Abstract
Bacillus subtilis is a widely distributed aerobic Gram-positive species of bacteria. As a tool in the lab, it has the advantages of nonpathogenicity and limited likelihood of becoming drug resistant. It is a probiotic strain that can be directly used in humans and animals. It can be induced to produce spores under nutrient deficiency or other adverse conditions. B. subtilis spores have unique physical, chemical, and biochemical characteristics. Expression of heterologous antigens or proteins on the surface of B. subtilis spores has been successfully performed for over a decade. As an update and supplement to previously published research, this paper reviews the latest research on spore surface display technology using B. subtilis. We have mainly focused on the regulation of spore coat protein expression, display and application of exogenous proteins, and identification of developing research areas of spore surface display technology.
Keywords: Bacillus subtilis, Spores, Surface display, Coat protein
Introduction
Bacillus subtilis is an important industrial microorganism. Its genetics and physiology have been studied intensively. Among bacteria, the understanding of its genetic background and physiology is second only to Escherichia coli (Kunst et al. 1997; Sonenshein et al. 2002). Spore surface display is a method of anchoring exogenous functional proteins on the surface of spores by means of a special structure (Zhang et al. 2019). B. subtilis spore surface display has many advantages. First, spores are resistant to harsh environmental conditions, and this is conducive to the use and stability of exogenous proteins in complex environments (Wang et al. 2011). Second, spores are synthesized in the cytoplasm of bacterial cells, so any heterologous protein to be anchored on the spore surface does not need to cross any membrane (Kim and Schumann 2009). Third, molecular chaperone in the cytoplasm of B. subtilis can appropriately promote the secretion and expression of foreign proteins (Muller et al. 2000). The first spore display system was established by Isticato et al. (2001), using CotB as an anchor protein to display tetanus toxin (TTFC) on the surface of B. subtilis spores. With growing knowledge of the B. subtilis genome and proteome, spore surface display has now been successfully applied in many fields, including oral vaccine development, antibody production, biocatalysis, bioremediation, and creating of diagnostic tools (Fig. 1) (Georgiou et al. 1997; Li et al. 2019).
Fig. 1.
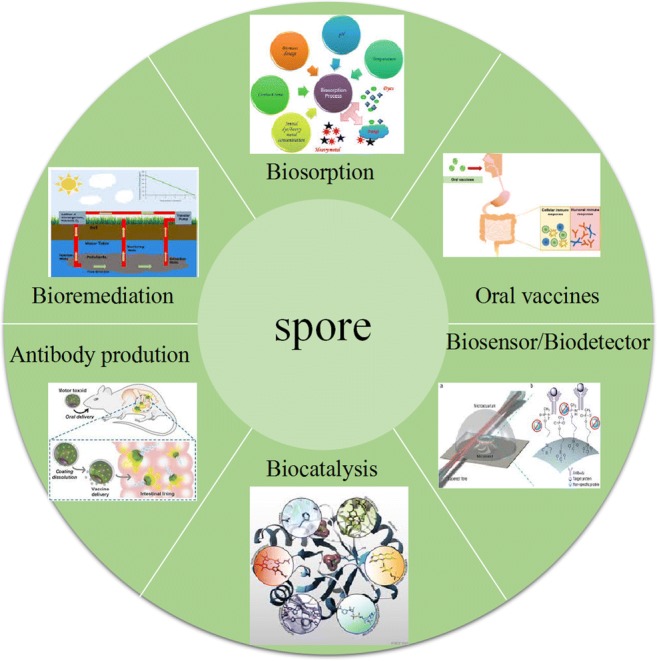
Applications of B. subtilis spore surface display
B. subtilis spore surface display follows two main approaches: a recombinant approach and a nonrecombinant approach (Isticato and Ricca 2014; Ricca et al. 2014). The recombinant approach requires modification of the bacterial genome to express a protein of interest as a fusion with spore coat protein (Hinc et al. 2013; Isticato and Ricca 2014), and the nonrecombinant approach is based on the direct adsorption of heterologous proteins on the spore surface or anchoring exogenous proteins on the spore surface with a cross-linking agent (Isticato et al. 2019; Ricca et al. 2014). The display by recombination approach avoids the isolation and purification steps of foreign proteins, the production process is simple, and it is the mainstream of B. subtilis spore surface display technology (Chen et al. 2017b; Kim and Schumann 2009). In this review, we summarize the application of genetic recombination-based spore surface display technology in many fields, discuss new and developing research, and determine the future prospects of the technology.
Formation and structure of B. subtilis spores
Bacteria have many strategies to cope with the challenges of their environment (Tasaki et al. 2017). These strategies often involve rapid changes in gene expression, which temporarily alter the phenotype of cells and allow them to survive. A more complex and persistent example of stress response is sporulation, in which the bacterial genome is isolated in a protected space (spore) until environmental conditions improve, at which point spores will germinate to form vegetative cells with reproductive capacity (Setlow 2014). Among Gram-positive bacteria, B. subtilis and a few similar species are the most commonly used experimental systems, and many studies have been conducted to assess the process and morphology of sporulation (Higgins and Dworkin 2012).
Formation of B. subtilis spores
It is challenging for B. subtilis to form spores; their formation is controlled by a series of regulatory and structural genes whose expressions themselves are tightly regulated (Bejerano-Sagie et al. 2006). When nutrients are depleted, sporulation is triggered by the activation of histidine sensor kinases, including KinA, KinB, and KinC, which shuttle phosphate through an extended phosphorelay, resulting in phosphorylation of the master regulator of sporulation,transcription factor Spo0A (Molle et al. 2003). KinA is the major kinase responsible for initiation of sporulation and KinA (or KinB) overexpression during exponential growth is sufficient to induce entry into sporulation (Fujita and Losick 2005). In fact, inducing KinA synthesis beyond a certain level leads toentry into sporulation regardless of nutrient availability (Eswaramoorthy et al. 2010). The effect of a kinC mutation on sporulation is weaker than that of kinA or kinB (Lopez et al. 2009). Phosphorylated Spo0A can directly activate or inhibit the transcription of many genes; it indirectly controls genes involved in asymmetric cell division and those involved in the activation of sporulation-specific sigma factors and ultimately promotes spore formation (Hilbert and Piggot 2004).
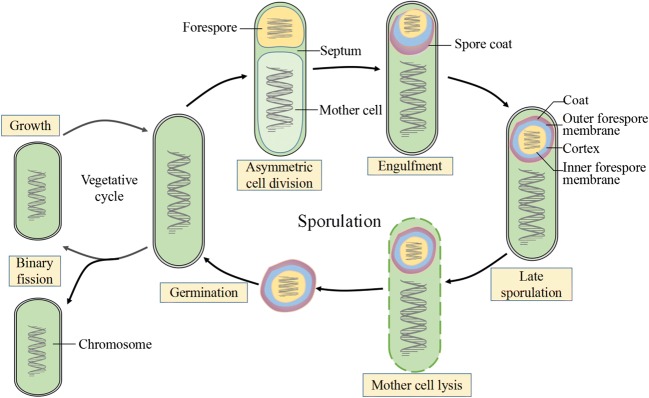
The formation of spores can be roughly divided into the following steps (Eichenberger et al. 2003; Higgins and Dworkin 2012): In harsh environments, B. subtilis cells begin to form dormant spores that resist adverse environments, and the activity of σH begins to increase. Cells then divide unequally using specific asymmetric septum to form large mother cells and a small forespore. The mother cell is necessary for spore formation, but it is eventually lysis and the prospore eventually produces a mature spore. Mother cell and prospore express different σ factors; σE factors are expressed in mother cells, while σF factors are expressed in prospores (Losick and Stragier 1992), and phosphorylated Spo0A can induce the activation of σE and σF factors (Wang et al. 2006). After unequal division is completed, the maternal plasma membrane gradually encapsulates the forespore, so the outer membrane of the forespore encapsulates two layers of membrane structure. After that, the activated or synthesized σG and σK begin to induce gene expression in the forespore and mother cell. Lastly, specific structures such as spore crust, cortex, and spore coat are gradually synthesized. The cortex is composed of peptidoglycan (PG), which is located between the inner and outer membrane of spore, and spore PG precursors are synthesized in the mother cell (Popham 2002). The spore coat is formed in the mother cell and covers the outer surface of the prospore (Henriques and Moran 2007; Kim et al. 2006). Dipicolinic acid (DPA) synthesized in the mother cell gradually fills the forespores, which could help the forespores dehydrate continuously, the mother cell lyses, and mature spore is generated (Fig. 2) (McKenney et al. 2013).
Fig. 2.
The sporulation and germination cycle in B. subtilis. Adapted from McKenney et al. (2013)
Structure of B. subtilis spores
The B. subtilis spore is a complex structure. The spore core contains the chromosomal DNA that is maintained in a compact state by small acid-soluble proteins (SASPs). The original membrane that surrounded the forespore surrounds the core and the peptidoglycan rich cortex surrounds this membrane. Surrounding the cortex, the spore coat consists of about 80 proteins deposited by the mother cell arranged in inner and outer layers (Fig. 3) (Liu et al. 2016; McKenney et al. 2013).
Fig. 3.
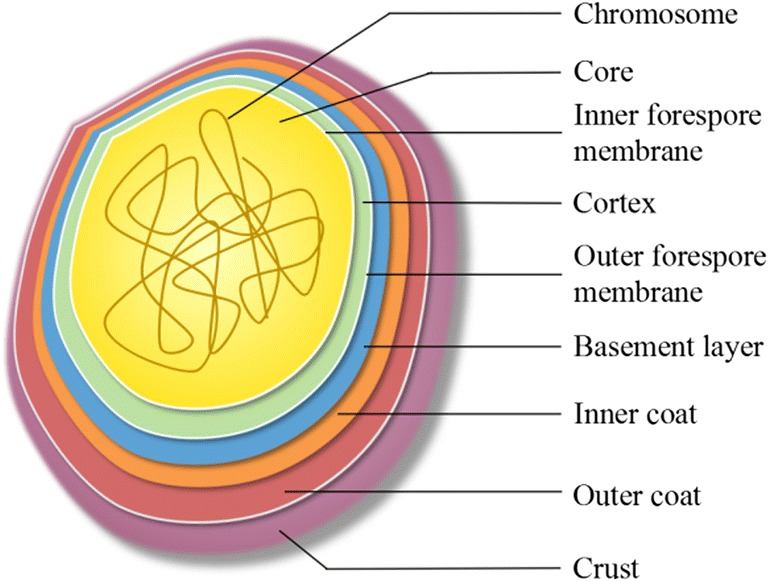
Spore structure
Spore coat
The assembling coat is synthesized in the mother cell and is targeted to the outer forespore membrane by SpoIVA (Wang et al. 2009). SpoIVA binds and hydrolyzes ATP, allowing it to self-assemble into cable-like structures (Setlow 2012; Ramamurthi and Losick 2008) that form a basement layer that serves as a platform for coat assembly. Other proteins involved in assembly are SpoVID that directly interacts with SpoIVA (Mullerova et al. 2009; Wang et al. 2009) and SafA, which is necessary for the encasement of the spore (Mullerova et al. 2009). SafA was found to affect the localization of about 16 inner coat protein fusions (McKenney et al. 2010) substantiating its central role in coat assembly. Three layers of the B. subtilis spore coat are observed in thin-section electron microscopy: an inner coat, an outer coat, and crust (Warth et al. 1963; McKenney et al. 2010). The outer coat is indispensable for spore formation, yet its specific functions remain unclear. Compared with the outer coat of spores, the inner coat is a selective permeability barrier that protects spore DNA from being destroyed by some chemical agents (McKenney et al. 2013). The spore coat makes spores resistant to chemical reagents and external lysozymes, and prevents the nuclei from being degraded or ingested by protozoa (Setlow 2006); however, the resistance to heat, radiation, and some other chemical reagents is poorly understood (Borch-Pedersen et al. 2016).
Spore cortex
The cortex and core of spores are the key structures in the formation and maintenance of dormant spores. The spore cortex is thick, mainly composed of PG, which can reach 10% of the total dry weight of the spore. The structure of the PG is similar to that in vegetative cell wall, but in cortex, the structure is relatively loose, which is extremely important for maintaining spore resistance and dormancy (Aguilar et al. 2007; Higgins and Dworkin 2012). Because some amino acid residues of N-acetylmuramic acid of PG in the spore cortex are replaced by short peptides, the degree of cross-linking of PG in cortex is lower than that of PG in vegetative cell wall (Popham 2002). In addition, B. subtilis spore-cortex PG was found to be O-acetylated, a common PG modification that reduces sensitivity to the innate immune anti-microbial lysozyme (Laaberki et al. 2011). However, since lysozyme is unable to penetrate the outer coat (Driks 1999), this modification would not appear to be useful.
Spore core
The innermost layer of spores is the core, it is surrounded by the inner forespore membrane, germ cell wall (McKenney et al. 2013). The inner forespore membrane is located in the inner of cortex, it It has extremely low permeability, and small molecular substances are difficult to penetrate, which can prevent DNA-damaging substances from penetrating the inner forespore membrane to cause damage to the spore core DNA (Setlow 2006). Spore core contains most of its enzymes, chromosomal DNA, ribosomes, and tRNA, it also contains the special small molecule, DPA, it is chelated with Ca2+ in the spore core, exists as a calcium salt, CaDPA, which can promote dehydration of the spore core and increase the thermal resistance of the spore (Higgins and Dworkin 2012; McKenney et al. 2013). SASPs are tightly bound to the core DNA of the spore, which can make the spore tolerate damage from UA radiation, drying, and high temperature, and can be used as a carbon source and energy source during spore germination (Setlow 2007).
Regulation of B. subtilis spore coat protein expression
The anchoring proteins used in B. subtilis spore surface display can be linked to exogenous proteins through their C- or N-termini. The correct selection of anchoring proteins is key to successfully displaying exogenous proteins on the spore surface. A suitable anchoring protein needs to meet the following requirements: (1) it must have a strong anchoring domain to ensure that foreign proteins can be immobilized on the spore surface (Potocki et al. 2017); (2) they must be compatible with foreign proteins, be able to form fusion proteins, and should not be able to interact with each other (Lee et al. 2003); and (3) anchored proteins must be resistant to protease hydrolysis (Lee et al. 2003; Potocki et al. 2017). To date, various spore coat proteins, such as CotB, CotC, CotE, CotG, CotX, CotY, CotZ, CgeA, and OxdD, have been successfully used as the anchoring proteins to display exogenous proteins on the spore surface of B. subtilis.
Spore surface display of B. subtilis using CotB as an anchoring protein
CotB was the first spore coat protein to be used in B. subtilis spore surface display. Its expression and assembly require the assistance of a variety of regulatory factors and proteins (Kodama et al. 2011; Zilhao et al. 2004). The expression of cotB is regulated by the maternal cell-specific sigma factors and transcription regulators GerE and GerR (Cangiano et al. 2010). CotB has a strongly hydrophilic C-terminus, which is composed of three serine-rich repeats; the serine residues accounts for more than 50% of the CotB C-terminus. Some studies have shown that CotB modification requires the involvement of CotG and CotH (Zilhao et al. 2004), and CotG is known to interact directly with CotB. Mutation of cotG results in the accumulation of a 46-kDa CotB protein in cells, but the specific mechanisms for this remain unclear. CotH, or proteins regulated by CotH, can prevent CotG from being hydrolyzed by proteases in the cell before assembling into spores, and it has an indirect regulatory effect on CotB (Nguyen et al. 2016). Isticato et al. deleted the amino acid residue in position 105 of the CotB C-terminus (CotBΔ105), used CotBΔ105 as an anchoring protein, and integrated the tetanus toxin gene into the amylase gene locus of the cotB-deleted B. subtilis genome. It was found that exogenous protein could not be expressed in spores, which proved that the fusion protein could not be assembled on the surface of spores in the absence of the original cotB gene (Henriques and Moran 2007). Therefore, the cotB, cotG, and cotH genes of B. subtilis should be retained when CotB is being used as an anchor to display exogenous proteins.
Spore surface display of B. subtilis using CotC as an anchoring protein
CotC is an abundant, 66-amino-acid protein known to assemble in the outer coat in various forms: a monomer of 12 kDa, a homodimer of 21 kDa, and two less abundant forms of 12.5 and 30 kDa, probably due to posttranslational modifications of CotC (Isticato et al. 2010; Isticato et al. 2008). Assembly of CotC requires expression of both cotH and cotE, but CotC does not accumulate in the mother cell compartment when its assembly is prevented by mutation of CotH (Isticato et al. 2004). In contrast, overexpression of cotH allows the accumulation of CotC in the mother cell compartment, suggesting that CotH, or a CotH-dependent factor, acts to prevent degradation of CotC in the mother cell and then allows its assembly within the coat (Baccigalupi et al. 2004). The mechanism of assembly of CotC is of interest, as the abundant CotC protein has been used as a vehicle for the display of heterologous proteins at the spore surface (Isticato et al. 2007). At present, heat-labile enterotoxin B subunit, urea, ethanol dehydrogenase, β-galactosidase, proinsulin, enolase, and trehalose synthase have all been successfully displayed on the spore surface using CotC as molecular carrier, which improves their tolerance to harsh environments (Hinc et al. 2010b; Romero et al. 2007).
Spore surface display of B. subtilis using CotG as an anchoring protein
CotG is a 24-kDa protein regulated by mother cell RNA polymerase σK and transcription regulator GerE. Like CotC, the expression of CotG is also indirectly regulated by GerR because GerR can activate SpoVIF, which plays an active role in GerE and GerE-dependent genes (Cangiano et al. 2010). The assembly of CotG on spore surfaces is mainly as 32- and 36-kDa proteins. Thirty-two kilodalton CotG may be formed by abnormal migration of unmodified initial proteins. Thirty-six kilodalton CotG may be produced by extensive cross-linking of proteins when proteins are assembled into the spore coat (Eichenberger et al. 2004). Like CotB and CotC, CotG assembly also requires cotH expression. CotH protects CotG from protease hydrolysis before sporulation, which is essential for the formation and assembly of CotG (Naclerio et al. 1996; Zilhao et al. 2004). Therefore, cotH should be retained when CotG is used as anchoring protein to display exogenous proteins (Saggese et al. 2014).
Spore surface display of B. subtilis using other anchoring proteins
OxdD is a secondary component of the spore shell and has oxalate decarboxylase activity. It can catalyze the conversion of oxalate into formate and CO2. Its molecular weight is approximately 43 kDa (Garcia-Ramon et al. 2017). oxdD gene is transcribed by a σK-recognized promoter during sporulation and is negatively regulated by GerE. Therefore, OxdD is produced in the mother cell chamber of sporangia and depends on SafA assembly in the coat. Genetic and cytobiological analyses have shown that OxdD is located in the outer layer of the spore. As an anchoring protein, OxdD could encapsulate the exogenous proteins under the spore surface, providing more effective protection for the exogenous proteins and reducing effects on spore formation (Romero et al. 2007).
CotH is an intermediate morphogenetic protein that plays a role in the assembly of the spore shell, but differs from CotG. CotH, as an inner layer protein of 42.8 kDa, has a strong correlation with CotB and CotG. The expression of cotH is regulated by σK. As mentioned earlier, the assembly of spore coat proteins CotB, CotC, and CotG in CotH-mutant strains also has multiple validity defect, indicating that the inner and outer layers of the spore coat require CotH function (Isticato et al. 2015).
CotZ, a key component of the crust belongs to the last encasement class and is more abundant at the mother cell proximal pole of the forespore (Imamura et al. 2010). It is dependent on σE, σK and the transcription factor GerE for expression (McKenney et al. 2013). CotZ is a 16-kDa protein, and it has been found to act as a new anchoring motif for the efficient display of UreA of Helicobacter acinonychis on the spores (Imamura et al. 2011; Hinc et al. 2013). In the case of the CotZ-UreA fusion protein, the calculated number of recombinant protein molecules is 2.5 × 102 from a single spore. This fusion protein is more effective in stimulating immunological response than other antigens in mice.
Similar to CotZ, CgeA is another 14 kDa crust protein. CgeA is dependent on σK and the transcription factor GerE for expression (Imamura et al. 2011). Iwanicki et al. (2014) described an example application of presented vector system to display CagA protein of Helicobacter pylori in fusion with CgeA spore coat protein.
Applications of B. subtilis spore surface display
B. subtilis has a well-established fermentation and production technology, and spores are resistant to harsh environmental conditions (Wang et al. 2011), so the application of spore surface display technology is very extensive. To date, this technology has been used in the production of multimeric proteins, oral vaccine preparations, and industrial enzyme production (Guoyan et al. 2019). Table 1 summarizes the related applications of spore surface display of foreign proteins based on recombinant approach in previous studies.
Table 1.
List of fusion and target proteins, used vectors, and application of Bacillus subtilis spore surface-displayed proteins based on recombinant approach
| Fusion protein | Bacterial strain | Target protein | Used vector | Substrate/antibody | Product | Application | Reference |
|---|---|---|---|---|---|---|---|
| CotB | B. subtilis PY79 | TTFC | pGEM | Anti-TTFC | ―* | Oral vaccination | (Isticato et al. 2001) |
| B. subtilis PY79 and RH103 | TTFC | pET28b | Anti-TTFC | ― | Oral vaccination | (Duc et al. 2003) | |
| B. subtilis PY79 and PP108 | TcdA | ― | Anti-TcdA | ― | Oral vaccination | (Hong et al. 2017) | |
| B. subtilis DB403 | Tm1350 | pHS | p-Nitrophenyl butyrate | p-Nitrophenyl | Industrial biocatalysis | (Chen et al. 2015a) | |
| B. subtilis DB403 | DSM | pHS | p-Nitrophenyl butyrate | p-Nitrophenyl | Industrial biocatalysis | (Chen et al. 2015b) | |
| B. subtilis PY79 | VP28 | pDG364 | White Spot Syndrome virus | ― | Vaccine for shrimps | (Nguyen et al. 2014; Pham et al. 2016) | |
| B. subtilis PY79 | UreA | pGEM | Anti-UreA | ― | Anti-Helicobacter vaccine | (Hinc et al. 2010b) | |
| B. subtilis HU58 | MPT64 | pcotVac | Anti-MPT64 | ― | Vaccine against tuberculosis | (Sibley et al. 2014) | |
| B. subtilis PY79 | RSM2e3 | pDG1664 | Anti-RSM2e3 | ― | Influenza vaccine | (Zhao et al. 2014) | |
| B. subtilis 168 | FliD | pDL | Anti-FliD | ― | C. difficile oral vaccines | (Negri et al. 2013) | |
| B. subtilis PY79 | GST-Cpa247-370 | pDG1664 | Anti-Cpa247–370 | ― | Vaccine against necrotic enteritis | (Hoang et al. 2008) | |
| B. subtilis PY79 | PA | pDG364 | Anti-PA | ― | Anthrax vaccine | (Le et al. 2007) | |
| B. subtilis PY79 and RH201 | pDHAFB | Anti-His | ― | Bioremediation | (Hinc et al. 2010a) | ||
| CotC | B. subtilis PY79 (Spo+) | TTFC and LTB | pRH22 and pIM51 | Anti-TTFC and anti-LTB | ― | Clostridium tetani and E. coli vaccine | (Mauriello et al. 2004) |
| B subtilis 168 (trp−) | BmADH | pJS700 | Ethanol and NAD+ | Acetaldehyde and NADH | Industrial biocatalysis | (Wang et al. 2011) | |
| B. subtilis PY79 and PP108 | TcdA | ― | Anti-TcdA | ― | Oral vaccination | (Hong et al. 2017) | |
| B subtilis 168 (trp−) | OmpC | pDG364 | ― | ― | Vaccine against Salmonella | (Dai et al. 2018) | |
| B. subtilis DB431 and BB80 | VP28 and VP26 | pDG1662 | Anti-Vp28 and anti-Vp26 | ― | Oral vaccination | (Valdez et al. 2014) | |
| B. subtilis WB600 | Urease B and CTB | pUS186 | Rat anti UreB serum | ― | Oral vaccine for H. pylori | (Zhou et al. 2017) | |
| B. subtilis WB600 | CsCP | pEB03 | Rat anti-rCsCP serum | ― | Vaccine against Clonorchis sinensis | (Tang et al. 2016; Tang et al. 2017) | |
| B. subtilis WB600 | TP20.8 | pGEX | TP20.8-specific antibody | ― | Vaccine against Clonorchis sinensis | (Zhou et al. 2008b) | |
| B. subtilis WB600 | CsPmy | PEB03 | Rat anti-rCsPmy serum | ― | Vaccine against Clonorchis sinensis | (Sun et al. 2018) | |
| B. subtilis WB600 | CsTP22.3 | pGEX | Rat anti-TP22.3 sera | ― | Vaccine against Clonorchis sinensis | (Zhou et al. 2008a) | |
| B. subtilis WB600 | CsLAP2 | PEB03 | Rat anti-CsLAP2 serum | ― | Vaccine against Clonorchis sinensis | (Qu et al. 2014) | |
| B. subtilis WB800N | TreS | pDG1730 | D-maltosee | D-trehalose | Industrial biocatalysis | (Liu et al. 2019) | |
| B. subtilis WB600 | VP4 | pEB03 | Rabbit anti-rVP4 serum | ― | Vaccine against grass carp reovirus | (Jiang et al. 2018) | |
| B subtilis 168 (trp−) | hGH | pJS700 | Anti-hGH | ― | Oral vaccination | (Lian et al. 2014) | |
| B. subtilis PY79 | UreA | pGEM | Anti-UreA | ― | Anti-Helicobacter vaccine | (Hinc et al. 2010b) | |
| B subtilis 168 | β-galactosidase | pKH40 | ONPG | ONP | Industrial biocatalysis | (Tavassoli et al. 2013) | |
| B. subtilis 168 (trp−) | GP64 | pJS700 | GP64-specific antibody | ― | Vaccine against Bombyx mori Nucleopolyhedrovirus | (Li et al. 2011) | |
| B. subtilis 168 (trp−) | HSA | pJS700 | HSA-specific antibody | ― | Oral vaccination | (Mao et al. 2012) | |
| CotE | B. subtilis DB104 | Tyrosinase | pCSK1 | L-tyrosine | ― | Industrial, medical, and environmental applications | (Hosseini-Abari et al. 2016) |
| B. subtilis DB104 | β-galactosidase | pDG1728 | Anti β-galactosidase, antibody mouse IgM | ― | Industrial biocatalysis | (Hwang et al. 2013) | |
| B. subtilis DB104 | Lipase A and Lipase B | pHPS9 | pNPP | ― | Industrial biocatalysis | (Kim 2017) | |
| CotG | B.subtilis DB403 | Nitrilase | pHS | Tomalononitrile, Succinonitrile, Glutaronitrile | 2-cyanoacetic acid, 3-cyanopropionic acid, 4-cyanobutyric acid | Industrial biocatalysis | (Chen et al. 2015c) |
| B. subtilis DB104 | DhaA | pHY300PLK | 2-CEES | Chloride | Bioremediation | (Wang et al. 2019) | |
| B. subtilis DB104 | β-galactosidase | pDG1728 | Anti β-galactosidase, antibody mouse IgM | ― | Industrial biocatalysis | (Hwang et al. 2013) | |
| B. subtilis DB104 | ω-transaminase | pHPS9 | (S)-α-methylbenzylamine and pyruvate | Acetophenone | Industrial biocatalysis | (Bum-Yeol et al. 2011) | |
| B. subtilis DB104 | GFPUV | pCSK1 | ― | ― | Diagnosis | (Kim et al. 2007) | |
| B. subtilis MI111 | Phytase | pHT304 | Sodium phytate | Inorganic phosphate | Industrial biocatalysis and animal probiosis | (Mingmongkolchai and Panbangred 2018) | |
| B. subtilis WB800N | TreS | pDG1730 | D-Maltosee | D-Trehalose | Industrial biocatalysis | (Liu et al. 2019) | |
| B. subtilis 168 c-trp | ChiS | pDHAFB | Chitin | 3,5-dinitrosalicylic acids and N-acetyl glucosamine | Biopesticide | (Rostami et al. 2017) | |
| B. subtilis DB403 | Nitrilase | pHS. | 3-Cyanopyridine | 3-Carboxypyridine | Industrial biocatalysis | (Chen et al. 2017a) | |
| B. subtilis DB403 | L-arabinose isomerase | pHS | D-galactose | D-tagatose | Industrial biocatalysis | (Qi et al. 2018) | |
| B. subtilis WB600 | NanA | pEASY | Pyruvate | Neu5Ac | Industrial biocatalysis | (Xu et al. 2011) | |
| B. subtilis DB104 | Streptavidin | pHPS9 | Anti-streptavidin, Antibody | ― | Biological diagnosis | (Kim et al. 2005) | |
| CotX | B. subtilis 168 (trpC2) | β-galactosidase | pJS700a | ONPG | ONP | Industrial biocatalysis | (He et al. 2015; Wang et al. 2016) |
| CotY | B. subtilis 168(trp−) | β-galactosidase | pJS700a | ONPG | ONP | Industrial biocatalysis | (He et al. 2015) |
| CotZ | B. subtilis 168(trp−) | β-galactosidase | pJS700a | ONPG | ONP | Industrial biocatalysis | (He et al. 2015) |
| B. subtilis WB800(trp-) | DPEase | pET22b(+) | D-fructose | D-allulose | Industrial biocatalysis | (He et al. 2016) | |
| CgeA | B. subtilis 168 | CagA | pMUTIN4 | Mouse anti-CagA antibody | ― | Vaccine formulation | (Iwanicki et al. 2014) |
| OxdD | B. subtilis PY79 | Phytase | pDG364 | Sodium phytate | Inorganic phosphate | Industrial biocatalysis and animal probiosis | (Potot et al. 2010) |
*Not available
Application in polyprotein production
B. subtilis can spontaneously form spores in harsh or nutrient poor environments. Spores have strong resistance to adverse environments, such as high temperature, chemical reagents, ultraviolet rays, and lysozymes. The spore coat is a complex structure comprising at least 70 different proteins. Spore surface display requires the expression of exogenous proteins fused with coat proteins, so that the exogenous proteins are assembled on the spore surface directly without transmembrane localization after synthesis in the mother cell. The fusion proteins can be immobilized by spore surface display, which improves the stability of the protein and makes isolation and purification easier. Liu H et al. fused trehalose synthase with spore-anchoring proteins CotC and CotG for display on the surface of B. subtilis spores, and immunofluorescence, Western blot analysis, and enzyme activity assays showed that trehalose synthase was indeed present on the spore surface. The trehalose synthase on the surface of the recombinant spore can react with maltose as a substrate to form trehalose, after reused four times, the recombinant spore retained most of the enzymatic activity. (Liu et al. 2019). β-Galactosidase is a high molecular weight protein (116 kDa). It is active in a tetramer state and can affect the structure of host cells in general surface display systems. To date, this protein has been displayed on the spore surface using the B. subtilis spore coat proteins CotC, CotE, CotG, CotX, CotY, and CotZ, the enzyme expressed on the surface of the spore still retains its activity (He et al. 2015). The active polyproteins were anchored on the spore surface by spore surface display technology, which demonstrates that this technology represents a new method for the production of polyproteins.
Application in preparation of oral vaccine
CotB and CotC were selected as anchoring proteins to display antigens on the surface of B. subtilis spores. Since the first successful display of surface antigens, the list of displayed antigens has grown steadily (Amuguni and Tzipori 2012; Rosales-Mendoza and Angulo 2015). Spores have good resistance to stress; therefore, vaccines developed with this method can tolerate the acidic environment of the gastrointestinal tract and have a long shelf life (Zhou et al. 2008a). They can pass through the gastrointestinal mucosa smoothly and quickly induce the body to produce a protective immune response. In addition, the use of spores as vaccine carriers can improve the efficiency of the immune response (Batista et al. 2014; Vogt et al. 2016).
In recent years, Clonorchiasis sinensis, caused by Clonorchis sinensis, has become increasingly prevalent. Effective prevention strategies are urgently needed to control this food-borne infectious disease. Previous studies have shown that C. sinensis paramyosin (CsPmy) functions as a preferred vaccine. Sun et al. (2018) displayed CsPmy on the spore surface using CotC as anchoring protein. The expression of CsPmy on the spore surface was analyzed by SDS-PAGE, Western blot analysis, and immunofluorescence assay, and the results showed that CsPmy was successfully expressed on spore surfaces and the fusion protein had good thermostability. Specific IgGs in sera and intestinal mucosa were increased after intraperitoneal and intragastrical immunization. Oral immunization with B. subtilis spore expressing CsPmy on the surface was a promising, safe, and needle-free vaccination strategy against clonorchiasis (Mingmongkolchai and Panbangred 2018). In addition, CsPmy, CsCP, TP20.8, CsTP22.3, and CsLAP2 have also been successfully displayed on spore surfaces for immunization against clonorchiasis sinensis (Tang et al. 2017; Zhou et al. 2008a). Salmonellosis is a major public health problem throughout the world. Dai et al. have assessed the potential use of B. subtilis spores for the expression of a major protective antigen of Salmonella serovar pullorum, OmpC. Mice immunized with recombinant spores expressing the OmpC antigen presented significant higher levels of OmpC-specific serum IgG and mucosal SIgA antibodies than mice immunized with nonrecombinant spores (p < 0.01) (Dai et al. 2018). These results indicate that B. subtilis spores have broad applicability in vaccine development.
Application in the production of industrial enzymes
Industrial enzymes are at the core of the biocatalysis and biotransformation industries. They are characterized by high catalytic efficiency, high specificity, and low pollution in the production process. It can be difficult to separate enzymes from substrates, and the reaction conditions are usually strictly controlled. This leads the enzymes to be easily inactivated and makes their reuse difficult. However, enzymes can be easily separated from their substrates by displaying them on the surface of spores. The excellent stress resistance of spores can enhance the stability of enzymes in complex environments and promote the reuseabilty of enzymes. He et al. produced D-allulose by using D-psicose 3-epimerase (DPEase) expressed and displayed on the surface of B. subtilis spores. DPEase was fused at the C-terminus of the anchoring protein, CotZ, via a peptide linker, and trophic genes were used as selection markers during chromosomal integration. The optimal temperature and pH of the fusion protein CotZ-DPEase were 55 °C and pH 7.5–8.0, respectively, and the anchored DPEase exhibited high thermostability. Under optimal conditions, 60% of the yield was maintained after five cycles of utilization. Therefore, this biocatalyst system, capable of expressing and immobilizing DPEase on the spore surface of B. subtilis, was an appropriate alternative for D-allulose production (He et al. 2016).
Lipases expressed in microbial hosts have great commercial value, but their applications are restricted by the high costs of production and harsh conditions used in industrial processes. Chen et al. successfully displayed the thermophilic lipase Tm1350 on the B. subtilis spore surface. The results showed that spore surface-displayed Tm1350 had more stable enzyme activity than free enzyme. Meanwhile, recycling experiments showed that the recombinant spores could be used for up to three reaction cycles without a significant decrease in catalytic rate (84%) (Chen et al. 2015a). These studies have played a positive role in development of the application of spore surface-displayed enzymes in the industrial field.
Application in the field of biological control of environmental pollution
Enzymatic technology has been applied to the treatment of environmental pollution due to its advantages of stability against environmental stress and high catalytic efficiency. Tyrosinases, which are copper-containing monooxygenases, could be used for bioremediation of phenol-polluted environments and production of L-DOPA and melanin from L-tyrosine, are widely used for environmental applications (Sok and Fragoso 2018). Hosseini-Abari et al. displayed tyrosinase on spore surfaces using CotE as a molecular carrier. Tyrosinase activity on spores was monitored in the presence of L-tyrosine and CuSO4. Recombinant spores could be used repeatedly, with 62% of enzymatic activity remaining after washing six times with Tris-HCl buffer (Hosseini-Abari et al. 2016).
Chitinase is a hydrolytic enzyme that has the specific function of hydrolyzing chitin into chitosan or N-acetylglucosamine. Chitinase is mainly used to control pests in agriculture. It can be used alone as an insecticide or used in conjunction with other microorganisms to control pests (Rishad et al. 2016). Rostami et al. fused chitinase with CotG and successfully displayed it on the surface of B. subtilis spores. Enzyme activity assays showed that the surface-displayed chitinase was active and was also able to inhibit the growth of Rhizoctonia solani and Trichoderma harzianum fungi (Rostami et al. 2017) This suggests a new bioremediation method to treat the problem of residual organophosphorus pesticides in the environment.
Application in animal feed preparation
Feed enzymes must remain active under the harsh conditions of feed preparation and the gastrointestinal tract. The strong stress resistance of spores enables them to be used as new tools for improving bioactive molecular preparations. E. coli phytase (AppA) has been widely used as an exogenous feed enzyme for monogastric animals. Sirima et al. displayed AppA on the spore surface of B. subtilis using spore coat protein CotG as an anchoring protein. AppA was successfully produced on the spore surface as verified by Western blot analysis and phytase activity assays. The highest enzyme activity was observed at 55 °C and thermal stability measurements demonstrated that more than 30% activity remained after 30 min incubated at 60 °C (Mingmongkolchai and Panbangred 2018).
Research hotspots on surface display of B. subtilis spores
As mentioned above, many B. subtilis spore surface display systems have been developed. However, up to now, most of these studies have been confined to the laboratory. Therefore, research on how to scale up the production of target proteins has become an active area of research. Strategies include introducing linker peptide chains (Huang et al. 2015), using multiple anchoring proteins to display exogenous proteins at the same time (Iwanicki et al. 2014), and increasing the number of copies of exogenous genes (Xu et al. 2011). Research on improving the sporulation efficiency of B. subtilis is another recent approach to optimizing spore surface display (Devi et al. 2015; Tojo et al. 2013).
An appropriate intermediate ligand can improve the folding efficiency of foreign target proteins and anchoring proteins. It can also change the interactions between foreign target proteins and anchoring proteins, as well as between target foreign proteins and the cell surface. Strauss et al. found that the activity of lipase on the spore surface was positively correlated with the length of the intermediate. Lipase activity increased from 0.8 to 83 U/mg when the length of the intermediate increased from 10 to 92 amino acids (Strauss and Götz 1996). Hinc et al. found that the binding mode of anchoring proteins and foreign target proteins was the key factor for the success of spore display. The conformation of linker peptides could affect the results of spore surface display, and alpha helices have shown to be most effective under some conditions (Hinc et al. 2013).
Using multiple anchoring proteins to display exogenous proteins at the same time can also improve spore display efficiency. The structure of B. subtilis spores is complex and contains dozens of different proteins. The number of potential anchoring proteins in spores is an important factor that restricts display efficiency. Therefore, the simultaneous display of various exogenous proteins by multiple anchoring proteins has become a hot area of research (Liu et al. 2019). At present, the chromosome insertion sites selected by the researchers are all the growth non-essential amyE gene. Iwanicki et al. constructed spore surface display integrative vectors using the non-essential genes lacA and pyrD as insertion sites, and using CotC and CotG as anchoring proteins, thus creating a multi-anchoring protein display system (Iwanicki et al. 2014).
Conclusions and future perspectives
B. subtilis spore surface display technology has developed rapidly over the past decade, and many coat proteins, including CotB, CotC, CotE, CotG, CotZ, CgeA, and OxdD, have been successfully used to display exogenous proteins or polypeptides on the spore surface. The nonpathogenicity of B. subtilis make this technology applicable to food and biological industries. The resistance of spores to stress makes industrial enzymes displayed on their surface more stable, and also provides the advantages of easy purification and recycling of immobilized enzyme, which can greatly reduce the cost of industrialization. B. subtilis spore surface display provides feasible avenues to improve industrial production efficiency, while providing for food and biological safety. At the same time, there are still some problems in spore surface display, such as the limited number of anchoring proteins on the spore surface, which is not conducive to a large number of exogenous proteins. Further, the success of surface display on spores depends on the fusion of anchored and target proteins, so it is critical to choose the correct fusion and anchor partner (Hinc et al. 2010b).
B. subtilis spore surface display technology has shown great promise for use in vaccine and drug preparation, enzymatic catalysis, biological detection, and other areas because of its unique advantages. It is believed that with further research on surface display using B. subtilis, this technology will play an important role in even more fields in the future.
Funding information
This work was supported by the National Natural Science Foundation of China (31501413); Shandong key project of Research & Development plan (2017GSF221019); Young doctorate Cooperation Fund Project, QiLu University of Technology (Shandong Academy of Sciences) (2017BSHZ021); State Key Laboratory of Biobased Material and Green Papermaking, Qilu University of Technology, Shandong Academy of Sciences (ZZ20190314); Natural Science Foundation of Shandong Province (ZR2017BC072, ZR2019PC060); The Dragon City Excellent Researcher Award Program from Zhucheng and Taishan Industry Leading Talents Program; and A Project of Shandong Province Higher Educational Science and Technology Program (A18KA116).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ping Lin and Haibo Yuan contributed equally to this work.
Contributor Information
Hongling Liu, Email: lhl0538@163.com.
Tengfei Wang, Email: wangtengfeiSCI@163.com.
References
- Aguilar C, Vlamakis H, Losick R, Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol. 2007;10(6):638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amuguni H, Tzipori S. Bacillus subtilis: a temperature resistant and needle free delivery system of immunogens. Hum Vaccin Immunother. 2012;8(7):979–986. doi: 10.4161/hv.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccigalupi L, Castaldo G, Cangiano G, Isticato R, Marasco R, De Felice M, Ricca E. GerE-independent expression of cotH leads to CotC accumulation in the mother cell compartment during Bacillus subtilis sporulation. Microbiology (Reading, England) 2004;150(Pt 10):3441–3449. doi: 10.1099/mic.0.27356-0. [DOI] [PubMed] [Google Scholar]
- Batista MT, Souza RD, Paccez JD, Luiz WB, Ferreira EL, Cavalcante RC, Ferreira RC, Ferreira LC. Gut adhesive Bacillus subtilis spores as a platform for mucosal delivery of antigens. Infect Immun. 2014;82(4):1414–1423. doi: 10.1128/iai.01255-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125(4):679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Borch-Pedersen K, Lindbäck T, Madslien EH, Kidd SW, O'Sullivan K, Granum PE, Aspholm M. The cooperative and interdependent roles of GerA, GerK, and Ynd in germination of Bacillus licheniformis spores. Appl Environ Microbiol. 2016;82(14):4279–4287. doi: 10.1128/AEM.00594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bum-Yeol H, Byung-Gee K, June-Hyung K. Bacterial surface display of a co-factor containing enzyme, ω-transaminase from Vibrio fluvialis using the Bacillus subtilis spore display system. Biosci Biotechnol Biochem. 2011;75(9):1862–1865. doi: 10.1271/bbb.110307. [DOI] [PubMed] [Google Scholar]
- Cangiano G., Mazzone A., Baccigalupi L., Isticato R., Eichenberger P., De Felice M., Ricca E. Direct and Indirect Control of Late Sporulation Genes by GerR of Bacillus subtilis. Journal of Bacteriology. 2010;192(13):3406–3413. doi: 10.1128/JB.00329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian R, Ni Z, Zhang Q, Zhang T, Chen Z, Chen K, Yang S. Surface display of the thermophilic lipase Tm1350 on the spore of Bacillus subtilis by the CotB anchor protein. Extremophiles. 2015;19(4):799–808. doi: 10.1007/s00792-015-0755-0. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang T, Jia J, Vastermark A, Tian R, Ni Z, Chen Z, Chen K, Yang S. Expression and display of a novel thermostable esterase from Clostridium thermocellum on the surface of Bacillus subtilis using the CotB anchor protein. J Ind Microbiol Biotechnol. 2015;42(11):1439–1448. doi: 10.1007/s10295-015-1676-8. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang T, Sun T, Ni Z, Le Y, Tian R, Chen Z, Zhang C. Clostridium thermocellum nitrilase expression and surface display on Bacillus subtilis spores. J Ind Microbiol Biotechnol. 2015;25(6):381–387. doi: 10.1159/000441642. [DOI] [PubMed] [Google Scholar]
- Chen H, Chen Z, Wu B, Ullah J, Zhang T, Jia J, Wang H, Tan T. Influences of various peptide linkers on the Thermotoga maritima MSB8 Nitrilase displayed on the spore surface of Bacillus subtilis. J Mol Microbiol Biotechnol. 2017;27(1):64–71. doi: 10.1159/000454813. [DOI] [PubMed] [Google Scholar]
- Chen H, Ullah J, Jia J. Progress in Bacillus subtilis spore surface display technology towards environment, vaccine development, and biocatalysis. J Mol Microbiol Biotechnol. 2017;27(3):159–167. doi: 10.1159/000475177. [DOI] [PubMed] [Google Scholar]
- Dai X, Liu M, Pan K, Yang J. Surface display of OmpC of Salmonella serovar Pullorum on Bacillus subtilis spores. PLoS One. 2018;13(1):e0191627. doi: 10.1371/journal.pone.0191627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SN, Kiehler B, Haggett L, Fujita M. Evidence that autophosphorylation of the major sporulation kinase in Bacillus subtilis is able to occur in trans. J Bacteriol. 2015;197(16):2675–2684. doi: 10.1128/JB.00257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63(1):1–20. doi: 10.1128/MMBR.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc LH, Hong HA, Fairweather N, Ricca E, Cutting SM. Bacterial spores as vaccine vehicles. Infect Immun. 2003;71(5):2810–2818. doi: 10.1128/iai.71.5.2810-2818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, Van Ooij C, Silvaggi J, Gonzalez-Pastor J-E, Fujita M, Ben-Yehuda S, Stragier P, Liu JS. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol. 2003;327(5):945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2(10):e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J Bacteriol. 2010;192(15):3870–3882. doi: 10.1128/jb.00466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19(18):2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramon DC, Berry C, Tse C, Fernández-Fernández A, Osuna A, Vílchez S. The parasporal crystals of Bacillus pumilus strain 15.1: a potential virulence factor? Microb Biotechnol. 2017;11(2):302–316. doi: 10.1111/1751-7915.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou George, Stathopoulos Christos, Daugherty Patrick S., Nayak Amiya R., Iverson Brent L., III Roy Curtiss. Display of heterologous proteins on the surface of microorganisms: From the screening of combinatorial libraries to live recombinant vaccines. Nature Biotechnology. 1997;15(1):29–34. doi: 10.1038/nbt0197-29. [DOI] [PubMed] [Google Scholar]
- Guoyan Z, Yingfeng A, Zabed H, Qi G, Yang M, Jiao Y, Li W, Wenjing S, Xianghui Q. Bacillus subtilis spore surface display technology: a review of its development and applications. J Microbiol Biotechnol. 2019;29(2):179–190. doi: 10.4014/jmb.1807.06066. [DOI] [PubMed] [Google Scholar]
- Wang He, Yang Ruijin, Hua Xiao, Zhao Wei, Zhang Wenbin. Functional display of active β-galactosidase on Bacillus subtilis spores using crust proteins as carriers. Food Science and Biotechnology. 2015;24(5):1755–1759. doi: 10.1007/s10068-015-0228-3. [DOI] [Google Scholar]
- He W, Jiang B, Mu W, Zhang T. Production of d-Allulose with d-Psicose 3-Epimerase expressed and displayed on the surface of Bacillus subtilis spores. J Agric Food Chem. 2016;64(38):7201–7207. doi: 10.1021/acs.jafc.6b03347. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Moran CP. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68(2):234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinc K, Ghandili S, Karbalaee G, Shali A, Noghabi KA, Ricca E, Ahmadian G. Efficient binding of nickel ions to recombinant Bacillus subtilis spores. Res Microbiol. 2010;161(9):757–764. doi: 10.1016/j.resmic.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Hinc K, Isticato R, Dembek M, Karczewska J, Iwanicki A, Peszyńska-Sularz G, De Felice M, Obuchowski M, Ricca E. Expression and display of UreA of Helicobacter acinonychis on the surface of Bacillus subtilis spores. Microb Cell Factories. 2010;9(1):2. doi: 10.1186/1475-2859-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinc K, Iwanicki A, Obuchowski M. New stable anchor protein and peptide linker suitable for successful spore surface display in B. subtilis. Microb Cell Factories. 2013;12(1):22. doi: 10.1186/1475-2859-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TH, Hong HA, Clark GC, Titball RW, Cutting SM. Recombinant Bacillus subtilis expressing the Clostridium perfringens alpha toxoid is a candidate orally delivered vaccine against necrotic enteritis. Infect Immun. 2008;76(11):5257–5265. doi: 10.1128/IAI.00686-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HA, Hitri K, Hosseini S, Kotowicz N, Bryan D, Mawas F, Wilkinson AJ, van Broekhoven A, Kearsey J, Cutting SM. Mucosal antibodies to the C terminus of toxin A prevent colonization of Clostridium difficile. Infect Immun. 2017;85(4):e01060–e01016. doi: 10.1128/iai.01060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini-Abari A, Kim BG, Lee SH, Emtiazi G, Kim W, Kim JH. Surface display of bacterial tyrosinase on spores of Bacillus subtilis using CotE as an anchor protein. J Basic Microbiol. 2016;56(12):1331–1337. doi: 10.1002/jobm.201600203. [DOI] [PubMed] [Google Scholar]
- Huang Z, Li G, Zhang C, Xing X-H. A study on the effects of linker flexibility on acid phosphatase PhoC-GFP fusion protein using a novel linker library. Enzym Microb Technol. 2015;83:1–6. doi: 10.1016/j.enzmictec.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Hwang B-Y, Pan J-G, Kim B-G, Kim J-H. Functional display of active tetrameric β-galactosidase using Bacillus subtilis spore display system. J Nanosci Nanotechnol. 2013;13(3):2313–2319. doi: 10.1166/jnn.2013.6889. [DOI] [PubMed] [Google Scholar]
- Imamura D, Kuwana R, Takamatsu H, Watabe K. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J Bacteriol. 2010;192(2):518–524. doi: 10.1128/jb.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D, Kuwana R, Takamatsu H, Watabe K. Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J Bacteriol. 2011;193(16):4075–4080. doi: 10.1128/jb.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Ricca E. Spore surface display. Microbiol Spectr. 2014;2(5):TBS-0011-2012. doi: 10.1128/microbiolspec.TBS-0011-2012. [DOI] [PubMed] [Google Scholar]
- Isticato R, Cangiano G, Tran HT, Ciabattini A, Medaglini D, Oggioni MR, De Felice M, Pozzi G, Ricca E. Surface display of recombinant proteins on Bacillus subtilis spores. J Bacteriol. 2001;183(21):6294–6301. doi: 10.1128/JB.183.21.6294-6301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Esposito G, Zilhao R, Nolasco S, Cangiano G, De Felice M, Henriques AO, Ricca E. Assembly of multiple CotC forms into the Bacillus subtilis spore coat. J Bacteriol. 2004;186(4):1129–1135. doi: 10.1128/jb.186.4.1129-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato Rachele, Scotto Di Mase Donatella, Mauriello Emilia M.F., De Felice Maurilio, Ricca Ezio. Amino terminal fusion of heterologous proteins to CotC increases display efficiencies in the Bacillus subtilis spore system. BioTechniques. 2007;42(2):151–156. doi: 10.2144/000112329. [DOI] [PubMed] [Google Scholar]
- Isticato R, Pelosi A, Zilhao R, Baccigalupi L, Henriques AO, De Felice M, Ricca E. CotC-CotU heterodimerization during assembly of the Bacillus subtilis spore coat. J Bacteriol. 2008;190(4):1267–1275. doi: 10.1128/jb.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Pelosi A, De Felice M, Ricca E. CotE binds to CotC and CotU and mediates their interaction during spore coat formation in Bacillus subtilis. J Bacteriol. 2010;192(4):949–954. doi: 10.1128/jb.01408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Sirec T, Vecchione S, Crispino A, Saggese A, Baccigalupi L, Notomista E, Driks A, Ricca E. The direct interaction between two morphogenetic proteins is essential for spore coat formation in Bacillus subtilis. PLoS One. 2015;10(10):e0141040. doi: 10.1371/journal.pone.0141040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isticato R, Ricca E, Baccigalupi L. Spore adsorption as a nonrecombinant display system for enzymes and antigens. J Vis Exp. 2019;19(145):e59102. doi: 10.3791/59102. [DOI] [PubMed] [Google Scholar]
- Iwanicki A, Piątek I, Stasiłojć M, Grela A, Łęga T, Obuchowski M, Hinc K. A system of vectors for Bacillus subtilis spore surface display. Microb Cell Factories. 2014;13(1):30. doi: 10.1186/1475-2859-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Bian Q, Zeng W, Ren P, Sun H, Lin Z, Tang Z, Zhou X, Wang Q, Wang Y. Oral delivery of Bacillus subtilis spores expressing grass carp reovirus VP4 protein produces protection against grass carp reovirus infection. Fish Shellfish Immunol. 2018;84:768–780. doi: 10.1016/j.fsi.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Kim J. Surface display of lipolytic enzyme, lipase A and lipase B of Bacillus subtilis on the Bacillus subtilis spore. Biotechnol Bioproc E. 2017;22(4):462–468. doi: 10.1007/s12257-017-0205-1. [DOI] [Google Scholar]
- Kim J, Schumann W. Display of proteins on Bacillus subtilis endospores. Cell Mol Life Sci. 2009;66(19):3127–3136. doi: 10.1007/s00018-009-0067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee CS, Kim BG. Spore-displayed streptavidin: a live diagnostic tool in biotechnology. Biochem Biophys Res Commun. 2005;331(1):210–214. doi: 10.1016/j.bbrc.2005.03.144. [DOI] [PubMed] [Google Scholar]
- Kim H, Hahn M, Grabowski P, McPherson DC, Otte MM, Wang R, Ferguson CC, Eichenberger P, Driks A. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59(2):487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Roh C, Lee CW, Kyung D, Choi SK, Jung HC, Pan JG, Kim BG. Bacterial surface display of GFPUV on Bacillus subtilis spores. J Microbiol Biotechnol. 2007;17(4):677–680. doi: 10.1007/s10295-006-0202-4. [DOI] [PubMed] [Google Scholar]
- Kodama T, Matsubayashi T, Yanagihara T, Komoto H, Ara K, Ozaki K, Kuwana R, Imamura D, Takamatsu H, Watabe K, Sekiguchi J. A novel small protein of Bacillus subtilis involved in spore germination and spore coat assembly. Biosci Biotechnol Biochem. 2011;75(6):1119–1128. doi: 10.1271/bbb.110029. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini A, Alloni G, Azevedo V, Bertero M, Bessieres P, Bolotin A, Borchert S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Laaberki MH, Pfeffer J, Clarke AJ, Dworkin J. O-acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J Biol Chem. 2011;286(7):5278–5288. doi: 10.1074/jbc.M110.183236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HD, Hong HA, Atkins HS, Flick-Smith HC, Durrani Z, Rijpkema S, Titball RW, Cutting SM. Immunization against anthrax using Bacillus subtilis spores expressing the anthrax protective antigen. Vaccine. 2007;25(2):346–355. doi: 10.1016/j.vaccine.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21(1):45–52. doi: 10.1016/S0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Li G, Tang Q, Chen H, Yao Q, Ning D, Chen K. Display of Bombyx mori nucleopolyhedrovirus GP64 on the Bacillus subtilis spore coat. Curr Microbiol. 2011;62(5):1368–1373. doi: 10.1007/s00284-011-9867-7. [DOI] [PubMed] [Google Scholar]
- Li Wanqiang, Feng Jie, Li Jiajun, Li Jianzhen, Wang Zhenhua, Khalique Abdul, Yang Miao, Ni Xueqin, Zeng Dong, Zhang Dongmei, Jing Bo, Luo Qihui, Pan Kangcheng. Surface Display of Antigen Protein VP8* of Porcine Rotavirus on Bacillus Subtilis Spores Using CotB as a Fusion Partner. Molecules. 2019;24(20):3793. doi: 10.3390/molecules24203793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C, Zhou Y, Feng F, Chen L, Tang Q, Yao Q, Chen K. Surface display of human growth hormone on Bacillus subtilis spores for Oral administration. Curr Microbiol. 2014;68(4):463–471. doi: 10.1007/s00284-013-0500-9. [DOI] [PubMed] [Google Scholar]
- Liu H, Qiao H, Krajcikova D, Zhang Z, Wang H, Barak I, Tang J. Physical interaction and assembly of Bacillus subtilis spore coat proteins CotE and CotZ studied by atomic force microscopy. J Struct Biol. 2016;195(2):245–251. doi: 10.1016/j.jsb.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Liu H, Yang S, Wang X, Wang T. Production of trehalose with trehalose synthase expressed and displayed on the surface of Bacillus subtilis spores. Microb Cell Factories. 2019;18(1):100. doi: 10.1186/s12934-019-1152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33(1):152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355(6361):601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Mao L, Jiang S, Li G, He Y, Chen L, Yao Q, Chen K. Surface display of human serum albumin on Bacillus subtilis spores for oral administration. Curr Microbiol. 2012;64(6):545–551. doi: 10.1007/s00284-012-0109-4. [DOI] [PubMed] [Google Scholar]
- Mauriello EM, Duc LH, Isticato R, Cangiano G, Hong HA, De Felice M, Ricca E, Cutting SM. Display of heterologous antigens on the Bacillus subtilis spore coat using CotC as a fusion partner. Vaccine. 2004;22(9–10):1177–1187. doi: 10.1016/j.vaccine.2003.09.031. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol : CB. 2010;20(10):934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11(1):33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingmongkolchai S, Panbangred W. Display of Escherichia coli phytase on the surface of Bacillus subtilis spore using CotG as an anchor protein. Appl Biochem Biotechnol. 2018;187(3):838–855. doi: 10.1007/s12010-018-2855-7. [DOI] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, González-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50(5):1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Muller JP, Ozegowski J, Vettermann S, Swaving J, van Wely KHM, Driessen AJM. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem J. 2000;348(2):367–373. doi: 10.1042/0264-6021:3480367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullerova D, Krajcikova D, Barak I. Interactions between Bacillus subtilis early spore coat morphogenetic proteins. FEMS Microbiol Lett. 2009;299(1):74–85. doi: 10.1111/j.1574-6968.2009.01737.x. [DOI] [PubMed] [Google Scholar]
- Naclerio G, Baccigalupi L, Zilhao R, De Felice M, Ricca E. Bacillus subtilis spore coat assembly requires cotH gene expression. J Bacteriol. 1996;178(15):4375–4380. doi: 10.1128/jb.178.15.4375-4380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri A, Potocki W, Iwanicki A, Obuchowski M, Hinc K. Expression and display of Clostridium difficile protein FliD on the surface of Bacillus subtilis spores. J Med Microbiol. 2013;62(9):1379–1385. doi: 10.1099/jmm.0.057372-0. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Pham CK, Pham HT, Pham HL, Nguyen AH, Dang LT, Huynh HA, Cutting SM, Phan T-N. Bacillus subtilis spores expressing the VP28 antigen: a potential oral treatment to protect Litopenaeus vannamei against white spot syndrome. FEMS Microbiol Lett. 2014;358(2):202–208. doi: 10.1111/1574-6968.12546. [DOI] [PubMed] [Google Scholar]
- Nguyen KB, Sreelatha A, Durrant ES, Lopez-Garrido J, Muszewska A, Dudkiewicz M, Grynberg M, Yee S, Pogliano K, Tomchick DR. Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc Natl Acad Sci U S A. 2016;113(25):E3482–E3491. doi: 10.1073/pnas.1605917113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham KC, Tran H, Van Doan C, Le P, Van Nguyen A, Nguyen H, Hong H, Cutting S, Phan TN. Protection of Penaeus monodon against white spot syndrome by continuous oral administration of a low concentration of Bacillus subtilis spores expressing the VP 28 antigen. Lett Appl Microbiol. 2016;64(3):184–191. doi: 10.1111/lam.12708. [DOI] [PubMed] [Google Scholar]
- Popham DL. Specialized peptidoglycan of the bacterial endospore: the inner wall of the lockbox. Cell Mol Life Sci. 2002;59(3):426–433. doi: 10.1007/s00018-002-8435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki W, Negri A, Peszyńska-Sularz G, Hinc K, Obuchowski M, Iwanicki A. The combination of recombinant and non-recombinant Bacillus subtilis spore display technology for presentation of antigen and adjuvant on single spore. Microb Cell Factories. 2017;16(1):151. doi: 10.1186/s12934-017-0765-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potot S, Serra CR, Henriques AO, Schyns G, Microbiology E. Display of recombinant proteins on Bacillus subtilis spores, using a coat-associated enzyme as the carrier. Appl Environ Microbiol. 2010;76(17):5926–5933. doi: 10.1128/AEM.01103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G, An Y, Yun J, Yang M, Qi X. Enhanced D-tagatose production by spore surface-displayed L-arabinose isomerase from isolated Lactobacillus brevis PC16 and biotransformation. Bioresour Technol. 2018;247:940–946. doi: 10.1016/j.biortech.2017.09.187. [DOI] [PubMed] [Google Scholar]
- Qu H, Xu Y, Sun H, Lin J, Yu J, Tang Z, Shen J, Liang C, Li S, Chen W. Systemic and local mucosal immune responses induced by orally delivered Bacillus subtilis spore expressing leucine aminopeptidase 2 of Clonorchis sinensis. Parasitol Res. 2014;113(8):3095–3103. doi: 10.1007/s00436-014-3975-9. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31(3):406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricca E, Baccigalupi L, Cangiano G, De Felice M, Isticato R. Mucosal vaccine delivery by non-recombinant spores of Bacillus subtilis. Microb Cell Factories. 2014;13(1):115. doi: 10.1186/s12934-014-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishad KS, Rebello S, Shabanamol PS, Jisha MS. Biocontrol potential of Halotolerant bacterial chitinase from high yielding novel Bacillus pumilus MCB-7 autochthonous to mangrove ecosystem. Pestic Biochem Physiol. 2016;137:36–41. doi: 10.1016/j.pestbp.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Romero S, Merino E, Bolívar F, Gosset G, Martinez A. Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl Environ Microbiol. 2007;73(16):5190–5198. doi: 10.1128/AEM.00625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Mendoza S, Angulo C. Bacillus subtilis comes of age as a vaccine production host and delivery vehicle. Expert Rev Vaccines. 2015;14(8):1135–1148. doi: 10.1586/14760584.2015.1051469. [DOI] [PubMed] [Google Scholar]
- Rostami A, Hinc K, Goshadrou F, Shali A, Bayat M, Hassanzadeh M, Amanlou M, Eslahi N, Ahmadian G. Display of B. pumilus chitinase on the surface of B. subtilis spore as a potential biopesticide. Pestic Biochem Physiol. 2017;140:17–23. doi: 10.1016/j.pestbp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Saggese A, Scamardella V, Sirec T, Cangiano G, Isticato R, Pane F, Amoresano A, Ricca E, Baccigalupi L. Antagonistic role of CotG and CotH on spore germination and coat formation in Bacillus subtilis. PLoS One. 2014;9(8):e104900. doi: 10.1371/journal.pone.0104900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101(3):514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15(4):172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Setlow P. Dynamics of the assembly of a complex macromolecular structure-the coat of spores of the bacterium Bacillus subtilis. Mol Microbiol. 2012;83(2):241–244. doi: 10.1111/j.1365-2958.2011.07948.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol. 2014;196(7):1297–1305. doi: 10.1128/jb.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley L, Reljic R, Radford DS, Huang JM, Hong HA, Cranenburgh RM, Cutting SM. Recombinant Bacillus subtilis spores expressing MPT64 evaluated as a vaccine against tuberculosis in the murine model. FEMS Microbiol Lett. 2014;358(2):170–179. doi: 10.1111/1574-6968.12525. [DOI] [PubMed] [Google Scholar]
- Sok V, Fragoso A. Kinetic, spectroscopic and computational docking study of the inhibitory effect of the pesticides 2,4,5-T, 2,4-D and glyphosate on the diphenolase activity of mushroom tyrosinase. Int J Biol Macromol. 2018;118(Pt A):427–435. doi: 10.1016/j.ijbiomac.2018.06.098. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL, Hoch JA, Losick R (2002) Bacillus subtilis and its closest relatives: from genes to cells. Am Soc Microbiol. 10.1128/9781555817992.ch36
- Strauss A, Götz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21(3):491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- Sun H, Lin Z, Zhao L, Chen T, Shang M, Jiang H, Tang Z, Zhou X, Shi M, Zhou L, Ren P, Qu H, Lin J, Li X, Xu J, Huang Y, Yu X. Bacillus subtilis spore with surface display of paramyosin from Clonorchis sinensis potentializes a promising oral vaccine candidate. Parasit Vectors. 2018;11(1):156. doi: 10.1186/s13071-018-2757-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shang M, Chen T, Ren P, Sun H, Qu H, Lin Z, Zhou L, Yu J, Jiang H. The immunological characteristics and probiotic function of recombinant Bacillus subtilis spore expressing Clonorchis sinensis cysteine protease. Parasit Vectors. 2016;9(1):648. doi: 10.1186/s13071-016-1928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Sun H, Chen T, Lin Z, Jiang H, Zhou X, Shi C, Pan H, Chang O, Ren P, Immunology S. Oral delivery of Bacillus subtilis spores expressing cysteine protease of Clonorchis sinensis to grass carp (Ctenopharyngodon idellus): induces immune responses and has no damage on liver and intestine function. Fish Shellfish Immunol. 2017;64:287–296. doi: 10.1016/j.fsi.2017.03.030. [DOI] [PubMed] [Google Scholar]
- Tasaki S, Nakayama M, Shoji W. Morphologies of Bacillus subtilis communities responding to environmental variation. Develop Growth Differ. 2017;59(5):369–378. doi: 10.1111/dgd.12383. [DOI] [PubMed] [Google Scholar]
- Tavassoli S, Hinc K, Iwanicki A, Obuchowski M, Ahmadian G. Investigation of spore coat display of Bacillus subtilis β-galactosidase for developing of whole cell biocatalyst. Arch Microbiol. 2013;195(3):197–202. doi: 10.1007/s00203-013-0867-9. [DOI] [PubMed] [Google Scholar]
- Tojo S, Hirooka K, Fujita Y. Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control. J Bacteriol. 2013;195(8):1656–1665. doi: 10.1128/JB.02131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez A, Yepiz-Plascencia G, Ricca E, Olmos J. First Litopenaeus vannamei WSSV 100% oral vaccination protection using CotC:: Vp26 fusion protein displayed on Bacillus subtilis spores surface. J Appl Microbiol. 2014;117(2):347–357. doi: 10.1111/jam.12550. [DOI] [PubMed] [Google Scholar]
- Vogt CM, Schraner EM, Aguilar C, Eichwald C. Heterologous expression of antigenic peptides in Bacillus subtilis biofilms. Microb Cell Factories. 2016;15(1):137. doi: 10.1186/s12934-016-0532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358(1):16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, Grabowski P, Chua MH, Barry SN, Guan M, Bonneau R, Henriques AO, Eichenberger P. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol. 2009;74(3):634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Chang C, Yao Q, Li G, Qin L, Chen L, Chen K. Display of Bombyx mori alcohol dehydrogenases on the Bacillus subtilis spore surface to enhance enzymatic activity under adverse conditions. PLoS One. 2011;6(6):e21454. doi: 10.1371/journal.pone.0021454/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang R, Hua X, Zhang W, Zhao W. An approach for lactulose production using the CotX-mediated spore-displayed β-galactosidase as a biocatalyst. J Microbiol Biotechnol. 2016;26:1267–1277. doi: 10.4014/jmb.1602.02036. [DOI] [PubMed] [Google Scholar]
- Wang F, Song T, Jiang H, Pei C, Huang Q, Xi H. Bacillus subtilis spore surface display of haloalkane dehalogenase DhaA. Curr Microbiol. 2019;76(10):1161–1167. doi: 10.1007/s00284-019-01723-7. [DOI] [PubMed] [Google Scholar]
- Warth AD, Ohye DF, Murrell WG. The composition and structure of bacterial spores. J Cell Biol. 1963;16:579–592. doi: 10.1083/jcb.16.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gao C, Zhang X, Che B, Ma C, Qiu J, Tao F, Xu P. Production of N-acetyl-D-neuraminic acid by use of an efficient spore surface display system. Appl Environ Microbiol. 2011;77(10):3197–3201. doi: 10.1128/AEM.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, An Y, Zabed HM, Guo Q, Yang M, Yuan J, Li W, Sun W, Qi X. Bacillus subtilis spore surface display technology: a review of its development and applications. J Microbiol Biotechnol. 2019;29(2):179–190. doi: 10.4014/jmb.1807.06066. [DOI] [PubMed] [Google Scholar]
- Zhao G, Miao Y, Guo Y, Qiu H, Sun S, Kou Z, Yu H, Li J, Chen Y, Jiang S. Development of a heat-stable and orally delivered recombinant M2e-expressing B. subtilis spore-based influenza vaccine. Hum Vaccin Immunother. 2014;10(12):3649–3658. doi: 10.4161/hv.36122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Xia H, Hu X, Huang Y, Li Y, Li L, Ma C, Chen X, Hu F, Xu J. Oral administration of a Bacillus subtilis spore-based vaccine expressing Clonorchis sinensis tegumental protein 22.3 kDa confers protection against Clonorchis sinensis. Vaccine. 2008;26(15):1817–1825. doi: 10.1016/j.vaccine.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Xia H, Hu X, Huang Y, Ma C, Chen X, Hu F, Xu J, Lu F, Wu Z. Immunogenicity of recombinant Bacillus subtilis spores expressing Clonorchis sinensis tegumental protein. Parasitol Res. 2008;102(2):293–297. doi: 10.1007/s00436-007-0762-x. [DOI] [PubMed] [Google Scholar]
- Zhou Zhenwen, Dong Hui, Huang Yanmei, Yao Shuwen, Liang Bingshao, Xie Yongqiang, Long Yan, Mai Jialiang, Gong Sitang. Recombinant Bacillus subtilis spores expressing cholera toxin B subunit and Helicobacter pylori urease B confer protection against H. pylori in mice. Journal of Medical Microbiology. 2017;66(1):83–89. doi: 10.1099/jmm.0.000404. [DOI] [PubMed] [Google Scholar]
- Zilhao R, Serrano M, Isticato R, Ricca E, Moran CP, Jr, Henriques AO. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J Bacteriol. 2004;186(4):1110–1119. doi: 10.1128/jb.186.4.1110-1119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]