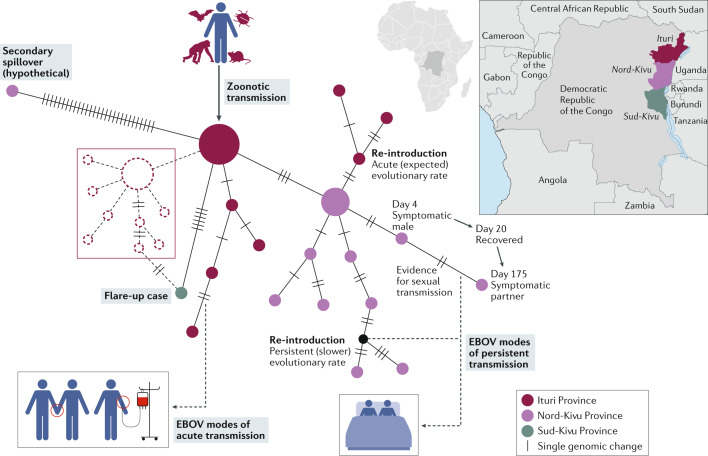
Fig. 3. Ebola virus transmission.
Following a zoonotic transmission event, sequencing the Ebola virus (EBOV) genome in real-time can help epidemiologists pinpoint the likely route of infection and of transmission during an outbreak of Ebola virus disease (EVD). A theoretical median-joining haplotype network of the Nord-Kivu/Sud-Kivu/Ituri Province EVD outbreak caused by EBOV/“Itu” in the Democratic Republic of the Congo (provinces are indicated on the map) provides different hypotheses that may explain person-to-person transmission events. A consistent number of genomic changes occurring in a specific window of time can indicate acute transmission events (that is, transmission via close contact with an individual with EVD, bottom left). The detection of transmission depends on diagnostics and sampling frequency. When sampling frequency is low, flare-up cases of EVD can appear with limited epidemiological data and with an unexpectedly large number of differences to earlier sampled haplotypes, which may point to undetected transmission networks (middle left; indicated by dashed lines). When viral diversity cannot be explained by spillover and spatial–temporal estimations, a secondary spillover may be possible (top left). Persistent infections through sexual transmission present with low genetic diversity (that is, with a slow evolutionary rate) over periods (bottom right) that are much longer than expected for acute reintroduction at the expected evolutionary rate (top right). A similar analysis was performed during the first discovery of sexual transmission during the 2013–2016 Western African EVD outbreak73, and a theoretical example is shown (bottom right). This example indicates the number of days after the initial presentation of symptoms at which an acutely infected male is sampled (day 4) and the day at which he recovers (day 20). On day 175, the sexual partner of this male becomes symptomatic owing to a very similar EBOV genotype, confirmed with epidemiological information and visualized using a median-joining haplotype network.
Virus persistence
EBOV initially infects monocyte-derived macrophages and dendritic cells, which disseminate the virus through the circulatory system to all main target organs, including the liver, spleen and kidneys. After infecting and damaging the vascular endothelia, EBOV infiltrates the parenchymata of these organs, resulting in focal necroses and inflammation. Such damage can eventually lead to multi-organ dysfunction syndrome and ultimately death76,77. However, in some cases, the intrinsic, innate and adaptive immune responses can contain viral replication and dissemination, resulting in the survival of the patient78.
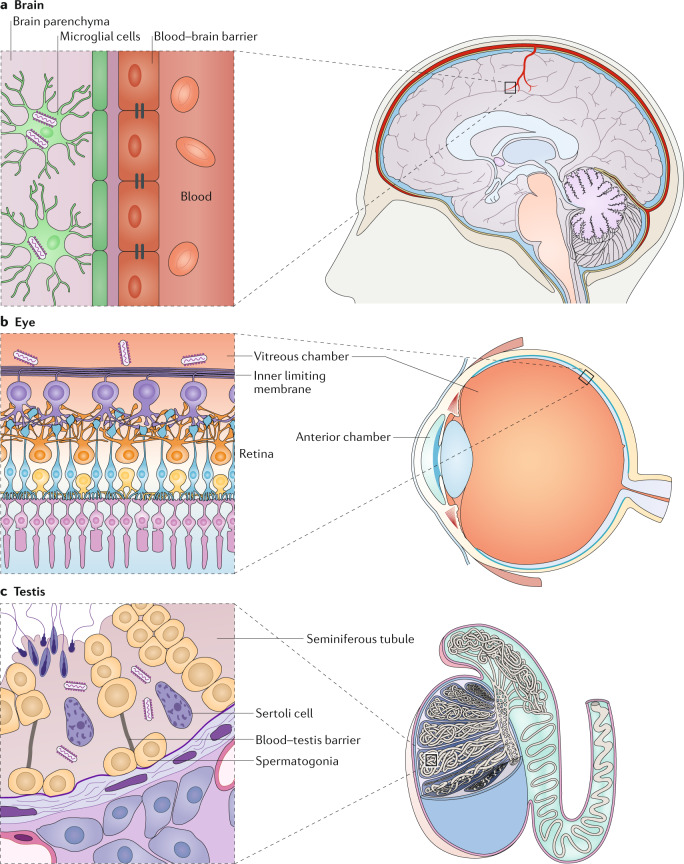
Until recently, it was hypothesized that survivors of filovirus diseases effectively abolished filovirus infection. However, new evidence indicates that EBOV can persist in certain sites of the body in the absence of viraemia and that this persistence could cause disease flare-ups (for example, see ref.73). Some of these sites, including the brain, eyes and testes, are immune-privileged because they are physically separated from tissues and cells that are under immune surveillance (Fig. 4). Thus, foreign antigens such as EBOV particles are tolerated within these sites without eliciting an inflammatory immune response79.
Fig. 4. Persistent Ebola virus infection in immune-privileged sites.
a | Ebola virus (EBOV) can break down the blood–brain barrier and persist in microglial cells. b | EBOV has been detected in aqueous humour extracted from the anterior chamber of the eye of human survivors. However, EBOV is mainly detected in the vitreous chamber and in the inner limiting membrane of the retina to which it is adjacent in rhesus monkeys that survive experimental infection with EBOV. c | The blood–testis barrier created by tight junctions between Sertoli cells protects sperm from immune recognition in seminiferous tubules, the main site of EBOV persistence in testis. Sertoli cells are the main cellular reservoir of EBOV persistence. Image courtesy of W. Discher.
Prior to the outbreak of EVD in Western Africa, evidence of persistent EBOV infection had been sparse probably owing to the small number of spillover events arising from this persistence (Fig. 1). Nevertheless, infectious EBOV and MARV and/or filoviral RNA had been detected in the eyes and semen of convalescent survivors prior to this outbreak80–83.
The large pool of survivors following the 2013‒2016 outbreak of EVD (specifically, 17,323 individuals) was different. Disease flare-ups or re-emergences were reported in at least nine individuals and attributed to sexual transmission or breast-feeding84. Sexual transmission of persistent EBOV was implicated in the initiation of new EBOV transmission chains49,73,85–87. Genomic analyses revealed that the evolutionary rate of EBOV persisting in testes during convalescence is reduced relative to the rates of EBOV persisting in blood and plasma85,88. All these observations prompted a notable number of studies of the long-term effects of persistent EBOV infection in EVD survivors. Indeed, EBOV persistence can be accompanied by various sequelae, colloquially often referred to as ‘post-Ebola [virus] syndrome’89. Studies of EBOV persistence in humans and experimentally in NHPs have revolutionized our understanding of EBOV infection and changed the guidelines of clinical operation as well as the recommendations of the World Health Organization for EVD survivors.
The impact of genomics on understanding the persistence of EBOV is broader than next-generation sequencing; advances have also revolutionized the field of pathology by allowing the rapid exploration of transcriptional expression in a chosen site of filovirus infection. The development of multi-labelled and targeted ‘probes’ that allow multiplex immunopathological hybridizations in sites of interest have also boosted our knowledge of EBOV persistence. Indeed, studies using novel histopathological tools have benefited genomic research in immune-privileged and non-immune-privileged sites.
Immune-privileged sites
Various neurological complications have been noted in survivors of EVD90. In experimentally infected rhesus monkeys (Macaca mulatta), encephalitic EBOV persistence is always accompanied by various degrees of encephalitis or meningoencephalitis91. Persistent infection of EBOV in the brain may lead to EVD relapse and late-onset encephalitis in human survivors several months after acute disease92,93. In experimentally infected NHPs, EBOV enters the brain by breaking down the blood–brain barrier by directly infecting and damaging endothelial cells91 (Fig. 4a). Interestingly, EBOV primarily infects and persists in microglia91.
Ocular complications, including uveitis, are some of the most common findings during EVD convalescence94, and persistent EBOV and persistent MARV have been isolated from the aqueous humour of human survivors with uveitis80,95. In experimentally infected NHPs, EBOV infects blood vessels during the acute phase of infection and later infects parenchymal eye tissues. However, in rhesus monkeys surviving the experimental infection of EBOV with various degrees of uveitis, retinitis and vitritis, EBOV only persisted in CD68+ cells (monocytes or macrophages) in the vitreous chamber and in the inner limiting membrane of the retina to which it is adjacent91 (Fig. 4b). Whether EBOV isolated from the aqueous humour of human survivors originates from the vitreous chamber and its adjacent structures, as appears to be the case in NHPs, remains unknown96.
The first recorded sexual transmission of a filovirus occurred in 1967, when a male survivor of Marburg virus disease transmitted MARV to his wife81. EBOV genomic RNA was repeatedly detected in the semen of EVD survivors up to 40 months after acute infection82,85,87,97–99, and infectious EBOV was isolated from a few semen samples100. EBOV infects the seminiferous tubules of both human and NHPs (Fig. 4c), which are the immune-privileged sites of sperm production, during the acute phase of disease101. Persistent EBOV infection was detected in the epididymis of a single rhesus monkey survivor with epididymitis, whereas MARV persistence in seminiferous tubules was multifocal in 22 of 73 crab-eating macaques that survived91,102. Recent studies indicate that testicular persistence is not restricted to filoviruses, as potential cases of the sexual transmission of Crimean-Congo haemorrhagic fever virus (CCHFV; an orthonairovirus of the family Nairoviridae) and Lassa virus (LASV; a mammarenavirus of the family Arenaviridae) have been reported103–105. Sertoli cells, the supporting cells of spermatogenesis, are the main cellular reservoir of testicular MARV and CCHFV persistence in experimentally infected NHPs102,106, and they may also be the reservoir of testicular LASV.
Non-immune-privileged sites
The majority of cases of filovirus persistence have been associated with immune-privileged sites. However, in several EVD flare-ups originating from asymptomatic survivors of EVD that were persistently infected with EBOV, identifying the exact sites of viral persistence in the index cases was not possible84,107. Thus, it is expected that sites of EBOV persistence that are not immune-privileged will be discovered. EBOV can be detected in breastmilk of female survivors of EVD and in various tissues, including the blood and liver, of laboratory mice that have been experimentally infected with EBOV and have partial immunity up to 150 days post-exposure108,109. Persistence in sites that are not immune-privileged has also been recently reported for CCHFV106 and LASV110,111. Interestingly, CCHFV persists within granulomas of NHP survivors with latent tuberculosis106. LASV persists in the smooth muscle cells of blood vessels with vasculitis in both crab-eating macaque and domesticated guinea pig survivors110,111, suggesting that a local altered immunological environment may sustain viral persistence.
Genomics of persistent infection
Genomics is uniquely suited to study the pattern of transmission from patients who are asymptomatic or paucisymptomatic. Genomic studies of persistent infections revealed distinct evolutionary dynamics that might result in patterns that can help identify the source of the flare-ups100. During the 2013‒2016 EVD outbreak, an unexpectedly low genetic diversity and complementary epidemiological data provided the first evidence of EBOV sexual transmission49,73. Recent pathogenesis studies elucidated the persistent infection of EBOV and MARV in seminiferous tubules101,102 and epididymides55, but niche-specific genomic studies (or single-cell sequencing efforts) have yet to be reported. Certain EBOV mutations may be tissue-specific and may therefore be required to establish persistence; genomics will be key in identifying these mutations. For instance, the sequencing of EBOV genomes present in the plasma and cerebrospinal fluid of a patient with EVD during a relapse revealed that only two non-coding changes to the genome had occurred compared to the originally obtained EBOV genomes sequenced from plasma during the acute phase of disease93. Multiple or even single nucleotide changes may permit EBOV to transverse the blood–brain barrier by directly infecting endothelial cells and microglia cells and establishing a persistent infection91. Although EBOV and MARV have been detected and isolated from the aqueous humour of EVD survivors with uveitis80,95 and in breastmilk of survivors109, genomic studies have not yet investigated the evolutionary dynamics in these niches.
Host responses
In nature, diverse biotic communities constantly interact and evolve within an ecosystem in response to environmental factors. Microorganisms, pathogenic or not, participate in these relationships and constantly react and evolve to changes within the environment. In highly complex organisms such as mammals, immune responses against viruses are inherently complex and involve humoural and intracellular mediators and diverse cell types. In turn, viruses such as EBOV evolve sophisticated and diverse strategies to evade the host immune response. By studying host–pathogen interactions, we may improve our understanding of the mechanisms that govern infection, immunity and immune evasion.
Even though EBOV is highly virulent and lethal in humans, some individuals survive infection. Furthermore, some individuals who were exposed to EBOV or who tested seropositive for the virus never reported disease. The seroprevalence rate of these individuals, categorized as asymptomatic or paucisymptomatic, has been reported to be >1% throughout Africa112–117. Transcriptomics offer insight as to why certain factors, such as the source, viral load, and infectivity of EBOV or host genetics, contribute to the range of disease severity and survivorship in patients. Host ‘immune gene signatures’ in patients infected with EBOV/Mak have been associated with clinical prognosis8,118. Indeed, data generated from 112 patients infected with EBOV/Mak (24 survivors and 88 fatal cases) revealed that interferon response-related genes and acute-phase responses were dysregulated in patients who did not survive8. However, natural killer cell populations were increased in EVD survivors, suggesting a crucial role for natural killer cells in controlling EBOV infection. Furthermore, low levels of inflammation and robust T cell responses with an upregulation of cytotoxic T lymphocyte-associated protein 4 (CTLA4) and programmed cell death 1 (PD1) expression in T cells also correlated with survival from EVD118. Additional transcriptomic studies focusing on the population dynamics of EBOV in infected humans and possibly in naive NHPs will complement earlier characterization of host responses to EVD and provide a greater understanding of the mechanisms of EVD pathogenesis and disease outcome. Bringing transcriptomics tools to the field, facilitated by our ability to perform genomic sequencing in outbreak areas, will allow the promises of precision medicine to be realized in an outbreak setting.
Genomics in threat characterization
The genomic characterization of an outbreak pathogen is only the first step in characterizing a threat, a process that can be facilitated by reverse functional genomics. Classical reverse genetics has focused on the virus rescue of filoviruses based on cloning a reference (that is, a consensus) filovirus sequence or using a replicating filovirus isolate to clone filovirus sequences61,62. However, synthetic reverse genomics can rapidly rescue individual virus haplotypes or genotypes from a virus population in the absence of replicating isolates, using gene and genome synthesis based on sequence information alone. These rescued viruses can be used to evaluate the functional aspects of individual filovirus genome mutations. Thus, functional genomics can facilitate the rapid and precise functional characterization of a newly emerging filovirus or filovirus mutant. Given that natural filovirus isolates must be studied while fulfilling strict biosafety and biosecurity requirements that are frequently not available in areas of filovirus disease outbreaks, the availability of in-country and field-deployable sequencing platforms increases the ability of researchers to gather crucial information about the virus without the need to export biological specimens across national borders. To succeed, field sequencing needs to produce highly accurate finished filovirus genome sequences (for example, sequences that include the complete genomic leader and trailer regions of the virus) from a variety of sample sources31,42,72. Filoviruses rescued from synthetic reverse genetics systems can then be used to study host adaptation and attenuation as well as the efficacy of therapeutics118–121. The de novo generation of EBOV from the ongoing EVD outbreak in the Democratic Republic of the Congo (that is, of EBOV/“Itu”) demonstrates the value of synthetic reverse genetics9 (Fig. 5). After the development of modular reverse genetic systems that are even more efficient than those available today, even high-throughput phenotypical characterizations of large numbers of minimally divergent filoviruses are likely to be possible even in the absence of a biological isolate.
Fig. 5. Reverse functional genomics for characterizing authentic Ebola virus isolates.
Reverse functional genomics facilitates external support in the response to outbreaks of Ebola virus disease (EVD) that occur in remote areas that lack in-house resources to test available medical countermeasures. Rapid, high accuracy, complete genome sequences determined in-country are shared with out-of-country collaborators to evaluate key genomic and proteomic changes in Ebola virus (EBOV) that may affect the efficacy of available therapeutics (part a). For example, changes in the double-stranded RNA (dsRNA)-binding site of the EBOV polymerase cofactor viral protein 35 (VP35), a protein targeted by several therapeutics, can be identified from the sequencing data obtained for a new EBOV isolate. Changes in this region of VP35 may compromise the efficacy of treatments. Indeed, if two available therapeutic agents (hypothetical treatment A and hypothetical treatment B) target VP35, reverse genetic systems can produce replicative EBOVs de novo that contain the changes identified in VP35 (part b). These replicative EBOVs can then be used for the in vitro and in vivo therapeutic evaluation of both hypothetical treatments; data obtained using this approach can inform on which treatment is potentially more efficacious against the EBOV isolate causing the outbreak.
Genomics in prevention and therapy
MCMs and EBOV population dynamics
Multiple MCMs against EBOV infection have been developed, including small molecules, monoclonal antibodies (mAbs), antibody cocktails and vaccines122, and clinical trials have yielded promising results. However, the population dynamics and evolution of EBOV are influenced by mutations in RNA, RNA recombination rates, virus population bottlenecks, natural selection and fitness (including diversifying and purifying selection), host range and mode of transmission123. These traits re-adjust depending on the environment in a natural host, a naive accidental host, a previously exposed accidental host (owing to infection or vaccination) or an accidental host that received an MCM. Therapeutic pressures will force the selection of individual EBOV genotypes, and thereby adaption, to ensure virus survival or persistence. Highly accurate genomic data can be used to evaluate available MCMs in silico prior to in vitro and in vivo testing and to characterize host responses to specific therapeutic interventions in real time. For example, accurate sequencing data from EBOV/“Itu” allowed for the rapid in silico assessment of the ability of the mAb114 (National Institutes of Health) and ZMapp (Mapp Biopharmaceutical) mAbs to bind to the receptor-binding domain of the EBOV spike glycoprotein GP1,2, predicting that these mAbs should be effective against circulating EBOV/“Itu”42. Using genomic approaches to understand how MCMs influence EBOV population dynamics and to identify mutant viruses that escape MCMs could augment future therapeutic designs and filovirus-targeting strategies.
Vaccinating against EBOV
Correlates of protection for different vaccines and vaccination regimens against EBOV have not been fully determined. However, both humoural and cellular immunity independently correlate with the protection of NHPs from EBOV infection and disease124,125. The use of a recombinant vesicular stomatitis Indiana virus vaccine expressing EBOV GP1,2, rVSVΔG-ZEBOV-GP (sold under the brand name Ervebo), the first EBOV vaccine to be approved by the FDA126,127, diminished EVD case fatality rates in a limited ring-vaccination trial performed in Guinea in 2015 (ref.128) and it is currently administered to people living in the area of the current EVD outbreak. However, unfortunately, some vaccinated individuals still present with clinical signs of mild EVD. The exact reasons for these ‘breakthrough cases’ are not completely understood; however, they are thought to result from EBOV infection within the first 10 days after vaccination. A breakthrough could be due to the individual having an insufficient immune response to control the virus (that is, when the vaccine induces a low titre of anti-EBOV antibodies and/or scarce effector cells) or to EBOV adapting to escape the immunological pressures produced by the vaccine. The characterization of the genomic EBOV population in patients who developed EVD after vaccination could help address these questions.
Immunotherapy against EBOV
Immunotherapy has also been successfully used as a therapy for EVD, most notably during the ongoing outbreak in the Democratic Republic of the Congo129. The Pamoja Tulinde Maisha (PALM) randomized controlled trial recently evaluated four investigational therapeutics (ZMapp, remdesivir (Gilead Sciences, Inc.), mAb114 and REGN-EB3 (Regeneron Pharmaceuticals)) in the treatment of EVD130. mAb114 (a single mAb) and REGN-EB3 (a cocktail of three mAbs) were significantly more effective than ZMapp (a cocktail of three mAbs) in treating EVD (P = 0.007 and P = 0.002, respectively) and were also more effective than remdesivir (a nucleoside analogue); the effect of ZMapp and remdesivir on EVD was not significantly different. mAb114, obtained from a 1995 EVD survivor from the Democratic Republic of the Congo, is one of several recently identified ebolavirus-neutralizing antibodies that are being developed as therapeutics for EVD129–132. This mAb protected NHPs from EBOV-induced disease and death, even when administered 5 days after inoculation with EBOV133. As mAb114 binds to the EBOV glycoprotein GP1,2, the risk of escape mutants arising might be constrained by viral fitness. REGN-EB3 also showed promising results in EBOV-exposed NHPs, and no mutant viruses were reported to escape this MCM134,135. By contrast, EBOV mutants that escape ZMapp have been characterized136. A mutation in amino acid residue 508 of EBOV GP1,2 abrogated its binding to two of the mAbs within ZMapp, and a mutation in amino acid residue 273 of EBOV GP1,2 abrogated its binding to the third mAb137. Moreover, epitope mapping of EBOV GP1,2 using an alanine-scanning assay138 identified several residues that are crucial for antibodies to bind to EBOV GP1,2, although none of these mutations was tested for fitness.
In silico modelling using the three-dimensional structure of EBOV GP1,2 was used to predict which mutations in EBOV would enable it to escape antibody binding139,140. Initially, mAb KZ52 was analysed, followed by mAb100, mAb114 and mAb 13F6–1-2. A list of 34 mutations predicted to interfere with KZ52–EBOV GP1,2 binding was expanded to 127 mutations predicted to disrupt the binding of other mAbs to EBOV GP1,2. Three of the predicted mutations were found in EBOVs infecting humans or experimental animals34,141, showing that viruses may escape current MCMs under investigation.
Despite the proven efficacy of two therapeutics assessed in the PALM trial, namely mAb114 and REGN-EB3, none of the treatments achieved complete protection against EVD129. We hypothesize that the characterization of the EBOV population in individuals who did not benefit from treatment in the PALM trial might help elucidate the reasons for this failure. A clear demonstration of the power of genomics for this purpose was the investigation of two NHPs that succumbed to experimental EBOV exposure despite experimental treatment with MB-003, a cocktail of three mAbs. Two clusters of genomic mutations (a cluster of five non-synonymous changes in one animal and a cluster of four non-synonymous changes in the other) reduced the binding of the three mAbs to EBOV GP1,2 (Fig. 6). Viral isolates obtained after MB-003 treatment recapitulated the observed ‘escape’ phenotype. The mutations, all located in the EBOV glycoprotein (GP) gene, abrogated antibody binding to GP1,2. Although this study was restricted in size, it highlights mechanisms by which EBOV can evade immunotherapies. First, adaptation to a selection pressure can occur rapidly. Second, methods to monitor the appearance and phasing of viral mutants that can escape immunotherapies are crucial. Third, with the use of reverse genetics systems, scientists can acquire pertinent information on the therapeutic efficacy of sequence-based MCMs during an ongoing outbreak of EVD9 (Fig. 5). For example, phenotypical changes resulting from clade-defining non-synonymous mutations (for example, alanine to histidine substitution at residue 547 in nucleoprotein or isoleucine to methionine substitution at residue 78 in viral protein 35, both in EBOV/“Itu”) or positively selected non-synonymous mutations identified in the ongoing outbreak can be investigated.
Fig. 6. Therapeutic and phylodynamic perspectives of the emergence of treatment-resistant Ebola virus mutants.
a | During an Ebola virus (EBOV) infection, the low-fidelity viral RNA-directed RNA polymerase creates replication errors that lead to increasing intra-host variation through successive replications (that is, genetic drift occurs). Therapeutic monoclonal antibodies, such as MB-003, neutralize EBOV, and infected patients subsequently recover. However, such treatment can also create a ‘therapeutic bottleneck’ in which intra-host diversity alters to select genotypes that only harbour the escape variant that continues to replicate in current and future hosts. b | The evolution of EBOV intra-host populations over time, pre-treatment and post-treatment, and the genomic bottleneck, can be visualized by estimating a phylogenetic tree (notable events along the tree are indicated by colourless circles). Samples can be characterized using next-generation sequencing (NGS) at various time points. Each coloured circle represents a finished EBOV genome from an individual patient. After an inter-host bottleneck, viral diversity should be low in the earliest phase of infection (1). As EBOV continually replicates, expanding intra-host diversity can create heterogeneous subpopulations (2, blue circles). Once exposed to treatment (3, dashed line), EBOV intra-host populations may be countered and eliminated, demonstrating an effective treatment without further intra-host replication or diversification (green circles). However, a small subpopulation surviving the therapeutic bottleneck may allow escape mutants to subvert treatment (4) and continue diversifying into novel, treatment-resistant genotypes (5, red circles).
Resistance of EBOV to vaccines and MCMs
Unfortunately, not many studies have investigated the resistance of EBOV to candidate vaccines or to MCM. Cost is a consideration, but the limited number of experimental animals typically available for therapeutic investigations (~5–10) reduces the analytical power of comparative virus–host interactions and inter-host population-level genomics. In addition, experiments aimed at understanding the appearance of antiviral resistance are restrictive; precise protocols to measure and evaluate EBOV and the host immune response to infection without violating established policies are urgently required. Furthermore, the possibility that a more virulent or less treatable virus may evolve during such experiments and be accidently released is a concern, which is why Institutional Biosafety Committees are cautious in approving experiments aimed at creating such viruses. Additionally, concerns may arise from other nations that are parties of the Biological Weapons and Toxins Convention. Convincing national rivals that experiments resulting in EBOV strains that are resistant to MCMs were actually intended to strengthen MCMs, and not to deliberately create biological weapons, is not straightforward.
Conclusions
The advancement of targeted and unbiased genomic sequencing has profoundly impacted our biological knowledge of, epidemiological preparedness for, and response strategies to known and unknown high-consequence filovirus infections. Greater investments in in-country genomic infrastructure and mobile sequencing platforms have reduced the time from sample to viral genome sequence and simultaneously informed local clinicians, epidemiologists and public health officials of genome-guided information on the virus responsible for a current outbreak. Advances in large-scale and timely data generation encourage future genomic studies to go beyond consensus-level pathogen genome sequencing during an outbreak. For example, persistent infections in immune-privileged sites of survivors can be characterized using single-cell sequencing technologies. Additionally, an increased focus on host responses in an infected individual and the contribution of host genome characteristics to disease outcome and transmission would potentially further benefit the development of MCMs and improve the outbreak response. Complete pathogen–host genomic investigations could be applied to infected individuals, NHPs and known natural host reservoirs of filoviruses. The successful integration of current and future genomic tools will rely on the establishment of new long-term partnerships between government, academia and public health agencies and on maintaining in-country genomic capabilities where the threat of filovirus outbreaks is imminent.
Acknowledgements
We thank Laura Bollinger (NIH/NIAID Integrated Research Facility at Fort Detrick, Frederick, MD, USA) for critically editing the manuscript and William Discher (USAMRIID, Fort Detrick, Frederick, MD, USA) for the diagram used in Fig. 4. This work was supported in part through the prime contract of Laulima Government Solutions, LLC, with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under contract no. HHSN272201800013C and Battelle Memorial Institute’s former prime contract with NIAID under contract no. HHSN272200700016I. J.H.K. performed this work as a former employee of Battelle Memorial Institute and a current employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under contract no. HHSN272201800013C. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Defense, US Department of Health and Human Services, US Department of the Army, or of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication.
Glossary
- Next-generation sequencing
A collection of continuously evolving technologies and techniques that allow for the digitalization of genomic material.
- Metagenomic sequencing
The sequencing of genetic material recovered directly from an environmental or clinical sample that allows the identification of all organisms and mobile genetic elements represented in the sample.
- Genetic drift
The change in the frequency of an existing gene variant (allele) in a population owing to the occurrence of random mutations.
- Exaptation
The novel use of an evolved trait for a different function.
- Median-joining haplotype network establishment
A minimum spanning tree analysis of recombinant-free genomic sequences that infers ancestry–descendant relationships using haploid genotypes that can be visualized in a single unrooted, reticulate network.
- Phylodynamic inferences
The study of how evolutionary processes interact with epidemiological and immunological factors to influence phylogenetic estimations.
- Uveitis
Inflammation of the uvea (the pigmented layer between retina and the fibrous layer composed of sclera and cornea of the eye).
- Population bottlenecks
The erosion of genetic diversity and concomitant reduction in individual fitness and evolutionary potential.
Author contributions
These authors researched data for the article, contributed to the writing and discussion of the content, and reviewed and edited the manuscript prior to submission, focusing on the following sections: introduction (N.D., J.H.K. and G.P.); identifying filovirus reservoirs (J.H.K.); identifying and characterizing outbreaks (N.D., J.H.K., G. P. and X.Z.); genomics in threat characterization (N.D. and G.P.); and genomics in prevention and therapy (M.S.-L. and G.P.). All of the authors contributed to the conclusions.
Competing interests
All authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Microbiology thanks Miles Carroll and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
FDA news release on the first FDA-approved vaccine for the prevention of Ebola virus disease: https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health
References
- 1.Kuhn JH. Filoviruses. A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch. Virol. Suppl. 2008;20:13–360. doi: 10.1007/978-3-211-69495-4_1. [DOI] [PubMed] [Google Scholar]
- 2.National Center for Biotechnology Information. GenBankhttps://www.ncbi.nlm.nih.gov/genbank/ (2019).
- 3.Kuhn, J. H., Amarasinghe, G. & Perry, D. L. in Fields Virology: Emerging Viruses Ch. 12 (eds Peter Howley, M. Knipe, David M. & Whelan, Sean P. J.) (Wolters Kluwer, 2020).
- 4.World Health Organization. Situation report. Ebola Virus Disease. https://apps.who.int/iris/bitstream/handle/10665/208883/ebolasitrep_10Jun2016_eng.pdf;jsessionid=4F7DFD58B3A8FDF69F885F6DF11F4E60?sequence=1 (2016).
- 5.World Health Organization. Ebola in the Democratic Republic of Congo. Health Emergency Update. https://www.who.int/emergencies/diseases/ebola/drc-2019 (2020).
- 6.Whitley R. The new age of molecular diagnostics for microbial agents. N. Engl. J. Med. 2008;358:988–989. doi: 10.1056/NEJMp0708085. [DOI] [PubMed] [Google Scholar]
- 7.Palacios G, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, et al. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol. 2017;18:4. doi: 10.1186/s13059-016-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMullan LK, et al. Characterisation of infectious Ebola virus from the ongoing outbreak to guide response activities in the Democratic Republic of the Congo: a phylogenetic and in vitro analysis. Lancet Infect. Dis. 2019;19:1023–1032. doi: 10.1016/S1473-3099(19)30291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demetria C, et al. Reemergence of Reston ebolavirus in cynomolgus monkeys, the Philippines, 2015. Emerg. Infect. Dis. 2018;24:1285–1291. doi: 10.3201/eid2407.171234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisbert TW, Jahrling PB. Use of immunoelectron microscopy to show Ebola virus during the 1989 United States epizootic. J. Clin. Pathol. 1990;43:813–816. doi: 10.1136/jcp.43.10.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda MEG, Miranda NLJ. Reston ebolavirus in humans and animals in the Philippines: a review. J. Infect. Dis. 2011;204(Suppl. 3):S757–S760. doi: 10.1093/infdis/jir296. [DOI] [PubMed] [Google Scholar]
- 13.Albariño CG, et al. Insights into Reston virus spillovers and adaption from virus whole genome sequences. PLoS One. 2017;12:e0178224. doi: 10.1371/journal.pone.0178224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amman BR, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawęska JT, et al. Marburg virus infection in Egyptian rousette bats, South Africa, 2013-2014. Emerg. Infect. Dis. 2018;24:1134–1137. doi: 10.3201/eid2406.172165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towner JS, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amman BR, et al. Isolation of Angola-like Marburg virus from Egyptian rousette bats from West Africa. Nat. Commun. 2020;11:510. doi: 10.1038/s41467-020-14327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanepoel R, et al. Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 2007;13:1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajihara M, et al. Marburgvirus in Egyptian fruit bats, Zambia. Emerg. Infect. Dis. 2019;25:1577–1580. doi: 10.3201/eid2508.190268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrette RW, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 21.Pan Y, et al. Reston virus in domestic pigs in China. Arch. Virol. 2014;159:1129–1132. doi: 10.1007/s00705-012-1477-6. [DOI] [PubMed] [Google Scholar]
- 22.Jayme SI, et al. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol. J. 2015;12:107. doi: 10.1186/s12985-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemenesi G, et al. Re-emergence of Lloviu virus in Miniopterus schreibersii bats, Hungary, 2016. Emerg. Microbes Infect. 2018;7:66. doi: 10.1038/s41426-018-0067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negredo A, et al. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog. 2011;7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes KM, et al. Bombali virus in Mops condylurus Bat, Kenya. Emerg. Infect. Dis. 2019;25:955–957. doi: 10.3201/eid2505.181666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein T, et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 2018;3:1084–1089. doi: 10.1038/s41564-018-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karan LS, et al. Bombali virus in Mops condylurus bats, Guinea. Emerg. Infect. Dis. 2019;25:1774–1775. doi: 10.3201/eid2509.190581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He B, et al. Filovirus RNA in fruit bats, China. Emerg. Infect. Dis. 2015;21:1675–1677. doi: 10.3201/eid2109.150260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X-L, et al. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat. Microbiol. 2019;4:390–395. doi: 10.1038/s41564-018-0328-y. [DOI] [PubMed] [Google Scholar]
- 30.Yang X-L, et al. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg. Infect. Dis. 2017;23:482–486. doi: 10.3201/eid2303.161119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi M, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 32.Leroy EM, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 33.Bermejo M, et al. Ebola outbreak killed 5000 gorillas. Science. 2006;314:1564. doi: 10.1126/science.1133105. [DOI] [PubMed] [Google Scholar]
- 34.Leroy EM, et al. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–390. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 35.Wittmann TJ, et al. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc. Natl Acad. Sci. USA. 2007;104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes EC, Dudas G, Rambaut A, Andersen KG. The evolution of Ebola virus: insights from the 2013-2016 epidemic. Nature. 2016;538:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baize S, et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 38.Gire SK, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arias A, et al. Rapid outbreak sequencing of Ebola virus in Sierra Leone identifies transmission chains linked to sporadic cases. Virus Evol. 2016;2:vew016. doi: 10.1093/ve/vew016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kugelman JR, et al. Monitoring of Ebola virus Makona evolution through establishment of advanced genomic capability in Liberia. Emerg. Infect. Dis. 2015;21:1135–1143. doi: 10.3201/eid2107.150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mbala-Kingebeni P, et al. Medical countermeasures during the 2018 Ebola virus disease outbreak in the North Kivu and Ituri Provinces of the Democratic Republic of the Congo: a rapid genomic assessment. Lancet Infect. Dis. 2019;19:648–657. doi: 10.1016/S1473-3099(19)30118-5. [DOI] [PubMed] [Google Scholar]
- 43.Mbala-Kingebeni P, et al. 2018 Ebola virus disease outbreak in Équateur Province, Democratic Republic of the Congo: a retrospective genomic characterisation. Lancet Infect. Dis. 2019;19:641–647. doi: 10.1016/S1473-3099(19)30124-0. [DOI] [PubMed] [Google Scholar]
- 44.Mbala-Kingebeni P, et al. Rapid confirmation of the Zaire Ebola virus in the outbreak of the Equateur Province in the Democratic Republic of Congo: implications for public health interventions. Clin. Infect. Dis. 2019;68:330–333. doi: 10.1093/cid/ciy527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grubaugh ND, et al. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 47.Park DJ, et al. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sozhamannan S, et al. Evaluation of signature erosion in Ebola virus due to genomic drift and its impact on the performance of diagnostic assays. Viruses. 2015;7:3130–3154. doi: 10.3390/v7062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blackley DJ, et al. Reduced evolutionary rate in reemerged Ebola virus transmission chains. Sci. Adv. 2016;2:e1600378. doi: 10.1126/sciadv.1600378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Vega M-A, et al. Ebola viral load at diagnosis associates with patient outcome and outbreak evolution. J. Clin. Invest. 2015;125:4421–4428. doi: 10.1172/JCI83162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dudas G, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers L, Frawley T, Goss S, Kang C. Ebola virus outbreak 2014: clinical review for emergency physicians. Ann. Emerg. Med. 2015;65:101–108. doi: 10.1016/j.annemergmed.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 53.Urbanowicz RA, et al. Human adaptation of Ebola virus during the West African outbreak. Cell. 2016;167:1079–1087. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll MW, et al. Temporal and spatial analysis of the 2014-2015 Ebola virus outbreak in West Africa. Nature. 2015;524:97–101. doi: 10.1038/nature14594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nkoghé D, et al. Cas isolé de fièvre hémorragique survenu au Gabon en 2002 pendant l’épidémie d’Ebola mais distant des régions épidémiques [French] Méd. Trop. 2005;65:349–354. [PubMed] [Google Scholar]
- 56.Kerber R, et al. Analysis of diagnostic findings from the European mobile laboratory in Gueckedou, Guinea, March 2014 through March 2015. J. Infect. Dis. 2016;214:S250–S257. doi: 10.1093/infdis/jiw269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lauck M, et al. GB virus C coinfections in west African Ebola patients. J. Virol. 2015;89:2425–2429. doi: 10.1128/JVI.02752-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li T, et al. Metagenomic next-generation sequencing of the 2014 Ebola virus disease outbreak in the Democratic Republic of the Congo. J. Clin. Microbiol. 2019;57:e00827–19. doi: 10.1128/JCM.00827-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreuels B, et al. A case of severe Ebola virus infection complicated by gram-negative septicemia. N. Engl. J. Med. 2014;371:2394–2401. doi: 10.1056/NEJMoa1411677. [DOI] [PubMed] [Google Scholar]
- 60.Wolf T, et al. Severe Ebola virus disease with vascular leakage and multiorgan failure: treatment of a patient in intensive care. Lancet. 2015;385:1428–1435. doi: 10.1016/S0140-6736(14)62384-9. [DOI] [PubMed] [Google Scholar]
- 61.Carroll MW, et al. Deep sequencing of RNA from blood and oral swab samples reveals the presence of nucleic acid from a number of pathogens in patients with acute Ebola virus disease and is consistent with bacterial translocation across the gut. mSphere. 2017;2:e00325–17. doi: 10.1128/mSphereDirect.00325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.di Paola N, et al. An outbreak of human parvovirus B19 hidden by dengue fever. Clin. Infect. Dis. 2019;68:810–817. doi: 10.1093/cid/ciy630. [DOI] [PubMed] [Google Scholar]
- 63.Delwart EL. Viral metagenomics. Rev. Med. Virol. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houldcroft CJ, Beale MA, Breuer J. Clinical and biological insights from viral genome sequencing. Nat. Rev. Microbiol. 2017;15:183–192. doi: 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palacios G, et al. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briese T, et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6:e01491–15. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chalkias S, et al. ViroFind: a novel target-enrichment deep-sequencing platform reveals a complex JC virus population in the brain of PML patients. PLoS One. 2018;13:e0186945. doi: 10.1371/journal.pone.0186945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kamperschroer C, Gosink MM, Kumpf SW, O’Donnell LM, Tartaro KR. The genomic sequence of lymphocryptovirus from cynomolgus macaque. Virology. 2016;488:28–36. doi: 10.1016/j.virol.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 69.O’Flaherty BM, et al. Comprehensive viral enrichment enables sensitive respiratory virus genomic identification and analysis by next generation sequencing. Genome Res. 2018;28:869–877. doi: 10.1101/gr.226316.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kafetzopoulou LE, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 2019;363:74–77. doi: 10.1126/science.aau9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ladner JT, et al. Evolution and spread of Ebola virus in Liberia, 2014-2015. Cell Host Microbe. 2015;18:659–669. doi: 10.1016/j.chom.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mate SE, et al. Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cottam EM, et al. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J. Virol. 2006;80:11274–11282. doi: 10.1128/JVI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Worby CJ, Lipsitch M, Hanage WP. Shared genomic variants: identification of transmission routes using pathogen deep-sequence data. Am. J. Epidemiol. 2017;186:1209–1216. doi: 10.1093/aje/kwx182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baseler L, Chertow DS, Johnson KM, Feldmann H, Morens DM. The pathogenesis of Ebola virus disease. Annu. Rev. Pathol. 2017;12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- 77.Messaoudi I, Amarasinghe GK, Basler CF. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat. Rev. Microbiol. 2015;13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong G, Kobinger GP, Qiu X. Characterization of host immune responses in Ebola virus infections. Expert Rev. Clin. Immunol. 2014;10:781–790. doi: 10.1586/1744666X.2014.908705. [DOI] [PubMed] [Google Scholar]
- 79.Matzinger P, Kamala T. Tissue-based class control: the other side of tolerance. Nat. Rev. Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 80.Kuming BS, Kokoris N. Uveal involvement in Marburg virus disease. Br. J. Ophthalmol. 1977;61:265–266. doi: 10.1136/bjo.61.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martini GA, Schmidt HA. Spermatogene Übertragung des „Virus Marburg“ (Erreger der „Marburger Affenkrankheit“) [German] Klin. Wochenschr. 1968;46:398–400. doi: 10.1007/BF01734141. [DOI] [PubMed] [Google Scholar]
- 82.Rodriguez LL, et al. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 1999;179:S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- 83.Smith DH, et al. Marburg-virus disease in Kenya. Lancet. 1982;1:816–820. doi: 10.1016/S0140-6736(82)91871-2. [DOI] [PubMed] [Google Scholar]
- 84.Subissi L, et al. Ebola virus transmission caused by persistently infected survivors of the 2014-2016 outbreak in West Africa. J. Infect. Dis. 2018;218:S287–S291. doi: 10.1093/infdis/jiy280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diallo B, et al. Resurgence of Ebola virus disease in Guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days. Clin. Infect. Dis. 2016;63:1353–1356. doi: 10.1093/cid/ciw601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sow MS, et al. New evidence of long-lasting persistence of Ebola virus genetic material in semen of survivors. J. Infect. Dis. 2016;214:1475–1476. doi: 10.1093/infdis/jiw078. [DOI] [PubMed] [Google Scholar]
- 87.Uyeki TM, et al. Ebola virus persistence in semen of male survivors. Clin. Infect. Dis. 2016;62:1552–1555. doi: 10.1093/cid/ciw202. [DOI] [PubMed] [Google Scholar]
- 88.Whitmer SLM, et al. Active Ebola virus replication and heterogeneous evolutionary rates in EVD survivors. Cell Rep. 2018;22:1159–1168. doi: 10.1016/j.celrep.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott JT, et al. Post-Ebola syndrome, Sierra Leone. Emerg. Infect. Dis. 2016;22:641–646. doi: 10.3201/eid2204.151302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vetter P, Kaiser L, Schibler M, Ciglenecki I, Bausch DG. Sequelae of Ebola virus disease: the emergency within the emergency. Lancet Infect. Dis. 2016;16:e82–e91. doi: 10.1016/S1473-3099(16)00077-3. [DOI] [PubMed] [Google Scholar]
- 91.Zeng X, et al. Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat. Microbiol. 2017;2:17113. doi: 10.1038/nmicrobiol.2017.113. [DOI] [PubMed] [Google Scholar]
- 92.Howlett P, et al. Ebola virus disease complicated by late-onset encephalitis and polyarthritis, Sierra Leone. Emerg. Infect. Dis. 2016;22:150–152. doi: 10.3201/eid2201.151212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacobs M, et al. Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet. 2016;388:498–503. doi: 10.1016/S0140-6736(16)30386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shantha JG, Crozier I, Yeh S. An update on ocular complications of Ebola virus disease. Curr. Opin. Ophthalmol. 2017;28:600–606. doi: 10.1097/ICU.0000000000000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Varkey JB, et al. Persistence of Ebola virus in ocular fluid during convalescence. N. Engl. J. Med. 2015;372:2423–2427. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor AW. Ocular immune privilege and transplantation. Front. Immunol. 2016;7:37. doi: 10.3389/fimmu.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deen GF, et al. Ebola RNA persistence in semen of Ebola virus disease survivors - final report. N. Engl. J. Med. 2017;377:1428–1437. doi: 10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rowe AK, et al. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. J. Infect. Dis. 1999;179:S28–S35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- 99.The PREVAIL III Study Group A longitudinal study of Ebola sequelae in Liberia. N. Engl. J. Med. 2019;380:924–934. doi: 10.1056/NEJMoa1805435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schindell BG, Webb AL, Kindrachuk J. Persistence and sexual transmission of filoviruses. Viruses. 2018;10:683. doi: 10.3390/v10120683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J. Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 102.Coffin KM, et al. Persistent Marburg virus infection in the testes of nonhuman primate survivors. Cell Host Microbe. 2018;24:405–416.e3. doi: 10.1016/j.chom.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 103.McElroy AK, et al. A case of human Lassa virus infection with robust acute T-cell activation and long-term virus-specific T-cell responses. J. Infect. Dis. 2017;215:1862–1872. doi: 10.1093/infdis/jix201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pshenichnaya NY, Sydenko IS, Klinovaya EP, Romanova EB, Zhuravlev AS. Possible sexual transmission of Crimean-Congo hemorrhagic fever. Int. J. Infect. Dis. 2016;45:109–111. doi: 10.1016/j.ijid.2016.02.1008. [DOI] [PubMed] [Google Scholar]
- 105.Richmond JK, Baglole DJ. Lassa fever: epidemiology, clinical features, and social consequences. BMJ. 2003;327:1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith DR, et al. Persistent Crimean-Congo hemorrhagic fever virus infection in the testes and within granulomas of non-human primates with latent tuberculosis. PLoS Pathog. 2019;15:e1008050. doi: 10.1371/journal.ppat.1008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dokubo EK, et al. Persistence of Ebola virus after the end of widespread transmission in Liberia: an outbreak report. Lancet Infect. Dis. 2018;18:1015–1024. doi: 10.1016/S1473-3099(18)30417-1. [DOI] [PubMed] [Google Scholar]
- 108.Gupta M, et al. Persistent infection with Ebola virus under conditions of partial immunity. J. Virol. 2004;78:958–967. doi: 10.1128/JVI.78.2.958-967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sissoko D, et al. Ebola virus persistence in breast milk after no reported illness: a likely source of virus transmission from mother to child. Clin. Infect. Dis. 2017;64:513–516. doi: 10.1093/cid/ciw793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cashman KA, et al. Immune-mediated systemic vasculitis as the proposed cause of sudden-onset sensorineural hearing loss following Lassa virus exposure in cynomolgus macaques. mBio. 2018;9:e01896–18. doi: 10.1128/mBio.01896-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu DX, et al. Persistence of Lassa virus associated with severe systemic arteritis in convalescing guinea pigs (Cavia porcellus) J. Infect. Dis. 2019;219:1818–1822. doi: 10.1093/infdis/jiy641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Becquart P, et al. High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One. 2010;5:e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diallo MSK, et al. Prevalence of infection among asymptomatic and paucisymptomatic contact persons exposed to Ebola virus in Guinea: a retrospective, cross-sectional observational study. Lancet Infect. Dis. 2019;19:308–316. doi: 10.1016/S1473-3099(18)30649-2. [DOI] [PubMed] [Google Scholar]
- 114.Glynn JR, et al. Asymptomatic infection and unrecognised Ebola virus disease in Ebola-affected households in Sierra Leone: a cross-sectional study using a new non-invasive assay for antibodies to Ebola virus. Lancet Infect. Dis. 2017;17:645–653. doi: 10.1016/S1473-3099(17)30111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Leroy EM, et al. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. doi: 10.1016/S0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- 116.Moyen N, et al. Risk factors associated with Ebola and Marburg viruses seroprevalence in blood donors in the Republic of Congo. PLoS Negl. Trop. Dis. 2015;9:e0003833. doi: 10.1371/journal.pntd.0003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nkoghe D, et al. Risk factors for Zaire ebolavirus-specific IgG in rural Gabonese populations. J. Infect. Dis. 2011;204 (Suppl. 3):S768–S775. doi: 10.1093/infdis/jir344. [DOI] [PubMed] [Google Scholar]
- 118.Ruibal P, et al. Unique human immune signature of Ebola virus disease in Guinea. Nature. 2016;533:100–104. doi: 10.1038/nature17949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 2002;76:406–410. doi: 10.1128/JVI.76.1.406-410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Volchkov VE, et al. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291:1965–1969. doi: 10.1126/science.1057269. [DOI] [PubMed] [Google Scholar]
- 121.Cross RW, Geisbert TW. Use of reverse genetics to inform Ebola outbreak responses. Lancet Infect. Dis. 2019;19:925–927. doi: 10.1016/S1473-3099(19)30346-9. [DOI] [PubMed] [Google Scholar]
- 122.Hoenen T, Groseth A, Feldmann H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 2019;17:593–606. doi: 10.1038/s41579-019-0233-2. [DOI] [PubMed] [Google Scholar]
- 123.Chisholm PJ, Busch JW, Crowder DW. Effects of life history and ecology on virus evolutionary potential. Virus Res. 2019;265:1–9. doi: 10.1016/j.virusres.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 124.Warfield KL, et al. Role of antibodies in protection against Ebola virus in nonhuman primates immunized with three vaccine platforms. J. Infect. Dis. 2018;218:S553–S564. doi: 10.1093/infdis/jiy316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wong G, et al. Immune parameters correlate with protection against Ebola virus infection in rodents and nonhuman primates. Sci. Transl Med. 2012;4:158ra146. doi: 10.1126/scitranslmed.3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garbutt M, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 2004;78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang Z-Y, et al. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78654. [DOI] [PubMed] [Google Scholar]
- 128.Henao-Restrepo AM, et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.National Institute of Allergy and Infectious Diseases. Independent Monitoring Board Recommends Early Termination of Ebola Therapeutics Trial in DRC Because of Favorable Results with Two of Four Candidates. https://www.niaid.nih.gov/news-events/independent-monitoring-board-recommends-early-termination-ebola-therapeutics-trial-drc (2019).
- 130.Mulangu S, et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Corti D, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 132.Wec AZ, et al. Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell. 2017;169:878–890.e15. doi: 10.1016/j.cell.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gaudinski MR, et al. Safety, tolerability, pharmacokinetics, and immunogenicity of the therapeutic monoclonal antibody mAb114 targeting Ebola virus glycoprotein (VRC 608): an open-label phase 1 study. Lancet. 2019;393:889–898. doi: 10.1016/S0140-6736(19)30036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Misasi J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pascal KE, et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis. 2018;218 (Suppl. 5):S612–S626. doi: 10.1093/infdis/jiy285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qiu X, et al. Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci. Transl Med. 2012;4:138ra81. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 137.Audet J, et al. Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus. Sci. Rep. 2014;4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davidson E, et al. Mechanism of binding to Ebola virus glycoprotein by the ZMapp, ZMAb, and MB-003 cocktail antibodies. J. Virol. 2015;89:10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miller CR, et al. Initiating a watch list for Ebola virus antibody escape mutations. PeerJ. 2016;4:e1674. doi: 10.7717/peerj.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Patel JS, Quates CJ, Johnson EL, Ytreberg FM. Expanding the watch list for potential Ebola virus antibody escape mutations. PLoS One. 2019;14:e0211093. doi: 10.1371/journal.pone.0211093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kugelman JR, et al. Emergence of Ebola virus escape variants in infected nonhuman primates treated with the MB-003 antibody cocktail. Cell Rep. 2015;12:2111–2120. doi: 10.1016/j.celrep.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 142.Johnson WE. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019;17:355–370. doi: 10.1038/s41579-019-0189-2. [DOI] [PubMed] [Google Scholar]
- 143.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 144.Kryukov K, Ueda MT, Imanishi T, Nakagawa S. Systematic survey of non-retroviral virus-like elements in eukaryotic genomes. Virus Res. 2019;262:30–36. doi: 10.1016/j.virusres.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 145.Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient Bornavirus and Ebolavirus/Marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Horie M, Tomonaga K. Paleovirology of Bornaviruses: what can be learned from molecular fossils of Bornaviruses. Virus Res. 2019;262:2–9. doi: 10.1016/j.virusres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 147.Chiba S, et al. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011;7:e1002146. doi: 10.1371/journal.ppat.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.ter Horst AM, Nigg JC, Dekker FM, Falk BW. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-interacting RNAs. J. Virol. 2019;93:e02124–18. doi: 10.1128/JVI.02124-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Taylor DJ, Dittmar K, Ballinger MJ, Bruenn JA. Evolutionary maintenance of filovirus-like genes in bat genomes. BMC Evol. Biol. 2011;11:336. doi: 10.1186/1471-2148-11-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taylor DJ, Leach RW, Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 2010;10:193. doi: 10.1186/1471-2148-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 2019;14:319–338. doi: 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Faria NR, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]