Abstract
Lassa virus (LASV) is responsible for a type of acute viral haemorrhagic fever referred to as Lassa fever. Lack of adequate treatment and preventive measures against LASV resulted in a high mortality rate in its endemic regions. In this study, a multi-epitope vaccine was designed using immunoinformatics as a prophylactic agent against the virus. Following a rigorous assessment, the vaccine was built using T-cell (NCTL = 8 and NHTL = 6) and B-cell (NLBL = 4) epitopes from each LASV-derived protein in addition with suitable linkers and adjuvant. The physicochemistry, immunogenic potency and safeness of the designed vaccine (~ 68 kDa) were assessed. In addition, chosen CTL and HTL epitopes of our vaccine showed 97.37% worldwide population coverage. Besides, disulphide engineering also improved the stability of the chimeric vaccine. Molecular docking of our vaccine protein with toll-like receptor 2 (TLR2) showed binding efficiency followed by dynamics simulation for stable interaction. Furthermore, higher levels of cell-mediated immunity and rapid antigen clearance were suggested by immune simulation and repeated-exposure simulation, respectively. Finally, the optimized codons were used in in silico cloning to ensure higher expression within E. coli K12 bacterium. With further assessment both in vitro and in vivo, we believe that our proposed peptide-vaccine would be potential immunogen against Lassa fever.
Electronic supplementary material
The online version of this article (10.1007/s10989-019-10003-8) contains supplementary material, which is available to authorized users.
Keywords: Lassa fever, Immunoinformatics, Peptide vaccine, Molecular docking, Immune simulation
Introduction
Lassa virus (LASV) is an emerging viral pathogen belongs to the Arenaviridae family and can cause severe viral haemorrhagic fever which is known as Lassa fever, with a 20% fatality rate (Charrel and De Lamballerie 2003). Lassa fever is a viral acute zoonotic disease due to its capability to affect the highest number of people (~ 500,000) and causes 5000 annual deaths in Western Africa (Ogbu et al. 2007). Also, the people of Ghana (~ 10%), Côte d’Ivoire (~ 30), Nigeria (~ 40%), Guinea (~ 50%), Sierra- Leone and Liberia (~ 80%) and a few areas of Mali are assumed to be affected by Lassa fever (Fichet-Calvet and Rogers 2009; Safronetz et al. 2010). About 200 million people of West African regions (i.e., Nigeria and Senegal) are at high risk of LASV outbreak (Charrel and De Lamballerie 2003). Moreover, it also affects many areas of Europe such as the United Kingdom (Kitching et al. 2009), Netherlands (WHO 2000) and Germany (Haas et al. 2003). Though it has been revealed that the virus primarily targets antigen-presenting cells (mainly dendritic cells and macrophages) and endothelial cells and interferes with their complete maturation and activation, but the pathogenesis of Lassa fever is yet not clearly understood (Hallam et al. 2018; Oti 2018). Given the high annual incidence and mortality rate, however, the development of an effective LASV vaccine is an urgent necessity.
LASV is endemic to West Africa, and the genomic organization of the Lassa virus is an enveloped, ambisense and has a bisegmented, negative sense and single-stranded RNA genome consisting of large (L) segments and small (S) segments (Oti 2018). The large or L segment of the RNA encodes the 200 kDa RNA polymerase (L) protein and the small ring-finger protein (matrix protein or Z-protein, 11 kDa) that regulate replication and transcription (Cornu and de la Torre 2001; Djavani et al. 1997). The small segments encode the surface glycoprotein precursor (GP, 75 kDa) and the nucleoprotein (NP, 63 kDa), which is proteolytically cleaved into GP1 and GP2 (envelope glycoprotein) that bind to the alpha-dystroglycan receptor and mediate entering into the host cell (Cao et al. 1998; Oti 2018).
LASV is transmitted to the human being through the rodent reservoir Mastomys natalensis, a typical African rat lurking in village houses (Bonner et al. 2007). Recent evidence, however, indicates that other rodent species may also be LASV recipients, like, African wood mouse Hylomyscus pamfi (Nigeria), and Guinea mouse M. erythroleucus (Nigeria and Guinea) (Hallam et al. 2018). Exchange of LASV occurs when a healthy individual comes in contact with the blood, secretion, tissue or excretion of any infected personal or by food contaminated with the host excreta. However, skin to skin contact without exchange of blood fluid cannot transmit the virus (Keenlyside et al. 1983). Children under ten years old are considered as the most vulnerable to LASV. For instance, a study showed 15% seropositivity in the under-aged population in West Africa (Kernéis et al. 2009). Besides, pregnant patients with Lassa fever results in spontaneous abortions (Price et al. 1988). Ribavirin, an antiviral drug, is found to be effective at the initial phase of Lassa fever and can reduce the fatality rate (Jahrling et al. 1980; McCormick et al. 1986). However, the development of potential toxicity and teratogenicity when used in the later stage of disease drives us to think that Ribavirin is not a potent therapeutic against Lassa fever (Fisher-Hoch et al. 1992; Kochhar 1990). Peptide vaccines are immune stimulants where fragments of virus-derived proteins mimic natural pathogens; hence, more influential in terms of safety, efficacy and specificity (Skwarczynski and Toth 2016). In 1985, the first epitope-based vaccine was developed using cholera toxin against E. coli (Jacob et al. 1985). Furthermore, peptide vaccines against many pathogenic agents (i.e., HIV, malaria, swine fever, influenza, anthrax, etc.) are promptly under development (Li et al. 2014).
Present study demonstrates the screening of whole LASV proteome followed by the grouping of viral proteins. Each protein group was evaluated separately for the identification of T-cell and B-cell epitopes along with their respective MHC alleles using vaccinomics. Subsequently, a vaccine was designed using the most persuasive epitopes from each protein with suitable adjuvant and linkers. The primary sequence was used for physicochemical analysis and immunogenic profiling, followed by the secondary and tertiary structure predictions. The predicted three-dimensional (3D) structure was applied for refinement and validation. Besides, disulphide bridging was done to improve structural stability. The binding affinity and interactions between the vaccine protein and the receptor were calculated by molecular docking and dynamics simulation, respectively. Codon optimization and in silico cloning were taken care of for the evaluation of the expression of chimeric protein within the appropriate host. Finally, an immune simulation was performed to estimate the immunogenic potency in real-life.
Methodology
Proteome Retrieval and Antigenicity Prediction
The whole proteome of the Lassa mammarenavirus was retrieved from the ViPR database (Virus Pathogen Database and Analysis Resource), an integrated robust database for several virus families and their respective species (Pickett et al. 2012). Initially, the proteins from LASV proteome were isolated and classified as glycoprotein, L-protein, matrix protein, nucleocapsid protein, nucleoprotein, polymerase, ring-finger protein, Z-protein. As a measure of an immune response, the structural proteins were then applied for antigenicity prediction using the Vaxijen v2.0 server with a 0.5 threshold value (Doytchinova and Flower 2007). This server uses auto and cross-covariance (ACC) transformation method to maintain 70–89% prediction accuracy. A protein with the best antigenic score was chosen from each class of structural proteins.
Prediction of Cytotoxic T-Lymphocyte (CTL) Epitopes and MHC-I Binding Alleles
The selected protein was submitted to the NetCTL v1.2 server for the prediction of CTL epitopes (9-mer) for 12 supertypes (i.e., A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, and B62). This server predicts CTL epitopes based on three criteria, namely, MHC-I binding peptides, C-terminal cleavage, and TAP transport efficiency. MHC-I binding and C-terminal cleavage are obtained by using artificial neural networks whereas TAP transporter efficiency is evaluated by the weight matrix (Larsen et al. 2007). In this study, the threshold was set to 0.5 which has a sensitivity and specificity of 0.89 and 0.94, respectively. Furthermore, MHC-I binding alleles for each CTL epitope were predicted using the consensus method in the IEDB analysis tool (Moutaftsi et al. 2006). The human was selected as the source species, and percentile rank ≤ 5 was considered since lower score indicates higher affinity.
Assessment of CTL Epitopes for Immunogenicity, Allergenicity and Toxicity
Firstly, selected CTL epitopes were rechecked for antigenicity to ensure their ability to induce immune response with VaxiJen v2.0 server. Further, they were also evaluated for immunogenicity with MHC-I immunogenicity tool of IEDB server (Calis et al. 2013). Vaccine components should be free from an allergic reaction. So, AllergenFP v1.0 server was used for allergenicity prediction. This server can recognize both allergens and non-allergens with 88% accuracy (Dimitrov et al. 2014b). Furthermore, toxic epitopes should be eliminated as they could compromise the functionality of the vaccine construct. Therefore, we used ToxinPred server to sort out the toxic CTL epitopes (Gupta et al. 2013).
Prediction of Helper T-Lymphocyte (HTL) Epitopes and MHC-II Binding Alleles
Helper T-lymphocyte (HTL) responses play an essential role in the induction of both humoral and cellular immune responses. Therefore, HTL epitopes are likely to be a significant element of preventive and immunotherapeutic vaccines. The IEDB MHC-II binding tool was applied to predict 15 amino acid long HTL epitopes using NN-align method (Nielsen and Lund 2009). A percentile rank was generated by comparing peptide’s binding affinity with a comprehensive set of randomly selected peptides from the Swiss-Prot database. Percentile rank ≤ 5 was also considered for this analysis (Paul et al. 2016).
Identification of Cytokine-Inducing HTL Epitopes
Innate immune system, B-lymphocytes, cytotoxic T-cells and other immune cells are activated by the help of helper T-cells which further releases different types of cytokines, i.e., interferon-gamma (IFN-γ), interleukin-4 (IL-4) and interleukin-10 (IL-10), HTL epitopes have the ability to overcome proinflammatory response and thus diminish tissue damage (Luckheeram et al. 2012). Therefore, cytokine-inducing HTL epitopes are essential in vaccine development. So, we used IFNepitope server for the prediction of IFN-γ inducing HTL epitopes using a hybrid method (Motif and SVM) along with IFN-gamma versus Non-IFN-gamma model (Dhanda et al. 2013a, b). In addition to IFN-gamma, IL-4 and IL-10 properties were also evaluated with IL4pred and IL10pred servers, respectively (Dhanda et al. 2013a, b; Nagpal et al. 2017).
Prediction and Assessment of Linear B-lymphocyte (LBL) Epitopes
Identification of B-cell epitopes (BCEs) is a fundamental step for epitope-based vaccine development as it produces antibodies which provide humoral immunity. Therefore, LBL epitopes were predicted using iBCE-EL server. It uses a method that combines gradient boosting algorithms with an extremely randomized tree method (Manavalan et al. 2018). The predicted LBL epitopes were then further assessed to check their antigenicity, allergenicity and toxicity with VaxiJen v2.0, AllergenFP v1.0 and ToxinPred server, respectively.
Estimation of Population Coverage
Different HLA alleles, as well as their expression, are sensationally distributed at various frequencies in diverse ethnicities (Adhikari and Rahman 2017). Hence, the HLA-alleles distribution among the world population is crucial for successful multi-epitope vaccine development. In this study, the IEDB population coverage analysis tool was used for the analysis of the population coverage of the potential CTL and HTL epitopes and their MHC binding alleles (Bui et al. 2006).
Designing of Multi-epitope Vaccine Construct
To construct multi-epitope vaccine, finally selected CTL, HTL, and LBL epitopes were linked together with the help of AAY, GPGPG and KK linkers, respectively (Gu et al. 2017; Nain et al. 2019). Peptides used for the vaccine construction are generally poorly immunogenic when used alone, therefore, requires adjuvants to boost up the immune response (Li et al. 2014). Consequently, the amino acid sequence of OmpA protein (GenBank: AFS89615.1) was chosen as an adjuvant and linked ahead of the first CTL epitope through EAAAK linker (Arai et al. 2001). Using linker for joining of two epitopes is required for the effective functioning of each epitope (Nezafat et al. 2014).
Antigenicity, Allergenicity and Physicochemical Evaluation
As vaccine protein should be highly antigenic, antigenicity prediction is necessary. The designed vaccine construct was analysed using Vaxijen v2.0 (Doytchinova and Flower 2007) and cross-checked with ANTIGENpro server (Magnan et al. 2010). Vaxijen is free from any alignment and work based on various physicochemical properties of the protein, whereas ANTIGENpro server has been developed based on microarray analysis data. Screening for allergenicity is important as it indicates the potentiality of a vaccine construct to cause sensitization and allergic reaction. We used AllergenFP v1.0 (Dimitrov et al. 2014b) and AllerTOP v2.0 (Dimitrov et al. 2014a) server to check the allergenicity of the vaccine protein. Inducing an immune response after injecting the vaccine into the body is the sole purpose of vaccination. Therefore, the assessment of various physicochemical properties of the chimera protein is essential. The primary protein sequence of the vaccine was used to predict the various physiochemical features through ProtParam (Wilkins et al. 1999) web-server. Furthermore, the solubility of the vaccine protein upon overexpression in E. coli was predicted by SOLpro (Magnan et al. 2009) tool in the SCRATCH suite.
Secondary and Tertiary Structure Prediction of the Vaccine Construct
PSIPRED v4.0 server was used for the prediction of vaccine’s secondary structure. Two feed-forward neural networks are the basis of PSIPRED, which process the PSI-BLAST (Position-Specific Iterated-Blast) to predict a secondary structure from an amino acid sequence as an input (Buchan et al. 2013).The tertiary structure of the final subunit vaccine was predicted using the RaptorX server, which is based on three steps; specifically single-template threading, multiple-template threading and alignment quality prediction. The server predicts 3D protein model and provides some confidence scores to evaluate the quality of predicted models. P-value, GDT, uGDT, modelling error at each residue etc. are different confidence scores that provide a clear indication to the relative good model (Källberg et al. 2017).
Refinement and Validation of 3D Vaccine Construct
The predicted model of the vaccine protein was refined through GalaxyRefine web-server. This server initially rebuilds sidechains, then sidechain repacking and consequently uses the molecular dynamics simulation for overall structural relaxation (Heo et al. 2013). Structural validation is a process to identify potential errors in the predicted tertiary structure (Khatoon et al. 2017). Therefore, ProSA-web was used for structural validation, which provides an overall quality score for the input structure. If the calculated score falls outside the range characteristics of the native protein, then the structure contains errors (Wiederstein and Sippl 2007). The ERRAT server was also used to analyse non-bonded atom–atom interactions (Colovos and Yeates 1993). Finally, Ramachandran plot was obtained using PROCHECK server. The Ramachandran plot is a way to visualize energetically allowed and disallowed dihedral angles psi (ψ) and phi (ϕ) of amino acid and is calculated based on van der Waal radius of the side chain. The result from PROCHECK include the percentage and number of residues in most favoured, additional allowed, generously allowed, and disallowed region, which defines the quality of modelled structure (Laskowski et al. 1993).
Conformational B-cell Epitope Prediction from Vaccine Construct
The conformational B-cell epitope is the collection of amino acid residues on the 3-dimensional geometry of the vaccine protein which interacts directly to the immune receptor. Therefore, ElliPro tool of IEDB server was used to determine the presence of conformational B-cell epitopes in the validated tertiary structure. ElliPro uses three algorithms relying on their protrusion index (PI) values to approximate the protein structure as an ellipsoid, calculate the residue PI, and adjacent cluster residues. ElliPro provides an average PI value over each epitope residue for each generated epitope. For each epitope residue, the PI value is calculated based on the residue mass centre outside the largest possible ellipsoid (Ponomarenko et al. 2008).
Disulphide Engineering of Final Vaccine Construct
Disulphide bonds are covalent interactions that stabilizes molecular interactions and provide considerable stability by confirming precise geometric conformations. Disulphide engineering is a novel approach for creating disulphide bonds into the target protein structure. Therefore, disulphide engineering was executed with Disulphide by Design v2.12 web tool. Initially, the refined protein model was uploaded and run for the residue-pair search that can be used for the disulphide engineering purpose. Potential residue pairs were selected for mutation, and cysteine residue was used as a final target for disulphide engineering (Craig and Dombkowski 2013).
In Silico Codon Adaptation and Cloning
Codon optimization is essential as unadapted codon may lead to the minor expression rate in the host. Codon optimization was performed using Java Codon Adaptation Tool (JCat) server with a view to improving the translational efficiency in E. coli K12 strain (Grote et al. 2005). Three additional options were selected to avoid the rho-independent transcription termination, prokaryote ribosome binding site, and restriction enzymes cleavage sites. Codon adaptation index (CAI) value and GC content of the adapted sequence was obtained and compared with the ideal range (Sharpl and Li 1987). Consequently, the received nucleotide sequence was cloned into the E. coli pET28a(+) vector by using SnapGene v4.2 tool.
Molecular Docking Between the Vaccine and TLR2 Receptor
Molecular docking is a computational method which involves the interaction between a ligand molecule and the receptor molecule to provide a stable adduct. Also, a calculated score was provided as a measure of the degree of binding interaction (Lengauer and Rarey 1996). Toll-like receptor 2 (TLR2) can mediate high proinflammatory responses against LASV infection (Hayes and Salvato 2012). Therefore, TLR2 structure was used as the receptor (PDB ID: 3A7B) and the refined vaccine protein as the ligand (Berman et al. 2002). Finally, the binding affinity between the multi-epitope vaccine and TLR2 receptor was calculated through the ClusPro v2.0 server (Kozakov et al. 2017). This server completed the task in three consecutive steps such as rigid body docking, clustering of lowest energy structure, and structural refinement by energy minimization. The best-docked complex was selected based on the lowest energy scoring and docking efficiency.
Molecular Dynamics Simulation
Molecular dynamics study is critically essential for checking the stability of the protein–protein complex in any in silico analysis. Protein stability can be determined by comparing essential protein dynamics to their normal modes (Van Aalten et al. 1997; Wüthrich et al. 1980). The iMODS server was used to explain the collective protein motion in the internal coordinates through normal mode analysis (NMA) (López-Blanco et al. 2014). The server estimated the direction and extent of the immanent motions of the complex in terms of deformability, eigenvalues, B-factors, and covariance. The deformability of the main chain depends on whether a specified molecule can deform at each of its residues. The eigenvalue of each normal mode describes the rigidity of motion. This value is related directly to the energy required for the structural deformation, and the deformation is much easier if the eigenvalue is low.
In Silico Evaluation of Immune Response
To estimate the immunogenic potential of the final vaccine, in silico immune simulations were conducted using the C-ImmSim server. This immune simulator uses a position-specific scoring matrix (PSSM) and machine learning techniques for the prediction of epitope prediction and immune interactions, respectively (Rapin et al. 2010). The minimum suggested interval between dose 1 and dose 2 is 4 weeks, according to most vaccines in current use (Castiglione et al. 2012). All parameters were set at default with time steps set at 1, 84, and 168 where each time step is equal to 8 h. Therefore, three injections were given four weeks apart. Moreover, six doses of injections of the designed vaccine were given in the same manner to simulate repeated exposure interaction to the antigen seen in a typical endemic area to probe for clonal selection.
Results
Protein Retrieval and Highest Antigenic Protein Selection
For the construction of candidate vaccine, a total of 1665 protein sequences of different structural (glycoprotein, matrix protein, nucleocapsid protein, nucleoprotein, ring-finger protein and Z-protein) and non-structural (L-protein, Polymerase, RNA directed RNA-polymerase) protein sequences of the LASV were retrieved from ViPR database. Structural protein enables viruses to invade and assemble viral particles in the host, while non-structural proteins secrete various enzymes that assist in viral replication and development of structural proteins. Vaxijen v2.0 server revealed the highest antigenic protein from each type and as we set a threshold of 0.5, only glycoprotein, matrix protein, ring-finger protein and Z-protein showed antigenicity (Table 1).
Table 1.
The most antigenic protein of each type, along with their GenBank accession ID, antigenicity score, and length
| Proteins name | GenBank accession | Length | Antigenicity score |
|---|---|---|---|
| Z-protein | AAT48997.1 | 99 | 0.7645 |
| Matrix protein | ANH09745.1 | 99 | 0.7368 |
| Glycoprotein | AAT49014.1 | 254 | 0.6956 |
| Ring-finger protein | AAO59514.1 | 99 | 0.5849 |
Prediction and Appraisal of CTL, HTL and LBL Epitopes
A total of 180 unique CTL epitopes (9-mer) were predicted from the four LASV highest antigenic proteins. Here 97, 22, 32 and 29 CTL epitopes were predicted from the glycoprotein, matrix protein, ring-finger protein and Z-protein, respectively, using the NetCTL v1.2 server. Among them, only 42 epitopes were found as antigenic, immunogenic, and non-toxic (Supplementary Table 1). Of the 42 epitopes, 22 were found to be non-allergenic. These non-allergenic epitopes were further used to predict their MHC-I binding alleles using MHC-I allele prediction tool of the IEDB server. Similarly, a total of 72 unique HTL epitopes (15-mer) and their MHC-II binding molecules were predicted using the IEDB MHC-II prediction tool. The cytokine (i.e., IFN-γ, IL-4 and IL-10) inducing ability of these HTL epitopes were also evaluated (Supplementary Table 2). B-cell epitopes are antigenic regions of a protein that can trigger antibody formation. The iBCE-EL tool was used to predict linear B lymphocyte (LBL) from the LASV proteins. We found a total of 101 LBL epitopes and after evaluation, only 37 unique epitopes (14, 5, 9 and 9 LBL epitopes were found from the glycoprotein, matrix protein, ring-finger protein and Z-protein, respectively) were found to be non-allergenic and non-toxic and considered for vaccine construction (Supplementary Table 3).
Construction of Multiepitope Vaccine
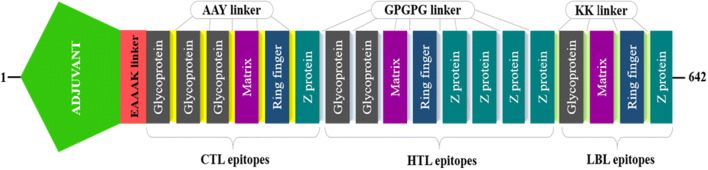
For multi-epitope vaccine designing, we have considered highly antigenic CTL epitopes from every type of proteins that are immunogenic, non-allergenic and non-toxic (Table 2). On the other hand, at HTL epitope selection, we screened cytokine-inducing properties and found that only two epitopes from the glycoprotein have the capacity to induce all three type of cytokine, while epitopes from the other proteins were positive for a maximum of two cytokines. Hence, we selected two epitopes from the glycoprotein and epitopes having inducing feature for at least two cytokines were selected in case of other proteins (Table 3). As we get a numerous number of B-cell epitopes with higher antigenicity, non-toxicity and non-allergenicity, we took the epitope with the best probability score (obtained from iBCE-EL tool) from each type of protein (Table 4). Therefore, 6 CTL, 8 HTL and 4 LBL epitopes are merged by AAY, GPGPG and KK linkers, respectively (Fig. 1). OmpA agonist (GenBank ID: AFS89615.1), which is 352 amino acid residues long, was used as an adjuvant for TLR2 receptor using EAAAK linker. The final vaccine construct comprises 642 amino acid residues.
Table 2.
Finally selected cytotoxic T-lymphocyte (CTL) epitopes for multi-epitope vaccine construction
| Protein | CTL epitopes | Position | Combined score | Antigenicity | Immunogenicity | Allergenicity | Toxicity |
|---|---|---|---|---|---|---|---|
| Glycoprotein | RPSPIGYLG | 234–242 | 0.6122 | 2.3297 | 0.08704 | No | Non-toxic |
| CGIIGLVAF | 41–49 | 0.5494 | 1.4004 | 0.25738 | No | Non-toxic | |
| ATCGIIGLV | 39–47 | 0.7889 | 1.3749 | 0.2936 | No | Non-toxic | |
| Matrix | APPARTGEN | 83–91 | 0.527 | 1.1219 | 0.20981 | No | Non-toxic |
| Ring-finger | FENRGLVEC | 36–44 | 0.7557 | 1.228 | 0.16588 | No | Non-toxic |
| Z-protein | GDRTRPPPY | 89–97 | 0.6183 | 1.4294 | 0.07998 | No | Non-toxic |
Table 3.
Finally selected helper T-lymphocyte (HTL) epitopes with their cytokine inducing properties
| Protein | HTL epitopes | Position | Antigenicity | IL4pred | IL10pred | IFNepitope |
|---|---|---|---|---|---|---|
| Glycoprotein | GYLGLLSQRTRDIYI | 239–253 | 1.8077 | IL4 inducer | IL10 inducer | Positive |
| GVLQTFMRMAWGGRY | 185–199 | 0.4769 | IL4 inducer | IL10 inducer | Positive | |
| Matrix | NCLTLLLSVSSRCPI | 52–66 | 0.6165 | IL4 inducer | IL10 inducer | Negative |
| Ring-finger | NCLSLLLSVSSRCPI | 52–66 | 0.7793 | IL4 inducer | IL10 inducer | Negative |
| Z-protein | LTLLLSVSNRCPICK | 54–68 | 0.2654 | IL4 inducer | IL10 inducer | Negative |
| NCLTLLLSVSNRCPI | 52–66 | 0.7145 | IL4 inducer | IL10 inducer | Negative | |
| CLTLLLSVSNRCPIC | 53–67 | 0.8906 | IL4 inducer | IL10 inducer | Negative | |
| CLNCLTLLLSVSNRC | 50–64 | 0.9551 | Non IL4 inducer | IL10 inducer | Positive |
Table 4.
Finally selected linear B-lymphocyte (LBL) epitopes for multi-epitope vaccine construction
| Protein | Peptide sequence | Position | Probability | Allergenicity | Antigenicity | Toxicity |
|---|---|---|---|---|---|---|
| Glycoprotein | GKSCSLTLKGGY | 54–65 | 0.8007 | Non-allergen | Antigen | Non-toxic |
| Matrix | KQTKAPEVEDGP | 4–15 | 0.7259 | Non-allergen | Antigen | Non-toxic |
| Ring-finger | WFENRGLVECNN | 35–46 | 0.7579 | Non-allergen | Antigen | Non-toxic |
| Z-protein | APETKNSPRASL | 8–19 | 0.7927 | Non-allergen | Antigen | Non-toxic |
Fig. 1.
Graphical presentation of the multi-epitope vaccine construct. A 642 amino acid residues long vaccine construct consisting of an adjuvant (green) at the N-terminal end is linked with the whole multi-epitope sequence through EAAAK linker (red). CTL, HTL and BL epitopes are fused with the help of AAY (yellow), GPGPG (light blue) and KK (light green) linkers, respectively (Color figure online)
Population Coverage Analysis
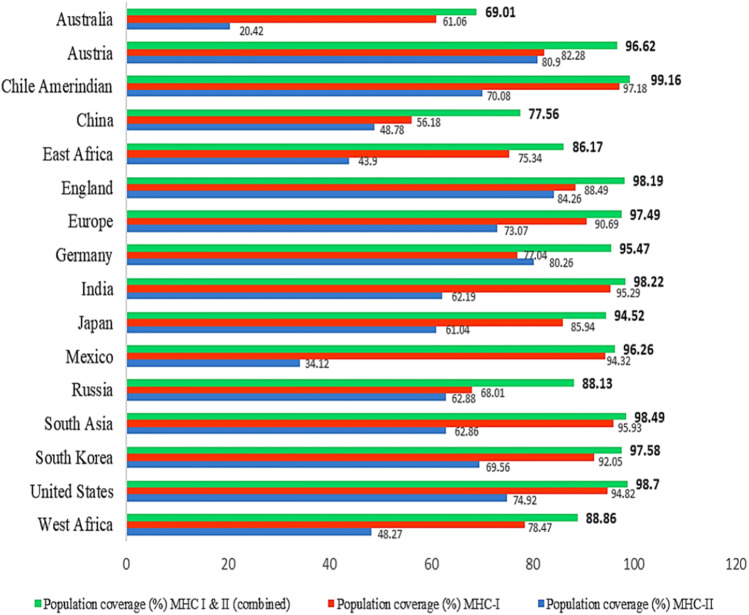
The distribution of HLA allele varies between different geographical and ethnic regions around the globe. Therefore, population coverage during the development of an efficient vaccine must be taken into account. In this study, selected CTL and HTL epitopes, which were used to construct the vaccine and their corresponding HLA alleles (Supplementary Tables 4 and 5) were obtained for population coverage analysis both individually and in combination. Our selected CTL and HTL epitopes were found to cover 91.74% and 68.15% of the world population, respectively. Importantly, CTL and HTL epitopes showed 97.37% population coverage worldwide when used in combination. The highest population coverage was found to be 99.16% in the South American country Chile Amerindian. In West Africa, where the bacteria first appeared and had several outbreaks, the population coverage was 88.86%. LASV caused several epidemics in different countries of the world, especially in England, Germany, and the Netherlands where the population coverage was found to be 98.19%, 95.47%, and 69.80%, respectively (Supplementary Table 6). In addition, the population coverage in China, Europe, France, India, Japan, South Korea, Russia, South Asia, and United States were found to be 77.56%, 97.49%, 94.65%, 98.22%, 94.52%, 97.58%, 88.13%, 98.49% and 98.70%, respectively (Fig. 2).
Fig. 2.
Population coverage of the selected T-cell epitopes and their respective HLA alleles. Regions of particular importance were considered in this graph with their MHC-I (red), MHC-II (blue) and combined (green) coverage rate (Color figure online)
Physiochemical Analysis and Solubility Prediction
Multiple physicochemical properties were calculated from the ProtParam server by inserting the whole vaccine construct as an amino acid sequence. The molecular weight of the construct was calculated as ~ 68 kDa and the antigenicity prediction showed that the construct has good antigenic properties. The analysis shows 9.31 pI (Isoelectric point) value which indicates the vaccine construct is basic in nature. The instability index (II) was computed to be 29.05, which implies that the sequence of the construct will remain stable after expression. The aliphatic index was calculated as 84.98 which indicates the construct’s thermostability. The grand average of hydropathicity (GRAVY) was calculated to be negative (− 0.118). This negative value indicates the hydrophilic nature of protein; therefore this protein tends to have better interaction with other proteins. Estimated half-life in mammalian reticulocyte (in vitro) was found to be 30 h, while in yeast and Escherichia coli the estimated half-life (in vivo) are > 20 h and > 10 h, respectively. The immunogenic appraisal also revealed that our vaccine construct is highly antigenic, non-allergenic and showed higher solubility rate calculated through SOLpro server. These evaluations suggested that our vaccine construct might be an ideal vaccine against LASV (Table 5).
Table 5.
Antigenic, allergenic and physiochemical assessments of the primary sequence of final vaccine protein
| Features | Assessment |
|---|---|
| Number of amino acids | 642 |
| Molecular weight | 67,909.01 Dalton |
| Chemical formula | C3018H4809N845O886S25 |
| Theoretical pI | 9.31 |
| Total number of negatively charged residues (Asp + Glu) | 49 |
| Total number of positively charged residues (Arg + Lys) | 76 |
| Total number of atoms | 9583 |
| Extinction coefficient (at 280 nm in H2O) | 64,970 M−1 cm−1 |
| Instability index | 29.05 |
| Aliphatic index | 84.98 |
| Grand average of hydropathicity (GRAVY) | − 0.118 |
| Antigenicity | 0.904775 (AntigenPro) |
| 0.7223 (Vaxijen v2.0) | |
| Allergenicity | Probable non-allergen (AllergenFP v1.0) |
| Probable non-allergen (AllerTOP v2.0) | |
| Solubility | 0.990675 |
Secondary Structural Feature Prediction
PSIPRED v4.0 workbench was used to predict the secondary structure of the final vaccine construct. The final vaccine construct (642 amino acid long) was analysed in which 352 amino acids involved in random coil formation while 149 amino acids involved in α-helix creation and β-strands are formed only by 141 amino acids. So, overall secondary structural feature prediction results indicate 54.83% are random coils, 23.21% forms α-helix and 21.96% are β-strands (Fig. 3).
Fig. 3.
Graphical representation of secondary structure prediction of the multi-epitope vaccine. Here, the β-strands, α-helix and random coils are indicated by yellow, pink and blue color, respectively
Tertiary Structure Modelling and Refinement
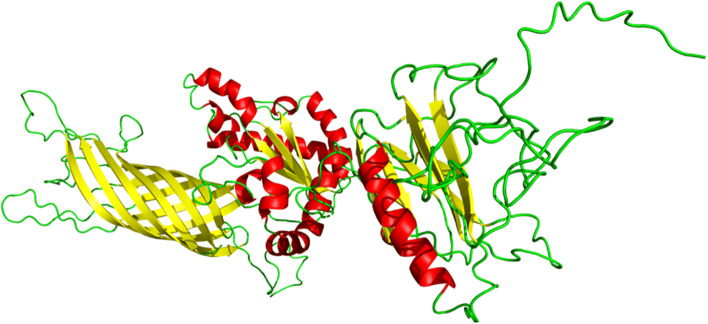
The tertiary structure of the multiepitope vaccine was modelled from the RaptorX server. In RaptorX server, 100% (642) amino acid residues were modelled as six domains. The protein structure (PDB ID: 1bxwA) was used as the best template for modelling. The relative quality of the modelled structure was evaluated by P-value. Calculated P-value for the modelled structure was 5.39 × 10−06 which is very low. Here, lower the P-value higher the quality of the model. Furthermore, uGDT is also used as a parameter for 3D structure evaluation and a construct with > 100 residues, uGDT > 50 is a good indicator. As we got 354 as uGDT score, it indicates the tertiary model as acceptable protein model for further analysis (Supplementary Table 7). The predicted tertiary (3D) structure was further refined using GalaxyRefine server, leads to an increase in the number of residues in the favoured region, generated five refined models. The refined best model showed 93.6% residues in the most favoured region in the Ramachandran plot, GDT-HA score 0.9276, RMSD 0.490, MolProbity 2.052, Clash score 13.2 and Poor rotamers 0.8 (Supplementary Table 8), which indicate the quality of the refined model among all the five models after comparison and finally helped us to select the model for further studies (Fig. 4).
Fig. 4.
Tertiary structure of the multi-epitope vaccine after refinement indicating α-helix (red), β-strand (yellow) and random coil (green) in colour (Color figure online)
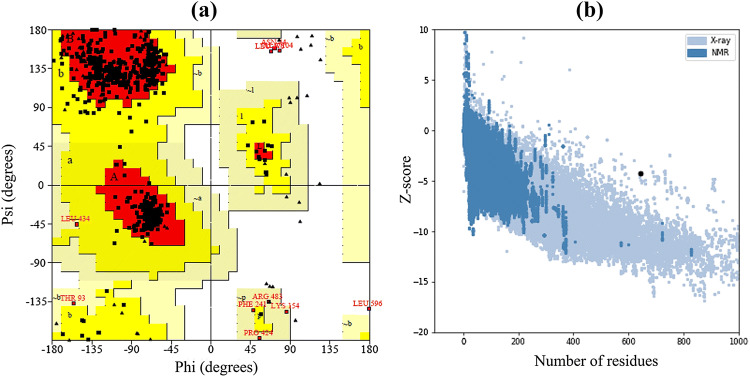
Validation of Refined Tertiary Structure
Validation of the refined tertiary structure was checked by using PROCHECK, ProSA-Web and ERRAT server. Ramachandran plot analysis of the crude structure, by PROCHECK server, revealed that 85.9% of the structure was located in the most favoured region, 11.1% in additional allowed regions, 2.0% in generously allowed regions and 1.0% of the residues were the in disallowed regions. Whereas, after refinement PROCHECK generated a better result, 89.7% of residues were located in the most favoured regions, 8.6% in additionally allowed, 1.0% in generously allowed and 0.8% of residues were found in the disallowed region (Fig. 5a). ProSA-web and ERRAT verified the quality and potential errors in a crude 3D model. The selected best model after refinement had an overall quality factor of 78.2% with ERRAT while ProSA-web gave a Z-score of − 4.23 for the input vaccine protein model, indicating the model is slightly in the range of native protein conformation (Fig. 5b).
Fig. 5.
Structural validation of the tertiary structure of the vaccine construct. a Represents the Ramachandran plot of the refined model where most favoured, allowed and disallowed regions are 89.7%, 9.6% and 0.8% respectively. b Indicates ProSA-web validation of 3D structure showing Z-score (− 4.23)
Conformational B-cell Epitope Prediction
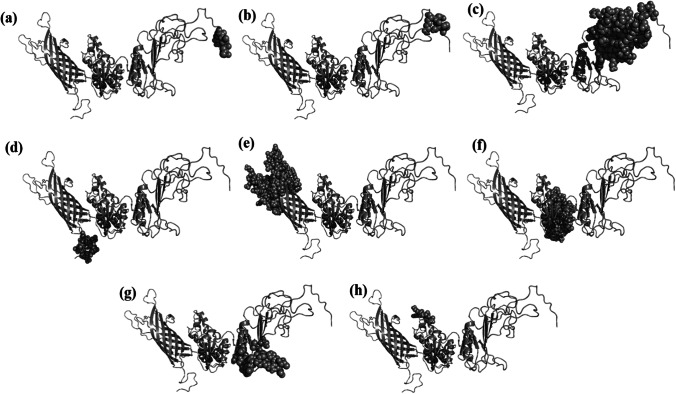
Conformational B-cell epitopes were predicted using ElliPro, an online web server that predicts epitope based on the tertiary structure. A total of 305 residues with scores varying from 0.566 to 0.944 were predicted to be located in eight conformational B-cell epitopes (Fig. 6, Table 6). The epitopes ranged in size from 4 to 93 residues.
Fig. 6.
Graphical presentation of eight conformational B-cell epitopes of the peptide vaccine. Here the predicted epitopes are indicated by cyan colour (spheres) and the rest of the residues are in grey (cartoon) (Color figure online)
Table 6.
Predicted conformational B-cell epitopes of the peptide vaccine
| No. | Residues | No. of residues | Score | Figure |
|---|---|---|---|---|
| 1 | R639, A640, S641, L642 | 4 | 0.944 | 6 (a) |
| 2 | K635, N636, S637, P638 | 4 | 0.937 | 6 (b) |
| 3 | I486, G487, P488, G489, P490, G491, N492, C493, L494, G531, V558, S559, N560, R561, C562, P563, I564, C565, K566, G567, P568, G569, P570, G571, N572, C573, L574, T575, L576, L577, L578, S579, V580, S581, N582, R583, C584, P585, I586, K587, K588, G589, K590, S591, C592, S593, L594, T595, L596, K597, G598, G599, Y600, K602, Q604, T605, K606, A607, P608, E609, V610, E611, D612, G613, P614, K615, K616, W617, F618, E619, N620, R621, G622, L623, V624, E625, C626, N627, N628, K629, K630, A631, P632, E633, T634 | 85 | 0.773 | 6 (c) |
| 4 | K3, T4, A5, I6, A7, L8, T9, I10, A11, A12, L13, A14, A15, A16, S17, V18 | 16 | 0.771 | 6 (d) |
| 5 | F39, H40, D41, G42, L43, N44, Q45, I46, E47, K48, I49, E50, G51, P52, I53, S54, F55, G56, F57, K58, R59, R87, A88, K89, V90, R91, A92, T93, D94, P95, K96, T97, K98, E99, T100, V101, G102, E103, A104, K105, H106, T107, L135, V136, R137, S138, D139, Y140, K141, V142, Y143, D144, K145, E146, L147, A148, D149, L150, S151, F152, L153, K154, R155, T156, H157, S158, T159, Q160, V161, S162, P163, F165, W185, L186, N187, N188, V189, G190, K191, L192, K193, D194, A195, K196, G197, E198, R199, V200, D201, Y202, P204, D205, I206 | 93 | 0.759 | 6 (e) |
| 6 | V227, L233, N234, S235, D236, V237, T238, F239, G240, F241, D242, K243, A244, D245, L246, K247, P248, A249, A250, Q251, N252, V253, L254, D255, G256, I257, Y258, G259, E260, I261, A262, Q263, L264, K265, S266, A267, S268, V269, R291, A292, D293, T294, V295, A296, N297, Y298, L299, V300, A301, K302, G303, V304, A305, Q306, N307, A308, I309, S310, R342, V348, G350, N351, K352, E353, A354, A355, A356 | 67 | 0.72 | 6 (f) |
| 7 | S499, V500, S501, S502, R503, C504, P505, I506, G507, P508, G509, P510, G511, C512, L513, N514, C515, S540, N541, R542, C543, P544, I545, C546, G547, P548, G549, P550, G551 | 29 | 0.644 | 6 (g) |
| 8 | V328, K329, G330, R331, A333, I335, A336 | 7 | 0.566 | 6 (h) |
Disulphide Bridging for Vaccine Stability
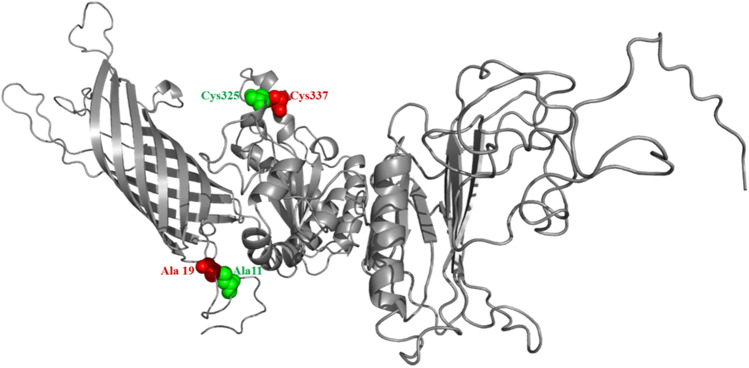
Disulphide engineering was performed using Disulfide by Design v2.12 to stabilize the modelled structure of the final vaccine construct. In total, 54 pairs of residues could be used in disulphide engineering have been discovered (Supplementary Table 9). However, only two pairs of residuals have been concluded after the evaluation of other parameters such as energy score and χ3 angle, as their value falls below the allowed range, i.e. energy should be less than 2.2 kcal/mol, and χ3 angles are expected to be between − 87 and + 97° (Craig and Dombkowski 2013). Therefore, a total of four mutations were generated on the residue pairs. For Ala11–Ala19 residual pairs, the energy score is 0.98 kcal/mol, and the χ3 angle is 85.64 degree. Whereas, for Cys325–Cys337, the χ3 angle and the energy were − 74.33° and 1.93 kcal/mol (Fig. 7).
Fig. 7.
Disuphide engineering of the vaccine construct to improve stability. Two mutated pairs are shown in green and red colour which are selected based on their energy, χ3 value, and B-factor (Color figure online)
Codon Optimization and In Silico Cloning
Expressing the LASV-derived vaccine protein epitope into the E. coli expression system was the primary purpose of in silico cloning. Therefore, according to the codon usage of the E. coli expression system, it was necessary to adapt the codon respectively to the subunit vaccine construct. To optimize codon usage of the vaccine construct in E. coli (strain K12) for maximal protein expression, the Java Codon Adaptation Tool (JCat) was used. The length of the optimized codon sequence was 1926 nucleotides. Codon optimization evaluates the sequence and tells about GC content of the cDNA sequence and codon adaptive index (CAI) where GC content was calculated as 53.63% which lies in the optimum range of (30–70) %. CAI was calculated as 0.98, which also lies in the range (0.8–1.0), which indicates the possibility of good expression of the vaccine candidate in the E. coli host. XhoI (158) and NdeI (238) restriction sites were later created and cloned using SnapGene software into the pET28a (+) vector (Fig. 8). Thus, the total length of the clone was 7.22 kbp.
Fig. 8.
In silico cloning of the final vaccine construct into pET28a(+) expression vector where the red part indicates the coding gene for the vaccine surrounded between XhoI (158) and NdeI (2091) while the vector backbone has shown in a black circle (Color figure online)
Molecular Docking of Vaccine with Immune Receptor
To assess the interaction between the refined model and the TLR2 (PDB ID-3A7B) immune receptor, molecular docking was performed by using online server ClusPro v2.0 and a total of 30 models were generated (Supplementary Table 10). Among them, only that model was selected, which occupied the receptor properly and having the lowest energy score. Since model number 1 fulfils the desired criteria, therefore, was chosen as the best-docked complex (Fig. 9). The energy score obtained for the model 1 was found to be − 1406, which is lowest among all other predicted docked complex confirming the highest binding affinity.
Fig. 9.
The docked complex of vaccine protein and TLR-2 (PDB ID- 3A7B) receptor. Here the receptor is represented in green colour whereas the vaccine protein is in red (Color figure online)
Molecular Dynamics Simulation of the Vaccine-TLR2 Complex
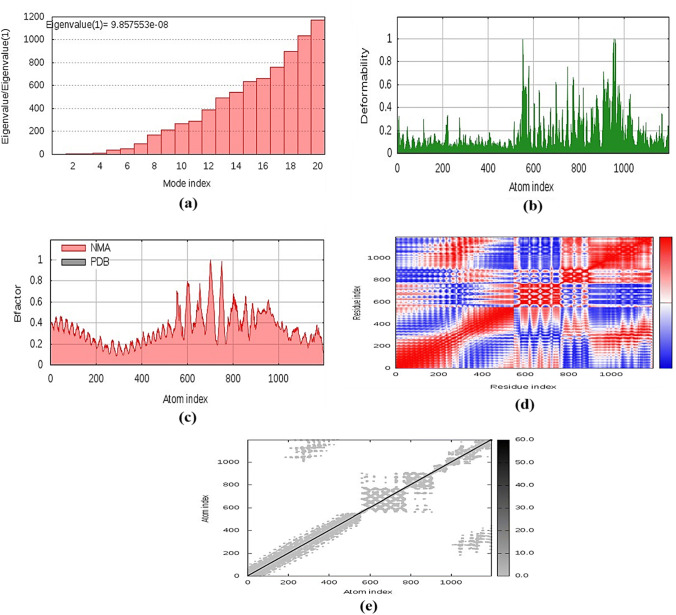
Normal mode analysis (NMA) was conducted to scrutinize protein stabilization and their large-scale mobility. This assessment was conducted by iMODS server depending on the internal coordinates of the docked complex. The complex’s deformability depends on the individual distortion of each residue, depicted by chain hinges (Fig. 10b). The eigenvalue found for the complex was 9.857553e−08 (Fig. 10a). The variance correlated with each normal mode was inverted to the eigenvalue (Kovacs et al. 2004). The B-factor values generated from normal mode analysis were proportional to RMS (Fig. 10c). Covariance matrix showing various pairs of related, anti-correlated or uncorrelated motions represented by red, blue and white colours, has stated the coupling of pairs of residues, respectively (Fig. 10d). The result also provided an elastic model of the network that distinguished the pairs of atoms linked through springs (Fig. 10e). Each dot in the diagram showing one spring, coloured by the degree of stiffness, between the corresponding atom pairs. The darker the greys, the more rigid the springs were.
Fig. 10.
Molecular dynamics simulation of the vaccine-TLR2 complex, showing a eigenvalue; b deformability; c B-factor; d covariance matrix; and e elastic network analysis
In Silico Immune Simulation
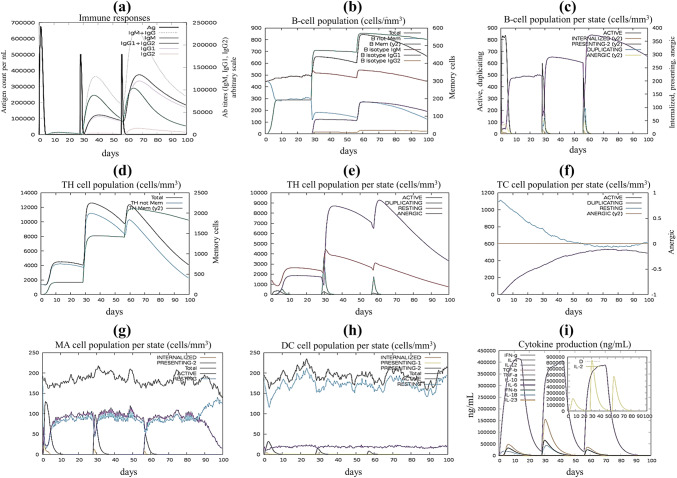
The simulated immune response was compatible with actual immune responses (Fig. 11). For instance, the secondary and tertiary responses were higher than the primary response. High concentrations of IgM was characterized at the primary response. In both the secondary and tertiary reactions, the typical high levels of immunoglobulin activities (i.e., IgG1 + IgG2, IgM, and IgG + IgM antibodies) were evident with concomitant antigen reduction (Fig. 11a). This indicates the emergence of immune memory and thus increased antigen clearance upon subsequent exposures (Fig. 11e). Furthermore, several long-lasting B-cell isotypes were observed, suggesting the potential for isotype switching and memory formation (Fig. 11b, c). In the TH (helper) and TC (cytotoxic) cell populations with the respective memory development, a similarly elevated response was noticed (Fig. 11d–f). During exposure, increased macrophage activity was demonstrated, with continuously proliferating dendritic cells (Fig. 11g, h). High levels of IFN-γ and IL-2 were also evident. Besides, a lower Simpson index (D) indicates greater diversity (Fig. 11i). Moreover, multiple exposure (n = 6) simulation as an encounter to endemic regions led to an increase of concentrations of IgG1 and decreasing IgM and IgG2 levels, while maintaining a high concentration of IFN-γ, IL-2, TC, and TH cell populations. (Supplementary Fig. 1). This profile suggests immune memory development and, therefore, natural immune protection against the virus in question.
Fig. 11.
C-ImmSim presentation of an in silico simulation of immune response using the chimeric peptide as antigen, showing a Immunoglobulin production in response to antigen injection, b B cell population after three injections, c B cell population per state, d The evolution of T-helper cell, e T-helper cell population per state, f Cytotoxic T-cell population per state, g Macrophages population per state, h Dendritic cell population per state, and i Production of cytokine and interleukins with Simpson index (d)
Discussion
LASV is a virus with a higher mortality rate and has the potential to bring upon catastrophe among the endemic region like West Africa. So, for developing a prevention method to fight against LASV is an obligation. Nowadays, vaccination is the most dynamic approach to improve the immunity system to fight against infectious diseases. Efficient development and manufacturing of live or attenuated vaccine, however, is expensive and can take years to complete. Nonetheless, the incorporation of excessive antigenic load in the attenuated vaccine appears not only to contribute little to the protective immune response but to complicate the state by causing allergic reactions (Li et al. 2014). Compared to traditional vaccines, multi-epitope vaccines decrease unwanted parts, which can either cause pathological immune responses or adverse effects (Zhang 2018). Increased safety, cost-effectiveness, the opportunity to rationally engineer the epitopes for increased potency and breadth, and the ability to focus immune responses on conserved epitopes also include potential benefits of epitope-based vaccines (Shey et al. 2019). For many years, researchers have sought to minimize the cost, time and side-effects of vaccine development. Different strategies are readily available at this moment for designing and developing efficient and competent new generation epitope-based vaccines depending on immunoinformatic approaches (María et al. 2017; Seib et al. 2012). Researchers also used immunoinformatics methods as a tool to provide futuristic models of multi-epitope driven vaccine against Ebola virus, Hepatitis C virus, Oropouche virus, Dengue virus, etc. (Adhikari et al. 2018; Ali et al. 2017; Dash et al. 2017; Ikram et al. 2018). Knowing all the pros of multiepitope based vaccine, our first and foremost concern was to construct a vaccine which will be able to elicit a robust immune response after vaccination. Though there had been a few attempts to suggest candidates for peptide vaccine against LASV, this is the very first approach recommending a fully functional multi-peptide based vaccine that has been evaluated by in silico approaches (Faisal et al. 2017; Hossain et al. 2018; Verma et al. 2015).
ViPR database was used to retrieve the whole complete sequence of LASV and after screening different types of protein, 4 protein sequences were selected due to their higher antigenicity. Through different servers and databases CTL, HTL and LBL epitopes were chosen as a vaccine candidate. An effective multi-epitope vaccine should be designed to include epitopes capable of producing CTL, HTL and B cells epitopes and inducing efficient reactions to a specific tumour or virus (Zhang 2018). Because of its function in inducing antibody manufacturing and mediating its effective features, we have been interested in incorporating B cell epitopes (Cooper and Nemerow 1984). Over time, however, the humoral response from memory B cells can easily be overcome by the emergence of antigens, whereas cell-mediated immunity (T-cell immunity) often leads to lifelong immunity (Bacchetta et al. 2005). CTL limits pathogen spread by identifying and destroying infected cells and by secreting unique antiviral cytokines (Garcia et al. 1999). Therefore, B and T-cell epitopes were predicted for the vaccine construct.
The vaccine candidates were selected from HTL, CTL and B-cell epitopes based on their antigenicity, allergenicity, immunogenicity and toxicity. Helper T-cells that release other types of cytokines such as interferon-gamma (IFN-γ), interleukin-4 (IL-4) and interleukin-10 (IL-10) have the potential to overcome pro-inflammatory response and therefore reduce tissue damage. The innate immune system, B-lymphocytes, cytotoxic T cells and other immune cells are activated with the help of helper T-cells. Thus, the cytokine (i.e., IFN-γ, IL-4 and IL-10) inducing ability of specific HTL epitopes were also evaluated for candidate choosing. The vaccine construction was completed after joining the CTL, HTL and B-cell epitopes with AAY, GPGPG and KK linkers, respectively. To enhance expression, folding and stabilization, linkers are implemented as an indispensable element in the development of vaccine protein (Shamriz et al. 2016). Furthermore, the OmpA agonist (GenBank ID: AFS89615.1) was used as a TLR2 adjuvant and joined to the first CTL epitope using EAAAK linker (Arai et al. 2001). When used alone, multiepitope-based vaccines are poorly immunogenic and require coupling to adjuvants (Meza et al. 2017). Adjuvants are ingredients added to vaccine formulations that affect particular immune responses to antigens, their development, stability, and longevity and are protective against infection (Lee and Nguyen 2015). Also, they gain great attention because the immune response to humoral and cell-mediated immune responses can be selectively modulated (Bonam et al. 2017). Though, the vaccine size is seemingly long as a peptide vaccine, several studies had been done where the vaccine length is even longer (Chatterjee et al. 2018; Kalita et al. 2019; Rahmani et al. 2019). Therefore, we think it won't be a problem in term of stability and expression. When assessing the vaccine construct, we observed that the non-adjuvant construct showed less antigenicity (0.624) than the adjuvant construct (0.7223) with the aid of Vaxijen server, which clearly states that the adjuvant is significant for the chimera.
The molecular weight of our vaccine candidate is ~ 68 kDa which is an average molecular weight for a multi-epitope vaccine. One of the fundamental requirements of many biochemical and functional analysis is the solubility of overexpressed recombinant protéin within the E. coli host (Khatoon et al. 2017). The constructed vaccine protein was found to be soluble which secured their easy access to the host. The basic nature of the vaccine is indicated by theoretical pI value. In addition, the predicted instability index shows that the protein will remain stable after expression, thus enhancing usage capacity further. The GRAVY score and aliphatic index depict the hydrophilicity and thermostability, respectively.
The 3D structure modelling includes sufficient information on the spatial arrangement of crucial protein components and excellent support in the investigation of protein function, dynamics, ligand interactions and other proteins. The desirable properties of the vaccine construct enhanced significantly after refinement. The Ramachandran plot demonstrates that most residues are discovered in the favored and allowed regions (99.2%) with very few residues in the disallowed region; which depicts that the quality of the overall model is satisfactory. Besides, GDT-HA, RMSD value, MolProbity, Clash Score and Poor Rotamers values indicate the good quality of our designed vaccine construct. Different structure validation tools were used to identify errors in the modelled vaccine construct. The Z-score (− 4.23) and ERRAT quality factor (78.2%) showed that the overall structure of the refined vaccine is adequate.
The HLA alleles maintain the response to T-cell epitopes, and in different ethnic communities, these alleles are highly polymorphic. The T-cell epitope should bind with more HLA alleles to obtain more population coverage. So, we selected the CTL and HTL epitopes with their respective HLA alleles to predict the allele distribution worldwide. The findings showed that the chosen epitopes and their individual alleles cover ideally in numerous geographic regions of the globe. The highest population coverage was recorded at 99.16% in Chile Amerindian, and those epitopes and their respective HLA alleles cover 97.37% of the world population when used in combination. In Western Africa, in particular, Nigeria, Senegal and Mali, the epidemic of the LASV happened in most significant measure. Therefore, in these geographical regions, vaccine candidates are essential to protect people against LASV infection. The population coverage was found to be 88.86% at West Africa, where the virus first appeared and had several outbreaks.
Data driven protein-receptor docking analysis and molecular dynamics simulation was carried out to evaluate a potential immune interaction and stability between TLR2 and the vaccine protein, considering the use of a TLR2 agonist as an adjuvant in the constructed chimera. Energy minimization was conducted to minimize the potential energy of the whole system for the complete conformational stabilization of the vaccine protein-TLR2 docked complex. The energy minimizes the inappropriate structural geometry by replacing individual protein atoms, thus making the structure more stable with adequate stereochemistry. The derived eigenvalue indicates the stiffness of motion and the required energy for the complex deformability.
The immunoreactivity testing through serological assessment is one of the first steps in validating a candidate vaccine (Gori et al. 2013). The expression of the recombinant protein in a suitable host is required. E. coli expression systems are determined for recombinant protein manufacturing (Chen 2012; Rosano and Ceccarelli 2014). Codon optimization had been performed with a view to achieving a high level of expression of our recombinant vaccine protein in E. coli K12. Both the codon adaptability index (0.98) and the GC content (53.63%) were promising for high-level protein expression in bacteria. Enhancing the stability of proteins is an indispensable objective in various biomedical and mechanical applications. In this study, we have introduced a disulphide bridging into the multi-epitope vaccine construct to improve protein thermostability, modify its practical features and assist in the analysis of genetic components.
The immune simulation revealed results consistent with typical immune responses. There was an overall increase in immune responses following repeated exposure of the antigen. The development of memory B-cells and T-cells were visible, with several months lasting memory B-cells. Another intriguing finding is that after the first injector concentrations of IFN-γ and IL-2 increased and were maintained at peaks after repeated antigen exposure. This finding shows elevated TH cell concentrations and therefore effective Ig production that support a humoral response. Both dendritic and macrophage cells activity were satisfactory in our study. Besides, components like epithelial cells of the innate immune system were active. The Simpson Index, D suggests a possibility of different immune responses for clonal specificity analysis.
Conclusion
Lassa virus is an emerging viral pathogen which is characterized by severe haemorrhagic fever with a higher mortality rate, hence, become an increasing concern. Though antiviral drug Ribavirin showed some promises earlier, excessive toxicity and teratogenicity rendered its effectivity questionable. Knowing all the merits that a peptide vaccine has to offer, immunoinformatics strategies have been taken into consideration for designing a multi-epitope vaccine. Both T-cell and B-cell epitopes derived from different LASV proteins were included in the vaccine to produce an effective immune response. We believe that our vaccine will hopefully generate cell-mediated and humoral immune responses. The binding potential and interaction between vaccine protein and receptor were higher and stable. Besides, effective immune responses in real life were observed in immune simulation. However, further investigations both in vitro and in vivo are warranted to ensure its true potential to fight against Lassa fever.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author Contributions
ZN and UKA conceived and designed the analysis; SBS and ZN performed immunoinformatic analyses; SBS prepared illustrations and wrote the manuscript; FA contributed in the dynamic simulation; SBS, SAK, and RT performed the antigenicity and epitope predictions; UKA and ZN contributed to the critical revision of the manuscript; UKA supervised the whole work; and all authors approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
All authors have declared that they have no conflict of interest.
Research Involving Human Participants and/or Animals
This article does not contain any studies involved with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adhikari UK, Rahman MM. Overlapping CD8 + and CD4 + T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift Valley fever virus using immunoinformatics approach. Infect Genet Evol. 2017;56(October):75–91. doi: 10.1016/j.meegid.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari UK, Tayebi M, Mizanur Rahman M. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging oropouche virus. J Immunol Res. 2018 doi: 10.1155/2018/6718083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-017-09199-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14(8):529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- Bacchetta R, Gregori S, Roncarolo MG. CD4+ regulatory T cells: mechanisms of induction and effector function. Autoimmun Rev. 2005;4(8):491–496. doi: 10.1016/j.autrev.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Berman HM, Bluhm WF, Philip E, Marvin J, Weissig H, John D. The protein data bank. Biol Crystallogr. 2002;D58:899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- Bonam SR, Partidos CD, Halmuthur SKM, Muller S. An overview of novel adjuvants designed for improving vaccine efficacy. Trends Pharmacol Sci. 2017;38(9):771–793. doi: 10.1016/j.tips.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Bonner PC, Schmidt WP, Belmain SR, Oshin B, Baglole D, Borchert M. Poor housing quality increases risk of rodent infestation and lassa fever in refugee camps of sierra leone. Am J Trop Med Hyg. 2007;77(1):169–175. doi: 10.4269/ajtmh.2007.77.169. [DOI] [PubMed] [Google Scholar]
- Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41(Web Server issue):349–357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf. 2006;7:1–5. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, et al. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science (New York, N.Y.) 1998;282(5396):2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- Castiglione F, Mantile F, De Berardinis P, Prisco A. How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Comput Math Methods Med. 2012 doi: 10.1155/2012/842329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, De Lamballerie X. Arenaviruses other than Lassa virus. Antiviral Res. 2003;57(1–2):89–100. doi: 10.1016/S0166-3542(02)00202-4. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Ojha R, Khatoon N, Prajapati VK. Scrutinizing Mycobacterium tuberculosis membrane and secretory proteins to formulate multiepitope subunit vaccine against pulmonary tuberculosis by utilizing immunoinformatic approaches. Int J Biol Macromol. 2018;118:180–188. doi: 10.1016/j.ijbiomac.2018.06.080. [DOI] [PubMed] [Google Scholar]
- Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv. 2012;30(5):1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper NR, Nemerow GR. The role of antibody and complement in the control of viral infections. J Investig Dermatol. 1984;83(1 SUPPL.):S121–S127. doi: 10.1038/jid.1984.33. [DOI] [PubMed] [Google Scholar]
- Cornu TI, de la Torre JC. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol. 2001;75(19):9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig DB, Dombkowski AA. Disulfide by design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinf. 2013 doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash R, Das R, Junaid M, Akash MFC, Islam A, Hosen SMZ. In silico-based vaccine design against Ebola virus glycoprotein. Adv Appl Bioinf Chem. 2017;10(1):11–28. doi: 10.2147/AABC.S115859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Gupta S, Vir P, Raghava GP. Prediction of IL4 inducing peptides. Clin Dev Immunol. 2013;2013:263952. doi: 10.1155/2013/263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8(1):1–15. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2—a server for in silico prediction of allergens. J Mol Model. 2014 doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- Djavani M, Lukashevich IS, Sanchez A, Nichol ST, Salvato MS. Completion of the Lassa fever virus sequence and identification of a RING finger open reading frame at the L RNA 5′ end. Virology. 1997;235(2):414–418. doi: 10.1006/viro.1997.8722. [DOI] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal A, Imtiaz SH, Zerin T, Rahman T, Shekhar HU. Computer aided epitope design as a peptide vaccine component against Lassa virus. Bioinformation. 2017;13(12):417–429. doi: 10.6026/97320630013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichet-Calvet E, Rogers DJ. Risk maps of lassa fever in West Africa. PLoS Neglect Trop Dis. 2009 doi: 10.1371/journal.pntd.0000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher-Hoch SP, Gborie S, Parker L, Huggins J. Unexpected adverse reactions during a clinical trial in rural West Africa. Antiviral Res. 1992;19(2):139–147. doi: 10.1016/0166-3542(92)90073-E. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17(1):369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- Gori A, Longhi R, Peri C, Colombo G. Peptides for immunological purposes: design, strategies and applications. Amino Acids. 2013;45(2):257–268. doi: 10.1007/s00726-013-1526-9. [DOI] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(SUPPL. 2):526–531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Sun X, Li B, Huang J, Zhan B, Zhu X. Vaccination with a paramyosin-based multi-epitope vaccine elicits significant protective immunity against Trichinella spiralis infection in mice. Front Microbiol. 2017;8(AUG):1–9. doi: 10.3389/fmicb.2017.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GPS. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013 doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas WH, Breuer T, Pfaff G, Schmitz H, Kohler P, Asper M, et al. Imported Lassa fever in Germany: surveillance and management of contact persons. Clin Infect Dis. 2003;36(10):1254–1258. doi: 10.1086/374853. [DOI] [PubMed] [Google Scholar]
- Hallam HJ, Hallam S, Rodriguez SE, Barrett ADT, Beasley DWC, Chua A, et al. Baseline mapping of Lassa fever virology, epidemiology and vaccine research and development review-article. NPJ Vaccines. 2018 doi: 10.1038/s41541-018-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes M, Salvato M. Arenavirus evasion of host anti-viral responses. Viruses. 2012;4(10):2182–2196. doi: 10.3390/v4102182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo L, Park H, Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:384–388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MU, Omar TM, Oany AR, Kibria KMK, Shibly AZ, Moniruzzaman M, et al. Design of peptide-based epitope vaccine and further binding site scrutiny led to groundswell in drug discovery against Lassa virus. 3 Biotech. 2018;8(2):1–14. doi: 10.1007/s13205-018-1106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram A, Zaheer T, Awan FM, Obaid A, Naz A, Hanif R, et al. Exploring NS3/4A, NS5A and NS5B proteins to design conserved subunit multi-epitope vaccine against HCV utilizing immunoinformatics approaches. Sci Rep. 2018;8(1):1–14. doi: 10.1038/s41598-018-34254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob CO, Leitner M, Zamir A, Salomon D, Arnon R. Priming immunization against cholera toxin and E. coli heat-labile toxin by a cholera toxin short peptide-beta-galactosidase hybrid synthesized in E. coli. EMBO J. 1985;4(12):3339–3343. doi: 10.1002/j.1460-2075.1985.tb04086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling PB, Hesse RA, Eddy GA, Johnson KM, Callis RT, Stephen EL. Lassa virus infection of rhesus monkeys: pathogenesis and treatment with ribavirin. J Infect Dis. 1980;141(5):580–589. doi: 10.1093/infdis/141.5.580. [DOI] [PubMed] [Google Scholar]
- Kalita P, Lyngdoh DL, Padhi AK, Shukla H, Tripathi T. Development of multi-epitope driven subunit vaccine against Fasciola gigantica using immunoinformatics approach. Int J Biol Macromol. 2019;138:224–233. doi: 10.1016/j.ijbiomac.2019.07.024. [DOI] [PubMed] [Google Scholar]
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. Template-based protein structure modeling using the RaptorX web server. Physiol Behav. 2017;176(8):139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenlyside RA, McCormick JB, Webb PA, Smith E, Elliott L, Johnson KM. Case-control study of mastomys natalensis and humans in Lassa virus-infected households in Sierra Leone. Am Soc Trop Med Hyg. 1983 doi: 10.4269/ajtmh.1983.32.829. [DOI] [PubMed] [Google Scholar]
- Kernéis S, Koivogui L, Magassouba N, Koulemou K, Lewis R, Aplogan A, et al. Prevalence and risk factors of lassa seropositivity in inhabitants of the Forest Region of Guinea: a cross-sectional study. PLoS Neglect Trop Dis. 2009 doi: 10.1371/journal.pntd.0000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon N, Pandey RK, Prajapati VK. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitching A, Addiman S, Cathcart S, Bishop L, Krahé D, Nicholas M, et al. A fatal case of Lassa fever in London, january 2009. Rapid Commun. 2009;14(6):12–14. [PubMed] [Google Scholar]
- Kochhar DM. Effects of exposure to high concentrations of ribavirin in devloping embryos. Pediatr Infect Dis J. 1990;9:S88–S90. doi: 10.1097/00006454-199009001-00008. [DOI] [PubMed] [Google Scholar]
- Kovacs JA, Chacón P, Abagyan R. Predictions of protein flexibility: first-order measures. Proteins. 2004;56(4):661–668. doi: 10.1002/prot.20151. [DOI] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12(2):255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf. 2007;8:1–12. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. doi: 10.1107/s0021889892009944. [DOI] [Google Scholar]
- Lee S, Nguyen MT. Recent advances of vaccine adjuvants for infectious diseases. Immune Netw. 2015;15(2):51. doi: 10.4110/in.2015.15.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer T, Rarey M. Computational methods for biomolecular docking. Curr Opin Struct Biol. 1996;6(402–406):402–405. doi: 10.1016/S0959-440X(96)80061-3. [DOI] [PubMed] [Google Scholar]
- Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide vaccine: progress and challenges. Vaccines. 2014;2(3):515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. IMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):271–276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckheeram RV, Zhou R, Verma AD, Xia B. CD4 +T cells: differentiation and functions. Clin Dev Immunol. 2012 doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan CN, Randall A, Baldi P. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25(17):2200–2207. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- Magnan CN, Zeller M, Kayala MA, Vigil A, Randall A, Felgner PL, Baldi P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(23):2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan B, Govindaraj RG, Shin TH, Kim MO, Lee G. iBCE-EL: a new ensemble learning framework for improved linear B-cell epitope prediction. Front Immunol. 2018;9(July):1695. doi: 10.3389/fimmu.2018.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- María RR, Arturo CJ, Alicia JA, Paulina MG, Gerardo AO (2017) The impact of bioinformatics on vaccine design and development. In Afrin F, Hemeg H, Ozbak H (eds) Vaccines. Intech, Rijeka
- McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Belmont-williams R. Effective therapy with Ribavirin. N Engl J Med. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- Meza B, Ascencio F, Sierra-Beltrán AP, Torres J, Angulo C. A novel design of a multi-antigenic, multistage and multi-epitope vaccine against Helicobacter pylori: an in silico approach. Infect Genet Evol. 2017;49:309–317. doi: 10.1016/j.meegid.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, et al. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat Biotechnol. 2006;24(7):817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- Nagpal G, Usmani SS, Dhanda SK, Kaur H, Singh S, Sharma M, Raghava GPS. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep. 2017;7(February):1–10. doi: 10.1038/srep42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nain Z, Abdullah F, Rahman MM, Karim MM, Khan MSA, Sayed SB, et al. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J Biomol Struct Dyn. 2019 doi: 10.1080/07391102.2019.1692072. [DOI] [PubMed] [Google Scholar]
- Nezafat N, Ghasemi Y, Javadi G, Khoshnoud MJ, Omidinia E. A novel multi-epitope peptide vaccine against cancer: an in silico approach. J Theor Biol. 2014;349:121–134. doi: 10.1016/j.jtbi.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinf. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbu O, Ajuluchukwu E, Uneke CJ. Lassa fever in West African sub-region: an overview. J Vector Borne Dis. 2007;44(1):1–11. [PubMed] [Google Scholar]
- Oti VB. A reemerging lassa virus: aspects of its structure, replication, pathogenicity and diagnosis. IntechOpen. 2018 doi: 10.5772/intechopen.79072. [DOI] [Google Scholar]
- Paul S, Sidney J, Sette A, Peters B. TepiTool: a pipeline for computational prediction of T cell epitope candidates. Curr Protoc Immunol. 2016;2016:18.19.1–18.19.24. doi: 10.1002/cpim.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(D1):593–598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Fisher-Hoch SP, Craven RB, McCormick JB. A prospective study of maternal and fetal outcome in acute Lassa fever infection during pregnancy. BMJ. 1988;297(September):584–587. doi: 10.1136/bmj.297.6648.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani A, Baee M, Rostamtabar M, Karkhah A, Alizadeh S, Tourani M, Reza H. Development of a conserved chimeric vaccine based on helper T-cell and CTL epitopes for induction of strong immune response against Schistosoma mansoni using immunoinformatics approaches. Int J Biol Macromol. 2019;141:125–136. doi: 10.1016/j.ijbiomac.2019.08.259. [DOI] [PubMed] [Google Scholar]
- Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010 doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5(APR):1–17. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D, Lopez JE, Sogoba N, Traore SF, Raffel SJ, Fischer ER, et al. Detection of lassa virus, mali. Emerg Infect Dis. 2010;16(7):1123–1126. doi: 10.3201/eid1607.100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Zhao X, Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin Microbiol Infect. 2012;18(SUPPL. 5):109–116. doi: 10.1111/j.1469-0691.2012.03939.x. [DOI] [PubMed] [Google Scholar]
- Shamriz S, Ofoghi H, Moazami N. Effect of linker length and residues on the structure and stability of a fusion protein with malaria vaccine application. Comput Biol Med. 2016;76:24–29. doi: 10.1016/j.compbiomed.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Sharpl PM, Li W. Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shey RA, Ghogomu SM, Esoh KK, Nebangwa ND, Shintouo CM, Nongley NF, et al. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019 doi: 10.1038/s41598-019-40833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarczynski M, Toth I. Peptide-based synthetic vaccines. Chem Sci. 2016;7(2):842–854. doi: 10.1039/c5sc03892h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aalten DMF, De Groot BL, Findlay JBC, Berendsen HJC, Amadei A. A comparison of techniques for calculating protein essential dynamics. J Comput Chem. 1997;18(2):169–181. doi: 10.1002/(SICI)1096-987X(19970130)18:2<169::AID-JCC3>3.0.CO;2-T. [DOI] [Google Scholar]
- Verma SK, Yadav S, Kumar A. In silico prediction of B- and T-cell epitope on Lassa virus proteins for peptide based subunit vaccine design. Adv Biomed Res. 2015;4:201. doi: 10.4103/2277-9175.166137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Who WER (2000) Imported case of Lassa fever in the Netherlands. 10.2807/esw.04.30.01557-en
- Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(SUPPL. 2):407–410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol (Clifton, N.J.) 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Wüthrich K, Wagner G, Richarz R, Braun W. Correlations between internal mobility and stability of globular proteins. Biophys J. 1980;32(1):549–560. doi: 10.1016/S0006-3495(80)84989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol. 2018;15(2):182–184. doi: 10.1038/cmi.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.