Abstract
Outbreaks of porcine epidemic diarrhea (PED) caused by porcine epidemic diarrhea virus (PEDV) infection have caused high mortality of piglets and significant economic losses to the Chinese swine industry. In the current study, 184 specimens from pigs with or without signs of diarrhea were collected from 39 farms across eight provinces, mainly around Hunan, People's Republic of China, in 2017 to 2018 in order to obtain epidemiological information on PEDV infections in these regions. The results indicated an average PEDV-positive rate of 38.04% (70/184) and more-pronounced disease severity in diarrheic pigs (48.76%; 59/121) than in non-diarrheic pigs (17.46%; 11/63). Phylogenetic and sequence analysis demonstrated that 14 representative PEDV strains from 14 swine farms belonged to the G2 group (G2-a and G2-b subgroups) and displayed a high degree of genetic variation. In particular, two out of the 14 PEDV strains were found to have unique indels in the S1 gene. The strain HN-SY-2017-Oct had a 9-nucleotide (T1152GAAGCCAAT1160T) insertion, and the strain ZJ-2018-May had a 3-nucleotide (AAA) deletion at position 1126 in the S1 gene. A three-dimensional structural prediction revealed that these unique insertions might lengthen the loop on the surface or increase the likelihood of the surface protein being phosphorylated at 388Y, thereby affecting the virulence or pathogenicity of PEDV. Collectively, the data show that PED remains a severe threat to the pig industry and that variant PEDV stains are circulating in China. The updated PEDV epidemiological data will facilitate the design of PEDV vaccines and the application of effective measures for PED prevention.
Introduction
Porcine epidemic diarrhea virus (PEDV) is an enveloped, single-stranded, positive-sense RNA virus that belongs to the genus Alphacoronavirus of the family of Coronaviridae [22]. The disease porcine epidemic diarrhea (PED) was first described in England in 1971, the causative agent, PEDV, was subsequently identified, isolated, and designated CV777 in Belgium in 1978 [20]. Since October 2010, a new variant of PEDV has emerged in over ten provinces of China. The main clinical manifestations of PED include watery diarrhea, vomiting, and dehydration, with high mortality (nearly 100%) of newborn piglets [4]. Outbreaks of PED on farms where pigs had been vaccinated with inactivated or attenuated CV777-based vaccines have been reported in recent years, suggesting that the present vaccines based on classical PEDV strains cannot completely protect pigs against the variant strains of PEDV that are circulating in China [32]. The first outbreak of PED in the USA was confirmed in April 2013, and cases were documented in over 30 US states in a short period of time. PEDV then swept across Canada and Mexico [26]. Variant PEDV strains with nucleotide insertions/deletions in the S gene, referred to as S INDEL strains, have been reported in the USA [27] and other countries, including Japan [24], China [30], and Germany [21], suggesting that PEDV has circulated all over the world and can be considered a global threat to the swine industry [16].
PEDV has an RNA genome that is ~ 28 kb in length and contains at least eight major open reading frames (ORFs) encoding four structural and three non-structural proteins [12, 23]. The spike (S) glycoprotein of PEDV, a major surface protein, is composed of two subunits containing conserved nonamer and GxCx motifs, namely the S1 and S2 proteins [4]. The S1 protein is essential for recognition of cellular receptors, binding, and virulence, while the S2 protein mainly mediates membrane fusion during virus infection [15, 25]. The S1 gene of most PEDV strains is more variable than the S2 gene and has therefore been extensively employed for analysis of genetic relatedness and evolution as well as vaccine development [6]. The ORF3 protein of PEDV, a non-structural protein, has been shown to be important for the virulence of PEDV strains [19]. Due to the deletion of 17 amino acids in the ORF3 of PEDV vaccine isolates, the ORF3 gene can be used as a molecular marker to distinguish between PEDV vaccine and non-vaccine strains and to analyze the molecular epidemiology of PEDV [19].
Though there have been reports describing the epidemiology and genetic variations of PEDV strains in China [11, 13, 22, 28, 30, 33–35], little information is available about current epidemic trends and the molecular characteristics of PEDV, particularly in Hunan province and its surrounding provinces. In the present study, we collected 184 samples (79 fecal samples and 105 small-intestinal tissues) from 39 swine farms in eight provinces including Hunan and neighboring provinces during 2017-2018 for the detection of PEDV using a reverse transcription polymerase chain reaction (RT-PCR) assay. We also tested for other diarrhea-causing pathogens, including TGEV (transmissible gastroenteritis virus) and PoRV (porcine rotavirus group A), but not PDCoV (porcine deltacoronavirus), in order to estimate the co-infection rate in these samples. Furthermore, the complete ORF3 and S1 genes of 14 representative PEDV strains from 14 swine farms in these regions were cloned, sequenced, and extensively analyzed.
Materials and methods
Collection and pre-treatment of specimens
Porcine specimens (fecal swabs and small intestine) were collected from 184 pigs with different clinical symptoms on 39 commercial farms across eight central provinces (Hunan, Hubei, Guangxi, Jiangxi, Guangdong, Henan, Gansu, and Zhejiang) (Fig. 1A) between 2017 and 2018. Of the 184 specimens, 121 were obtained from 4- to 70-day-old pigs exhibiting clinical signs of diarrhea (watery diarrhea, vomiting, and dehydration), and the remaining 63 samples were collected from 14- to 140-day-old pigs showing no diarrhea symptoms. The pigs on most of the swine farms tested had been immunized with a commercial vaccine based on PEDV strain CV777, but those on some farms had not been immunized (Table 1). All specimens were frozen with ice packs and delivered to the Key Laboratory of Animal Vaccine & Protein Engineering (Hunan province, China) for further processing. All specimens were prepared individually and mixed with sterile phosphate-buffered saline (PBS) in sterile 1.5-ml microcentrifuge tubes, frozen and thawed three times, and centrifuged at 12000 × g. The supernatants were then collected and stored at -80 °C for later testing.
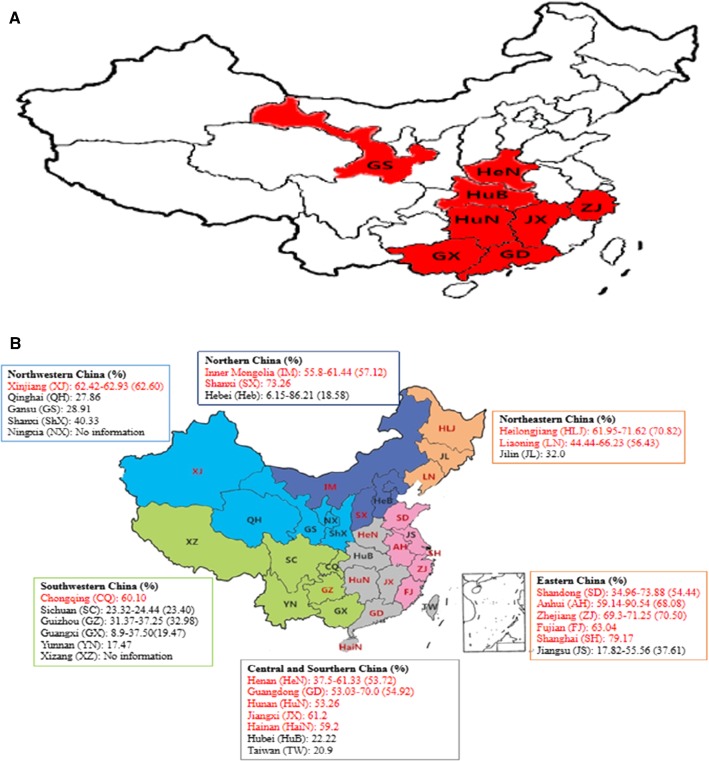
Fig. 1.
Summary of data on PEDV epidemics from the current study and previous studies. (A) Map of China with the eight provinces from which the 184 specimens were collected in the current study shown in red. (B) Results of representative epidemiological studies of PEDV infection conducted in China after 2010, collected from Chinese- and English-language publications, are summarized, analyzed, and displayed on a map of China. The data are divided into six groups according to region (boxes), and the range and average (in parentheses) PEDV infection rates are also indicated. The provinces with an average rate higher than 50% are highlighted in red
Table 1.
Information about specimens collected in the present study
| Province | Collection date | Region | Clinical diarrhea | Sample type (fecal swab [FS[ or intestine contents [IC]) |
Total number of samples | Number positive | Positive rate (%) |
Strain name | GenBank accession no. | Immunization | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF3 | S1 | ||||||||||
| Hunan | Oct-2017 | Changsha | Yes | IC | 12 | 0 | 0.0 | - | - | No | |
| Oct-2017 | Shaoyang | Yes | IC | 10 | 10 | 100.0 | HNSY/CH/2017/Oct | MK135454 | MK135449 | Yes | |
| Nov-2017 | Yueyang | Yes | IC | 5 | 5 | 100.0 | HNYY/CH/2017/Nov | MK135460 | MK135452 | Yes | |
| Dec-2017 | Changsha | Yes | IC | 2 | 2 | 100.0 | HNCS/CH/2017/Dec | Mk135458 | Mk135450 | No | |
| Dec-2017 | Loudi | No | IC | 2 | 2 | 100.0 | - | - | - | Yes | |
| Dec-2017 | Yueyang | No | IC | 4 | 1 | 25.0 | - | - | - | No | |
| Dec-2017 | Yueyang | No | IC | 3 | 2 | 66.67 | - | - | - | Yes | |
| Dec-2017 | Chengzhou | No | IC | 2 | 1 | 50.0 | - | - | - | Yes | |
| Jan-2018 | Loudi | Yes | IC | 2 | 0 | 0.0 | - | - | - | Yes | |
| Jan-2018 | Loudi | Yes | IC | 1 | 0 | 0.0 | - | - | - | Yes | |
| No | IC | 2 | 0 | 0.0 | - | - | - | ||||
| Jan-2018 | Hengyang | No | IC | 2 | 1 | 50.0 | - | - | - | No | |
| Mar-2018 | Xiangtan | Yes | FS | 3 | 1 | 33.33 | HNXT/CH/2018/Mar | MK135465 | MK135443 | Yes | |
| Mar-2018 | Xiangxi | Yes | IC | 3 | 1 | 33.33 | - | - | - | Yes | |
| Mar-2018 | Hengyang | No | IC | 4 | 0 | 0.0 | - | - | - | Yes | |
| Mar-2018 | Yueyang | Yes | IC | 3 | 3 | 100.0 | HNYY/CH/2018/Mar | MK135463 | MK135442 | Yes | |
| Mar-2018 | Yiyang | Yes | IC | 4 | 4 | 100.0 | HNYY/CH/2018/Mar | MK135467 | MK135446 | Yes | |
| Mar-2018 | Liuyang | Yes | IC | 4 | 4 | 100.0 | HNLY/CH/2018/Mar | Mk135466 | MK135445 | No | |
| Mar-2018 | Unknow | Yes | IC | 3 | 3 | 100.0 | - | - | - | Yes | |
| Apr-2018 | Zhuzhou | Yes | IC | 2 | 0 | 0.0 | - | - | - | Yes | |
| Apr-2018 | Xiangtan | Yes | IC | 2 | 2 | 100.0 | - | - | - | No | |
| Apr-2018 | Loudi | No | IC | 2 | 1 | 50.0 | - | - | - | No | |
| May-2018 | Xiangtan | Yes | IC | 4 | 1 | 25.0 | - | - | - | Yes | |
| Jun-2018 | Yongzhou | Yes | FS | 5 | 3 | 60.0 | HNYZ/HN/2018/Jue | MK135456 | MK135441 | Yes | |
| Jun-2018 | Zhangjiajie | Yes | IC | 3 | 0 | 0.0 | - | - | - | No | |
| Jun-2018 | Chenzhou | Yes | IC | 3 | 3 | 100.0 | HNCZ/CH/2018/Jue | MK135461 | MK135451 | Yes | |
| July-2018 | Hengyang | Yes | IC | 3 | 1 | 33.33 | - | - | - | No | |
| July-2018 | Xiangtan | Yes | IC | 7 | 2 | 28.57 | - | - | - | Yes | |
| July-2018 | Hengyang | Yes | IC | 11 | 4 | 36.37 | - | - | - | Yes | |
| Jiangxi | Dec-2017 | Nanchang | No | IC | 2 | 0 | 0.0 | - | - | - | Yes |
| Mar-2018 | Yingtan | Yes | IC | 1 | 0 | 0.0 | - | - | - | Yes | |
| No | IC | 10 | 0 | 0.0 | - | - | - | Yes | |||
| Guangxi | Jan-2018 | Hezhou | Yes | IC | 4 | 4 | 100.0 | GXHZ/CH/2018/Jan | MK135459 | MK135453 | No |
| Gansu | Mar-2018 | Jiuquan | Yes | FS | 10 | 4 | 40.0 | GSJQ/CH/2018/Mar | MK135464 | MK135440 | Yes |
| Guangdong | Mar-2018 | Maoming | Yes | FS | 5 | 0 | 0.0 | - | - | - | No |
| Mar-2018 | Maoming | Yes | FS | 5 | 0 | 0.0 | - | - | - | Yes | |
| Hubei | Mar-2018 | Wuhan | No | FS | 10 | 2 | 20.0 | HuBWH/CH/2018/Mar | MK135457 | MK135447 | Yes |
| Mar-2018 | Wuhan | No | FS | 10 | 0 | 0.0 | - | - | - | Yes | |
| Mar-2018 | Wuhan | No | FS | 10 | 1 | 10.0 | - | - | - | Yes | |
| Hebei | Mar-2018 | Zhangjiakou | Yes | FS | 3 | 1 | 33.33 | HebZJK/CH/2018/Mar | MK135462 | MK135444 | Yes |
| Zhejiang | May-2018 | Unknow | Yes | IC | 1 | 1 | 100.0 | ZJ/CH/2018/May | MK135455 | MK135448 | Yes |
-, not identified
Detection of viruses in clinical specimens
Trizol Reagent (Invitrogen, USA) was used to extract total viral RNA from the collected samples. About 1 μg of viral RNA was used to generate cDNA by reverse transcription (RT) using a Revert Aid first Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the protocols of the manufacturer. Three pairs of specific primers were designed for detecting the presence of PEDV (targeting the S gene), TGEV (targeting the S gene), and PoRV (targeting the nonstructural protein 4 gene), respectively, based on their reference sequences (Table 2). RT-PCR reactions were performed in a 20-μl mixture containing 10 μl of 2 × Taq PCR Master Mix (TsingKe Co, Ltd., People’s Republic of China), 1 μl of the cDNA sample, 1 μl of each primer (10 pmol), and sterilized water in a thermal cycler (Biometra), using the following program: 94 °C for 5 min (first denaturation), 35 cycles of 94 °C for 30 s (denaturation); 56 °C for 30 s (annealing), and 72 °C for 30 s (extension); followed by 7 min at 72 °C (final extension). The positive samples were confirmed to yield the expected DNA bands in 1% agarose gel electrophoresis (~ 500, 300, and 420 bp for PEDV, TGEV, and PoRV, respectively).
Table 2.
Primers employed in the present study
| Primer | Sequence | Binding position | Length | Purpose | Reference sequence |
|---|---|---|---|---|---|
| pPEDV-F-ZY171 | GATATGTTTGTAATGGTAACTC | 23063-23564 | 502 | Detection of PEDV (targeting the S gene) | MF807952 |
| pPEDV-R-ZY172 | AGCATAGCTAAAAGGCAATGC | ||||
| pTGEV-F-ZY173 | GATATGTTTGTAATGGTAACCC | 22946-23244 | 299 | Detection of TGEV (targeting the S gene) | KX900402 |
| pTGEV-R-ZY174 | CTCTATAGCTGAACGATACTTAC | ||||
| pPoRV-F-ZY175 | TTTACTCTACATAAAGCATCAAT | 171-586 | 416 | Detection of PORV (targeting the NSP4 gene) | KX453793 |
| pPoRV-R-ZY176 | GACGGCAACTCAACCTCTCACAT | ||||
| PEDV-ORF3-F | TCCTAGACTTCAACCTTACG | 24740-25570 | 831 | Sequencing | AF353511 |
| PEDV-ORF3-R | GGTGACAAGTGAAGCACAGA | ||||
| PEDV-S1-F | GGTAAGTTGCTAGTGCGTAA | 20570-23004 | 2435 | Sequencing | AF353511 |
| PEDV-S1-R | AATACTCATACTAAAGTTGGTGG |
Sequencing of the complete ORF3 and S1 genes
Two pairs of specific primers (Table 2) designed based on the complete genomic sequence of PEDV strain CV777 (AF353511) were used to amplify the whole ORF3 and S1 genes of the PEDV strains identified in this study. The PCR reactions were performed in a 50-μl mixture consisting of 25 μl of 2 × Taq PCR Master Mix (TsingKe Co, Ltd., People’s Republic of China), 2 μl of the cDNA sample, 2 μl of each primer (10 pmol), and sterilized water in a thermal cycler (Biometra) with the following program: 94 °C for 5 min (first denaturation), 35 cycles of 94 °C for 30 s (denaturation); 56 °C for 30 s (annealing), and 72 °C for 30 s or 2 min (for the complete ORF3 or S1 gene, respectively) (extension); followed by 7 min at 72 °C (final extension). The purified PCR products were cloned into the pUCm-T vector and then sequenced. The sequences of these novel PEDV strains were submitted to the NCBI database (GenBank accession numbers MK135440-MK135453 and MK135454-MK135467 for the S1 and ORF3 genes, respectively).
Bioinformatic analysis
The complete PEDV S1 and ORF3 gene sequences of 14 newly obtained PEDV strains from different regions and their corresponding reference strains were aligned using DNAStar version 7.10 (Lasergene) for comparison. Phylogenetic trees were constructed based on the complete PEDV ORF3 and S1 gene sequences determined in this study and those of several reference strains downloaded from the GenBank database, using the neighbor-joining (NJ) method in MEGA 7.0 software. The reliability of the trees was evaluated using 1000 bootstrap replicates. The S1 genes of two potential PEDV recombinant strains identified in this study were further analyzed using SimPlot version 3.5.1 recombination analysis software as described previously [31]. In addition, the potential effects of amino acid indels on the structure of the PEDV S1 protein were also analyzed. Briefly, three-dimensional structures of the S1 proteins of PEDV strains were predicted using SWISS-Model (https://swissmodel.expasy.org) [2] and displayed in PyMOL version 1.7.4.4 (www.pymol.org). The effects of amino acid indels on phosphorylation sites in the S1 protein were evaluated using NetPhos 2.0 software (http://www.cbs.dtu.dk/services/NetPhos/) [3].
Results
The epidemiology of PEDV and other viruses
RT-PCR analysis of all collected specimens (summarized in Table 1) revealed that 70 (38.04%) of 184 specimens from 28 (71.79%) of 39 swine farms were positive for PEDV and that the positive rate in pigs with diarrhea (48.76%, 59/121) was almost threefold higher than in those without clinical sign of diarrhea (17.46%, 11/63). Further classification by year indicated that 23 out of 42 specimens (54.76%, 23/42) from seven out of nine farms (77.78%, 7/9) in 2017 were positive for PEDV, while 47 (33.10%, 47/142) specimens from 21(70.0%, 21/30) farms in 2018 were PEDV positive. Classification by region demonstrated that the PEDV-positive rates were 50.44% (57/113), 100.0% (4/4), 40.0% (4/10), 33.33% (1/3), 100.0% (1/1), and 10.0% (3/30) for Hunan, Guangxi, Gansu, Hebei, Zhejiang, and Hubei provinces, respectively, while no samples from Guangdong (n = 10) or Jiangxi province (n = 13) were positive for PEDV. The positive rates for TGEV and PoRV were 3.26% (6/184) and 3.80% (7/184), respectively, and the co-infection rate for "PEDV + TGEV" and "PEDV + PoRV" was 1.09% (2/184) and 3.26% (6/184), respectively. No "TGEV + PoRV" or "PEDV + TGEV + PoRV" co-infections were found.
To provide an updated overview of the PEDV epidemic trends in China, we summarized the representative results from the main epidemiological studies of PEDV infection conducted in China after 2010 [1, 8, 17–22] and displayed them on a map of China according to region (Fig. 1B). Previous epidemical studies, together with the current one, collectively revealed that the average detection rates for PEDV infection range from 17.47 to 79.17%, and the PEDV-positive rate of 55.56% (15/27) provinces/regions (red marked) is higher than 50.0%, indicating a severe epidemic trend of PEDV in China.
Genetic characteristics of the newly identified PEDV strains
To analyze the genetic characteristics of these newly identified PEDV strains, the full-length ORF3 and S1 genes of 14 representative PEDV isolates were successfully amplified, sequenced, and submitted to the GenBank database (Table 1). The sequencing results showed that the S1 genes of 12 PEDV strains commonly had a length of 2376 nt (partial nsp13 gene + full-length S1 gene). Notably, the S1 gene of one strain (HN-SY-2017-Oct) from the city of Shaoyang in Hunan was 2385 nt long, which is 9 nt (T1152GAAGCCAAT1160T encoding three continuous amino acids [381KPI383] insertion) longer than those of the other 12 strains, while the S1 of another strain from Zhejiang (ZJ-2018-May) was 2373 nt in length with a 3-nt deletion (1126AAA1127) resulting in the deletion of an amino acid (372K) (Fig. 2A). For the ORF3 gene, no nucleotide insertions or deletions were found in any of the PEDV strains identified in the present study.
Fig. 2.
Comparison of S1 amino acid sequences of PEDV isolates from this study with those of reference strains (GenBank nos. AF353511.1, KT323979.1, KF272920.1, KC210145.1, and JX188454.1). (A) A 3-aa insertion (381KPI383) was found in the PEDV HN/SY/2017/Oct strain, and a 1-aa deletion (K) was found at position of 372 in the PEDV ZJ/2018/May strain. (B) Amino acid sequence variations in three epitope regions (aa 201-212, aa 748-755, and aa 764-771) in the S1 genes of 14 PEDV strains
Analysis of sequence variation in the new PEDV strains
To identify sequence variations, the nucleotide (nt) and deduced amino acid (aa) sequences of the whole S1 and ORF3 genes of the 14 new PEDV strains were aligned. The results indicated that they displayed high sequence similarity, with 97.2-100% identity at the nt level and 95.8-100% identity at the aa level for the S1 gene, and with 95.1-99.7% nt sequence identity and 96.4-99.6% aa sequence identity for the ORF3 gene. Notably, compared with the attenuated PEDV strain CV777 (KT323979.1), these 14 PEDV strains exhibited 90.7-91.6% nt and 89.1-90.0% aa sequence identity in the S1 gene and 95.9-97.2% and 93.8-96.4% in the ORF3 gene. Likewise, these strains had 90.7-91.6% nt and 88.5-89.1% aa sequence identity in the S1 gene and 95.9-97.3% nt and 85.9-89.1% aa sequence identity in the ORF3 gene to the PEDV strain CV777 (AF353511.1). However, all 14 PEDV strains showed high sequence similarity to the highly virulent Chinese PEDV strain AJ1102 (JX188454.1) and the highly pathogenic USA PEDV strain USA/Colorado/2013 (KF272920.1) (Table 3), which partly explains the immune failure of the traditional PEDV CV777 vaccine in pigs challenged with variant PEDV strains.
Table 3.
Percent sequence identity in the S1 and ORF3 genes of PEDV strains identified in the present study
| PEDV strains compared (GenBank accession no.) | nt/aa level | ORF3 | S1 |
|---|---|---|---|
| 14 PEDV strains identified in current study | nt | 95.1-99.7% | 97.2-100.0% |
| aa | 96.4-99.6% | 95.8-100.0% | |
| Comparison with the PEDV CV777 strain (AF353511.1) | nt | 95.9-97.2% | 91.4-92.3% |
| aa | 93.8-96.4% | 89.1-90.0% | |
| Comparison with the PEDV CV777 attenuated strain (KT323979.1) | nt | 93.8-97.1% | 90.7-91.6% |
| aa | 85.9-89.1% | 88.5-89.1% | |
| Comparison with the variant PEDV strain AJ1102 from Hubei province, China, in 2011 (JX188454.1) | nt | 95.7-99.3% | 97.3-98.7% |
| aa | 95.6-99.1% | 97.2-98.2% | |
| Comparison with the natural recombinant PEDV strain from Henan province, China, in 2016 (KR095279.1) | nt | 94.0-97.8% | 94.8-96.3% |
| aa | 93.5-96.8% | 92.5-94.5% | |
| Comparison with the PEDV prototype strain USA/Colorado/2013 (highly pathogenic strain) from the USA in 2013 (KF272920) | nt | 95.3-99.4% | 97.9-99.5% |
| aa | 96.0-99.1% | 97.2-99.2% | |
| Comparison with the PEDV S-INDEL strain OH851 (low-pathogenic strain) from the USA in 2014 (KJ399978) | nt | 95.6-99.7% | 91.9-92.9% |
| aa | 96.4-99.1% | 90.7-92.5% |
The molecular features of three major epitope regions (aa 201-212, aa 748-755, and aa 764-771) of the S1 gene from these 14 PEDV strains were further analyzed and characterized. Compared with the PEDV strain CV777 (AF353511.1), its attenuated variant (KT323979.1), and three variants in the G2 gene group (KF272920.1, KC210145.1, and JX188454.1), there were no amino acid mutations in the epitope region (Y748SNIGVCK755S) among the 14 S1 genes examined, while three amino acid mutations were found in one other epitope domain (C201AMQYVYTPTYYM212L), and one was found in another (P764SQYGQVK771I) (Fig. 2B).
Phylogenetic analysis of the novel PEDV strains
To determine the evolutionary position of each of the 14 PEDV strains, phylogenetic trees were constructed using the full-length S1 and ORF3 gene sequences of PEDV reference strains available in the GenBank database and the 14 newly identified PEDV strains. The phylogenetic tree based on the S1 gene sequences (Fig. 3A) showed that the 14 PEDV strains could be classified into two groups, with the lower group (G1) containing two major branches including the PEDV attenuated strain CV777 and its prototypical parent strain. The upper group consisted of two subgroups (named G2-a and G2-b), and 11 of 14 (78.57%) PEDV strains from Hunan (n = 8), Guangxi (n = 1), Hubei (n = 1), and Zhejiang (n = 1) provinces reported here were randomly distributed in the G2-a subgroup, forming five different clusters. Notably, among these strains, the Yongzhou isolate from Hunan province (HN-YZ) was the most closely related to the Chinese highly virulent PEDV strain AJ1102 (JX188454.1), which was isolated from Hubei province. In contrast, the other three (21.43%) PEDV strains belonged to the G2-b subgroup and were more closely related to the Chinese highly virulent strain AH2012 (KC210145.1), which was isolated from Anhui province, and the American highly pathogenic strain USA/Colorado/2013 (KF272920.1).
Fig. 3.
Phylogenetic trees based on the full-length S1 (A) and ORF3 (B) gene sequences of the 14 newly identified PEDV strains and reference strains, constructed by the neighbor-joining (NJ) method using MEGA 7.0 software (Kimura 2-parameter model; 1000 bootstrap replicates). Black squares and triangles represent PEDV strains from Hunan and other provinces, respectively, identified in this study
Phylogenetic analysis based on ORF3 gene sequences showed that 13 (92.85%) of the 14 PEDV strains obtained in this study, the Chinese highly virulent strain AH2012 (KC210145.1), and the American highly pathogenic strain USA/Colorado/2013 (KF272920.1) were in the G2-c subgroup, while another PEDV strain (HN-SY) (7.15%, 1 of 14) was in the G2-b subgroup and was closely related to the Chinese PEDV highly virulent strain AJ1102 (JX188454.1). Consistent with the results of phylogenetic analysis based on the S1 gene, all of the PEDV strains from this study had a distant relationship to the present PEDV vaccine strains CV777 and LZC) (Fig. 3B).
Recombination and three-dimensional structure analysis of the S1 genes of two novel PEDV strains
Recombination analysis of the S1 genes of two novel PEDV strains and their putative parental strains was performed using SimPlot software (version 3.5.1). The analysis identified potential breakpoints in the HN-SY strain at nt positions 968 and 1480 (position alignment based on the S1 gene) when compared with two parental PEDV strains (AJ1102 and HN-YY-CH-2018-Mar) (Fig. 4A). Another strain from Zhejiang province had potential breakpoints at nt 919 and 1582 relative to the AJ1102 and HN-SY strains (Fig. 4B). Three-dimensional structure modeling of the S1 proteins of two novel PEDV strains predicted that the unique insertion of three continuous amino acids (381KPI383) would affect the protein surface, lengthening a loop on the surface and slightly changing its conformation (Fig. 4C). Analysis using NetPhos 2.0 software indicated that this alteration might increase the likelihood of phosphorylation occurring at 388Y (score = .881 > 0.50). In contrast, the deletion of 376K does not appear to have an obvious impact on the surface conformation of the S1 protein or the potential shift of the phosphorylated site, as indicated in the center of Fig. 4D.
Fig. 4.
Recombination and 3D structures of the S1 proteins of the two novel PEDV strains predicted by SimPlot recombination analysis software (version 3.5.1) and structure-homology modeling, respectively. (A) The potential breakpoint positions for strain HN-SY are in the region of nt 968-1480 (position alignment based on the S1 gene) compared with the parental PEDV strains (AJ1102 and HN-YY-CH-2018-Mar). (B) The potential breakpoint positions for strain CH-ZJ are in the region of nt 968-1480 (position alignment based on the S1 gene) compared with the parental PEDV strains AJ1102 and HN-YY-CH-2018-Mar. Red vertical lines indicate the potential breakpoints. (C) Predicted 3D structure of the S1 protein of PEDV strain HN/SY/2017/Oct. The loop including the insertion of three continuous amino acids (381KPI383) is shown in red, and the same loop without this insertion is shown in cyan. (D) Predicted 3D structure of the S1 protein of PEDV strain ZJ/2018/May. The β-strand with one-amino-acid (376K) deletion is show in green, and the same β-strand without the deletion is shown in red. The right panels are enlarged images from the boxes in the left panels
Discussion
China is the largest swine-raising country in the world, and microbial infections pose a great threat to the porcine industry. Since the emergence of PEDV variant strains in China in 2010, PED has become a major factor contributing to diarrheal diseases in piglets, resulting in tremendous economic losses to the swine industry [8]. So far, most of the PEDV strains circulating on pig farms in China are variant ones, and the traditional PEDV vaccines based on virulent PEDV strains (CV777, LZC, etc.) belonging to the G1 group cannot provide complete protection against variant strains [30]. Therefore, it is essential and urgent to obtain accurate data on the prevalence and molecular characteristics of the currently circulating PEDV strains, which will facilitate the development of vaccines based on these newly identified PEDV variant strains.
Here, by examining 184 specimens from pigs with (n = 121) or without diarrhea (n = 63) on different farms in eight provinces around Hunan, China, several characteristics of the epidemiology of PED-related viruses could be summarized: a) A relatively high PEDV-positive rate (close to 40%) was found in over 70% of the pig farms where samples were collected. In particular, a nearly 50% PEDV-positive rate was found in pigs with diarrhea, which is consistent with a recent report [8]. This suggests that PEDV remains a major pathogenic agent in China, as was also confirmed by examining data from previous investigations of the PEDV epidemic conducted in China (Fig. 1B). Therefore, more efforts are required to prevent and control this virus in the future. b) The overall rate of PEDV infection in pigs without diarrhea in the present study was higher than that found in a recent investigation [30], suggesting that attention should also be paid to these pigs. c) Although the detection rates of TGEV (3.26%; 6/184) and PoRV (3.80%; 7/184) were relatively low, the presence and circulation of other diarrhea-causing pathogens, including intestinal parasites (Balantidium coli, Coccidia, etc.), bacteria (Escherichia coli, Salmonella, etc.), and novel viruses such as porcine deltacoronavirus [13], and porcine enteric alphacoronavirus [17], are possible on pig farms in China. In particular, the question of whether co-infections with multiple viruses occur in individual PEDV-positive pigs warrants further investigation.
The majority of the samples collected in this study were from Hunan province. Both the prevalence of PEDV and the PEDV-positive rate of pigs with diarrhea in this province exceeded 50%, which was higher than in most of the administrative regions in southwestern and northwestern China but lower than in eastern, northeastern, central and southern China (Fig. 1B). This suggests that, as in most provinces in China, the epidemic situation of PEDV in Hunan is serious, and close monitoring of PEDV infection on pig farms is needed. Consequently, further investigations are required to understand the pathogenicity and transmission of PEDV so that appropriate strategies can be applied to prevent PED. Notably, recent investigations have indicated that this viral pathogen can be transmitted through contaminated feed and appliances [9]. The airborne route is another main pathway for PEDV dissemination in addition to fecal-oral transmission [14]. These observations could partly explain why culture density, animal welfare, and climate collectively contribute to differences in prevalence of PEDV infection in these provinces.
There is substantial evidence that the S1 and ORF3 genes play an essential role in the virulence of PEDV [7, 25]. We therefore sequenced the whole S1 and ORF3 genes of 14 PEDV strains from 14 representative swine farms in order to obtain information about virulence determinants. We identified a series of single nucleotide substitutions located in the S1 (including two epitope domains) and ORF3 genes which might contribute to the variability of PEDV strains at both the nucleotide and amino acid level. Notably, these 14 PEDV strains share high nucleotide and amino acid sequence similarity in their S1 and ORF3 genes with each other but exhibit low nucleotide and amino acid sequence similarity to the typical PEDV strain CV777 and its attenuated variant. Two novel PEDV variants, one (HN-SY-2017-Oct) with a continuous amino acid (KPI) insertion at position 381 (381KPI), and the other (ZJ-2018-May) with an amino acid deletion (K) at position 372 (372K) were identified. Amino acid insertions and deletions in the S gene (including the S1 gene) have been shown to affect viral pathogenicity [10, 18]. Although attempts at direct isolation and propagation of these two novel PEDV strains in various cell lines failed, we are still able to speculate that these strains are virulent. For instance, the swine farm where the novel PEDV strain HN-SY-2017-Oct was obtained was documented to suffer from destructive economic losses, suggesting that this variant strain has enhanced virulence and that traditional vaccines failed to protect pigs against infection. Moreover, structural prediction analysis showed that the insertion of three continuous amino acids (381KPI383) would lengthen the loop domain on the surface and alter the surface structure of this variant S1 protein, thereby contributing to the phosphorylation at position of 388Y (score = 0.881 > 0.50, indicating a high probability of being phosphorylated). Accumulating evidence has shown that the phosphorylation of surface proteins affects viral pathogenicity by altering the corresponding signaling pathway in the host cell [1, 29]. Experimental studies are needed to verify that phosphorylation occurs at position 388Y and to examine its effects. These findings highlight the necessity to monitor currently circulating variant strains of PEDV.
Finally, phylogenetic analysis based on the S1 gene showed that all 14 PEDV strains are closely related to highly virulent PEDV strains isolated in the USA and China and belong to the G2 group (G2-a or G2-b subgroup), but they are less closely related to members of the G1 group, including the typical PEDV CV777 attenuated strain and its prototypical parent strain (Fig. 3A), suggesting that PEDV variant strains are the main subtypes circulating in China, which is consistent with recent reports [4, 5]. Similar to the results based on the S1 gene, phylogenetic analysis based on the ORF3 gene also revealed that all 14 PEDV strains belong to the G2 group and differ from the PEDV-vaccine-related strains in the G2-a and G1 subgroups (Fig. 3B). Therefore, the ORF3 gene of PEDV does not appear to be suitable for revealing the phylogenetic positions of classical virulent PEDV strains (CV777, LZC, etc.) and these newly identified strains belonging to G2 group (Fig. 3B), which is consistent with a recent report [22].
In the current study, 14 novel PEDV strains from 14 regions in six provinces, mainly surrounding Hunan province, were identified in the investigation of the severe epidemics of PED in China. All of these strains belonged to the G2 group (G2-a and G2-b subgroups) and displayed a high degree of genetic variation. Comparison of the current epidemic results with those in previous reports confirms the pathogenicity of PEDV variant strains in China. Moreover, the amino acids of two epitope domains (aa 201-212 and aa 764-771) in the S1 gene were variable among these PEDV strains. More importantly, two newly identified PEDV strains, one containing an insertion of unique three continuous amino acids (381KPI), and the other containing an amino acid deletion (372K) in the S1 gene, were predicted by structural modeling to have potential effects of on the virulence of PEDV. The information provided here will facilitate future investigations of how these amino acid sequence variations affect the virulence of PEDV variant strains.
Abbreviations
- PED
porcine epidemic diarrhea
- PEDV
porcine epidemic diarrhea virus
- ORF
open reading frame
- RT-PCR
reverse transcription polymerase chain reaction
- TGEV
transmissible gastroenteritis virus
- PoRV
porcine rotavirus group A
Funding
This work was supported by the General Program of National Natural Science Foundation of China (Grant nos. 31571432 and 31802252) and the Hunan Provincial Natural Science Foundation of China (Grant no. 2015 JC3097). Support was also provided by “Shennong” Scholar funding to Aibing Wang.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Backert S, Feller SM, Wessler S. Emerging roles of Abl family tyrosine kinases in microbial pathogenesis. Trends Biochem Sci. 2008;33:80–90. doi: 10.1016/j.tibs.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 4.Chen F, Ku X, Li Z, Memon AM, Ye S, Zhu Y, Zhou C, Yao L, Meng X, He Q. Genetic characteristics of porcine epidemic diarrhea virus in Chinese mainland, revealing genetic markers of classical and variant virulent parental/attenuated strains. Gene. 2016;588:95–102. doi: 10.1016/j.gene.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Wang K, Hou Y, Li H, Li X, Yu L, Jiang Y, Gao F, Tong W, Yu H, Yang Z, Tong G, Zhou Y. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011–2017. Infect Genet Evol. 2019;69:153–165. doi: 10.1016/j.meegid.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Zeng L, Yang J, Yu F, Ge J, Guo Q, Gao X, Song T. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010–2012. Viruses. 2013;5:2375–2383. doi: 10.3390/v5102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Zhang XX, Li C, Wang H, Wang H, Meng XZ, Ma J, Ni HB, Zhang X, Qi Y, Sun D. Epidemiology of porcine epidemic diarrhea virus among Chinese pig populations: a meta-analysis. Microb Pathog. 2019;129:43–49. doi: 10.1016/j.micpath.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Dee SA, Bauermann FV, Niederwerder MC, Singrey A, Clement T, de Lima M, Long C, Patterson G, Sheahan MA, Stoian AMM, Petrovan V, Jones CK, De Jong J, Ji J, Spronk GD, Minion L, Christopher-Hennings J, Zimmerman JJ, Rowland RRR, Nelson E, Sundberg P, Diel DG. Survival of viral pathogens in animal feed ingredients under transboundary shipping models. PLoS One. 2018;13:e0194509. doi: 10.1371/journal.pone.0194509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan B, Jiao D, Zhao X, Pang F, Xiao Q, Yu Z, Mao A, Guo R, Yuan W, Zhao P, He K, Li B. Characterization of chinese porcine epidemic diarrhea virus with novel insertions and deletions in genome. Sci Rep. 2017;7:44209. doi: 10.1038/srep44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge FF, Yang DQ, Ju HB, Wang J, Liu J, Liu PH, Zhou JP. Epidemiological survey of porcine epidemic diarrhea virus in swine farms in Shanghai, China. Arch Virol. 2013;158:2227–2231. doi: 10.1007/s00705-013-1722-7. [DOI] [PubMed] [Google Scholar]
- 12.Lara-Romero R, Gómez-Núñez L, Cerriteño-Sánchez JL, Márquez-Valdelamar L, Mendoza-Elvira S, Ramírez-Mendoza H, Rivera-Benítez JF. Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013–2016. Virus Genes. 2017;54:215–224. doi: 10.1007/s11262-017-1528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Feng H, Liu Y, Chen Y, Wei Q, Wang J, Liu D, Huang H, Su Y, Wang D, Cui Y, Zhang G. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res Vet Sci. 2018;120:63–69. doi: 10.1016/j.rvsc.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Wu Q, Huang L, Yuan C, Wang J, Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat Commun. 2018;9:3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L, Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Zhang Q, Zhang L, Zhou P, Yang J, Fang Y, Dong Z, Zhao D, Li W, Feng J, Cui B, Zhang Y, Wang Y. A newly isolated Chinese virulent genotype GIIb porcine epidemic diarrhea virus strain: biological characteristics, pathogenicity and immune protective effects as an inactivated vaccine candidate. Virus Res. 2018;259:18–27. doi: 10.1016/j.virusres.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S, Kim S, Song D, Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg Infect Dis. 2014;20:2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SJ, Moon HJ, Luo Y, Kim HK, Kim EM, Yang JS, Song DS, Kang BK, Lee CS, Park BK. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stadler J, Zoels S, Fux R, Hanke D, Pohlmann A, Blome S, Weissenböck H, Weissenbacher-Lang C, Ritzmann M, Ladinig A. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Y, Liu Y, Chen Y, Zhao B, Ji P, Xing G, Jiang D, Liu C, Song Y, Wang G, Li D, Deng R, Zhang G. Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene. Virol J. 2016;13:192. doi: 10.1186/s12985-016-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M, Ma J, Wang Y, Wang M, Song W, Zhang W, Lu C, Yao H. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J Clin Microbiol. 2015;53:1484–1492. doi: 10.1128/JCM.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Murakami S, Takahashi O, Kodera A, Masuda T, Itoh S, Miyazaki A, Ohashi S, Tsutsui T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect Genet Evol. 2015;36:363–368. doi: 10.1016/j.meegid.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki T, Terada Y, Enjuanes L, Ohashi S, Kamitani W. S1 subunit of spike protein from a current highly virulent porcine epidemic diarrhea virus is an important determinant of virulence in piglets. Viruses. 2018;10:467. doi: 10.3390/v10090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Diep N, Sueyoshi M, Norimine J, Hirai T, Myint O, Teh APP, Izzati UZ, Fuke N, Yamaguchi R. Molecular characterization of US-like and Asian non-S INDEL strains of porcine epidemic diarrhea virus (PEDV) that circulated in Japan during 2013–2016 and PEDVs collected from recurrent outbreaks. BMC Vet Res. 2018;14:96. doi: 10.1186/s12917-018-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Wang Y, Baloch AR, Pan Y, Tian L, Xu F, Shivaramu S, Chen S, Zeng Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai–Tibet Plateau of China. Transbound Emerg Dis. 2018;25:363–369. doi: 10.1111/tbed.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Zhan Y, Wang A, Zhang L, Khayat R, Yang Y. In silico analysis of surface structure variation of PCV2 capsid resulted from loops mutation of its capsid protein (Cap) J Gen Virol. 2016;97:3331–3344. doi: 10.1099/jgv.0.000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Chai X, Cheng Y, Xing G, Liao A, Du L, Wang Y, Lei J, Gu J, Zhou J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018;247:47–54. doi: 10.1016/j.virusres.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhan Y, Wang N, Zhu Z, Wang Z, Wang A, Deng Z, Yang Y. In silico analyses of antigenicity and surface structure variation of an emerging porcine circovirus genotype 2b mutant, prevalent in southern China from 2013 to 2015. J Gen Virol. 2016;97:922–933. doi: 10.1099/jgv.0.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Liu X, Zhang Q, Zhou P, Fang Y, Dong Z, Zhao D, Li W, Feng J, Zhang Y, Wang Y. Biological characterization and pathogenicity of a newly isolated Chinese highly virulent genotype GIIa porcine epidemic diarrhea virus strain. Arch Virol. 2019;164:1287–1295. doi: 10.1007/s00705-019-04167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Liu X, Fang Y, Zhou P, Wang Y, Zhang Y. Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016–2017. Virol J. 2017;14:194. doi: 10.1186/s12985-017-0860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou L, Sun Y, Lan T, Wu RT, Chen JW, Wu ZX, Xie QM, Zhang XB, Ma JY. Retrospective detection and phylogenetic analysis of swine acute diarrhea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis. 2018;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo Q, Zhao R, Liu J, Zhao Q, Zhu L, Zhang B, Bi J, Yang G, Liu J, Yin G. Epidemiology and phylogeny of spike gene of porcine epidemic diarrhea virus from Yunnan, China. Virus Res. 2018;249:45–51. doi: 10.1016/j.virusres.2018.03.008. [DOI] [PubMed] [Google Scholar]