Abstract
Chronic hepatitis B is caused by prolonged infection with the hepatitis B virus (HBV), which can substantially increase the risk of developing liver disease. Despite the development of preventive vaccines against HBV, a therapeutic vaccine inducing an effective antibody response still remains elusive. The preS1 domain of the large HBV surface protein is the major viral attachment site on hepatocytes and thus offers a therapeutic target; however, its poor immunogenicity limits clinical translation. Here, we design a ferritin nanoparticle vaccine that can deliver preS1 to specific myeloid cells, including SIGNR1+ dendritic cells (which activate T follicular helper cells) and lymphatic sinus-associated SIGNR1+ macrophages (which can activate B cells). This nanoparticle vaccine induces a high-level and persistent anti-preS1 response that results in efficient viral clearance and partial serological conversion in a chronic HBV mouse model, offering a promising translatable vaccination strategy for the functional cure of chronic hepatitis B.
Subject terms: Nanobiotechnology, Nanoparticles, Biotechnology
A ferritin nanoparticle that delivers the preS1 domain of the large hepatitis B surface protein to two specific myeloid cell populations provides a therapeutic vaccination strategy for the treatment of chronic hepatitis B.
Main
Hepatitis B is a major global health issue caused by the infection of hepatitis B virus (HBV), which attacks the liver and can cause both acute and chronic diseases1. It is estimated that 257 million people worldwide are HBV chronic carriers and have a high risk of developing liver disease, cirrhosis and hepatocellular carcinoma. Approximately 1 million chronic hepatitis B (CHB) patients die from these late complications each year. Conventional hepatitis B surface antigen (HBsAg) vaccination induces a protective antibody in most healthy vaccinated populations and has efficiently decreased the incidence rate of new HBV infections2. However, HBsAg fails to induce an effective antibody response in either preclinical animal models or patients with CHB infection, probably due to antigen-specific immune tolerance induced by the high load of viral antigens3. Therefore, an effective treatment strategy to eliminate and eradicate hepatitis B is highly desired.
Compared with HBsAg, the preS1 domain of the HBV large surface antigen has been suggested to be an ideal immunogen candidate for therapeutic vaccine development. PreS1 plays a pivotal role in HBV entry by interacting with sodium taurocholate cotransporting polypeptide (NTCP), identified as the cellular receptor for HBV4,5. Monoclonal antibodies against preS1 (MA18/7 and KR127) have been demonstrated to potently inhibit HBV infection in vivo and in vitro6,7. In addition, anti-preS1 can also eliminate HBV+ cells via antibody-dependent cell-mediated cytotoxicity and phagocytosis8. Clinically, anti-preS1 is an early serum marker for HBV clearance9,10. All these data suggest that anti-preS1 response might be beneficial for CHB treatment. Moreover, in CHB patients, immune tolerance to preS1, if any, appears to be much lower than that to HBsAg3,11, as it only occurs in a trace amount compared with the latter in CHB patients12,13, which favours antibody response induction. In fact, we recently showed that preS1 antigen vaccination, but not HBsAg, induced a neutralizing therapeutic antibody response in HBV carrier mice11. Nonetheless, preS1 appears not to be an efficient antibody inducer compared with HBsAg. This is reflected under both natural infections and active vaccinations11,14,15. Therefore, how to improve the immunogenicity of preS1 for enhanced therapeutic antibody response is currently a key issue for preS1-based therapeutic vaccine development.
Nanoparticles (NPs) have emerged as an important platform for vaccine development. Lymph node (LN) antigen-presenting cell (APC) targeting has been frequently considered to be a valuable feature for many NP-based vaccines16. For preS1 vaccine, HBV core NPs have also been used7,17. However, given the complex composition of APCs and diversified functions of different APC subsets, the specific roles of different APC subsets during NP vaccine immunization seem to be vaguely understood18,19. Moreover, it has been unknown how to coordinately target them for an efficient adaptive immune response.
Ferritin self-assembling NPs have recently become widely appreciated for vaccine design20–24. However, the underlying immunological mechanism is still not fully understood. In the current study, we rationally designed a ferritin NP–preS1 vaccine. This vaccine demonstrates impressive preventive and therapeutic effects in mouse models. Mechanistically, the ferritin NPs are discovered to coordinately target distinct SIGNR1+ APCs for T follicular helper (Tfh) cell induction and B cell activation, leading to enhanced antibody production. Importantly, we have also uncovered a previously unrecognized mechanism: that macrophages capture NP–antigens for B cell activation in a CXCR5-dependent manner.
Ferritin NP–preS1 induces efficient antibody response
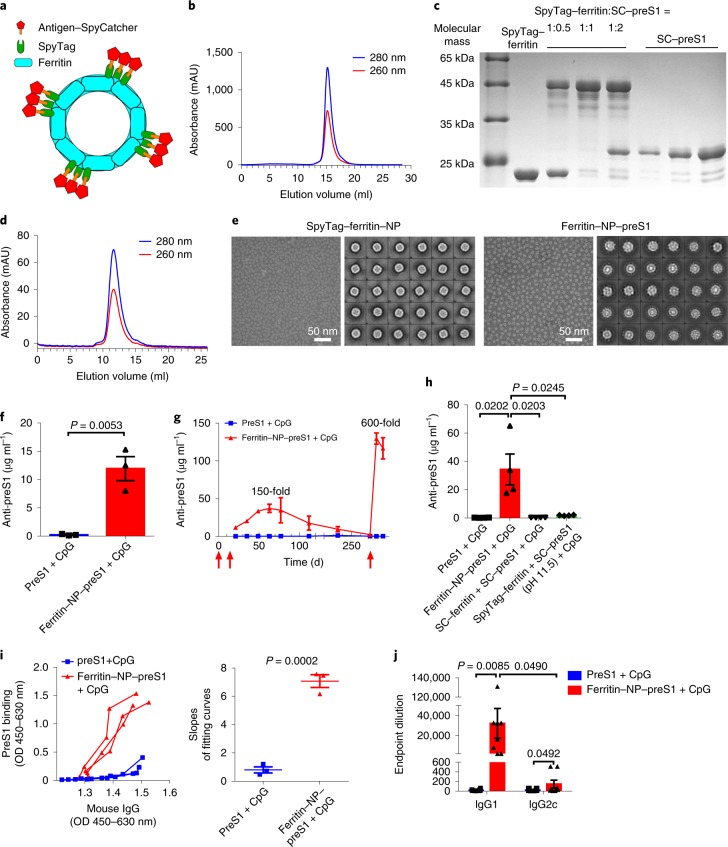
Ferritin NP–preS1 was generated taking advantage of a modified SpyTag/SpyCatcher technique25,26 (Fig. 1a). SpyTag–ferritin and ΔN SpyCatcher-fused preS1 (SC–preS1) were both expressed in Escherichia coli. The self-assembly of SpyTag–ferritin NPs was suggested by size exclusion chromatography (Fig. 1b). SpyTag- and SpyCatcher-mediated covalent conjugation was confirmed by SDS–polyacrylamide gel electrophoresis (Fig. 1c). The ferritin NP–preS1 was further purified by size exclusion chromatography, and its slightly larger diameter was reflected by its earlier elution (Fig. 1d). The self-assembled NPs were confirmed using transmission electron microscopy (TEM) (Fig. 1e).
Fig. 1. Molecular design and characterization of ferritin NP vaccine.
a, Schematic illustration of ferritin-based vaccine delivery vehicle. b, Size exclusion chromatography of SpyTag–ferritin NPs. The ultraviolet absorptions at 280 nm and 260 nm are shown. AU, absorbance unit. c, SDS–polyacrylamide gel electrophoresis analysis of the covalent ligation of SpyTag–ferritin (~22.4 kDa) and SC–preS1 (~25.4 kDa) at 1:0.5, 1:1 and 1:2 mole ratios at 4 °C overnight. d, Size exclusion chromatography of ferritin NP–preS1. e, TEM images and two-dimensional reconstruction of SpyTag–ferritin NP and ferritin NP–preS1. Panels b–e show representative results of at least three independent experiments. f, Anti-preS1 response at vaccine immunization on day 21 was analysed using enzyme-linked immunosorbent assay (ELISA) (n = 3 mice per group). Data are representative of five independent experiments. g, Kinetics of anti-preS1 levels after immunization (n = 3). The red arrows indicate the immunization times. h, Comparison of conjugated and unconjugated vaccines on anti-preS1 response. i, Anti-preS1 IgG avidity on vaccine immunization was detected with ELISA. Each curve represents an individual serum sample (n = 3) (left). The slopes of each fitting curve were calculated (right). OD, optical density. j, Titres of preS1-specific IgG1 and IgG2c in the sera from ferritin NP–preS1 vaccine (n = 8) or SC–preS1 (n = 14) immunized mice on day 21. g–j show representative results of three independent experiments. In f–j, data are shown as mean ± s.e.m.; statistical significance was determined using an unpaired two-tailed t-test. .
To assess the immunogenicity of the ferritin NP–preS1, naïve wild-type (WT) C57BL/6 mice were immunized as described in Methods. Ferritin NP–preS1 vaccine induced a 50 times higher antibody level than soluble SC–preS1 at day 21 (Fig. 1f). The anti-preS1 level continued to increase in the ferritin NP–preS1 immunization group and reached a peak around day 60, when an approximately 150 times higher antibody level was measured compared with the SC–preS1 control vaccine group, in which the antibody level still remained low. The antibody response gradually decreased but remained detectable at day 235. Furthermore, a boost immunization elicited a much stronger antibody response in the ferritin NP–preS1 group compared with the SC–preS1 control group (approximately 600-fold) (Fig. 1g). The enhanced immunogenicity was also seen in the absence of CpG-1826 adjuvant (Supplementary Fig. 1a), suggesting an intrinsic feature of ferritin NPs, probably due to repeated antigen display and an NP carrier effect16. However, in the presence of CpG, the enhancement becomes more marked, suggesting a coordination between a ferritin NP intrinsic feature and an adjuvant-induced innate response.
The markedly enhanced antibody response is not a dose-dependent effect (Supplementary Fig. 1b). The SC portion itself that was fused to preS1 has no effect on the immunogenicity of SC–preS1 (Supplementary Fig. 1c). The foreignness of the bacterial ferritin is also not responsible for the enhanced antibody response, as preS1 displayed by a mouse ferritin H chain generated a comparable antibody level to bacterial ferritin (Supplementary Fig. 1d). The efficient antibody response is also not restricted to mouse strains (Supplementary Fig. 1e). In addition, physically mixing preS1 antigens with ferritin NPs instead of covalent binding could not elicit a strong anti-preS1 response, suggesting the necessity of antigen display on the ferritin NPs (Fig. 1h). Thus, preS1 conjugated to ferritin NPs generally enhanced the immunogenicity of preS1 antigen and elicited an efficient antibody response.
Further characterization showed that anti-preS1 antibody induced by ferritin NP–preS1 vaccination has higher avidity than that induced by SC–preS1 (Fig. 1i). IgG1, and not IgG2c, was the dominant anti-preS1 isotype after ferritin NP–preS1 immunization (Fig. 1j). Regarding the safety perspective, no cross-reactive antibodies to mouse ferritin were induced after Pyrococcus furiosus (Pf) ferritin NP–preS1 immunization in mice, although anti-Pf ferritin was readily induced (Supplementary Fig. 1f). In addition, the serum levels of both glutamic–pyruvic transaminase and glutamic–oxaloacetic transaminase were normal (Supplementary Fig. 1g). The minimal iron content of ferritin NPs, compared with the iron storage capacity of 2,700 Fe per 24-mer for Pf ferritin27, was unlikely to affect iron homoeostasis in vivo (Supplementary Fig. 1h)20. Taken together, these results demonstrate notable immunogenicity and biosafety for the ferritin-NP-based preS1 vaccine.
Protective and therapeutic effect of ferritin NP–preS1
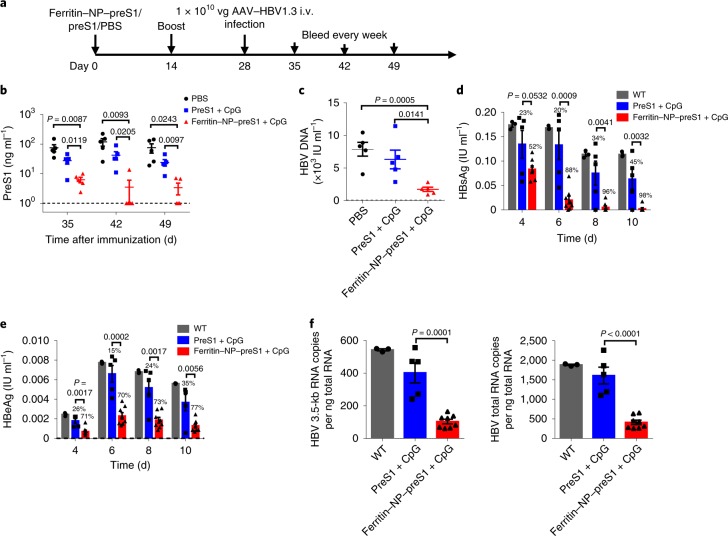
To test whether preS1 antisera induced by the ferritin NP–preS1 vaccine could prevent HBV infection, vaccine-immunized mice were challenged with adeno-associated virus (AAV)–HBV1.3 two weeks after the second immunization (Fig. 2a). PreS1 antigen and HBV DNA in the sera were greatly inhibited in the ferritin NP–preS1 vaccine group compared with the control groups (Fig. 2b,c), indicating effective clearance of infectious HBV particles in vivo, probably due to rapid elimination.
Fig. 2. Protective immunity of ferritin NP–preS1 vaccine against HBV.
a, Naïve WT C57BL/6 mice (n = 5) were subcutaneously immunized with 200 pmol ferritin NP–preS1 vaccine or equimolar SC–preS1 soluble antigen with 30 μg CpG-1826 on days 0 and 14. On day 28, mice were infected with 1 × 1010 viral genome equivalents (vg) AAV–HBV1.3 virus intravenously (i.v.). Blood samples were collected every week from day 35. b, The serum level of preS1 after AAV–HBV1.3 infection was detected using ELISA. c, The serum HBV DNA level on day 35 was quantified by real-time PCR. b and c show representative results of three independent experiments. d–f, HepG2-hNTCP cells were incubated with 1 × 107 vg HBV in the presence of immune sera collected from ferritin NP–preS1 (n = 8) or SC–preS1 (n = 5) immunized mice or WT mice (n = 3). After 24 h incubation, HepG2-hNTCP cells were washed and cultured in maintenance medium for 10 d. The culture supernatant was changed and collected every 2 d. d,e, Levels of HBsAg (d) and HBeAg (e) in the supernatant were measured using ELISA. On day 10, HepG2-hNTCP cells were collected in TRIzol and RNA was extracted. f, HBV 3.5-kb RNA and HBV total RNA were quantified using real-time PCR. The detection limits in b–f are shown as dashed lines. d–f show representative results of two independent experiments. In b–f, data are shown as mean ± s.e.m.; statistical significance was determined using an unpaired two-tailed t-test.
To evaluate the neutralizing capability of the antisera, HepG2-hNTCP cells were infected with HBV in the presence of sera from different immunization groups as described previously28. The levels of HBsAg and HBeAg in cell culture media were significantly inhibited by the sera from ferritin NP–preS1-immunized mice, in stark contrast with the preS1-immunization group (Fig. 2d,e). To more accurately measure HBV infection inhibition, the levels of HBV 3.5-kb RNA and HBV total RNA, indicating HBV viral replication in HepG2-hNTCP cells, were also determined, and significant inhibition by the sera of the ferritin NP–preS1 group but not the control groups was found (Fig. 2f). These results confirm that the antisera to preS1 elicited by the ferritin NP–preS1 vaccine can effectively prevent HBV infection of hepatocytes in vitro. Thus, both in vivo and in vitro results suggest that the ferritin NP–preS1 vaccine is an effective HBV prophylactic vaccine.
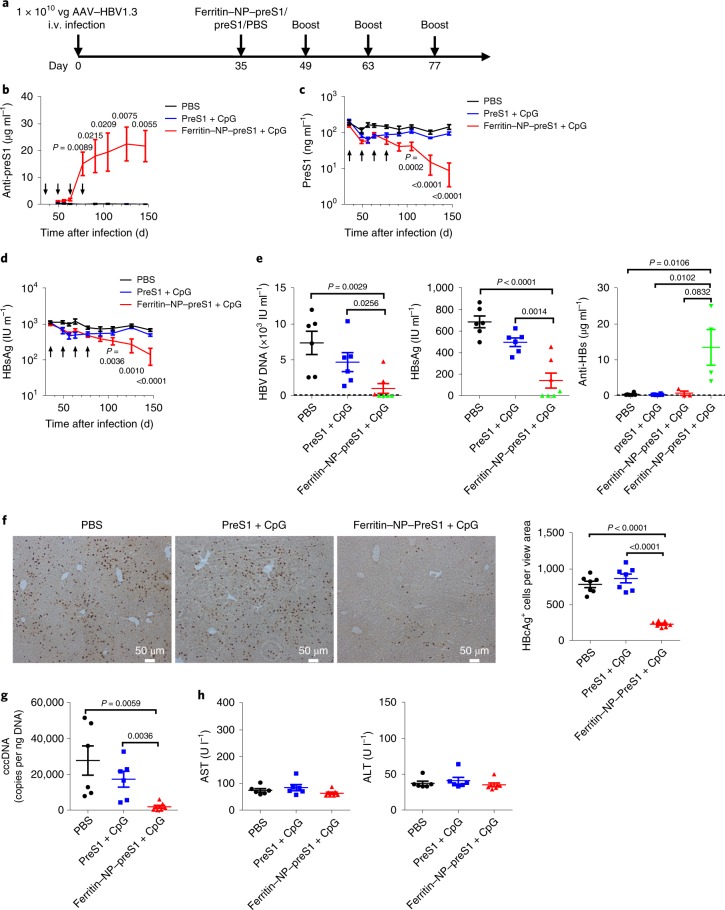
To determine the therapeutic effect of the ferritin NP–preS1 vaccine, HBV carrier mice were established11, and immunized with equimolar ferritin NP–preS1 vaccine or SC–preS1 four times every two weeks (Fig. 3a). There is no detectable anti-preS1 response in the established HBV carrier mice, consistent with the clinical observation29,30. However, after the second boost on day 63, the ferritin NP–preS1 vaccine induced a significant amount of anti-preS1, which was barely detectable in the SC–preS1 group (Fig. 3b). Meanwhile, preS1 antigen and HBsAg in the sera declined significantly with the induction of anti-preS1 (Fig. 3c,d). On day 147, about 10 weeks after the last immunization, serum HBV DNA was also significantly reduced in the ferritin NP–preS1 group but not in the control groups (Fig. 3e). Moreover, in four out of seven mice in the ferritin NP–preS1 group, serum HBV DNA dropped to undetectable levels, also showing barely detectable HBsAg and significant amounts of anti-HBs (Fig. 3e), suggesting potential for a functional cure. In addition, the HBV core antigen as detected by immunological histological chemistry staining (Fig. 3f) and covalently closed circular DNA (cccDNA) as detected by quantitative real-time PCR (Fig. 3g) were also significantly diminished in the liver of the ferritin NP–preS1 group. The clearance of HBV core antigen in the liver was not accompanied by liver injury (Fig. 3h). Thus, ferritin–preS1 NPs plus CpG functioned as an effective and non-pathogenic therapeutic vaccine in the mouse model of protracted HBV infection.
Fig. 3. Ferritin NP–preS1 as therapeutic vaccine.
a, C57BL/6 mice were inoculated with 1 × 1010 vg AAV–HBV1.3 virus intravenously. After 5 weeks, stable HBV carrier mice were vaccinated with 500 pmol ferritin NP–preS1 vaccine (n = 7) or equimolar SC–preS1 soluble antigen (n = 6) with 30 μg CpG-1826 adjuvant four times every 2 weeks. Sera were collected every 2 weeks. b–d, Serum levels of anti-preS1 (b), preS1 antigen (c) and HBsAg (d) were determined using ELISA. There is no difference between PBS (n = 6) and SC–preS1, and the P values shown in the figure are statistical significances between PBS and ferritin NP–preS1. IU, international units. e, Serum levels of HBV DNA, HBsAg and anti-HBs on day 147. Green dots represent the same mice across the three panels with undetectable HBV DNA. f, Immunological histological chemistry staining for hepatitis B core antigen (HBcAg) in hepatocytes on day 147. Positive cells were counted using ImageJ software in each section field (n = 7 from three mice). g, HBV cccDNA in the liver quantified using real-time PCR on day 147. h, Glutamic–pyruvic transaminase (ALT) and glutamic–oxaloacetic transaminase (AST) measured for the sera collected on day 147. b–h show representative results of four independent experiments. Data are shown as mean ± s.e.m.; statistical significance was determined using an unpaired two-tailed t-test.
Interferon-γ (IFN-γ) is an important activator for antibody-dependent cell-mediated cytotoxicity and phagocytosis31, which have been suggested to mediate anti-preS1 for therapeutic effect8. On ferritin NP–preS1 vaccination, significant numbers of preS1-specific IFN-γ-producing T cells were detected using enzyme-linked immune absorbent spot (ELISpot) assay in LNs, spleens and liver tissues (Supplementary Fig. 2a). Furthermore, IFN-γ blockade with a blocking antibody (XMG1.2) almost completely abolished the therapeutic effect on HBV core antigen clearance (Supplementary Fig. 2b,c). These data suggest that the Th1 response induced by ferritin NP–preS1 vaccine might play an important role in achieving therapeutic efficacy: enhancing IFN-γ response may synergize with ferritin NP–preS1 vaccine.
Ferritin NP vaccine targets distinct APCs
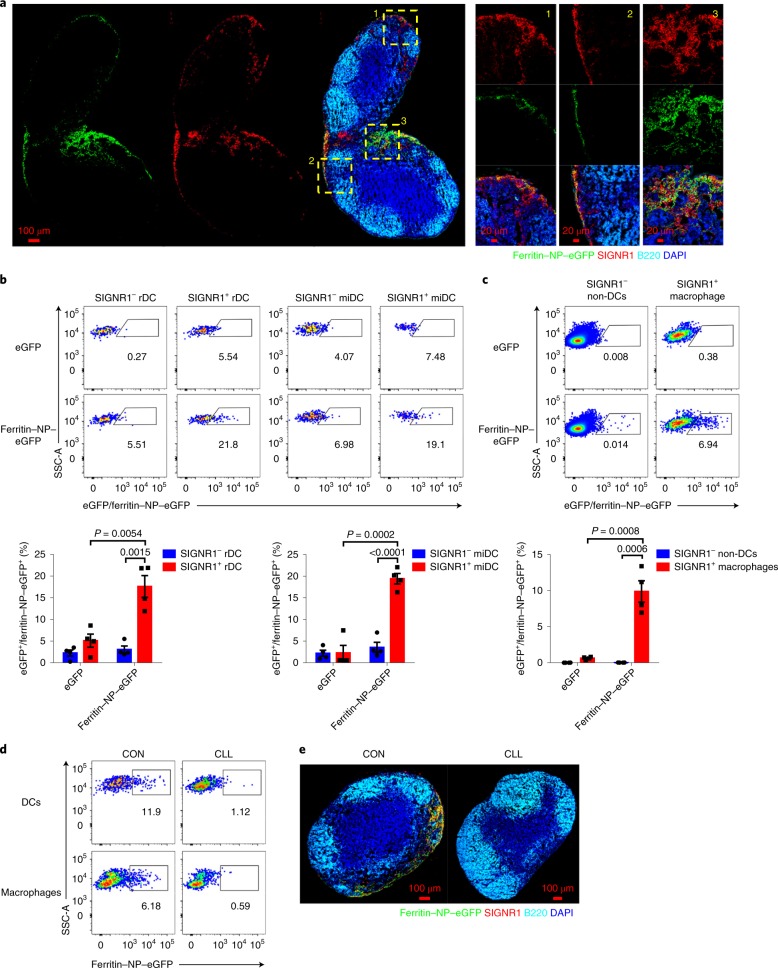
To understand how the ferritin NP vaccine is recognized and processed by the host immune system, fluorescein-labelled ferritin NPs were generated to trace their path. On subcutaneous injection, ferritin NP–eGFP (enhanced green fluorescent protein) was abundantly located in the medullary regions and also but to a lesser extent in the interfollicular region (Fig. 4a). Interestingly, ferritin NP–eGFP was highly colocalized with cells with SIGNR1 expression. SIGNR1 mediates the recognition of several virus particles32,33. This finding prompted us to hypothesize that ferritin NP–eGFP was actively captured by SIGNR1+ cells during their passive transportation and retained in LNs. To further confirm this and examine the ferritin NP cell-targeting property in more detail, ferritin NP capture by phagocytic cells was determined by flow cytometry. Supportively, SIGNR1+ resident dendritic cells (rDCs) and migratory DCs (miDCs) (Supplementary Fig. 3a) showed significantly higher GFP immunofluorescence intensity than their SIGNR1− counterparts, indicating the preferential capture of ferritin NPs by SIGNR1+ DCs (Fig. 4b). Similarly, in non-DC cells, the SIGNR1+ population, which was mostly CD169+ macrophages, also showed preferential capture of ferritin NPs (Fig. 4c). As control, the soluble eGFP was almost undetectable in either SIGNR1+ or SIGNR1− subsets. Similar results were found when fluorescein isothiocyanate (FITC) was used to label ferritin NP–preS1 or SC–preS1 (Supplementary Fig. 3b–d). The CpG-1826 adjuvant had no influence on these captures (Supplementary Fig. 4). Therefore, ferritin NPs appear to be predominantly captured by both SIGNR1+ DCs and SIGNR1+ macrophages in the draining LNs (dLNs).
Fig. 4. Ferritin NP vaccine targets distinct APCs.
a, C57BL/6 mice were subcutaneously injected with 2 nmol ferritin NP–eGFP. 4 h after injection, cryosections of inguinal LNs (iLNs) were obtained. The section was stained with anti-GFP, anti-SIGNR1, anti-B220 and 4,6-diamidino-2-phenylindole (DAPI). Three representative areas of interfollicular region and medulla are shown in magnification. Representative results of four independent experiments. b,c, C57BL/6 mice were subcutaneously injected with 2 nmol ferritin NP–eGFP (n = 4) or equimolar eGFP–SpyTag (n = 4). 4 h after injection, iLNs were digested into single cells and the capture of ferritin NP–eGFP or eGFP–SpyTag by the indicated DCs (b) or non-DCs (c) was presented and statistically analysed. Numbers adjacent to the outlined areas indicate the percentage of each gate. Representative results of three independent experiments. Data are shown as mean ± s.e.m.; statistical significance was determined using an unpaired two-tailed t-test. SSC-A, side scatter area. d, 10 d after CLL or control liposome (CON) f.p. injection, the capture of ferritin NP–eGFP by DCs or CD11c−CD11b+ macrophages was analysed. e, 10 d after CLL or CON treatment, ferritin NP–eGFP accumulation in pLNs 2 h after f,p. injection was determined by immunofluorescence microscopy. d and e show representative results of three independent experiments.
To further evaluate the role of SIGNR1+ cells in ferritin NP–eGFP capture, clodronate liposome (CLL) was injected into the footpads (f.p.) to eliminate APCs from popliteal LNs (pLNs)34. Both SIGNR1+ DCs and SIGNR1+ macrophages were efficiently depleted (Supplementary Fig. 3e). The capture of ferritin NP–eGFP was almost completely lost after CLL treatment (Fig. 4d,e), suggesting an essential role of SIGNR1+ cells in ferritin NP retention. This preferential capture by SIGNR1+ APCs is not only because of their location-determined accessibility to injected ferritin NPs. In fact, in vitro coculture of digested single-cell suspensions from LNs with ferritin NPs demonstrated similar preferential capture by SIGNR1+ APCs (Supplementary Fig. 5), suggesting an intrinsic feature of ferritin NPs or SIGNR1+ APCs.
Although SIGNR1 expression is highly positively correlated with ferritin NP capture, it does not seem to be essential, at least on its own, for ferritin NP capture, as shown in the data obtained from Signr1-deficient mice (Supplementary Fig. 6). Given the presence of several other orthologues of SIGNR1 in mouse35,36, the exact function of SIGNR1 and this gene family in ferritin NP capture remains to be determined in the future.
To further determine whether this preferential targeting is transferable to the human situation, we have tested its capture using digested single cells from human LNs. The human homologue of murine SIGNR1 is DC-SIGN (CD209), not only in terms of genomic structure, but also for cell distribution and function in LNs37,38. Flow cytometric analysis showed that ferritin NPs were indeed preferentially captured by DC-SIGN+ macrophages compared with DC-SIGN− macrophages (Supplementary Fig. 7a), probably supporting the feasibility of clinical translation.
Coordination of SIGNR1+ DCs and macrophages
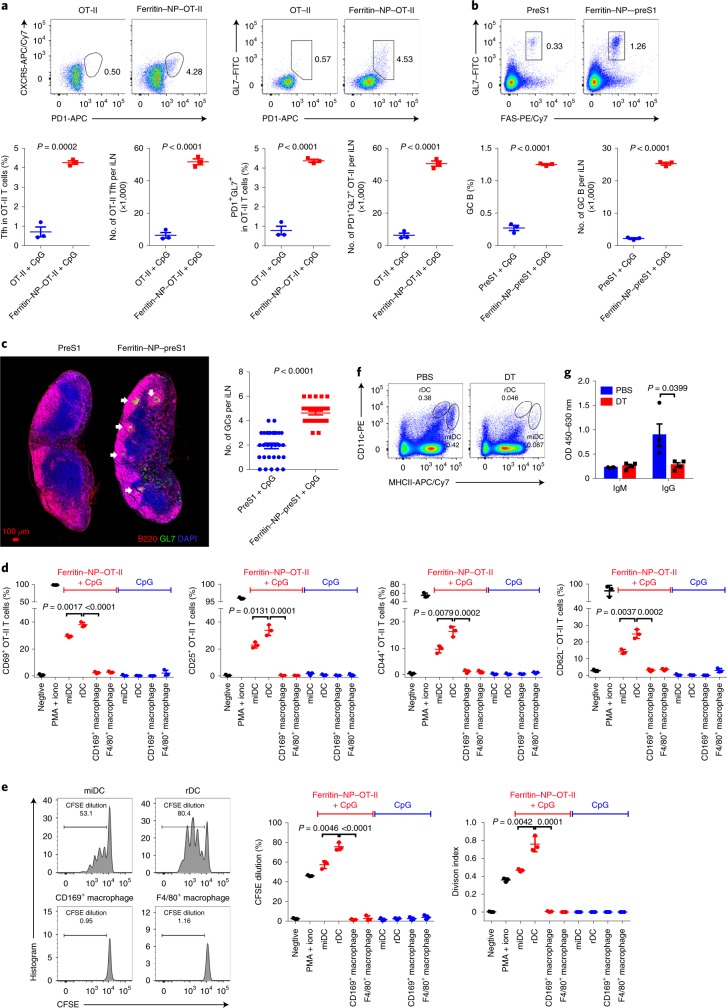
Germinal centre (GC) formation is essential for a strong antibody response, and the affinity selection of GC B cells is dependent on CD4+ Tfh cells39. Employing the well established OVA-OT-II antigen-specific CD4+ T cell system, we found that ferritin NP–OT-II induced about eight times as many CXCR5+PD1+ and PD1+GL7+ Tfh cells compared with control SC–OT-II 6 d after immunization (Fig. 5a). Furthermore, about 11 times more GC B (FAS+GL7+) cells were detected in the ferritin NP–preS1 vaccine group compared with the SC–preS1 control group (Fig. 5b). Histological analysis revealed that GC numbers in ferritin NP–preS1-immunized dLNs were also significantly higher than those in the SC–preS1 group (Fig. 5c). Therefore, ferritin NP vaccine is an efficient antigen-delivery platform to enhance Tfh and GC B responses.
Fig. 5. Ferritin NP vaccine induces efficient Tfh and GC responses.
a, Naïve WT Thy1.2 C57BL/6 mice (n = 3) were adoptively transferred with 1 × 107 Thy1.1+ OT-II splenocytes and immunized. On day 6, CD4+Thy1.1+TCR Vα2+ OT-II Tfh cells from LNs were analysed by flow cytometry (TCR, T-cell antigen receptor). The percentage and quantification of OT-II Tfh cells are shown. b, On day 63 of immunization (peak anti-preS1 response), GC B cells from iLNs were analysed by flow cytometry. a and b show representative results of three independent experiments. c, Cryosections of iLNs were stained with anti-B220, anti-GL7 and DAPI. The number of GCs was counted. Data were pooled from n = 30 slide sections from 10 mice in three independent experiments. d,e, 12 h after immunization, rDCs (CD103−), miDCs (CD103+), CD169+ macrophages (CD11c−CD11b+CD169+) and F4/80+ macrophages (CD11c−CD11b+CD169−F4/80+) were sorted from dLNs, and then incubated with naïve OT-II T cells (n = 3 culture wells). OT-II T cell activation (d) and carboxyfluorescein succinimidyl ester (CFSE)-labelled OT-II T cell proliferation (e) were determined by flow cytometry at 24 h and 72 h, respectively. Representative results of five independent experiments. iono, ionomycin. f, Flow cytometry analysis confirmed DC depletion in iLNs of CD11c-DTR bone marrow chimaeric mice treated with DT. Data show representative results of three independent experiments. g, PreS1-specific IgM and IgG responses were detected in CD11c-depleted mice on day 10 at immunization (n = 4). Data are representative of three independent experiments. In a–e and g, data are shown as mean ± s.e.m.; statistical significance was determined using an unpaired two-tailed t-test.
All rDCs, miDCs and SIGNR1+ (CD169+) macrophages efficiently capture ferritin NPs on immunization. To study which cells induce T cell activation, dLNs were collected 12 h after ferritin NP–OT-II vaccine immunization. Four populations of APCs were sorted according to the gating strategy (Supplementary Fig. 8) and cocultured with OT-II T cells. Macrophages did not induce a detectable level of OT-II T cell activation or proliferation (Fig. 5d,e). Interestingly, rDCs induced a significantly higher degree of T cell activation and proliferation than miDCs (Fig. 5d,e). Therefore, rDCs appear to play a major role in Tfh generation compared with miDCs.
For particulate antigen immunization, DCs can carry antigens to interfollicles for B cell activation32. To directly determine the role of DCs in B cell activation in the current scenario, rDCs and miDCs were depleted in CD11c–DTR (diphtheria toxin receptor) bone marrow chimaeric mice with diphtheria toxin (DT), with PBS-treated mice as controls (Fig. 5f). DC depletion resulted in a lower titre of IgG response, indicating that DCs played an essential role in T-cell-dependent antibody response (Fig. 5g). However, IgM antibodies, produced by early-activated B cells largely in a T-cell-independent manner32, were not affected (Fig. 5g). Thus, while DCs play a major role in T cell activation, they are dispensable for early B cell activation during ferritin NP vaccine immunization.
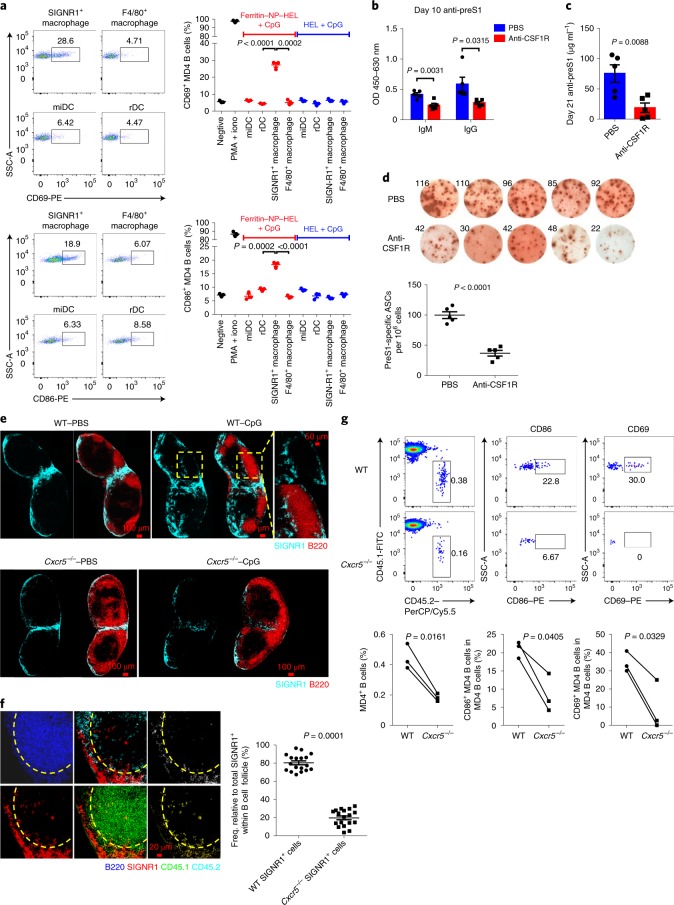
In addition to DCs, SIGNR1+ macrophages captured ferritin NP efficiently (Fig. 4c). SIGNR1+ macrophages showed markedly reduced transcript abundance for lysosome-associated membrane proteins-1/2, lysozyme and lysosomal proteases compared with SIGNR1−F4/80+ (abbreviated as F4/80+) macrophages (Supplementary Fig. 9a), indicating their poor degradation ability and potential role as antigen retainers for B cell activation. To directly test this, four subsets of APCs (miDCs, rDCs, SIGNR1+ macrophages and F4/80+ macrophages) were isolated as shown in Supplementary Fig. 8 from dLNs 3 d after ferritin NP–HEL (hen egg lysozyme) immunization and were incubated with MD4 B cells in vitro for 24 h. Except for SIGNR1+ macrophages, no other populations activated MD4 B cells as demonstrated by the upregulation of CD69 and CD86 (Fig. 6a), indicating a durable retention of antigen by SIGNR1+ macrophages. In vivo, a CSF1R-blocking antibody-mediated SIGNR1+ macrophage depletion (Supplementary Fig. 9b)33 resulted in significantly reduced anti-preS1 IgM and IgG 10 d after the first immunization (Fig. 6b). On day 21, significantly lower levels of anti-preS1 IgG and fewer preS1-specific IgG antibody-secreting cells (ASCs) were also observed in anti-CSF1R-treated mice (Fig. 6c,d). Thus, SIGNR1+ macrophages play a vital role in early B cell activation and IgG response.
Fig. 6. SIGNR1+ macrophages are required for B cell activation and antibody production.
a, On day 3 at f.p. immunization in WT mice, rDCs, miDCs, SIGNR1+ macrophages and F4/80+ macrophages were sorted from pLNs and incubated with MD4 B cells (n = 3 culture wells). Activation of MD4 B cells was detected 24 h later. Data are representative results of four independent experiments. b–d, WT mice (n = 5) were treated with PBS or anti-CSF1R before ferritin NP–preS1 vaccine immunization. b, On day 10, preS1-specific IgM and IgG were detected; c,d, on day 21, preS1-specific IgG (c) and ASCs (d) were detected. b–d show representative results of two independent experiments. e, Distribution of SIGNR1+ cells in iLNs of WT and Cxcr5−/− mice on day 3 at immunization. A representative B cell follicular region is shown in magnification (top right). Data show representative results of four independent experiments. f, Microscopy of WT SIGNR1+ cell (CD45.1) or Cxcr5−/− SIGNR1+ cell (CD45.2) distribution in iLNs 5 d after CpG-1826 immunization in WT:Cxcr5−/− mixed bone marrow chimaeric mice. The area of total SIGNR1+ cells (red), WT SIGNR1+ cells (yellow) or Cxcr5−/− SIGNR1+cells (white) within follicles (yellow dashed line) was analysed using ImageJ software. Data were pooled from n = 18 follicles from six mice in three independent experiments. Each symbol represents an individual follicle. g, SIGNR1+ macrophages were sorted from the spleen of vaccinated WT or Cxcr5−/− mice, injected f.p. in naive CD45.1 mice, who had received MD4 B cells (CD45.2) the day before. 1 d later, the percentage and activation of CD45.2 MD4 B cells were analysed. Data show representative results of three independent experiments. PerCP, peridinin-chlorophyll-protein; PE, phycoerythrin. In a–d and f, data are shown as mean ± s.e.m.; statistical significance was determined with an unpaired two-tailed t-test. In g, the data were analysed using a paired two-tailed t-test.
SIGNR1+ macrophages are mainly distributed in medulla and interfollicular regions in LNs. We wondered how they can transfer antigen for B cell activation. On CpG immunization, a cluster of SIGNR1+ cells emerged in the follicular regions of dLNs (Fig. 6e, top panel), which express CD169 rather than CD11c, indicating their macrophage identity (Supplementary Fig. 10a). This suggested that SIGNR1+ macrophages might directly carry antigens to follicles for B cell activation. To understand how this migration is regulated, we examined the CXCL13–CXCR5 axis, given the abundant expression of CXCL13 in B cell follicles and its important role for follicular migration40. In Cxcr5−/− mice, no SIGNR1+ cells could be found in B cell follicles on immunization, in stark contrast to WT mice (Fig. 6e, bottom panel). In supporting these data, flow cytometric analysis showed that SIGNR1+ macrophages, but not SIGNR1− macrophages or SIGNR1+ DCs, significantly upregulated CXCR5 on immunization (Supplementary Fig. 10b). The upregulation of CXCR5 appears to be a direct effect of CpG and unique to macrophages, as CpG alone induces CXCR5 expression on isolated primary SIGNR1+ macrophages but not DCs under in vitro culture (Supplementary Fig. 10c). Given the aberrant LN organization in Cxcr5−/− mice, WT:Cxcr5−/− mixed bone marrow chimaeric mice were generated and confirmed the macrophage cell intrinsic role of CXCR5 on their migration to B cell follicles (Fig. 6f).
To further evaluate the role of CXCR5 and follicular migration of SIGNR1+macrophages in B cell activation, we injected ferritin NP–HEL-carrying WT or Cxcr5−/− SIGNR1+ macrophages into the f.p. of naive CD45.1 mice, who had received MD4 B cells (CD45.2) the day before. 24 h later, MD4 B cell activation was determined by flow cytometry. The result showed that, while ferritin NP–HEL-carrying WT SIGNR1+ macrophages activated MD4 B cells significantly, Cxcr5−/− SIGNR1+ macrophages were unable to do so (Fig. 6g). Thus, these data together suggest a previously unrecognized mechanism for B cell activation via CXCR5-dependent SIGNR1+ macrophage follicular migration and antigen transportation.
Conclusions
CHB therapy has different levels of goals: complete sterilizing cure, functional cure and partial cure41,42. So far, although complete cccDNA elimination still appears a great challenge, functional cure, which is defined as sustained, undetectable serum HBsAg and HBV DNA with or without seroconversion to anti‐HBs after a finite course of therapy, but with the persistence of residual cccDNA, is more realistic. In our study, we have found significantly reduced serum HBsAg and HBV DNA after four vaccinations of ferritin NP–preS1. Furthermore, some mice in the ferritin NP–preS1 group showed undetectable HBsAg and HBV DNA in the sera, and more impressively these mice also showed significant amounts of anti-HBs. These data indicate that ferritin NP–preS1 vaccine may be a good candidate to achieve a functional cure. Moreover, encouragingly, ferritin NP–preS1 vaccine also significantly reduced HBV cccDNA in the liver. Thus, our study might provide a new strategy toward eliminating cccDNA that could be worthy of future evaluation when used either alone or in combination43.
Ferritin is a unique NP vector that recently drew a lot of attention and showed an impressive antibody-enhancing effect for several other important viral antigens20–24. However, the underlying immunological mechanism is still poorly understood, which might limit its further improvement. In the current study, we have revealed that ferritin NPs are primarily captured by SIGNR1+ cells at the lymphatic zone of the dLNs, where both resident DCs and macrophages are enriched19,44,45. The ferritin-capturing DCs and macrophages play different roles in antibody response. Resident DCs appear to be the most important APCs for Tfh induction, which is also supported by the recent finding that lymphatic-sinus-associated resident DCs are strategically localized to induce rapid T cell response on particulate antigen immunization45.
The role of LN macrophages in antibody response has been an intriguing question. In the previous studies34,46,47, subcapsular sinus macrophages were considered immotile and the particulate antigen was speculated to be transferred to B cells via transcytosis or cytoplasmic membrane movement from the subcapsular sinus side to the other side facing the B cell follicles19,46,48. In our study, we found that SIGNR1+ macrophages, but not other macrophages or DCs, can upregulate CXCR5 on immunization and migrate to the follicular regions. When CXCR5 is lacking, antigen-loaded macrophages fail to activate B cells, which might suggest a previously unrecognized means of particulate antigen transfer by migrating SIGNR1+ macrophages. Whether CXCR5 deficiency could impair other functions of macrophages, resulting in impaired B cell activation, remains to be studied in future. It should be noted that the upregulation of CXCR5 on macrophages is dependent on CpG instead of ferritin NPs themelves. How CpG stimulation upregulates macrophage CXCR5 expression is still unknown. Whether other adjuvants have similar effects also remains to be systemically investigated. Nonetheless, ferritin NPs seem to be able to employ this machinery for enhanced antibody response.
In summary, data presented here demonstrated that a ferritin-NP-based preS1 vaccine manifests an efficient antibody response that is both preventive and therapeutic in a mouse HBV model via coordinately dual-targeting SIGNR1+ resident DCs and macrophages in the LNs. This study might provide a good candidate and offer new insights for future translational study on CHB treatment.
Methods
Mice
Naïve WT C57BL/6 mice and BALB/c mice were obtained from Vital River. CD11c-DTR mice and OT-II TCR transgenic mice were purchased from The Jackson Laboratory. Signr1−/− mice were generated by Cyagen. MD4 transgenic mice were provided by B. Hou (Chinese Academy of Science Key Laboratory for Infection and Immunity, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). Cxcr5−/− mice were kindly provided by H. Qi (Institute for Immunology, School of Medicine, Tsinghua University, Beijing, China). All mice were housed under specific pathogen-free conditions in the animal care facilities at the Institute of Biophysics, Chinese Academy of Sciences. All animal experiments were performed in accordance with the guidelines of the Institute of Biophysics, Chinese Academy of Sciences, using protocols approved by the institutional laboratory animal care and use committee.
Human sample
Human fresh distal mesenteric LNs from neuroblastoma patients 2.5–14 years old were derived from the patients who underwent surgery at the Department of Paediatric Surgical Oncology, Children’s Hospital of Chongqing Medical University. The study was performed in accordance with the committee guidelines and regulations. Informed consent was obtained from all patients.
Cloning, expression and purification of fusion proteins
The gene coding for Pf ferritin (residues 5–174) was synthesized by GENEWIZ using E. coli preferred codons. SpyTag spaced with three G4S linkers was fused to the amino-terminus of Pf ferritin during PCR. The SpyTag–Pf ferritin gene was cloned into the pDEST14 prokaryotic expression plasmid using the NdeI and SacI sites. The SpyTag–mouse ferritin gene was cloned into pDEST14 in the same manner. For the cloning of ΔN SC–preS1, SC–eGFP, SC–OT-II and SC–HEL, the PCR product of SC–(G4S)3 was cloned into pDEST14 using the NdeI and EcoRI sites and the antigen gene was cloned using the EcoRI and SacI sites. For the cloning of preS1 without SC, preS1 was cloned into pDEST14 using the NdeI and SacI sites. The preS1 sequence consisting of the first 108 amino acids (ayw) of HBV large surface protein was synthesized by PCR from complementary DNA extracted from livers of mice infected with AAV–HBV1.3 (genotype D3, subtype ayw3). The pDEST14-SC-Pf ferritin clone was obtained in the same manner.
For SpyTag–Pf ferritin NP expression and purification, BL21 (DE3) competent E. coli cells were transformed with pDEST14–SpyTag–(G4S)3–Pf ferritin plasmid, and a single colony was picked and cultured in 5 ml lysogeny broth at 37 °C overnight. The cultures were then amplified in 400 ml lysogeny broth and induced by 0.3 mM isopropylthiogalactoside for 6 h. The bacterial cultures were harvested and lysed in Tris buffer (20 mM Tris, 50 mM NaCl, pH 7.5). The lysate supernatants were diluted to 1 mg ml−1 and heated to 70 °C for 15 min to precipitate most of the E. coli proteins. After centrifugation and concentration, the supernatants were loaded onto a Superose 6 Increase (GE Healthcare) size exclusion column that was pre-equilibrated with 20 mM Tris 50 mM NaCl buffer (pH 7.5) and eluted with the same buffer at a rate of 0.5 ml min−1. The total volume of the column (Vt) was 24 ml, the outer volume of the separation medium (Vo) was 8 ml and the elution volume (Ve) of SpyTag–ferritin NP was 15 ml. The expression and purification of SpyTag–mouse ferritin NPs and SC–Pf ferritin NPs were the same as for SpyTag–Pf ferritin NPs. The expression of SC–preS1 was the same as for SpyTag–Pf ferritin NPs. For SC–preS1 purification, the lysate supernatants were purified using a Ni–NTA agarose column (ComWin Biotech) according to the manufacturer’s protocol. SC–eGFP, SC–OT-II, SC–HEL and preS1 without SC were expressed and purified in the same manner as SC–preS1. Endotoxin was removed from all of the proteins with a ToxinEraser endotoxin removal kit (GenScript). The amount of endotoxin is less than 0.1 EU ml−1.
Generation and purification of ferritin-NP-based vaccine
The purified SC–preS1 was conjugated to SpyTag–ferritin NP in vitro to construct the ferritin NP–preS1 vaccine. To assay reconstitution, SpyTag–ferritin NP was reacted with SC–preS1 at molar ratios of 1:0.5, 1:1 and 1:2 at 4 °C overnight. SDS–PAGE was used to evaluate the reconstitution efficiency. For vaccine generation, SpyTag–ferritin NP was mixed with SC–preS1 at a molar ratio of 1:1.5 at 4 °C overnight. The conjugated ferritin NP–preS1 was then purified using a Superose 6 Increase size exclusion column to remove the unbound SC–preS1. Ferritin NP–preS1 was eluted in 11.5 ml (Vt = 24 ml, Vo = 8 ml). The generation and purification of mouse ferritin NP–preS1, ferritin NP–eGFP, ferritin NP–OT-II and ferritin NP–HEL were performed in the same manner. To abrogate reconstitution in the specific assay, SC–ferritin NP and SC–preS1 were mixed physically to avoid their interaction. In addition, when the pH of the reaction buffer reached 11.5, the combination between SC and SpyTag was almost lost. Endotoxin was removed from the vaccine proteins with the ToxinEraser endotoxin removal kit before immunization. The amount of endotoxin is less than 0.1 EU ml−1.
Negative-stain electron microscopy
Grids of ferritin NPs for negative-stain electron microscopy were prepared as described previously49. Briefly, 4 μl samples (0.15 mg ml−1) were applied to glow-discharge electron microscopy grids covered by a thin layer of continuous carbon film and stained with 2% (w/v) uranyl acetate. Negatively stained grids were imaged on a Talos L120C microscope (Thermo Fisher) operating at 120 kV. Images were recorded at a magnification of ×12,000 and a defocus set to −2 μm, using a 4k × 4k CCD (charge-coupled device) camera. The particles were manually picked and two-dimensional classification was performed with EMAN2.
Immunization
Female naïve WT C57BL/6 mice or BALB/c mice (8–9 weeks old) were subcutaneously immunized with 500 pmol (approximately 23.9 μg, as determined by single ferritin–preS1 subunit) ferritin NP–preS1 vaccine or 500 pmol (approximately 12.7 μg) SC–preS1 with 30 μg CpG-1826 (Generay), respectively, at the tail base at day 0 and day 14. For the memory response assay, another boost immunization was performed at day 270. For high-dose immunization, 4 nmol (approximately 191.3 μg) ferritin NP–preS1 vaccine or 4 nmol (approximately 101.8 μg) SC–preS1 with 30 μg CpG-1826 was used. For the HBV prevention assay in vivo, 200 pmol (approximately 9.6 μg) ferritin NP–preS1 vaccine or 200 pmol (approximately 5.1 μg) SC–preS1 with 30 μg CpG-1826, respectively, was administered. For the therapy assay in vivo, 500 pmol (approximately 23.9 μg) ferritin NP–preS1 vaccine or 500 pmol (approximately 12.7 μg) SC–preS1 with 30 μg CpG-1826 was administered four times on days 35, 49, 63 and 77. Mouse ferritin NP–preS1 vaccine and other soluble proteins were immunized with the indicated dose in the same manner. IFN-γ blockade was performed using an intraperitoneal injection of 200 μg anti-IFN-γ (XMG1.2, BioXcell) at indicated times.
ELISA
For the preS1-specific ELISA, 5 μg ml−1 preS1 protein produced in our laboratory was coated onto 96-well high-binding Costar assay plates (Corning) at 4 °C overnight. After blocking with a blocking buffer (PBS containing 5% fetal bovine serum, FBS), serum samples with different dilutions (1:10–1:10000) were added to the plates. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H + L) (1:5,000, ZSGB-BIO), goat anti-mouse IgG1 (1:5,000, ProteinTech), goat anti-mouse IgG2c (1:5,000, ProteinTech) and goat anti-mouse IgM (1:5,000, ProteinTech) were used to detect each isotype. The concentration of specific antibodies was measured using TMB substrate (SeraCare) and the absorbance at 450 nm–630 nm was detected using a microplate reader (Molecular Devices). The absolute concentration of anti-preS1 IgG in serum is calibrated using a mouse monoclonal anti-Hep B preS1 (KR127, Santa Cruz) standard. For the detection of anti-preS1 avidity, mouse sera were serially diluted on the basis of IgG concentration and drawn on the x axis, and anti-preS1 levels of diluted sera were measured and drawn on the y axis simultaneously. The slope of the fitting curve was calculated to reflect the avidity of the antisera. For the detection of mouse ferritin- or Pf ferritin-specific antibody, plates were coated with 5 μg ml−1 mouse ferritin or Pf ferritin protein produced in our laboratory, and goat anti-mouse IgG (H + L)–HRP antibodies were used as detection antibodies. For the ELISA to test preS1 antigen located on HBsAg or HBV particles, plates were coated with 5 μg ml−1 anti-preS1 monoclonal antibody XY007 (provided by Y.-X. Fu), and the detection antibody was anti-HBs–HRP (Shanghai Kehua Bio-engineering). For HBsAg, HBeAg and anti-HBs testing, commercial ELISA kits were used (Shanghai Kehua Bio-engineering).
ELISpot
For IgG ASC detection, 10 μg ml−1 preS1 protein was coated onto 96-well ELISpot plates (Merck Millipore) at 4 °C overnight. After blocking with a blocking buffer (RPMI 1640 medium containing 10% FBS), 1 × 106 lymphocytes from LNs in 100 μl complete RPMI 1640 medium were added to the plates. After a 5–6 h incubation at 37 °C, preS1-specific IgG ASCs were analysed using a biotinylated donkey anti-mouse IgG (CWbio) and streptavidin–HRP (BD Biosciences). The spots were visualized with an AEC substrate set (BD Biosciences) and quantified with an autoanalysis system. For IFN-γ-secreting cell detection, 5 × 105 lymphocytes isolated from LNs, spleen and livers were incubated for 48 h at 37 °C in complete RPMI 1640 medium containing 5 μg ml−1 full preS1 protein or control protein in an IFN-γ ELISpot plate (Merck Millipore).
AAV–HBV1.3 infection
The AAV–HBV1.3 virus was purchased from the Beijing FivePlus Molecular Medicine Institute. This recombinant virus carries 1.3 copies of the HBV genome (genotype D3, subtype ayw3) and is packaged in AAV serotype 8 capsids15. Male adult C57BL/6 mice were injected with 1 × 1010 vg recombinant virus in 100 μl saline by tail vein injection. For the therapeutic assay, stable HBV carrier mice were used for vaccination after 5 weeks. Mice were bled through the ophthalmic vein at the indicated times in the respective experiments to monitor preS1 antibody, HBsAg, preS1 antigen and HBV genomic DNA in serum.
HBV infection and inhibition assays in vitro
The HepG2-hNTCP cell line and HBV (genotype D3, subtype ayw3) virus were provided by W. Li. In vitro HBV infection and inhibition assays were performed as reported11,28. Briefly, 1 × 107copies of genome-equivalent HBV were inoculated into the culture medium of 1 × 105 HepG2-hNTCP cells in 48-well plates in the presence of anti-preS1 sera from different mice and incubated for 24 h. The cells were then washed with medium three times and maintained in primary hepatocyte maintenance medium (William’s E medium (Gibco) with 5 μg ml−1 transferrin, 5 ng ml−1 sodium selenite, 3 μg ml−1 insulin (insulin–transferrin–sodium selenite; Corning), 2 mM l-glutamine, 10 ng ml−1 epidermal growth factor (Sigma-Aldrich), 2% dimethyl sulfoxide, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin). The medium was changed every 2 days. Viral infection at different times was analysed by measuring HBsAg and HBeAg in culture medium and detecting HBV RNAs in HepG2-hNTCP cells.
Bone marrow chimaera
C57BL/6 mice were lethally irradiated with Co60 at 10 Gy. The next day, 5 × 106 bone marrow cells from donor mice (CD11c-DTR mice or Cxcr5−/− mice) were intravenously transferred. Mixed bone marrow chimaeras were generated by transplanting an equal number of 2.5 × 106 WT CD45.1 and 2.5 × 106 Cxcr5−/− CD45.2 bone marrow cells. The chimaeras were given prophylactic water containing antibiotics for 4 weeks following irradiation. Then, 8–10 weeks after bone marrow transplantation, the mice could be used for subsequent operations.
DC and macrophage depletion
For systemic DC depletion, 8–10 weeks after CD11c-DTR bone marrow transfer, the chimaeric mice were given DT (Sigma) intraperitoneally at a dose of 5 ng per g body weight 24 h before deletion efficiency detection or vaccine immunization. To maintain the depletion, DT was administered every two days.
For macrophage depletion in pLN, C57BL/6 mice were injected with 30–50 μl CLLs (FormuMax) or control liposomes (CON) f.p. 7–10 d before experimentation.
For systemic macrophage depletion, a blocking antibody to CSF1R (clone ASF98, BioXCell) was administered. 50 μg anti-CSF1R was injected via f.p. and 100 μg via intraperitoneal injection once a day for three consecutive days before experimentation.
LN digestion and flow cytometry
LNs were harvested and gently minced using scissors in FACS buffer (PBS containing 2% FBS). LNs were then digested with 0.5 mg ml−1 collagenase I (Sigma) and 0.04 mg ml−1 DNase I (Roche) at 37 °C for 1 h. For CXCR5 detection and in the ferritin–FITC binding assay, since CXCR5 and FITC were highly susceptible to collagenase digestion, CLSPA (Worthington) was used. LNs were digested with 100 U ml−1 CLSPA and 0.04 mg ml−1 DNase I (Roche) in 1640 medium at 37 °C for 1 h. The suspension was washed with cold FACS buffer, disaggregated by passing through a 70-μm cell strainer (Biologix Group) and centrifuged at 500 g for 5 min. Splenocytes were collected in the same manner, and red blood cells were removed with ammonium–chloride–potassium buffer.
For flow cytometric analysis, single cells were resuspended in an appropriate volume of FACS buffer (1–5 × 106 cells per 100 μl), blocked with anti-FcγR monoclonal antibody (clone 2.4G2) to block non-specific binding, and labelled with fluorescence-conjugated antibodies. For DC and macrophage analysis, the antibodies used included anti-CD19 (6D5, BioLegend), anti-B220 (RA3–6B2, eBioscience), anti-CD11c (N418, BioLegend), anti-I-A/I-E (M5/114.15.2, BioLegend), anti-CD11b (M1/70, eBioscience), anti-SIGNR1 (22D1, eBioscience), anti-CD169 (3D6.112, BioLegend), anti-F4/80 (BM8, BioLegend) and anti-CD103 (2E7, eBioscience). For T cell and B cell activation analysis, the antibodies used included anti-Thy1.1 (HIS51, eBioscience), anti-TCR Vα2 (B20.1, eBioscience), anti-CD4 (GK1.5, BioLegend), anti-PD1 (J43, eBioscience), anti-CXCR5 (SPRCL5, eBioscience), anti-GL7 (GL-7, eBioscience), anti-FAS (Jo2, BD), anti-CD69 (H1.2F3, eBioscience), anti-CD25 (PC61.5, eBioscience), anti-CD44 (IM7, BioLegend), anti-CD62L (MEL-14, eBioscience) and anti-CD86 (GL1, eBioscience). The working concentration of the antibodies is 2.5 μg ml−1. For human immune cell staining, the antibodies used included Human Trustain FcX (BioLegend), anti-human CD14 (M5E2, BioLegend), anti-human HLA-DR (L243, BioLegend) and anti-human CD209 (9E9A8, BioLegend). For the T cell proliferation assay, OT-II T cells were labelled with 2 mM carboxyfluorescein succinimidyl ester (Sigma) in PBS for 10 min at 37 °C, then washed with PBS containing 10% FBS at least twice. An LSRFortessa flow cytometer (BD Biosciences) and FlowJo software (Tree Star) were used for data collection and analysis, respectively. Cell sorting was performed on a FACSAria III (BD Biosciences).
Immunofluorescence microscopy
For the detection of ferritin NP–eGFP or FITC-labelled ferritin NPs, LNs were harvested and fixed with periodate–lysine–paraformaldehyde fixative buffer (PLP, PBS buffer containing 2 mg ml−1 NaIO4, 0.1 M l-lysine (pH 7.4) and 1% (w/v) PFA) for 12 h, then dehydrated in 30% sucrose twice before embedding in optimal cutting temperature compound (Sakura) and snap frozen in liquid nitrogen. Next, 10 μm-thick cryosections were prepared. For GC and SIGNR1+ cell staining, LNs were embedded in optimal cutting temperature compound and snap frozen in liquid nitrogen directly. After cryosectioning, the slides were air-dried for 1 h and fixed for 10 min in cold acetone.
The cryosections were blocked for 2 h at room temperature in FACS buffer containing 1 mg ml−1 anti-FcγR mAb (2.4G2). The cryosections were incubated overnight at 4 °C with the following antibodies at 2.5 μg ml−1 working concentrations: anti-SIGNR1 (22D1, eBioscience), anti-B220 (RA3–6B2, eBioscience), anti-CD45.1 (A20, eBioscience), anti-CD45.2 (104, eBioscience) and anti-GL7 (GL-7, eBioscience). For eGFP staining, anti-full length GFP polyclonal antibody (1:1,000, Clontech) was used overnight at 4 °C. Then, the unconjugated anti-GFP antibody was detected with an AlexaFluor488-conjugated goat anti-rabbit IgG secondary antibody (1:1,000, Zsbio) at room temperature for 1 h. Images were taken on a confocal microscope (Zeiss LSM-710) and analysed with ZEN 2012 software (Carl Zeiss) and Fiji ImageJ.
Quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using a RevertAid First Strand cDNA synthesis kit (Life Technologies). Quantitative real-time PCR was performed using SYBR Premix Ex Taq (Takara) on an ABI QuantStudio 7 Flex real-time PCR system. The primers used for quantitative real-time PCR are as follows: Hprt (forward, 5′-TCCTCCTCAGACCGCTTTT-3′; reverse, 5′-CCTGGTTCATCATCGCTAATC-3′), Lamp1 (forward, 5′-GTGGGAGTTGCGGTATCAAC-3′; reverse, 5′-TATTCAAGCGCACTCCTTGC-3′), Lamp2 (forward, 5′-CTAGGAGCCGTTCAGTCCAA-3′; reverse, 5′-CTTGCAGGTGAATACCCCAA-3′), Lyz1 (forward, 5′-GGACTCCTCCTGCTTTCTGT-3′; reverse, 5′-TAAACACACCCAGTCAGCCA-3′), Lyz2 (forward, 5′-GGACTCCTCCTGCTTTCTGT-3′; reverse, 5′-AGTCAGTGCTTTGGTCTCCA-3′), Ctsb (forward, 5′-TCTGAAGAAGCTGTGTGGCA-3′; reverse, 5′-TTGTTCCCGTGCATCAAAGG-3′) and Ctsc (forward, 5′-CTCGGTGATGGAAGCAACAG-3′; reverse, 5′-CTGATAGCTGTGTGGCCTCT-3′). The ΔΔCt method was used to calculate relative target gene expression to Hprt. For HBV RNA detection, the primers used are as follows: HBV 3.5 kb (forward, 5′-GAGTGTGGATTCGCACTCC-3′; reverse, 5′-GAGGCGAGGGAGTTCTTCT-3′) and HBV total (forward, 5′-TCACCAGCACCATGCAAC-3′; reverse, 5′-AAGCCACCCAAGGCACAG-3′). For HBV DNA detection in serum, HBV DNA was extracted from 200 µl of serum and measured following the manufacturer’s instructions (careHBV, Qiagen). The detection of HBV cccDNA was performed as reported50. After total DNA isolation from liver, a combination of digestion with XmaI and SacI restriction enzymes followed by T5 exonuclease was used to degrade HBV relaxed circular DNA and all the AAV DNA. The remaining undigested HBV cccDNA allows specific detection by quantitative real-time PCR. The primers used are forward, 5′ -TGCACTTCGCTTCACCT-3′; reverse, 5′ -AGGGGCATTTGGTGGTC-3′.
Statistical analysis
All analysis was performed using GraphPad Prism statistical software. All of the data were analysed using an unpaired two-tailed t-test or paired two-tailed t-test. The results are expressed as the mean ± s.e.m. A value of P < 0.05 was considered statistically significant.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41565-020-0648-y.
Supplementary information
Supplementary Figs. 1–10.
Acknowledgements
We thank H. Qi for Cxcr5−/− mice and B. Hou for ΜD4 transgenic mice. We thank K. Fan for technical consultation on ferritin characterization, and X. Shi for mouse breeding and healthcare services. This work was supported by grants from the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB29040202 to M.Z.), the National Natural Science Foundation of China (81991493 to Q.C.), the National Key R&D Programme of China (2018YF1313000 and 2018YF1313004 to S.W., 2019YFA0905903 to M.Z.) and the Science and Technology Service Network Initiative Programme of the Chinese Academy of Sciences (KFJ-STS-ZDTP-062 to M.Z.).
Source data
Author contributions
W.W. and M.Z. designed the experiments and analysed the data; W.W. and X.Z. conducted most experiments; Y.B., H.P. and Y.-X.F. helped with an AAV–HBV1.3 infection mouse model and in vitro HBV infection assay; S.W. and Z.W. provided human LN samples and helped in the human immune cell targeting assay; Q.C. helped with the immunofluorescence staining; Z.G. and P.Z. performed transmission electron micrography imaging; X.Y. provided technical advice on ferritin preparation and characterization and helped in the iron measuring assay; W.L. provided the HepG2-hNTCP cell line and HBV virus and helped with the in vitro infection assay; W.W. and M.Z. wrote the manuscript; M.Z. conceived and supervised the project.
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Nanotechnology thanks John Kehrl, Daniel Shouval and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41565-020-0648-y.
References
- 1.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev. Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 2.Gerlich WH. Prophylactic vaccination against hepatitis B: achievements, challenges and perspectives. Med. Microbiol. Immunol. 2015;204:39–55. doi: 10.1007/s00430-014-0373-y. [DOI] [PubMed] [Google Scholar]
- 3.Dembek C, Protzer U, Roggendorf M. Overcoming immune tolerance in chronic hepatitis B by therapeutic vaccination. Curr. Opin. Virol. 2018;30:58–67. doi: 10.1016/j.coviro.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Glebe D, et al. Pre-S1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J. Virol. 2003;77:9511–9521. doi: 10.1128/JVI.77.17.9511-9521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai D, et al. Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation. Antimicrob. Agents Chemother. 2012;56:4277–4288. doi: 10.1128/AAC.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeng CY, Ryu CJ, Gripon P, Guguen-Guillouzo C, Hong HJ. Fine mapping of virus-neutralizing epitopes on hepatitis B virus PreS1. Virology. 2000;270:9–16. doi: 10.1006/viro.2000.0250. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Li M, Le X, Ma W, Zhou B. Recombinant hepatitis B core antigen carrying preS1 epitopes induce immune response against chronic HBV infection. Vaccine. 2004;22:439–446. doi: 10.1016/j.vaccine.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Li D, et al. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. eLife. 2017;6:e26738. doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coursaget P, et al. Antibody response to preS1 in hepatitis-B-virus-induced liver disease and after immunization. Res. Virol. 1990;141:563–570. doi: 10.1016/0923-2516(90)90087-y. [DOI] [PubMed] [Google Scholar]
- 10.Deepen R, Heermann KH, Uy A, Thomssen R, Gerlich WH. Assay of preS epitopes and preS1 antibody in hepatitis B virus carriers and immune persons. Med. Microbiol. Immunol. 1990;179:49–60. doi: 10.1007/BF00190150. [DOI] [PubMed] [Google Scholar]
- 11.Bian Y, et al. Vaccines targeting preS1 domain overcome immune tolerance in hepatitis B virus carrier mice. Hepatology. 2017;66:1067–1082. doi: 10.1002/hep.29239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heermann KH, et al. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem D, Prince AM. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 14.Park J-h, Cho E-w, Lee Y-j, Shin SY, Kim KL. Determination of the protective effects of neutralizing anti-hepatitis B virus (HBV) immunoglobulins by epitope mapping with recombinant HBV surface-antigen proteins. Microbiol. Immunol. 2000;44:703–710. doi: 10.1111/j.1348-0421.2000.tb02552.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang D, et al. A mouse model for HBV immunotolerance and immunotherapy. Cell. Mol. Immunol. 2014;11:71–78. doi: 10.1038/cmi.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DM, Simon JK, Baker JR., Jr. Applications of nanotechnology for immunology. Nat. Rev. Immunol. 2013;13:592–605. doi: 10.1038/nri3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke BE, et al. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature. 1987;330:381–384. doi: 10.1038/330381a0. [DOI] [PubMed] [Google Scholar]
- 18.Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Gray EE, Cyster JG. Lymph node macrophages. J. Innate Immun. 2012;4:424–436. doi: 10.1159/000337007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanekiyo M, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yassine HM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 22.Kanekiyo M, et al. Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell. 2015;162:1090–1100. doi: 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bu W, et al. Immunization with components of the viral fusion apparatus elicits antibodies that neutralize Epstein-Barr virus in B cells and epithelial cells. Immunity. 2019;50:1305–1316.e1306. doi: 10.1016/j.immuni.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanekiyo M, et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl Acad. Sci. USA. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, et al. A novel method for synthetic vaccine construction based on protein assembly. Sci. Rep. 2014;4:7266. doi: 10.1038/srep07266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatur J, Hagedoorn PL, Overeijnder ML, Hagen WR. A highly thermostable ferritin from the hyperthermophilic archaeal anaerobe Pyrococcus furiosus. Extremophiles. 2006;10:139–148. doi: 10.1007/s00792-005-0484-x. [DOI] [PubMed] [Google Scholar]
- 28.Yan H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontisso P, et al. Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology. 1989;173:522–530. doi: 10.1016/0042-6822(89)90564-3. [DOI] [PubMed] [Google Scholar]
- 30.Mimms LT, et al. Discrimination of hepatitis B virus (HBV) subtypes using monoclonal antibodies to the PreS1 and PreS2 domains of the viral envelope. Virology. 1990;176:604–619. doi: 10.1016/0042-6822(90)90031-l. [DOI] [PubMed] [Google Scholar]
- 31.Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018;18:545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez SF, et al. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat. Immunol. 2010;11:427–434. doi: 10.1038/ni.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conde P, et al. DC-SIGN+ macrophages control the induction of transplantation tolerance. Immunity. 2015;42:1143–1158. doi: 10.1016/j.immuni.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Junt T, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 35.Park CG, et al. Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN. Int. Immunol. 2001;13:1283–1290. doi: 10.1093/intimm/13.10.1283. [DOI] [PubMed] [Google Scholar]
- 36.Powlesland AS, et al. Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J. Biol. Chem. 2006;281:20440–20449. doi: 10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- 37.Parent SA, et al. Molecular characterization of the murine SIGNR1 gene encoding a C-type lectin homologous to human DC-SIGN and DC-SIGNR. Gene. 2002;293:33–46. doi: 10.1016/s0378-1119(02)00722-9. [DOI] [PubMed] [Google Scholar]
- 38.Angel CE, et al. Distinctive localization of antigen-presenting cells in human lymph nodes. Blood. 2009;113:1257–1267. doi: 10.1182/blood-2008-06-165266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 40.Cyster JG, et al. Follicular stromal cells and lymphocyte homing to follicles. Immunological Rev. 2000;176:181–193. doi: 10.1034/j.1600-065x.2000.00618.x. [DOI] [PubMed] [Google Scholar]
- 41.Ning Q, et al. Roadmap to functional cure of chronic hepatitis B: an expert consensus. J. Viral Hepat. 2019;26:1146–1155. doi: 10.1111/jvh.13126. [DOI] [PubMed] [Google Scholar]
- 42.Tang LSY, Covert E, Wilson E, Kottilil S. Chronic hepatitis B infection: a review. J. Am. Med. Assoc. 2018;319:1802–1813. doi: 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 43.Revill PA, et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 2019;4:545–558. doi: 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerner MY, Torabi-Parizi P, Germain RN. Strategically localized dendritic cells promote rapid T cell responses to lymph-borne particulate antigens. Immunity. 2015;42:172–185. doi: 10.1016/j.immuni.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 46.Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 2009;10:786–793. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 48.Cyster JG. B cell follicles and antigen encounters of the third kind. Nat. Immunol. 2010;11:989–996. doi: 10.1038/ni.1946. [DOI] [PubMed] [Google Scholar]
- 49.Roux KH. Negative-stain immunoelectron-microscopic analysis of small macromolecules of immunologic significance. Methods. 1996;10:247–256. doi: 10.1006/meth.1996.0099. [DOI] [PubMed] [Google Scholar]
- 50.Lucifora J, et al. Detection of the hepatitis B virus (HBV) covalently-closed-circular DNA (cccDNA) in mice transduced with a recombinant AAV-HBV vector. Antivir. Res. 2017;145:14–19. doi: 10.1016/j.antiviral.2017.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–10.
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author on reasonable request.